Abstract
Malignant tumors reprogram cellular metabolism to support cancer cell proliferation and survival. Although most cancers depend on a high rate of aerobic glycolysis, many cancer cells also display addiction to glutamine. Mounting evidence indicates key roles of glutamine transporters and glutaminase activity in cancer glutamine metabolism. Here, we show that the EphA2 receptor tyrosine kinase activates YAP and TAZ (YAP/TAZ), transcriptional co-activators of the TEAD family of transcription factors, to promote glutamine metabolism in models of HER2-positive breast cancer. EphA2 overexpression induced nuclear accumulation of YAP and TAZ and increased expression of YAP/TAZ target genes. Inhibition of Rho or ROCK kinase abolished EphA2-dependent YAP/TAZ nuclear localization, suggesting Rho signaling as a critical intermediary. Silencing of YAP or TAZ significantly reduced intracellular glutamate, through differential regulation of SLC1A5 and GLS, respectively. Indeed, the regulatory DNA elements of both SLC1A5 and GLS contain TEAD binding sites and were bound by TEAD4 in an EphA2-dependent manner. In human breast cancer, EphA2 expression positively correlates with YAP and TAZ, as well as GLS and SLC1A5. While high expression of EphA2 predicts enhanced metastatic potential and poor survival, increased EphA2 expression rendered HER2-positive breast cancer cells more sensitive to glutaminase inhibition. Together, these findings define a novel mechanism of EphA2-mediated YAP/TAZ activation to promote glutaminolysis through upregulation of GLS and SLC1A5 in HER2+ breast cancer.
Introduction
Receptor tyrosine kinases (RTKs) serve as regulators of key signaling pathways that promote cell growth, proliferation, and migration and are commonly increased in abundance and/or activity in human cancers (1). As a member of the largest family of RTKs, the Eph receptors, EphA2 is frequently overexpressed in human cancer, with enhanced EphA2 activity correlating with tumor progression and reduced survival (2–5). However, unlike other RTKs, cumulative evidence supports two modes of EphA2 signaling. In the first mode, ligand-dependent EphA2 forward signaling in normal cells suppresses cell proliferation and invasiveness, often resulting from cell-cell interactions. In the second mode, due to reduced ligand binding or transactivation through another RTK, such as the Epidermal Growth Factor Receptor (EGFR) or HER2, ligand-independent EphA2 signaling promotes tumor malignancy in part through activation of the Rho-GTPase (3, 5–9). Indeed, EphA2 is overexpressed whereas its ligands, including ephrin-A1, are often less abundant in malignant human breast cancer specimens, correlating with poor survival and resistance to therapeutic drugs (3, 4, 10, 11). In parallel to the role of EphA2 in human breast cancer, ablation of EphA2 inhibits metastatic progression in the murine HER2-dependent mammary tumor model MMTV-Neu (5).
Similar to many RTK signaling pathways, the Hippo tumor suppressor pathway also controls cell proliferation, with dysregulation often contributing to breast tumorigenesis. Like EphA2, the Hippo pathway represents a complex signaling cascade, differentially responding to various signals. While cell-cell contact activates Hippo signaling to suppress cell growth and proliferation, growth factor signaling deactivates the Hippo pathway, supporting tumor growth and progression (12–15). Cell growth and proliferation result from increased transcription due to nuclear accumulation and activation of the Yes-associated protein (YAP) and WW domain containing Transcription Regulator 1 (WWTR1 or TAZ), transcriptional co-activators that act as the primary downstream effectors of the Hippo pathway. In the nucleus, YAP and TAZ primarily increase pro-proliferative gene expression through interaction with the TEAD family of transcription factors (16–19). Phosphorylation by the large tumor suppressor kinases 1 and 2 (LATS1/2) or additional unidentified kinases suppress YAP and TAZ activity through 14-3-3-dependent nuclear export (20–22), with YAP phosphorylation at Ser127 and Ser397 resulting in cytoplasmic retention and proteasomal degradation, respectively (23–25), while TAZ is primarily degraded upon phosphorylation at Ser89 (26). Growth factors and defects in Hippo signaling decrease phosphorylation and promote nuclear accumulation of YAP and TAZ in human breast cancer (27–30), supporting RTK signaling as potential regulators of Hippo and YAP/TAZ activity. Nonetheless, no specific RTK has been identified as an upstream mediator of YAP/TAZ activity.
