Abstract
On December 18, 2014, a yellow female fly quietly emerged from her pupal case. What made her unique was that she had only one parent carrying a mutant allele of this classic recessive locus. Then, one generation later, after mating with a wild-type male, all her offspring displayed the same recessive yellow phenotype. Further analysis of other such yellow females revealed that the construct causing the mutation was converting the opposing chromosome with 95% efficiency. These simple results, seen also in mosquitoes and yeast, open the door to a new era of genetics wherein the laws of traditional Mendelian inheritance can be bypassed for a broad variety of purposes. Here, we consider the implications of this fundamentally new form of “active genetics”, its applications for gene-drives, reversal and amplification strategies, its potential for contributing to cell and gene therapy strategies, and ethical/biosafety considerations associated with such active genetic elements.
Introduction
Classic rules of Mendelian inheritance impose several significant constraints on genetic manipulation of organisms (e.g., random segregation of distant loci and coinheritance of closely linked loci). In this review we consider how these “passive” rules of inheritance can in principle be superseded by a new form of “active genetics” based on a new CRISPR method referred to as the Mutagenic Chain Reaction (MCR). Although other forms of active genetics can also bypass Mendelian inheritance (e.g., transposable elements), MCR-related strategies offer an array of new programmable functions that we believe will fundamentally redefine the field of genetics.
CRISPR/Cas9 based genome editing
The CRISPR/Cas9 system (reviewed in1–4) has proven to be a highly effective genome editing tool in a wide variety of organisms including diverse animals, plants and yeast (reviewed in5–11) (Fig. 1A,B). Briefly, this system consists of two entities, the Cas9 endonuclease, which cleaves DNA templates on both strands, and a guide-RNA (gRNA), the first 20 nucleotides of which direct the Cas9 cleavage of a complementary target DNA at a site three nucleotides upstream the 3′-end of the gRNA target sequence (Fig. 1A). Following cleavage of a targeted genomic sequence by a Cas9/gRNA complex, one of two alternative DNA repair mechanisms restores chromosomal integrity: 1) non-homologous end joining (NHEJ)12,13, which typically generates insertions and/or deletions (indels) of a few base-pairs (bp) of DNA near the gRNA cut site, or 2) homology-directed repair (HDR)13,14, which can correct the lesion via a DNA template with sequence homology spanning the gRNA cut site (Fig. 1B). In Drosophila melanogaster, individuals carrying sources of genomically-encoded germline Cas9 and gRNAs15,16 (or embryos injected with plasmid encoded sources of gRNAs17–19) efficiently mutate the target sequence via NHEJ in the great majority of somatic and germline cells. In addition, when a DNA template containing homologous sequences is co-injected into the polar plasm, these standard CRISPR components can trigger HDR-mediated repair in the germline16,20,21. The autocatalytic mutagenesis method described below combines features of the CRISPR/Cas9 system in a novel configuration, exploiting the cell’s endogenous repair mechanism to generate self-homozygosing alleles.
Figure 1.
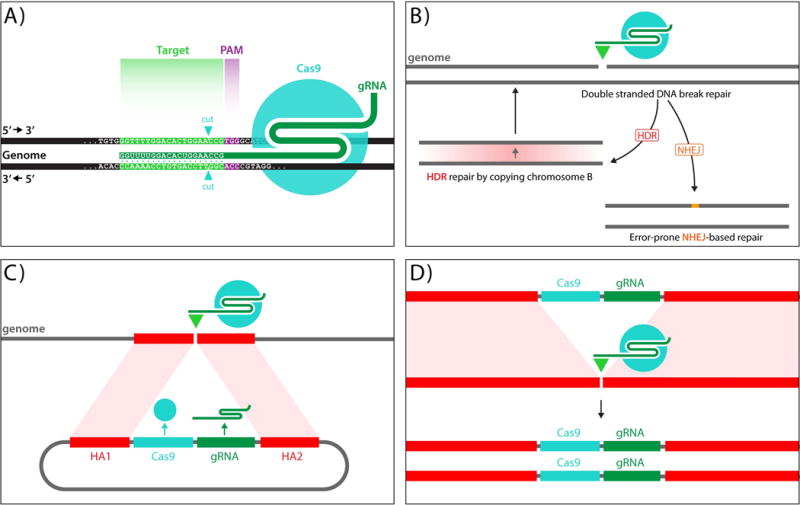
Outline of CRISPR and MCR methods. A) The CRISPR/Cas9 genome editing system consists of two elements, the Cas9 endonuclease, which generates blunt ended double stranded DNA breaks, and a 20 nucleotide guide RNA (gRNA) that binds to Cas9 and targets it to complementary genomic sequences, which in addition must have a so-called PAM sequence (NGG - violet type) recognized by Cas9 that lies immediately 3′ to the 20 nucleotides of gRNA match. B) Double stranded chromosomal breaks caused by targeted cas9/gRNA cleavage can be repaired by either the Rad51-dependent Homology Directed Repair (HDR) pathway, which faithfully copies information from the sister chromosome into the cut site, or the Ku70/80-dependent Non-homologous End-Joining (NHEJ) pathway, which typically results in short insertions/deletions (indels) at the cut site. C,D) MCR mutagenesis scheme: MCR elements (C) consist of three components: 1) a transgene encoding a nuclear targeted form of Cas9 endonuclease, 2) a gRNA directing cleavage to a desired genomic site, and 3) homology arms (HA1 and HA2) from the targeted locus that directly about the gRNA cut site. An injected MCR construct inserts into the genome at the site of gRNA directed cleavage. Once integrated into the genome (D), the MCR element acts on the opposing allele and inserts itself to generate a homozygous insertional mutation.
The Mutagenic Chain Reaction
Based on the CRISPR/Cas9 system summarized above, we reasoned that an autocatalytic genetic behavior could be achieved in which insertional mutants were generated by a construct having three components: 1) a central segment encoding Cas9 (expressed in both somatic cells and the germline), 2) a ubiquitously expressed gene encoding a gRNA targeted to a genomic sequence of interest, and 3) homology arms flanking the Cas9/gRNA cassette matching the two genomic sequences immediately adjacent to either side of the target cut site (Fig. 1C). Such a tripartite construct could result in Cas9 cutting the genomic target at the site determined by the gRNA followed by insertion of the Cas9/gRNA-bearing cassette into that locus via HDR directed by the flanking sequences. Expression of Cas9 and the gRNA from the insertion allele then should lead to cleavage of the opposing allele (Fig. 1D) followed by HDR-driven insertion of the Cas9/gRNA cassette into the companion chromosome. Analogously to the polymerase chain reaction (PCR), which doubles the number of DNA templates each cycle, we refer to this trans-acting autocatalytic mutagenesis scheme as the Mutagenic Chain Reaction (MCR), since it accomplishes the same end by in vivo DNA amplification each generation.
An MCR element efficiently converts its sister chromosome in fruit flies
We first tested the MCR concept in Drosophila22. As described below, we observed similar high frequencies of transmission of a significantly larger MCR in mosquitoes23 (also, related constructs efficiently bias inheritance in yeast24). In our initial study in flies, we used known efficient CRISPR/Cas9 components20,25, and two flanking homology arms of ~1 kb that precisely abut the gRNA-directed cut site (Fig. 2A). As mentioned above, this y-MCR element converted the opposing allele in the female germline ≈ 95% of the time, deviating significantly from the predicted 50% Mendelian transmission rate (Fig. 2B,C). In addition, somatic cells were converted to full body yellow mutant phenotype in the great majority of individuals (96%). PCR analysis confirmed the precise expected gRNA-driven genomic insertion of the y-MCR construct in such individuals indicating that the y-MCR element copied itself to the sister chromosome with high efficiency in the female germline. The yellow mutant phenotype was widespread in the vast majority of pigmented somatic cells of most individuals, however, molecular analysis revealed the presence of both MCR and wild-type size y locus PCR products in MCR females, indicating that allelic conversion was incomplete. Indeed, sequencing of the few exceptions in which the MCR did not convert or mutate the sister chromosome revealed NHEJ events. Rare non-converted y+ alleles had synonymous nucleotide changes at the gRNA directed cut site or small in-frame indels. Such mutations are potentially important in certain contexts (e.g., reducing the efficiency of gene-drives - see below) because they constitute ~ wild-type MCR-resistant alleles. We note that MCR alleles acting in both germline and in somatic cells (which may induce mutations via either HDR or NHEJ) can only be used to generate viable alleles. As described further below, however, targeting essential loci for the purposes of suppressive gene-drive systems is also possible if Cas9-dependent mutagenesis is strictly confined to the germline.
Figure 2.
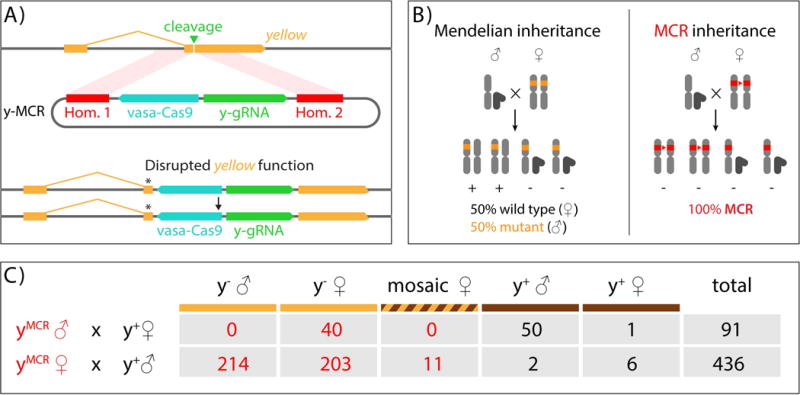
Efficient transmission of y-MCR element. A) Mendelian versus MCR inheritance of a yellow (y) allele. B) Structure of the y-MCR construct and its insertion into the genome at the yellow locus on the X chromosome. C) Summary of results of 8 crosses between F1 y- heterozygous flies and y+ flies (2 male MCR and 6 female MCR crosses) yielding a total of 527 F2 progeny. The MCR transmission rate in the experiments was 97%, which translates into a 95% rate of the MCR allele converting the opposite allele in the germline (conversion % = 2(X – 0.5N)/N where N = total number of flies and X = number of y flies with a y- phenotype or y mosaic phenotype).
Accessory elements can recall MCRs or expand their functionality
MCR elements carry both a source of Cas9 and a gRNA inserted at the gRNA cut site. We denote the MCR arrangement as: <cas9; gRNA> wherein the symbols <> represent the homology sequences flanking the gRNA cut site. Since persistent low-level Cas9 mutagenesis might reduce the fitness of individuals carrying such constructs and because it would be prudent to have methods for neutralizing MCRs, we have devised two types of constructs in which only the gRNA(s) are flanked by homology arms (denoted <gRNA> elements). Such <gRNA> constructs can only be copied to the other chromosome when a source of Cas9 is provided in trans. We consider here two categories of <gRNA> constructs that could be used in conjunction with MCRs: 1) ERACRs (Elements for Reversing the Autocatalytic Chain Reaction), which upon encountering an MCR deletes and replaces it (Fig. 3A), and 2) CHACRs (Constructs Hitchhiking on the Autocatalytic Chain Reaction) are targeted to other chromosomal loci and copied in parallel with the MCR. In the next section, we extend this strategy to devising active <gRNA> “copy-cat” cloning vectors.
Figure 3.
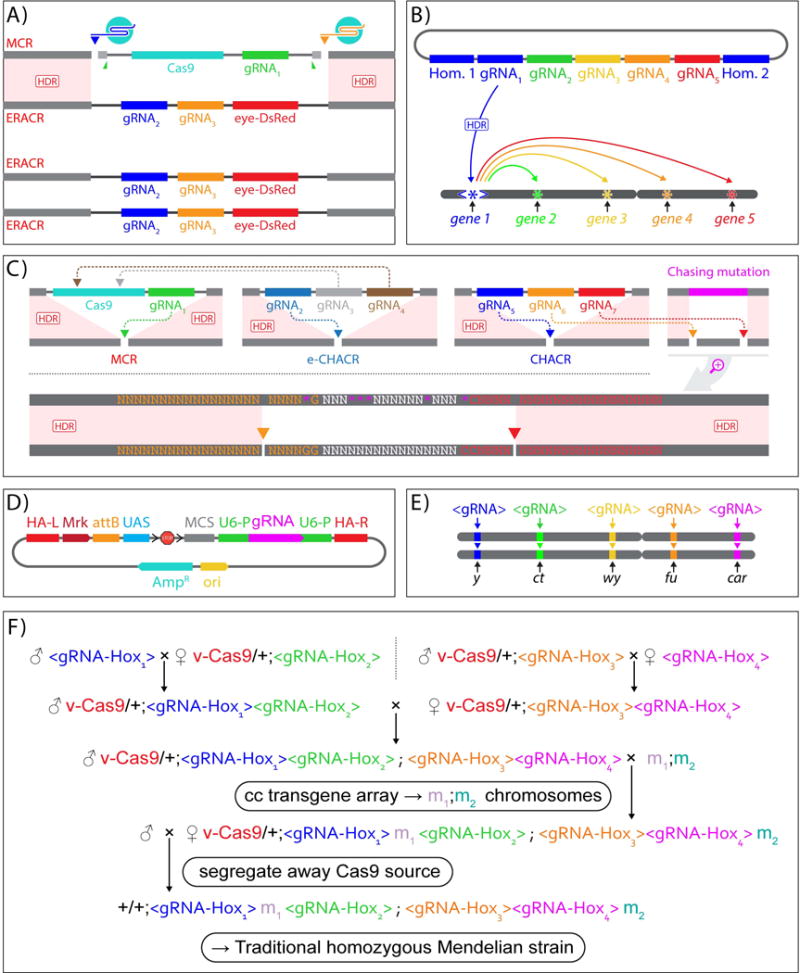
ERACRs, CHACRs, and “copy-cat” <gRNA> constructs: A) ERACRs: “Elements to Reverse the Autocatalytic Chain Reaction” delete MCR elements. In flies carrying both an MCR and an ERACR allele, Cas9 produced by the MCR cuts at sites directed by gRNA-2 and gRNA-3. eye-DsRed = dominant marker. The MCR inserted at a cut site determined by gRNA-1 lying within the deleted segment leading to the ERACR element becoming homozygous. B) CHACRs: “Constructs Hitchhiking on the Autocatalytic Chain Reaction” target other genomic targets. Shown here is an example in which a CHACR serves as a platform to launch an array of gRNAs to diverse targets where they induce standard NHEJ-dependent mutations. C) An MCR element (top left panel) could also be neutralized by CHACR elements used as second-site ERACRs (e-CHACR - inserted at site determined by gRNA2 - top second panel) that carries multiple gRNAs (gRNA3 - teal, gRNA4 - brown) targeting Cas9 in the MCR. Also, CHACRs could be used to drive the spread of unlinked auxiliary elements. Such a CHACR element is shown (top right panel) carrying 3 gRNAs inserted into the cut site of one of these gRNAs (gRNA5 - dark blue), which is in a different location in the genome than the MCR (inserted at a site defined by gRNA1 - green). Thus, like an ERACR, in the presence of an MCR carrying a Cas9 source, the CHACR cuts the opposing chromosome (via cleavage induced by gRNA5) and inserts itself into the resulting DNA gap. In addition, the depicted CHACR carries gRNA6 (orange) and gRNA7 (red), which cut at adjacent sites flanking a edited genomic locus (or existing natural allelic variant - top right panel). The resulting small deletion (region between the gRNA6 and gRNA7 cut sites) will then be repaired via HDR using the edited (pink) sequence. The lower panel shows a magnified view of the top right panel indicating the gene edited residues as pink asterisks and the two cleavage sites for gRNA6 (orange) and gRNA6 (red) relative to the sequences of perfect homology mediating HDR repair. D) “Copy-cat” or cc vectors allow the cloning of transgenes into multiple cloning sites (MCS) as well as matched sets of gRNA(s) flanked by both 5′ (U6p) and 3′ (U6-3′) U6-RNA regulatory elements, and homology arms (HA-L = left, HA-R = right), standard features of cloning vectors such as a bacterial origin of replication (Ori), a gene providing Ampicillin resistance (AmpR), as well as optional use cassettes such as a UAS promoter, an attBϕ31C recombinase donor site allowing for alternative recombinase-driven insertion of the construct into a genomic recipient site (attP), or instead, an attP recipient site to allow recombinase-mediated insertion into the genomically inserted copy-cat element, and an FRT-flanked transcriptional stop cassette (<Stop<). E) cc elements can insert at various loci along a chromosome (D. melanogaster X-chromosome shown as example) which are determined by their particular matched sets of gRNAs and homology arms. In the presence of a cas9 source, these elements will be copied to the sister chromosome, thereby homozygosing the element with the inserted transgene. F) Example of how copy-cat elements could be used in a model vertebrate organism such as a mouse or fish to create a cas9-dependent viable quadruple knock-out of a set of target genes (e.g., redundantly acting Hox gene paralogs). Not shown here for simplicity are various transgene constructs that also could be carried by each of the cc-elements (e.g., CRE/LOX components and fluorescent markers appropriate for expressing and analyzing the ability of a single Hox gene to substitute for the normal sets of genes in a given tissue). These cc elements/mutant alleles could be assembled in two generations. Next, in the maintained presence of cas9, they could be combined with two traditional Mendelian alleles (m1 and m2) by cc-ing the Hox mutant alleles into the mutant background. The source of cas9 then could be removed by segregation, resulting in the complex assembly of mutant alleles and transgenes which would now behave according to standard Mendelian rules.