Accumulating evidence supports a role of RTK signaling in regulating metabolic processes. While many tumors primarily depend on glycolysis to generate ATP and other biomolecules necessary for an enhanced proliferative phenotype under hypoxic conditions, breast cancer cells also utilize glutamine for biogenesis of nucleotides, lipids, and amino acids, generation of cellular anti-oxidant glutathione, as well as serving as a mitochondrial substrate (2, 31, 32). Increased glutamate concentrations in breast tumors are associated with invasiveness and drug resistance and correlate with increased risk of recurrence (33, 34), suggesting enhanced glutamine metabolism may support tumor survival and therapeutic evasion. The dependence of breast tumors on glutamine metabolism relies on glutamine uptake, primarily from the neutral amino acid transporter SLC1A5 (35, 36), as well as utilization of glutamine by glutaminase (GLS) through conversion to the glutamate metabolite (37). Among breast cancer subtypes, the HER2-positive subtype is one of the most reliant on glutamine metabolism, likely resulting from HER2-dependent upregulation of GLS and SLC1A5 (38, 39). We have previously shown that EphA2 promotes glutamine metabolism to support tumor growth in a HER2-positive breast cancer model (2); however, the mechanism of EphA2-dependent metabolism has not been elucidated.
Here, we demonstrate that the RTK EphA2 promotes glutamine metabolism by activating the transcriptional co-activators YAP and TAZ. First, we report that EphA2 overexpression increased nuclear localization and activation of YAP and TAZ in cells overexpressing HER2 or the rat ortholog Neu as well a HER2-positive breast cancer mouse model (MMTV-Neu). Next, we confirmed that active Rho and its downstream Rho-associated protein kinase, ROCK, were required for EphA2-mediated activation of YAP and TAZ (YAP/TAZ). Thirdly, YAP and TAZ are shown to enhance glutamine metabolism through differential transcriptional regulation of the glutaminolysis-related genes, SLC1A5 and GLS, respectively. Finally, data from human breast cancer samples indicated YAP, TAZ, SLC1A5, and GLS expression correlate with EphA2 expression, with greater activation of the EphA2-YAP/TAZ pathway associating with reduced survival, greater metastatic potential, and increased sensitivity to glutaminase inhibition. Together, these data define a novel mechanism of YAP and TAZ activation by the RTK EphA2 and describe a new transcriptional-dependent role of YAP and TAZ to enhance glutamine metabolism (GLS, SLC1A5) in a HER2-dependent breast cancer model.
Results
EphA2 activates YAP and TAZ in HER2-positive MCF10A cells and MMTV-NeuT mammary tumors
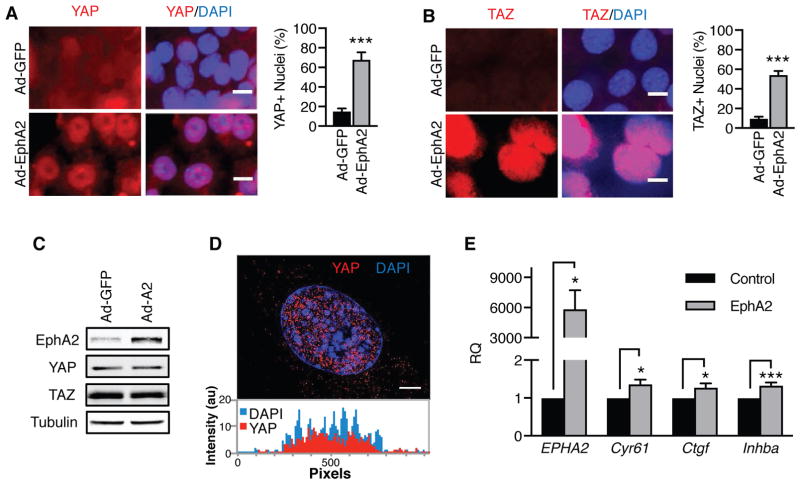
We have previously shown that EphA2 promotes glutamine metabolism through a RhoA-dependent pathway (2). Although Rho activation can lead to increased glutaminase activity (2, 40), the mechanism has not been elucidated. As RhoA can activate the transcriptional cofactors YAP and TAZ (41), we reasoned that increased glutaminolysis induced by EphA2 could be mediated by RhoA-dependent YAP and TAZ activation. To determine if the RTK EphA2 activates YAP and/or TAZ, MMTV-Neu cells were transduced with Ad-EphA2 or control Ad-GFP, and YAP or TAZ subcellular localization was assessed by immunofluorescence (Fig. 1A–B). Compared to control cells, both YAP and TAZ were more frequently co-localized with nuclear DAPI staining (Fig. 1A–B) in cells infected with Ad-EphA2, corresponding with increased EphA2 (Fig. 1C). To confirm the co-localization with DAPI was indeed indicative of intranuclear accumulation, cells were imaged by super-resolution microscopy, which collects multiple z-stack images of an individual cell. Intranuclear localization of YAP was observed in z-stack images upon EphA2 overexpression (Fig. 1D and Supplemental Video 1). Additionally, EphA2 was found to increase YAP and TAZ nuclear localization in the human MCF10A cell line overexpressing HER2 (MCF10A-HER2, Supplemental Fig. 1A–B), suggesting that YAP and TAZ are accumulated in the nucleus in response to EphA2 overexpression.