ERACR elements can recall MCRs
ERACRs are designed to delete and replace MCR elements, thereby eliminating cas9 from the genome. These elements carry two gRNAs that target sequences flanking the genomic integration site of a specific MCR element (denoted: <gRNA1; gRNA2>), but differ from other so-called reversal constructs that have been proposed26 in that they do not carry a source of Cas9 (Fig. 3A). ERACRs can be inserted into the genome by providing an exogenous source of Cas9 at the time of injection. When ERACR and MCR stocks are crossed, the gRNAs provided by the ERACR element combine with the Cas9 provided in trans by the MCR element to both delete the MCR and replace it with the ERACR. Importantly, ERACRs cannot spread through wild populations since they lack a Cas9 source, nor do they subject genomes to any Cas9-based mutagenesis. ERACRs also could include dominant markers (e.g., eye-DsRed), recoded gene cassettes that restore gene functions disrupted by insertion of the MCR element, or effector cassettes such as anti-malarial factors as discussed further below (these latter supplemental components are not depicted in Fig. 3A). ERACRs could be employed to eliminate an MCR that might inadvertently spread to an unintended population (e.g., from a pest population into a neighboring or distant indigenous population). In addition, the ability of ERACRs to delete the Cas9 source carried by MCRs would limit the accumulation of unwanted off-target mutations that might accompany the long term presence of an MCR in a population. One potential limitation of ERACRs, which like MCRs should generate a fraction of NHEJ generated lesions, is that some such events will destroy the gRNA cut site and hence prevent clean deletion of the MCR. Since subsequent HDR-mediated copying of the ERACR-resistant MCR could also include adjacent ERACR-induced mutations, such closely linked NHEJ mutations could spread along with MCR into the population. One way to circumvent this problem would be to target the ERACR cut sites far enough from the MCR (~ 1 kb) to prevent or greatly reduce HDR mediated copying of the ERACR-derived NHEJ mutations (e.g., via DNA resection, which typically extends < 500 bp from the double stranded break27).
MCRs can target secondary loci with CHACR elements
CHACRs are <gRNA> constructs that carry one or two gRNAs targeting non-MCR loci. For example, if a prominent off-target MCR site were identified in a population, a CHACR could be designed that clips out the mutation when crossed to the MCR and replaces the altered site with a recoded version of that sequence to repair damage caused by the MCR and to prevent subsequent mutagenesis at that site. After the CHACR had spread throughout the population and performed its reparative task, an ERACR could then be deployed to delete the MCR and restore the genome to a nearly wild-type condition. Although this chain of events may not always proceed with high frequencies (e.g., 95-100%), we note that correcting off-target effects and deleting MCR elements need occur in only a fraction of individuals in order to permit regeneration of a healthy population from a “rescued” minority sub-population via natural selection. CHACRs could also be used as second-site ERACRs by carrying gRNAs targeting Cas9 (Fig. 3C). An advantage of such elements is that they could be used to inactivate all MCRs carrying a given Cas9 isoform, in contrast to ERACRs described above, which are MCR-specific. Another variation would be to incorporate additional gRNAs in the MCR (or CHACR) that cut at sites where desired gene edits are to be made. One could then use such gRNAs to perform standard CRISPR edits of target genes in one strain and then cross it to the MCR strain. In subsequent generations, the gRNAs carried by the MCR (or CHACR) would cut the unedited alleles and HDR would efficiently repair the lesions using the edited (gRNA resistant) locus as a homology template (Fig. 3C). Such edits would then hitchhike with the MCR leading to their linked spread in the population. CHACRs could also carry a gRNA driving its insertion into a locus encoding a component in one pathway and a set of gRNAs targeting other genes acting redundantly in that same pathway or in parallel acting pathways to ensure that the desired process was inactivated (Fig. 3B).
Active genetics can enhance research in model and pioneer organisms
MCRs or split cas9; <gRNA> constructs could be used for a wide variety of applications in both traditional animal and plant model systems as well as in “pioneer organisms” currently lacking genetic tools. In addition, active genetic tools such as versatile “copy-cat” <gRNA> plasmid cloning vectors, which once inserted into the genome can be homozygosed in the presence of a separate cas9 source, should significantly accelerate the assembly of complex arrays of transgenes bypassing Mendelian rules of inheritance in well-developed models. These strategies should permit unprecedented genetic shortcuts enabling combinatorial genetic studies that are infeasible with currently available methods.
Active genetics provides an entry point for functional genomics in pioneer organisms
Our original motivation for conceiving the MCR was the unmet need to conduct functional genomic studies in pioneer organisms. The number of such species with sequenced genomes is growing exponentially, but development and mastery of genetic tools in some novel organisms remains difficult. Pioneer organisms are generally chosen for sequencing based on their informative position at phylogenetic nodes or because they offer particular advantages in a specific area of biology such as aging (e.g., African killifish), neurodegeneration, cancer, unique models for infectious disease (e.g., macaques for HIV, armadillos for leprosy, chinchillas or the hispid cotton rat for various viral infections), specialized behaviors (e.g., genetically-tractable primate models such as mouse lemurs, pigmy marmosets) or other adaptations.
Active genetic approaches offer an obvious avenue for gaining a genetic foothold in these species. Although in many cases it may be possible and more advisable to employ basic CRISPR/Cas9 technology to generate mutations in the germline of pioneer species, we have found that such endeavors can be very challenging, particularly in species without existing transgenesis methods. Thus, MCRs or split cas9; <gRNA> elements, offer potential advantages in generating identifiable homozygous mutations in G1 progeny. For full MCR elements, mutations could be generated in a single step bypassing any need for other transgenesis methods, while in the case of split cas9; <gRNA> configurations it would take two steps (i.e., first obtaining strains expressing a source of Cas9) and then injecting the <gRNA> construct into such backgrounds. The split cas9; <gRNA> option might be the best method to employ in species for which there is a serious concern of escape into wild populations (see Box 1). Another important advantage of mutations induced by split cas9; <gRNA> systems is that the <gRNA>-induced mutation could be segregated away from the cas9 transgene at which point it would behave as a simple Mendelian allele that could be used for traditional genetic studies. Full MCR elements may also create standard indel alleles of the locus at an appreciable rate (e.g., ~ 5%) via NHEJ22 that could similarly be segregated away from the MCR.
Box 1. Modeling and Safety considerations.
Modeling CRISPR-drives
As mentioned in the main text, introduction of a few MCR-bearing individuals into a wild-type population should initially result in doubling of the frequency the MCR allele at each generation (Fig. 4F). However, as this process continues, MCR individuals will begin mating with others carrying the allele, and the rate of increase will decline following a logistical growth curve (see figure legend for details). For an initial seeding frequency (c0) = 1%, an ideal MCR (100% allele conversion) could spread through a population in only 10 generations, increasing from 10% to 90% in just over four generations (Fig. 4G,H). Idealized ERACR elements would spread in exactly the same fashion within a uniform population of MCR bearing organisms, resulting in a concomitant reduction and elimination of the MCR (Fig. 4C).
Genomically encoded split cas9; <gRNA> configurations also create a gene-drive by virtue of the fact that the cas9 encoding gene cannot segregate away from the <gRNA>, assuming the gRNA is faithfully copied to the other allele 100% of the time (Fig. 4F). However, the reciprocal event will take place 50% of the time (i.e., one of the two <gRNA> copies will by necessity segregate from the cas9 source). The enforced association of cas9 with one copy of the <gRNA> results in a constant production of new <gRNA> alleles at each generation. We refer to such a copy-cat system as an “allelic pump”, since it pumps out a constant percentage of new alleles at each generation (see figure legend). For this scenario, an initial seeding at 1% would require more than 100 generations for mutant cas9/<gRNA> alleles to introgress completely into a population (Fig. 4G). If seeded at 10%, however, it could spread to ≈ 65% of the population in 10 generations (as compared to ≈ 4 generations for an MCR to spread through 90% of the population) (Fig. 4H). Thus, at high seeding frequencies allelic pumps could in principle spread significantly through a population if unopposed by any form of negative selection.
We note that even standard non-driving forms of genomically-encoded of Cas9 and gRNAs could result in a very weak mutational drive because each time the two elements encounter each other by random assortment, a new allele could be generated at the gRNA cut site (see figure legend). For initial seeding values c0 = g0 = 1%, this would amount to adding only 0.01% alleles/generation. However, if seeded at c0 = g0= 10% it would produce a drive of identical strength to an allelic pump seeded at c0 = g0 = 1% (compare red curve in Fig. 4G with green curve in Fig. 4H). Thus, it may make sense to consider coupled allelic pumps in the same general category as standard CRISPR mutagenesis configurations because they differ only in the effective seeding frequency, which is a quantitative not qualitative distinction. In contrast, MCRs or trans-complementing MCRs represent an entirely different category.
Responsible use of active genetic systems
Currently available sequence data are consistent with no bacterially-derived CRISPR system having ever been mobilized horizontally into a plant or animal genome in nature68. Thus, constructs such as MCRs that are inherently capable of rapid dispersion throughout naïve wild populations of plant and animal species are unprecedented. While it is clear that many other selfish genetic systems have appeared, driven themselves to fixation, and then responded as all fixed genes do to natural selection, prudence should obviously be employed to insure that Cas9-based drives do not gain access to wild populations of organisms that have never adapted to this system22,26,69,70. Recently, we and others published a consensus set of interim suggestions for safe use of active genetic elements in the laboratory71 in which we urge researchers to consider the implications of using active genetic elements and to use all reasonable precautions when embarking on their experiments (Fig. 6). Based on our own experience, we also urge researchers to work with their local Institutional Biosafety Committees (or equivalents) to obtain guidance for how to proceed safely with experiments. Moving forward, we believe it will also be helpful for researchers in specific areas to consult and make consensus recommendations available to bodies such as the National Academies of Sciences (USA) review panel that has been tasked with considering this issue (and related questions regarding the practical implementation of such technologies29,72) and to offer recommendations for appropriate regulation of CRISPR-based gene-drive systems to federal and local agencies. With the available resources and planning it should be possible to enthusiastically and responsibly engage in this exciting new field. Hopefully, the wisdom of the greater scientific community will help establish guidelines for active genetics that maintain a high level of safety while also affording flexibility to exploit the wealth of opportunities that hold such great promise to enhance human welfare.
“Copy-cat” transgenesis vectors can bypass constraints of Mendelian inheritance
The ability to homozygose <gRNA> constructs in a single step in the presence of a Cas9 source opens up fundamental new possibilities for genetically manipulating transgenic constructs and combining them with traditional Mendelian alleles. For example, one application would be to create a set of cloning vectors that we refer to as “copy-cat” (cc) elements. These vectors would harbor a gRNA and flanking homology sequences to guide its insertion into a desired chromosomal location and other standard features such as multiple cloning sites (MCS) and a dominant marker gene (Mrk) for identifying transgenic individuals (Fig. 3D). A modular kit of cc vectors could be generated for any given organism that targets sequences spaced along the various chromosomes to permit the flexible assembly of complex combinations of transgenes (Fig. 3E). cc elements could insert into coding regions of non-essential visible marker genes (e.g., pigment or bristle markers in Drosophila), into regulatory regions of essential genes that direct expression in a non-vital cell type (e.g., a wing-specific cis-regulatory sequence of an essential Drosophila gene), or into fitness neutral sites (e.g., rosa26 in mice) to avoid effects of the transgene insertion site on the sensitive biological systems (e.g., complex neuronal-based behaviors).
Short-cutting classical genetics in model organisms = Active Genetics
As described above, cc elements mobilized by Cas9 could insert a variety of different transgenes at defined loci (Fig. 3E), and then be combined by crossing strains carrying insertions at different sites. The progeny would inherit both transgenes, and then transmit them together to their progeny. cc-elements could also be tailored to insert into loci of interest and generate mutant phenotypes, combining transgenesis with mutagenesis. Once assembled, an array of cc-transgenic elements could be launched onto another set of chromosomes (e.g., that carried traditional sets of Mendelian alleles) in the maintained presence of a Cas9 source, by a process that could be referred to as cc-ing (e.g., example of targeting four Hox genes in Fig. 3F). One could then segregate away the cas9 transgene and exploit traditional stable Mendelian inheritance for experimental analysis of the resulting mutant phenotypes. This facilitated ability to assemble complex arrays of transgenic constructs and traditional alleles should greatly enable research in diverse fields (e.g., optogenetics in neuroscience or drought- or pest-resistance in polyploid crop plants). We note that for these types of applications cc elements would not have to be copied with exceptional efficiency as conversion rates greater than 50% should be more than adequate for recovering the desired allelic combinations.
Active genetics should also facilitate identification of modifier loci for a given trait or phenotype that encode missing components of a pathway. Such loci are typically identified in screens for dominant alleles that alter a reference homozygous mutant phenotype. Thus, a set of candidate interacting strains (which may have been generated in specific genetic backgrounds) could be crossed to the reference mutant to identify alterations (suppressed or enhanced) in that phenotype. If the reference homozygous mutant were generated using Cas9 and a <gRNA> allele in the gene of interest, it would be possible to screen F1 progeny directly for an altered mutant phenotype. To illustrate this strategy, one could cross a cas9; <gRNA> stock to a genome-wide collection of isogenic deletions and screen the progeny for alterations of the <gRNA> phenotype based on heterozygosity for the deleted interval. In contrast, existing genetic strategies would require intercrossing the F1 progeny to generate homozygous recessive mutants, which would necessarily assort the genetic background from the interacting strains in the ensuing F2 progeny, thus confounding analysis.
Employing MCR elements for genetic drives
An autosomal allele is defined as being under genetic drive if more than 50% of the progeny inherit the allele from an individual carrying a single copy of that allele28. A wide variety of genetic elements or symbiotic/parasitic organisms have been identified that generate drive, and are often referred to as selfish genes because they can spread through a population and become fixed (reviewed in29–33). Well-studied examples of such selfish elements or organisms include: chromosomal rearrangements28, transposons34, Medea elements35–37, homing endonuclease genes (HEGs)38–40, maternal-effect lethal underdominant elements41, and the bacterial endosymbiont/parasite Wolbachia42–44. CRISPR-based self-propagating elements such as MCRs22 or similar constructs24 are newcomers to this established selfish DNA realm.
Gene-converting drives can suppress or modify disease vector or pest populations
HEGs act much like MCRs in cutting chromosomes at a specific site and inserting themselves into the break via HDR. Austin Burt has proposed using HEGs as a potential drive mechanism for suppressing insect populations such as mosquitoes (Fig. 4A)45 that serve as vectors of diseases such as malaria, and dengue and chikungunya fevers. Indeed, based on data from the World Health Organization and other sources the Gates Foundation recently estimated that mosquitoes are responsible for more human deaths than any other animal. Burt and collaborators modeled the spread of HEGs targeting essential genes or various classes of genes required for fertility under conditions where the endonuclease was expressed in a germline-specific fashion. They showed that an HEG seeded at a frequency of 1% will rapidly spread through the population until it reaches a stable equilibrium in 12–14 generations. Thus, individuals carrying an HEG transgene targeting an essential locus will initially breed by chance most often with wild-type individuals. Their progeny will carry only a single targeted mutant allele in their somatic cells and hence will be viable. However, as the HEG allele frequency increases due to gene-drive HEG carriers will begin mating with each other. When such unions arise, a quarter of the offspring will inherit two mutant copies of the insertion and die. Eventually, a balance is struck between the HEG-mediated gene-drive and the fitness cost of carrying a lethal allele such that an equilibrium frequency for the HEG allele is reached equal to the efficiency (e) with which the HEG converts the opposing allele (Fig. 4A). If e is close enough to one, an HEG-drive can cause effective suppression (or in more extreme cases, elimination) of a population45.
Figure 4.