Figure 1. EphA2 activates the transcriptional co-activators YAP and TAZ in vitro.
A–B) Immunofluorescence of (A) YAP or (B) TAZ (both red) in MMTV-Neu cells infected with Ad-GFP or Ad-EphA2 for 72 hrs. DAPI-stained nuclei (blue) were counted in ImageJ, and YAP- or TAZ-positive nuclei were counted using three fields of view in three independent experiments. Error bars are SEM, and scale bars are 10 μm. ***p<0.005; Student’s t test. C) Western blot of EphA2, YAP, and TAZ in MMTV-Neu cells infected with Ad-GFP or Ad-EphA2. D) Super-resolution microscopy of YAP immunofluorescence from (A). Displayed image was compiled from ten consecutive z-stack images. Signal intensity of YAP (red) or DAPI (blue) was quantitated per pixel moving across the y-axis using ImageJ software. Scale bar is 10 μm. E) Relative mRNA expression was measured in MMTV-Neu cells retrovirally transduced with pBABE (“Control”) or pBABE-EphA2 (“EphA2”) by qRT-PCR from three independent experiments. Error bars are SEM. *p<0.05, ***p<0.005; Student’s t test.
Given the unique bidirectional nature of EphA2 signaling, we next assessed whether EphA2-mediated YAP and TAZ nuclear accumulation was due to EphA2-ephrinA1 interactions (Supplemental Fig. 2). Control or EphA2-overexpressing MMTV-Neu cells were treated with the soluble chimeric EphA2-Fc competitor, which binds ephrin-A1 to suppress ligand-dependent signaling (42), or the control Fc. While EphA2 overexpression increased YAP and TAZ nuclear accumulation, we did not observe any significant changes in the presence of soluble EphA2-Fc (Supplemental Fig. 2), suggesting a primarily receptor-driven role of EphA2 in YAP/TAZ activation.
Next, we sought to determine if EphA2-mediated nuclear localization of YAP and TAZ reflected a functional activation for the transcriptional co-activators. We examined the ability of EphA2 to enhance transcription of known YAP/TAZ-target genes, Cyr61, Ctgf, and Inhba (16, 19, 43–45) (Fig. 1E). Control or EphA2 overexpressing MMTV-Neu cells were subjected to quantitative real-time polymerase chain reaction (RT-PCR) (Fig. 1E). EphA2 overexpression resulted in an increase in mRNA expression of Cyr61, Ctgf, and Inhba expression (Fig. 1E). Similarly, EphA2 overexpression in MCF10A-HER2 cells (Supplemental Fig. 1C) also resulted in increased expression of the YAP/TAZ target genes, CYR61 and AXL (Supplemental Fig. 1D). Together, these findings support our hypothesis that EphA2 acts as a regulator of the transcriptional co-activators, YAP and TAZ.
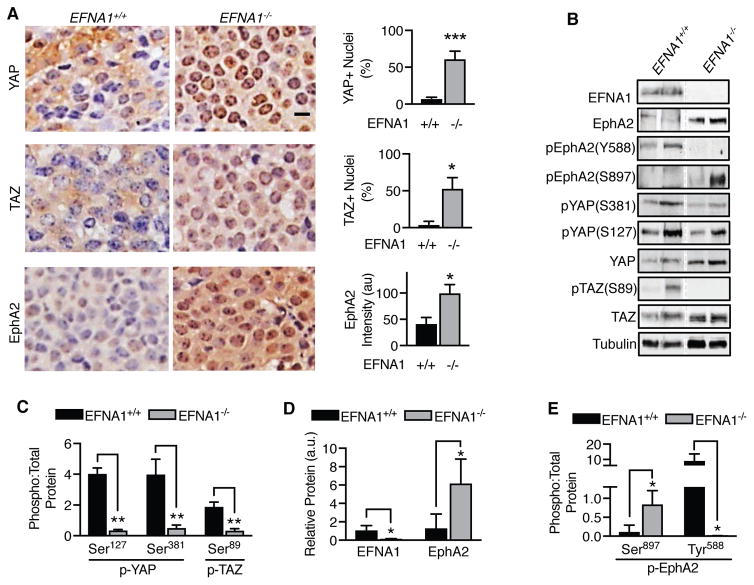
To determine if EphA2 increases YAP and TAZ activity in vivo, we utilized an ephrin-A1 (EFNA1) knockout MMTV-NeuT breast cancer mouse model, which has been demonstrated to overexpress EphA2 (2). Mammary tumors were collected from wild-type (EFNA1+/+) or ephrin-A1 knockout (EFNA1−/−) mice, and YAP and TAZ subcellular localization were determined by immunohistochemistry (Fig. 2A). As expected, EphA2 expression was significantly increased in ephrin-A1 knockout tumors (2). While YAP and TAZ were primarily observed in the cytoplasm of wild-type tumors, nuclear localization was significantly increased in ephrin-A1 knockout tumors (Fig. 2A). Analysis of tumor lysates by Western blot (Fig. 2B) indicated ephrin-A1 knockout tumors exhibited significantly lower phosphorylation of YAP at Ser381 (corresponding to Ser397 in humans) (Fig. 2B,C), which has been reported to mark YAP for degradation (23–25). A similar loss of TAZ phosphorylation at Ser89 was also observed (Fig. 2B, C). A robust inverse correlation between ephrin-A1 and EphA2 protein abundance was confirmed, with greater EphA2 protein detected in ephrin-A1 knockout tumors (Fig. 2B, D). Not surprisingly, loss of EFNA1 also reduced Tyr588 phosphorylation and increased Ser897 phosphorylation (Fig. 2B, E), indicating activation of a ligand-independent mode of EphA2 signaling (6, 46, 47). Although we cannot rule out the possibility that phenotype observed could be due to loss of ephrin-A1, increased EphA2, or both, together, our results strongly support that EphA2 overexpression and ligand-independent EphA2 signaling promotes YAP and TAZ activation.