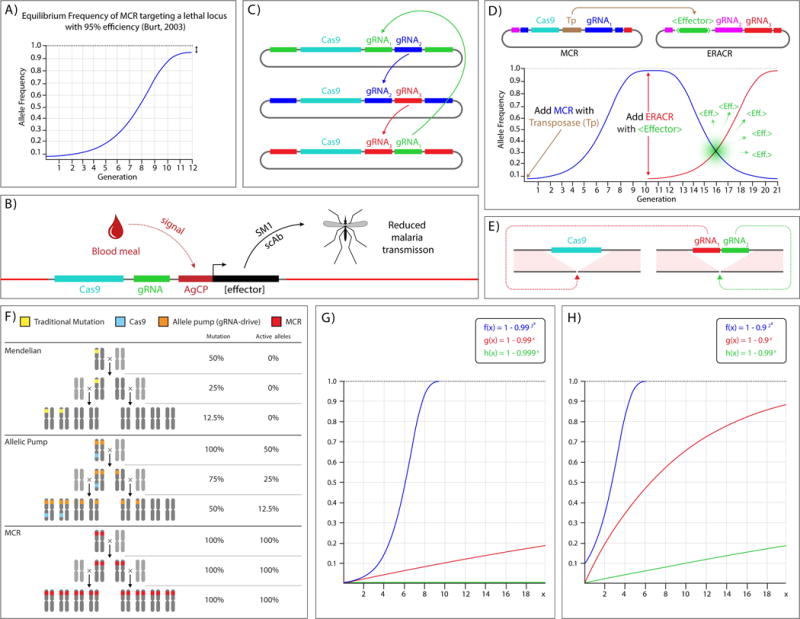
Modeling MCRs, ERACRs, and other <gRNA> elements. A) Modeling of an MCR powered by a germline specific source of cas9 that targets an essential gene based on the modeling of HEGs by Austin Burt. The example assumes that the MCR has a 95% efficiency of conversion (like the y-MCR in Drosophila) = the equilibrium frequency of the MCR allele in that population. B) Application of MCR to attenuate mosquito borne malaria in which an effector cassette encoding the SM1 peptide, which is conditionally activated by a blood meal (AgCP promoter) or a single chain antibody (scFvs) directed against the malarial agent P. falciparum50, is inserted along with core MCR components (Cas9 and gRNA) into a non-coding region of the mosquito genome. The SM1 peptide limits passage of P. falciparum through the gut, a required step in its exploitation of that vector host49. C) Reinforcing MCRs. A set of three mutually reinforcing MCRs. Each MCR carries two gRNAs, one targeting its own insertion site (color of gRNA matches color of homology arms) and a second gRNA targeting the cut site a companion MCR. If each of these elements behave as in the example shown in panel A, when integrated into the genome and released together they should create a sufficient genetic load to drive the population to extinction. Colors of flanking homology regions and gRNAs in the depicted plasmid constructs are matched to indicate which gRNAs direct cleavage at different genomic sites. Arrows summarize redundant patterns of gRNA cleavage that result in two gRNAs from different MCRs cutting at each chromosomal site. D) Top: A coupled pair of MCR and ERACR constructs designed to launch a transposon burst. The MCR carries a Transposase gene (Tp), while the ERACR carries an effector gene cassette <EF> flanked by inverted transposon ends. Bottom: The MCR (blue curve) seeded at 1:100 spreads through the target population following a logistic growth curve in ≈ 10 generations whereupon the ERACR is added. The ERACR (red curve) then spreads with the same dynamics through the MCR population. In individuals carrying both the MCR and ERACR (maximal in gray zone) the Transposase provided by the MCR mobilizes the transposon born effector cassette to new chromosomal sites. This mobilization is restricted to single generation since the ERACR also deletes the MCR. The result is an amplification of the number of effector cassettes in the population and their dispersion to potentially advantageous new genomic locations. E) Trans-complementing <cas9>; <gRNA> which together create a drive system equivalent to that of a single coupled <cas9; gRNA> MCR element. In this scheme, gRNA1 cleaves at the cas9 insertion site and gRNA2 cleaves at the <gRNA1,2> insertion site. F) Scheme depicting two generations of inheritance for a classic Mendelian allele (top), a copy-cat allelic pump consisting of a separated source of cas9 and a <gRNA> (middle), and an MCR (bottom). This logistic growth curve is defined by the second order recursion formula: fn+1 = fn +fn(1− fn) = 2fn − fn2, where fn is the frequency of the MCR in the population at generation n. This formula has the closed form solution f(n) = 1- (1- c0)(2n), where c0 = the seeding frequency of the MCR, which for low values of c0 can be approximated as expected, by the exponential equation f(n) = c02n. G) Time course of accumulated mutant alleles resulting from 1:100 seeding of an MCR (blue curve), a cas9; <gRNA> allelic pump (red curve), and a standard cas9; gRNA encoding transgenes green curve (buried in the baseline). The additive copy-cat drive can be modeled by the first order recursion formula: fn = fn-1 + c0(1-fn-1) where c0 = g0 (initial fractions of cas9 and gRNAs in the population). The closed form solution for this equation is f(n) = 1- (1- c0)n, which for low values of c0 = g0 can be approximated by the linear equation f(n) = c0(n). For comparison, the standard mutational drive can be represented by fn = fn-1 + c0g0(1-fn-1), which has the closed form solution f(n) = 1- (1- c0g0)n (≈ c0g0(n) for c0 and g0 <<1). H) Same as in panel G but with a seeding ratio of 1:10. Note that the allelic pump in G (red curve) has precisely the same behavior as the standard cas9; gRNA combination in H (green curve). Note that the growth curve for the copy-cat allelic pump seeding at c0 = g0 = 1% is identical to that of the standard non-drive mutagenesis scheme seeded at c0 = g0 = 10% (asterisks indicate equal endpoints).
Burt, colleagues, and others have since modeled a wide variety of scenarios for HEG-mediated gene-drives and reached several interesting conclusions, including: 1) targeting genes causing female sterility or grandchildless phenotypes is more effective than targeting essential genes for eradicating a population45,46, 2) targeting multiple sites with HEGs should provide more reliable suppression than a single element39, 3) low density populations are more prone to suppression than high density populations (fortunately many mosquito species carrying malaria are found in relatively low density)47, and 4) an aggressive HEG can lead to local elimination of an isolated pocket of a population before it can spread to the full population and thereby burn itself out47 (e.g., like highly-virulent forms of Ebola virus48).
Advantages of MCR-mediated gene-drives
An MCR element in which the cas9 source is expressed in a germline specific fashion should behave exactly as an HEG-drive. Thus, the pioneering modeling of HEG dynamics by Burt and colleagues can be directly applied to MCRs. The great benefit offered by an MCR is that it can be targeted to virtually any locus to generate either null or tissue-specific mutations in a target gene. In addition, one can select gRNA target sequences unique to a species within a closely related clade to greatly reduce the risk of inadvertent horizontal gene transfer. Since mosquitoes, like flies, are dipteran (two winged) insects, it is perhaps not surprising that MCRs can spread efficiently through mosquito populations23 (see below) as originally observed in Drosophila22. Likewise, MCRs would be expected to function efficiently in various invasive fly species to help restore ecologies to their native state and reduce associated agriculture damage.
In collaboration with Anthony James at UCI we recently tested the feasibility of a gene-drive strategy in mosquitoes23 by generating an MCR that carries one of several well-studied effector gene cassettes capable of blocking transmission of the malarial parasite Plasmodium falciparum49–51 (Fig. 4B). This kh-MCR targets insertion into an eye pigmentation locus (kynurenine hydroxylase = kh = cinnabar in Drosophila) in the Asian vector, Anopheles stephensi50. Previous studies by the James group have shown that the blood-meal inducible gene cassette carried by the ~ 17kb kh-MCR expresses two single-chain antibodies that block different steps of the parasite life cycle and are 100% effective in preventing propagation of P. falciparum in mosquitoes50,51. Since the kh-MCR propagates to 99.5% of progeny via both the male and female germline, a similar MCR targeted to one of several characterized fitness neutral loci50,51 should provide a potential strategy for sustainable malaria control. A potential add-on to this system would be for the MCR to carry an additional gRNA(s) targeting one of several host loci52–54 required for parasite transmission for either mutagenesis or editing (see above).
There are several advantages to using effector-bearing MCRs that target fitness neutral sites. First, such strategies should have the smallest possible ecological impact because their only effect is to block parasite transmission and not to harm the mosquito population. Second, the absence of a fitness handicap will allow isolated pockets of MCR mosquitoes to persist until they can disperse and mate with adjacent connected populations. Recall that modeling of lethal HEGs indicated that aggressive elements were subject to elimination in this type of scenario, particularly when the mosquito population density is low47. In contrast, fitness neutral MCR-modifier vectors, in principle, should be much more likely to spread smoothly through areas with uneven or locally disconnected population distributions.
Coupled MCR/ERACR/transposon systems could reinforce drive or amplify effector delivery
As Burt and colleagues pointed out with regard to population suppression with HEGs, deploying more than one such element can greatly increase the probability of success39. This same strategy could help overcome a potential weakness with MCRs such as described above that are designed to target fitness neutral loci because such elements are likely to generate MCR-resistant alleles via NHEJ at some frequency (~5% in our experiments with the y-MCR in Drosophila). Also, mutations could arise in MCR components that eliminate either Cas9 or gRNA function. One multiplicative strategy would be to generate a series of several reinforcing MCRs, each carrying two gRNAs: one that targets the site at which the MCR integrates and the other targeting the insertion site of a companion MCR (Fig. 4C). Such mutually reinforcing MCRs should virtually never fail to propagate through a population because at least one of them should be transmitted to nearly all progeny of an MCR parent mated to a wild-type individual. In addition, because of cross-reinforcement, such multiplicative MCRs should be relatively invulnerable to mutations in either cas9 (present at three different genomic sites) or the gRNAs (present at two distinct genomic sites).
It should also be possible to make use of a combination of MCRs, ERACRs, and transposons to broadly disseminate multiple copies of effector gene cassettes. For example, in the scheme depicted in Fig. 4D, an MCR carries a copy of a transposase gene (e.g., P-transposase Δ2–3) while a matched ERACR carries a desired effector cassette flanked by corresponding transposon ends. The idea would be first to release the MCR, and allow it to spread broadly throughout the population. These animals will not express the effector genes. Subsequently animals carrying the ERACR, which allows the expression of the effector gene, would be released. When an ERACR encounters an MCR the transposase encoded by the MCR should mobilize transposition of the effector cassette carried between the transposon ends. Because the ERACR also deletes the MCR, transposition should only take place for one generation, thereby creating a singular burst of transposon mobilization peaking at the point where the frequencies of the ERACR and MCR are equal. While this idealized scenario makes several assumptions, such as a higher relative rate of transposition versus deletion of the transposon55,56, in principle, it should increase the copy number of effector cassettes in the genome. In addition, transposon insertions could sample new loci for effective transgene expression, while deleterious insertions would be eliminated by natural selection.
Trans-complementing MCR-drives offer advantages over single-unit elements
Another variation on the theme of Cas9-drives, which offers potential husbandry advantages, is to have two separate trans-complementing drives for cas9 <cas9> and the gRNAs <gRNA1; gRNA2> wherein gRNA1 directs cleavage at the site of cas9 genomic insertion while gRNA2 cuts at the integration site of the <gRNA1; gRNA2> element (Fig. 4E). Since neither of the two constructs alone constitutes a drive, each single element could be propagated safely as a separate stock. When the two stocks are crossed (possibly after amplification of each of the stocks for release purposes) to test (or release) a full drive should result. In progeny of this cross the resulting <cas9>; <gRNA1; gRNA2> would combine to create a drive that should behave thereafter as a linked <cas9; gRNA> MCR. One additional advantage of such trans-complementing MCR-drives is that each of the two constructs could carry the same or different effector cassette, resulting in the former case to expression of four copies of a cassette, thereby doubling the levels of transgene expression as compared to that provided by a single cis-linked <cas9; gRNA> MCR element.
Potential applications of “active genetics” to human gene therapy
All of the examples of active genetics or gene-drives discussed above involve the spread of an MCR construct to offspring via the germline. The dissemination of MCR constructs might also be achieved between cells within an individual by coupling these elements to a viral delivery system. In such cases, the somatic spread of an MCR element could be exploited by targeting its insertion into such unique sequences. In principle, this approach might be used to fight any disease that results in specific alterations in genome sequence. We consider two such examples, using MCRs to target the HIV reservoir pool and selectively targeting cancer cells marked by distinguishing DNA sequence signatures.
An MCR strategy could be used to target the HIV reservoir
Retroviruses such as HIV insert into the host genome. Thus, it should be possible to engineer an MCR element that directs its insertion into the HIV Integrase gene and replaces its function with Cas9/gRNA-mediated insertion (Fig. 5A). If a construct of this kind were designed such that the Cas9 and gRNAs could be packaged within HIV viral particles, then the virus should be able to infect all CD4+ cells, but only integrate into those carrying an HIV provirus in the genome. Virus produced by such targeted MCR elements could then replicate and spread to other helper T-cells, but would only integrate into those with a proviral insertion. This process should continue until all cells carrying the provirus in their genome were neutralized. A caveat for this approach is that HIV reservoir cells are thought to be quiescent57 while HDR-mediated allelic conversion most likely requires DNA replication58–60. However, there are methods for inducing reservoir cells to re-enter the cell cycle61,62, which then may allow the conceptual chain of events described above to proceed. Another potential barrier to this type approach is that MCR mediated allelic conversion may be significantly less efficient in somatic cells than in the germline, as we believe to be the case in mosquitoes23. As mentioned above, NHEJ generated alleles once generated will often destroy the gRNA target site thereby precluding subsequent HDR-mediated gene conversion. Nonetheless, NHEJ generated mutations in an integrase gene would at least neutralize that particular proviral element. Efficient propagation of such viruses, however, may require development of methods to increase HDR-mediated gene copying such as suppression of NHEJ via silencing of key pathway components (e.g., Ku70-RNAi) or the use of alternative Cas9-related enzymes such as the recently characterized Cpf163, which cuts at a distance from its DNA recognition sequence thereby potentially permitting iterative rounds of NHEJ mutagenesis without destroying the gRNA-recognition sequence required for HDR.
Figure 5.
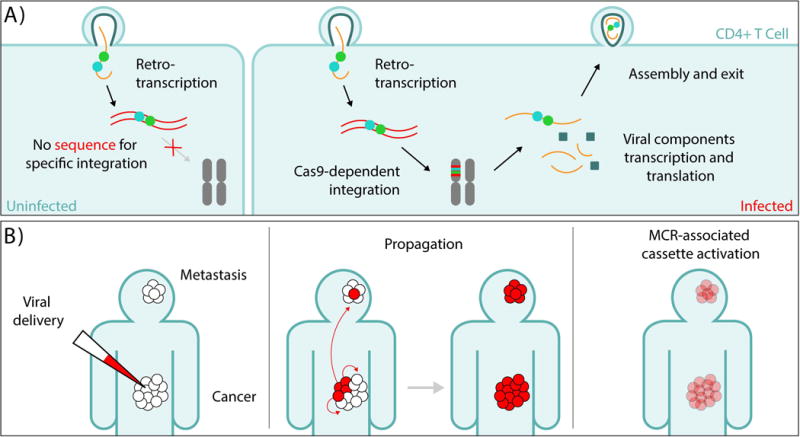
Potential applications of MCR technology to gene therapy. A) MCR-based spread of an Integrase-deficient Cas9/gRNA-dependent retroviral (e.g., HIV) construct directing its insertion into a chromosomal inserted provirus thereby rendering that proviral element inactive (e.g., reference73). Induction and maturation of such targeted proviruses should lead to the production of assembled viruses which could then infect all other CD4+ helper T-cells but only integrates into the genomes of cells carrying proviral insertions. This within-organism spread of the MCR construct could eventually incapacitate all proviruses leading to the eventual clearance of the HIV infection. B) An analogous retro-virally propagated MCR element directs its insertion into a cancer-specific genomic sequence. Infection and spread of this element throughout the body should lead to its selective insertion in cancer cells (in primary and metastatic tumors). When testing of patient cells indicates that the MCR has spread effectively to all cancer cells, an effector cassette carried by the MCR could be activated (e.g., by a hormone) to induce apoptosis or flag cells for destruction by the immune system.
MCR vectors might selectively target cancer cells
MCRs designed to spread among cells in the body might also be developed that target nucleotide differences between the cancer cell and normal cells, which can now be rapidly detected by deep sequencing64,65. While this approach may not be feasible for all types of cancer, those in which cancer-cell specific sequences can be identified, (e.g., chromosomal rearrangements) could be targeted by a construct comprising a cancer-specific gRNA carried by an MCR packaged in an Integrase-deficient retrovirus or adenovirus. Such an MCR-viral construct should infect both normal and cancer cells in the patient, but could only insert into the genome of cancer cells (Fig. 5B). If such an element were engineered to replicate and spread from cell-to-cell, an initial infection of only a small subset of cancer cells should result in spread of the MCR-virus until the great majority of cancer cells contained the construct even if the primary tumor had metastasized. Once MCR-viral delivery had become widespread among cancer cells, drug-inducible effectors (toxins, agents triggering apoptosis, or cellular antigens flagging cells for immune recognition) carried by the MCR could be activated. Again, as mentioned above, for these types of applications it may first be necessary to develop methods to increase the frequency of HDR-mediated gene copying in somatic cells. For such applications, it would also be important to use various means (e.g., careful gRNA target selection or nickase forms of Cas9) to reduce off-target effects to the lowest possible levels to avoid unintended secondary consequences of the therapy, particularly when the strategy is not to eliminate targeted cells.
Conclusions
We believe that the dawn of active genetic holds enormous promise to improving human welfare by accelerating research, combating disease, restoring the environment, and improving agriculture. At the same time, however, it is important to keep the unprecedented power of such systems on a tight leash.
The promise of active genetics
We expect that most applications of active genetic methods will employ various split cas9; <gRNA> copy-cat systems. These elements could be used for a broad variety of purposes such as: a novel system for transgenesis, inducing and combining mutations to test for cumulative or interacting effects, or assembling complex arrays of transgenes and traditional Mendelian alleles. Also, full MCR-related elements should serve as potent drive systems to disseminate effector transgenes through populations to combat insect-borne diseases or invasive species, and potentially allowing dispersal of gene therapy vectors throughout the human body targeting them to diseased cells. Given the explosive pace of CRISPR-related research in the last two years there will undoubtedly be continual waves of innovation that use these and new tools in yet unimagined ways.