Figure 2. EphA2 activates YAP and TAZ in an MMTV-Neu mouse model.
A) Immunohistochemistry of YAP, TAZ, and EphA2 (brown staining) in tumors collected from wild-type (EFNA1+/+) and EFNA1 knockout (EFNA1−/−) mice. Scale bar is 10 μm. Percentages of YAP- or TAZ-positive nuclei and EphA2 intensity/cell were calculated based on hematoxylin stained nuclei (blue) using CellProfiler software from four fields per sample (n=4). *p<0.05, ***p<0.005; Student’s t-test. B) Western blot of tumors collected from wild-type (EFNA1+/+) and EFNA1 knockout (EFNA1−/−) mice. The dotted line denotes non-contiguous lanes within a single blot. C–E) Quantitation of Western blot data shown in (B) from EFNA1+/+ (n=3) and EFNA1−/− (n=3) tumors. (C) Phospho-YAP (Ser127 or Ser381) to total-YAP, phospho-TAZ (Ser89) to total TAZ, (D) relative EFNA1 and EphA2 protein, and (E) phospho-EphA2 (Ser897 or Tyr588) are shown. Error bars are SEM. *p<0.05, **p<0.01; Student’s t-test.
EphA2 activates YAP and TAZ in a Rho-dependent mechanism
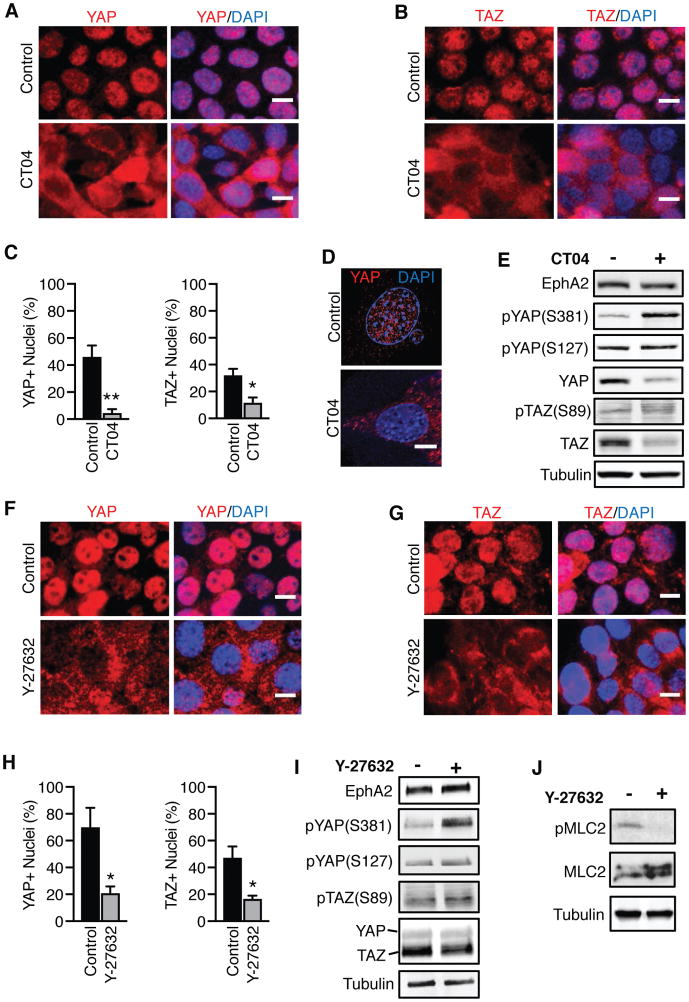
Overexpression of EphA2 has previously been shown to enhance RhoA signaling to support tumor malignancy (2, 5, 9, 48), and RhoA has also been implicated in the activation of YAP/TAZ (41), suggesting that the Rho GTPase may act as an intermediary in the EphA2-YAP/TAZ signaling axis. To examine if Rho is necessary to activate YAP and TAZ upon overexpression of EphA2, MMTV-Neu cells overexpressing EphA2 were treated with vehicle or the selective Rho inhibitor CT04, and YAP and TAZ subcellular localization were determined by immunofluorescence (Fig. 3A–D). In control cells, YAP and TAZ were localized to the nuclei, but CT04 treatment resulted in greater cytoplasmic localization (Fig. 3A–D and Supplemental Video 2), suggesting Rho inhibition blocks YAP and TAZ activity. CT04-treated cells also exhibited greater phosphorylation of YAP at Ser381 (but not Ser127), as well as reduced total YAP and TAZ protein (Fig. 3E), consistent with cytoplasmic retention and degradation of YAP and TAZ upon Rho inhibition. However, Rho inhibition did not increase LATS1 Ser909 phosphorylation (Supplemental Fig. 3A), a marker of LATS1 kinase activation, suggesting Rho may increase YAP and TAZ activity independent of the Hippo pathway.