Looking forward
How active genetics will evolve over the next decades is difficult to predict. In the immediate future, combining components from distinct CRISPR systems and other existing tools (e.g., transposons, ϕ31C, FLP/FRT, CRE/LOX, GAL4/UAS, LexA, Q-systems, and the wealth of such compounded tools in Drosophila66,67) should stimulate a flurry of innovation in genome engineering. Eventually, fusion of genome engineering with synthetic biology may allow transplantation or replacement of large chromosome segments from one organism into another. Such advances should result in a fuller understanding of developmental processes and how they evolve and ultimately permit the rational design of beneficial traits based on first principles. We hope that this exciting journey will be charted in a judicious and ethical fashion.
Figure 6.
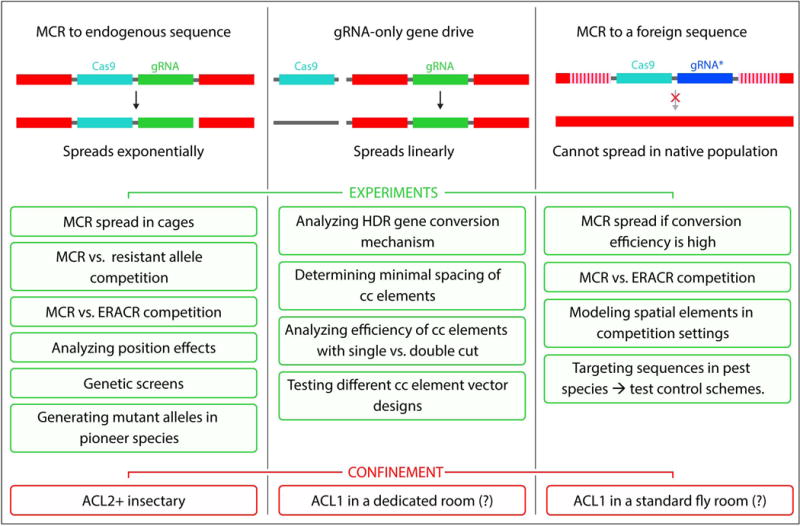
Biosafety options for sample experiments for different active genetic elements: Top: Schemes depicting an MCR targeting an endogenous sequence (left), a split cas9; <gRNA> allelic pump (middle), and an MCR targeting an exogenous sequence (right). Bottom: Types of experiments and recommended physical confinement strategies suitable for each type of active element. ACL = Arthropod Containment Levels. ACL1 corresponds to containment of arthropods judged to present a BioSafety Level 1 (BSL1) concern, which applies to standard laboratory organisms (e.g., flies or harmless strains of E. coli used for cloning) while ACL2 applies to insect vectors carrying BSL2 rated pathogens (e.g., mosquitoes carrying malarial parasites or tsetse flies carrying trypanosomes). For relevant current federal regulations on recombinant DNA and confinement procedures see references74–76. Question marks indicate tentative suggested levels of confinement for the different drive configurations.
Acknowledgments
We would like to thank Tony James, Hugo Bellen, Shinya Yamamoto, David Li-Kroeger, Fred Gould, Bill McGinnis, Steve Wasserman, Steve Hedrick, Steve Briggs, Marty Yanofsky, Annabel Guichard and the anonymous reviewers for helpful discussions and comments on the manuscript.
Glossary
- Active genetics
Genetic manipulations in which a genetic element is copied from one chromosome to the identical insertion site on the sister chromosome using Cas9 and gRNA elements (e.g., MCRs or split cas9; <gRNA> drives).
- Mutagenic Chain Reaction (MCR)
Method by which a cassette encoding Cas9 and a gRNA is inserted precisely into the gRNA cut site
- MCR elements
DNA constructs consisting of a Cas9/gRNA cassette flanked by homology arms that precisely abut the gRNA cut site. A shorthand for a given MCR can be denoted <cas9; gRNA> wherein the brackets denote the flanking homology arms.
- Element for Reversing the Autocatalytic Chain Reaction (ERACR)
A DNA construct consisting of two gRNAs that cut genomic sequences flanking an MCR construct. The gRNA construct is flanked by chromosomal homology arms that respectively abut the two gRNA cut sites. An important feature of the ERACR is that is does not carry a source of Cas9. When a stock carrying an ERACR is crossed to one carrying the targeted MCR, the Cas9 provided by the MCR results in the ERACR deleting the MCR and copying itself in place of the MCR. An ERACR could be denoted as <gRNA1; gRNA2> wherein gRNA1 cuts on one side of the MCR and gRNA2 cuts on the other side.
- Construct for Hitchhiking on the Autocatalytic Chain Reaction (CHACR)
A DNA construct similar to an ERACR in that it carries only gRNAs flanked by precisely abutting homology arms. It differs from an ERACR in that the gRNAs target insertion (a single gRNA) or insertion/deletion (two gRNAs) into a genomic site distinct from that of the MCR. In addition, CHACRs can carry gRNAs that drive edited genetic changes at a given genomic site or target loci for mutagenesis by NHEJ
- Split cas9; <gRNA>
A configuration in which a cas9 transgene inherited in a standard Mendelian fashion is combined with a gRNA flanked by homology arms (denoted as <gRNA>). In this situation, only the <gRNA> element is actively copied to the other chromosome
- Allelic pump
Since the combination of a traditional Mendelian source of cas9 and a <gRNA> results in the production of a constant new number of <gRNA> alleles at each generation we refer to this configuration as an allelic pump
- Copy-cat (cc) cloning vectors
Plasmid cloning vectors that in addition to having standard features (e.g., origin of replication, antibiotic resistance genes, multiple cloning sites) also carry a gRNA flanked by homology arms that direct insertion of the element into defined locations. Transgenes inserted into cc vectors can be readily rendered homozygous by providing a source of cas9 in trans
- Genetic drive
An allele of a diploid gene experiences genetic drive if it is inherited more than 50% of the time (i.e., more than by random chance alone)
- Effector gene cassette
A transgene encoding a protein that when expressed exerts a desired effect (e.g., anti-malarial peptides expressed following a blood meal in mosquitoes or a drug inducible cell lethal gene in a cancer cell)
References
- 1.Barrangou R. The roles of CRISPR-Cas systems in adaptive immunity and beyond. Curr Opin Immunol. 2015;32C:36–41. doi: 10.1016/j.coi.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci. 2014;71(3):449–65. doi: 10.1007/s00018-013-1438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12(5):317–26. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 4.Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–111. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassett AR, Liu JL. CRISPR/Cas9 and genome editing in Drosophila. J Genet Genomics. 2014;41(1):7–19. doi: 10.1016/j.jgg.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 2014;23(R1):R40–6. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 7.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sternberg SH, Doudna JA. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell. 2015;58(4):568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 10.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 11.Overcash JM, Aryan A, Myles KM, Adelman ZN. Understanding the DNA damage response in order to achieve desired gene editing outcomes in mosquitoes. Chromosome Res. 2015;23(1):31–42. doi: 10.1007/s10577-014-9450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5(5):a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aylon Y, Kupiec M. DSB repair: the yeast paradigm. DNA Repair (Amst) 2004;3(8–9):797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5(11):a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195(3):715–21. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A. 2014;111(29):E2967–76. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebo ZL, Lee HB, Peng Y, Guo Y. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly (Austin) 2014;8(1):52–7. doi: 10.4161/fly.26828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A. 2013;110(47):19012–7. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gratz SJ, Wildonger J, Harrison MM, O’Connor-Giles KM. CRISPR/Cas9-mediated genome engineering and the promise of designer flies on demand. Fly (Austin) 2013;7(4):249–55. doi: 10.4161/fly.26566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196(4):961–71. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Z, Chen H, Liu J, Zhang H, Yan Y, Zhu N, Guo Y, Yang B, Chang Y, Dai F, et al. Various applications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome. Biol Open. 2014;3(4):271–80. doi: 10.1242/bio.20147682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantz VM, Bier E. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science. 2015;348(6233):442–4. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gantz V, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito, Anohpeles stepensi. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1521077112. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. RNA-guided gene drives can efficiently bias inheritance in wild yeast. bioRxiv. 2015 [Google Scholar]
- 25.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4(1):220–8. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. Elife. 2014:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do AT, Brooks JT, Le Neveu MK, LaRocque JR. Double-strand break repair assays determine pathway choice and structure of gene conversion events in Drosophila melanogaster. G3 (Bethesda) 2014;4(3):425–32. doi: 10.1534/g3.113.010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtis CF. Possible use of translocations to fix desirable genes in insect pest populations. Nature. 1968;218(5139):368–9. doi: 10.1038/218368a0. [DOI] [PubMed] [Google Scholar]