Figure 3. EphA2 depends on Rho signaling to activate YAP and TAZ.
A,B,F,G) Immunofluorescence of (A,F) YAP or (B,G) TAZ (both red) and DAPI (blue) in MMTV-Neu cells infected with Ad-EphA2 and treated with PBS (control) or (A,B) Rho inhibitor (CT04, 3 μg/mL for 4 hrs) or ROCK inhibitor (Y-27632, 10 μM for 4 hrs). Scale bar is 10 μm. C,H). Quantification of immunofluorescence data in (C) (A,B) and (H) (F,G), respectively. DAPI-stained nuclei were counted in ImageJ, and YAP- or TAZ-positive nuclei were counted using three fields of view in three independent experiments. Error bars are SEM. *p<0.05, **p<0.01; Student’s t-test. D) Super-resolution microscopy of YAP immunofluorescence in control or CT04-treated cells from (A), compiled from ten consecutive z-stacks. Scale bar is 10 μm. E) Western blot analysis of cells in (A,B), treated with CT04 for 6 hrs. I) Western blot analysis of cells in (F,G), treated with Y-27632 (10 μM) for 6 hrs. J) Western blot analysis demonstrating ROCK inhibition by Y-27632.
Using a complementary genetic approach, YAP or TAZ subcellular localization was assessed by immunofluorescence in EphA2-overexpressed MMTV-Neu cells that were transduced with adenovirus to overexpress the control GFP, the dominant-negative Rho-T19N mutant, or the constitutively active Rho-Q63L mutant (Supplemental Figure 4). Compared to control cells, overexpression of the dominant-negative Rho mutant reduced the abundance of YAP and TAZ localizing to the nuclei, consistent with pharmacological Rho inhibition. Conversely, overexpression of the constitutively-active Rho-Q63L mutant maintained YAP and TAZ nuclear accumulation. Together, these data support the role of Rho catalytic activity in promoting EphA2-mediated nuclear accumulation of YAP and TAZ.
Next, we assessed how the downstream effector of Rho, ROCK, contributes to YAP/TAZ activation. Similar to treatment with CT04, a reduction in YAP and TAZ nuclear localization was observed in cells treated with the ROCK kinase inhibitor, Y-27632, compared to control (Fig. 3F–H). Likewise, inhibition of the ROCK kinase increased phosphorylation of YAP (Ser381 and Ser127) and TAZ (Ser89) (Fig. 3I), without affecting LATS1 Ser909 phosphorylation (Supplemental Fig. 3B). To confirm ROCK inhibition, we examined phosphorylation of myosin light chain 2 (MLC2) (Fig. 3J), a known substrate of the ROCK kinase (49). Compared to control cells, a substantial loss of MLC2 phosphorylation was observed in cells treated with Y-27632, confirming ROCK inhibition (50). Collectively, these results support the mechanism that YAP and TAZ are activated by EphA2 in a Rho-dependent manner.