- 29.James AA. Gene drive systems in mosquitoes: rules of the road. Trends Parasitol. 2005;21(2):64–7. doi: 10.1016/j.pt.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Burt A. Heritable strategies for controlling insect vectors of disease. Philos Trans R Soc Lond B Biol Sci. 2014;369(1645):20130432. doi: 10.1098/rstb.2013.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alphey L, McKemey A, Nimmo D, Neira Oviedo M, Lacroix R, Matzen K, Beech C. Genetic control of Aedes mosquitoes. Pathog Glob Health. 2013;107(4):170–9. doi: 10.1179/2047773213Y.0000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall JM. The effect of gene drive on containment of transgenic mosquitoes. J Theor Biol. 2009;258(2):250–65. doi: 10.1016/j.jtbi.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 33.Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7(6):427–35. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- 34.Skipper KA, Andersen PR, Sharma N, Mikkelsen JG. DNA transposon-based gene vehicles - scenes from an evolutionary drive. J Biomed Sci. 2013;20:92. doi: 10.1186/1423-0127-20-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward CM, Su JT, Huang Y, Lloyd AL, Gould F, Hay BA. Medea selfish genetic elements as tools for altering traits of wild populations: a theoretical analysis. Evolution. 2011;65(4):1149–62. doi: 10.1111/j.1558-5646.2010.01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CH, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BA. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316(5824):597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 37.Akbari OS, Chen CH, Marshall JM, Huang H, Antoshechkin I, Hay BA. Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth Biol. 2014;3(12):915–28. doi: 10.1021/sb300079h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alphey N, Bonsall MB. Interplay of population genetics and dynamics in the genetic control of mosquitoes. J R Soc Interface. 2014;11(93):20131071. doi: 10.1098/rsif.2013.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deredec A, Godfray HC, Burt A. Requirements for effective malaria control with homing endonuclease genes. Proc Natl Acad Sci U S A. 2011;108(43):E874–80. doi: 10.1073/pnas.1110717108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, Hovde BT, Baker D, Monnat RJ, Jr, Burt A, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473(7346):212–5. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akbari OS, Matzen KD, Marshall JM, Huang H, Ward CM, Hay BA. A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr Biol. 2013;23(8):671–7. doi: 10.1016/j.cub.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasgon J. Population replacement strategies for controlling vector populations and the use of Wolbachia pipientis for genetic drive. J Vis Exp. 2007;5:225. doi: 10.3791/225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, Magori K, Lloyd AL, Gould F. Introducing transgenes into insect populations using combined gene-drive strategies: modeling and analysis. Insect Biochem Mol Biol. 2007;37(10):1054–63. doi: 10.1016/j.ibmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinkins SP, Godfray HC. Use of Wolbachia to drive nuclear transgenes through insect populations. Proc Biol Sci. 2004;271(1546):1421–6. doi: 10.1098/rspb.2004.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003;270(1518):921–8. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deredec A, Burt A, Godfray HC. The population genetics of using homing endonuclease genes in vector and pest management. Genetics. 2008;179(4):2013–26. doi: 10.1534/genetics.108.089037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.North A, Burt A, Godfray HC, Buckley Y. Modelling the spatial spread of a homing endonuclease gene in a mosquito population. J Appl Ecol. 2013;50(5):1216–1225. doi: 10.1111/1365-2664.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345(6202):1369–72. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417(6887):452–5. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- 50.Isaacs AT, Jasinskiene N, Tretiakov M, Thiery I, Zettor A, Bourgouin C, James AA. Transgenic Anopheles stephensi coexpressing single-chain antibodies resist Plasmodium falciparum development. Proc Natl Acad Sci U S A. 2012;109(28):E1922–30. doi: 10.1073/pnas.1207738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isaacs AT, Li F, Jasinskiene N, Chen X, Nirmala X, Marinotti O, Vinetz JM, James AA. Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi. PLoS Pathog. 2011;7(4):e1002017. doi: 10.1371/journal.ppat.1002017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Wang X, Zhang G, Githure JI, Yan G, James AA. Genome-block expression-assisted association studies discover malaria resistance genes in Anopheles gambiae. Proc Natl Acad Sci U S A. 2013;110(51):20675–80. doi: 10.1073/pnas.1321024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bryant B, Raikhel AS. Programmed autophagy in the fat body of Aedes aegypti is required to maintain egg maturation cycles. PLoS One. 2011;6(11):e25502. doi: 10.1371/journal.pone.0025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corby-Harris V, Drexler A, Watkins de Jong L, Antonova Y, Pakpour N, Ziegler R, Ramberg F, Lewis EE, Brown JM, Luckhart S, et al. Activation of Akt signaling reduces the prevalence and intensity of malaria parasite infection and lifespan in Anopheles stephensi mosquitoes. PLoS Pathog. 2010;6(7):e1001003. doi: 10.1371/journal.ppat.1001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee YC, Langley CH. Transposable elements in natural populations of Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 2010;365(1544):1219–28. doi: 10.1098/rstb.2009.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maside X, Assimacopoulos S, Charlesworth B. Rates of movement of transposable elements on the second chromosome of Drosophila melanogaster. Genet Res. 2000;75(3):275–84. doi: 10.1017/s0016672399004474. [DOI] [PubMed] [Google Scholar]
- 57.Siliciano JD, Siliciano RF. HIV-1 eradication strategies: design and assessment. Curr Opin HIV AIDS. 2013;8(4):318–25. doi: 10.1097/COH.0b013e328361eaca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maher RL, Branagan AM, Morrical SW. Coordination of DNA replication and recombination activities in the maintenance of genome stability. J Cell Biochem. 2011;112(10):2672–82. doi: 10.1002/jcb.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431(7011):1011–7. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49(5):872–83. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Mousseau G, Mediouni S, Valente ST. Targeting HIV Transcription: The Quest for a Functional Cure. Curr Top Microbiol Immunol. 2015;389:121–45. doi: 10.1007/82_2015_435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sgarbanti M, Battistini A. Therapeutics for HIV-1 reactivation from latency. Curr Opin Virol. 2013;3(4):394–401. doi: 10.1016/j.coviro.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 63.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015 doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heuckmann JM, Thomas RK. A new generation of cancer genome diagnostics for routine clinical use: overcoming the roadblocks to personalized cancer medicine. Ann Oncol. 2015 doi: 10.1093/annonc/mdv184. [DOI] [PubMed] [Google Scholar]
- 65.Cronin M, Ross JS. Comprehensive next-generation cancer genome sequencing in the era of targeted therapy and personalized oncology. Biomark Med. 2011;5(3):293–305. doi: 10.2217/bmm.11.37. [DOI] [PubMed] [Google Scholar]
- 66.Venken KJ, Bellen HJ. Chemical mutagens, transposons, and transgenes to interrogate gene function in Drosophila melanogaster. Methods. 2014;68(1):15–28. doi: 10.1016/j.ymeth.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venken KJ, Bellen HJ. Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and PhiC31 integrase. Methods Mol Biol. 2012;859:203–28. doi: 10.1007/978-1-61779-603-6_12. [DOI] [PubMed] [Google Scholar]
- 68.Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42(4):2577–90. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oye KA, Esvelt K, Appleton E, Catteruccia F, Church G, Kuiken T, Lightfoot SB, McNamara J, Smidler A, Collins JP. Biotechnology. Regulating gene drives. Science. 2014;345(6197):626–8. doi: 10.1126/science.1254287. [DOI] [PubMed] [Google Scholar]
- 70.Lunshof J. Regulate gene editing in wild animals. Nature. 2015;521(7551):127. doi: 10.1038/521127a. [DOI] [PubMed] [Google Scholar]
- 71.Akbari BO, Bellen HJ, Bier E, Bullock SL, Burt A, Church GM, Cook KR, Duchek P, Edwards OR, Esvelt KM, et al. Safeguarding gene drive experiments in the laboratory. Science. 2015 doi: 10.1126/science.aac7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramsey JM, Bond JG, Macotela ME, Facchinelli L, Valerio L, Brown DM, Scott TW, James AA. A regulatory structure for working with genetically modified mosquitoes: lessons from Mexico. PLoS Negl Trop Dis. 2014;8(3):e2623. doi: 10.1371/journal.pntd.0002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Committee A-AD. Containment Guidelines (ver 3.1) The American Committee of Medical Entomology of the American Society for Tropical Medicine and Hygiene; 2001. [Google Scholar]
- 75.Health USDoHaHSPHSCfDCaPNIo. Biosafety in Microbiological and Biomedical Laboratories. 5th. 2009. (HHS Publication No. (CDC) 21-1112). [Google Scholar]
- 76.Services DoHaH. NIH guidelines for research involving recombinant or synthetic nucleic acid molecules. HHS; 2013. [Google Scholar]