YAP and TAZ enhance EphA2-mediated glutamine metabolism by regulating GLS and SLC1A5 expression
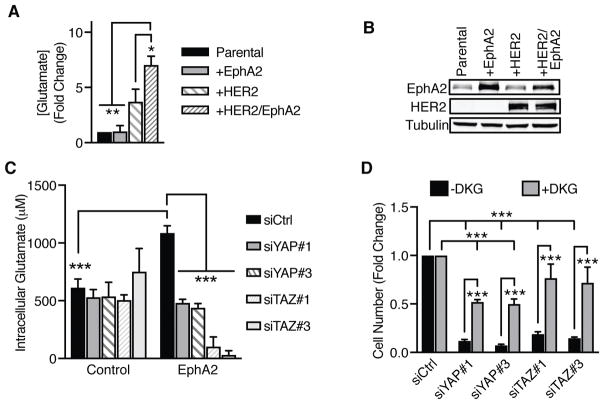
Overexpression of HER2 has been previously shown to promote “glutamine addiction” of breast tumors (38, 39). We reported that concurrent EphA2 overexpression in models of HER2-positive breast cancer promotes a similar increase of glutamine metabolism (2). To determine if crosstalk between EphA2 and HER2 are necessary to increase glutaminolysis, we examined the ability of MCF10A and MCF10A-HER2 cells with or without EphA2 overexpression to convert extracellular glutamine to intracellular glutamate following a period of glutamine stimulation of starved cells. Compared to parental cells, overexpression of EphA2 alone did not significantly increase intracellular glutamate concentration. However, concurrent overexpression of EphA2 along with HER2 significantly enhanced intracellular glutamate concentrations (Fig. 4A). A smaller increase in glutaminolysis was also observed with HER2 overexpression alone (Fig. 4B). Together, these results suggest that crosstalk with HER2 may promote EphA2-mediated glutamine metabolism in HER2-positive breast tumors.
Figure 4. YAP and TAZ promote glutamine metabolism in a HER2-positive breast cancer model.
A) Intracellular glutamate concentration was measured 20 minutes after addition of EGF (20 ng/mL) + L-glutamine (2.5 mM) in MCF10A (“Parental”), MCF10A-EphA2 (“+EphA2”), MCF10A-HER2 (“+HER2”), or MCF10A-HER2-EphA2 (“+HER2/EphA2”) cells. Data was calculated as fold change from parental cells. Error bars are SEM, calculated from three independent experiments. *p<0.05, ***p<0.005; one-way ANOVA; Tukey’s post hoc. B) Western blot of cells described in (A). C) Intracellular glutamate concentration (μM) was determined as described in (B) in MCF10A-HER2 (“Control”) or MCF10A-HER2-EphA2 (“EphA2”) cells with YAP or TAZ knockdown. Error bars are SEM, calculated from three independent experiments. ***p<0.005; two-factor ANOVA; Tukey’s post hoc. D) Growth assay of YAP or TAZ knockdown MCF10A-HER2-EphA2 cells treated with vehicle (-DKG) or dimethyl α-ketoglutarate (+DKG) for 3 days. Fold change in cell number was calculated based on controls in each respective treatment group from three independent experiments. Error bars are SEM. ***p<0.005; two-factor ANOVA; Tukey’s post hoc.
To determine whether YAP and TAZ can mediate EphA2-dependent glutaminolysis, YAP or TAZ was knocked down in MCF10A-HER2 or MCF10A-HER2-EphA2 cells by siRNA-mediated gene silencing (Fig. 4C), and intracellular glutamate concentrations were measured following a glutamine pulse under serum-starvation conditions. Consistent with previous results (Fig. 4A) (2), EphA2 overexpression significantly increased intracellular glutamate concentrations in MCF10A-HER2 cells. Knockdown of YAP or TAZ in control cells display no significant changes in glutamate concentrations (Fig. 4C). However, loss of YAP or TAZ significantly reduced intracellular glutamate concentrations in cells overexpressing EphA2 (Fig. 4C), indicating a role for YAP and TAZ in regulating EphA2-mediated glutamine metabolism. We observed a similar requirement for both YAP and TAZ in cells stimulated with both glutamine and serum (Supplemental Fig. 5), with loss of YAP (Supplemental Fig. 5A–B) or TAZ (Supplemental Fig. 5C–D) resulting in significantly reduced intracellular glutamate concentrations over time. The impact of YAP and TAZ on glutamine metabolism did not stem from off-target effects, since three independent siRNAs targeting YAP (Supplemental Fig. 5E–F) and TAZ (Supplemental Fig. 5G–H) resulted in significantly reduced intracellular glutamate compared to the control.
Since loss of YAP or TAZ has been associated with cellular growth defects (12–15), we next determined if restoration of α-ketoglutarate (α-KG), the TCA intermediate and downstream metabolite of glutamine uptake and conversion to glutamate, would rescue cell growth. MCF10A-HER2-EphA2 cells transfected with individual siRNAs to knockdown YAP or TAZ were treated with vehicle or dimethyl α-ketoglutarate (DKG), a cell-penetrable version of α-KG (51), and the number of cells were counted (Fig. 4D). Consistent with YAP and TAZ promoting cell growth, loss of YAP or TAZ significantly reduced cell number compared to the non-targeting control. However, addition of DKG partially rescued cell growth in cells with YAP or TAZ knockdown, supporting YAP and TAZ as regulators of glutamine metabolism.
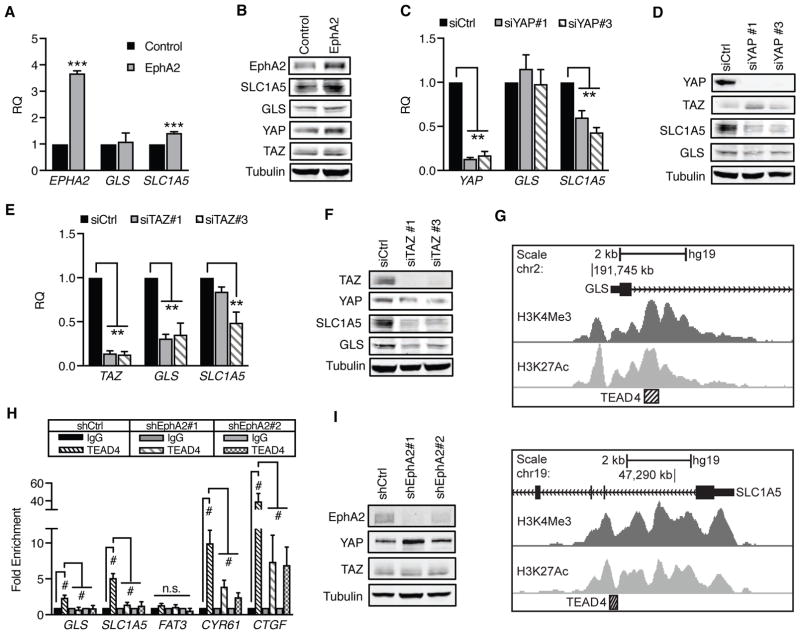
Next, we examined how glutamine metabolism is stimulated by YAP and TAZ. As regulators of transcription, we expected YAP and TAZ would promote expression of genes critical in glutamine metabolism, including the glutamine transporter, SLC1A5, and glutaminase, GLS, both of which are commonly overexpressed in human breast cancer, especially in the HER2-positive subtype (38, 39). We utilized quantitative RT-PCR to determine if EphA2, YAP, and TAZ promote SLC1A5 and GLS expression (Fig. 5). There are two alternatively-spliced isoforms of glutaminase encoded by GLS1, KGA and GAC (52). We focused on the more catalytically-active GAC isoform that is frequently overexpressed in human cancers, including breast cancer (52–54). Overexpression of EphA2 significantly increased SLC1A5 expression (Fig. 5A), an effect that was confirmed at the protein level (Fig. 5B). Surprisingly, expression of GLS was unaffected (Fig. 5A–B), possibly due to compensatory mechanisms of GLS regulation, such as MYC or NF-κB (39, 40, 55, 56). Interestingly, YAP and TAZ appear to differentially regulate glutamine metabolism genes. Loss of YAP significantly decreased expression of SLC1A5, but not GLS, both at the mRNA (Fig. 5C) and protein (Fig. 5D) levels. In contrast, loss of TAZ reduced both GLS and SLC1A5 expression (Fig. 5E–F).
Figure 5. GLS and SLC1A5 gene expression are enhanced by EphA2-YAP/TAZ-TEAD4 signaling.
A,C,E) Relative mRNA expression in (A) MCF10A-HER2 (“Control”) or MCF10A-HER2-EphA2 cells (“EphA2”) or (C,E) MCF10A-HER2-EphA2 cells transfected with control (siCtrl) or individual siRNAs targeting (C) YAP or (E) TAZ. (A) EphA2, (C) YAP, and (E) TAZ were included to demonstrate expression of target genes. Error bars are SEM, calculated from three independent experiments. **p<0.01, ***p<0.005, Student’s t-test for (A) or one-way ANOVA for (C) and (E); Dunnett’s post-hoc. B,D,F) Western blot of GLS and SLC1A5 in cells used in qRT-PCR. G) TEAD4 ChIP-seq at GLS (top) and SLC1A5 (bottom), downloaded from UCSC ENCODE Genome Browser. TEAD4-associated regions (hatched box) correlate with H3K4Me3 and H3K27Ac near exon 1 (black). H) Chromatin immunoprecipitation of MCF10A-HER2 cells transduced with shCtrl, shEphA2#1, or shEphA2#2. Relative immunoprecipitated genomic DNA was determined by qRT-PCR and normalized to IgG controls from four independent experiments. Error bars represent SEM. #p<0.005, two-way ANOVA, Tukey’s post hoc; non-significant (n.s.) comparisons are indicated. A) Western blot of EphA2 knockdown in cells used in (H).
To identify the mechanism by which YAP and TAZ promote GLS and SLC1A5 expression, the ENCODE database (57), which consolidates submitted ChIP-seq data, was searched to identify transcription factor binding sites near the promoters of genes critical in glutaminolysis (Fig. 5G). The TEAD family of transcription factors are necessary for YAP/TAZ functionality as a transcriptional regulator, supporting transcription of proliferative genes while suppressing pro-apoptotic gene expression (16–19). ChIP-seq data from ENCODE indicated that TEAD4 localizes to the promoters of both GLS and SLC1A5 (Fig. 5G), as demonstrated by enhanced histone H3 methylation (H3K4Me3) and acetylation (H3K27Ac) defining promoter areas and active enhancer elements, respectively (58, 59). ChIP-seq data for other members of the TEAD family of transcription factors was unavailable.(60, 61)
To investigate if YAP and TAZ may bind to GLS and SLC1A5 promoters through TEAD4 in HER-positive cancer cells, chromatin immunoprecipitation (ChIP) was performed in control or EphA2 knockdown cells using antibodies targeting IgG or TEAD4 (Fig. 5H–I). Specificity for TEAD4 was assessed using the positive controls CYR61 and CTGF, as well as the negative control FAT3, whose expression occurs either dependently (CYR61, CTGF) or independently (FAT3) of the YAP/TAZ/TEAD4 complex (16). Immunoprecipitated DNA for CYR61 and CTGF, but not FAT3, was enriched in TEAD4 immunocomplexes from control cells (Fig. 5H). Additionally, TEAD4-immunoprecipitated GLS and SLC1A5 DNA were significantly enriched over IgG controls in control cells. EphA2 knockdown significantly reduced this enrichment for CYR61, CTGF, GLS, and SLC1A5, suggesting that EphA2 is required to induce TEAD4 binding to GLS and SLC1A5 promoters. Like TEAD4, YAP was also found to be associated with GLS and SLC1A5 promoters (Supplemental Fig. 6). Together, these results support that EphA2 activates the YAP/TAZ/TEAD4 complex to promote glutamine metabolism through enhancement of GLS and SLC1A5 expression.
Hyperactivation of the EphA2-YAP/TAZ signaling axis correlates with increased glutamine metabolism and reduced survival in breast cancer patients
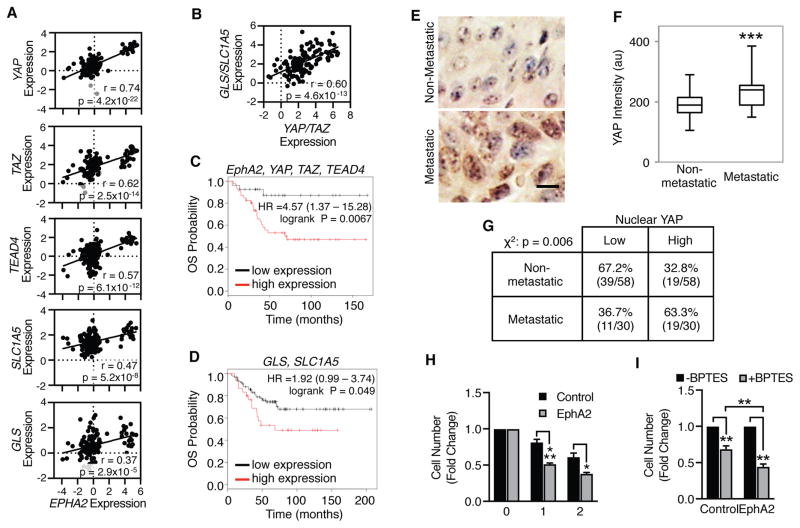
To determine if the EphA2-YAP/TAZ signaling axis is relevant to glutamine metabolism in human breast cancer, expression of EphA2, YAP, TAZ, TEAD4, GLS, and SLC1A5 from patient samples were analyzed in a previously published breast cancer database (62). Consistent with EphA2 overexpression promoting enhanced YAP/TAZ activity, strong correlations exist between expression of EphA2 and YAP as well as EphA2 and TAZ (Fig. 6A). Additional positive correlations between EphA2 and TEAD4, SLC1A5, and GLS were also observed (Fig. 6A). Furthermore, YAP/TAZ expression is positively correlative with GLS and SLC1A5 (Fig. 6B), supporting that YAP and TAZ regulate EphA2-mediated glutamine metabolism. To determine if the EphA2-YAP/TAZ-glutaminolysis signaling axis impact clinical outcomes, breast cancer databases from the Gene Expression Omnibus (GEO) (63) were examined for overall survival (OS) in patients with low or high combined expression of EphA2, YAP, TAZ, and TEAD4 (Fig. 6C) or GLS and SLC1A5 (Fig. 6D). High expression of genes involved in the EphA2-YAP/TAZ signaling axis significantly correlated with decreased overall survival in HER2-positive breast cancer patients (Fig. 6C). A similar trend was also observed with glutaminolysis genes, with high expression of GLS and SLC1A5 also strongly associated with decreased overall survival (Fig. 6D), strongly supporting the importance of EphA2, YAP/TAZ, and GLS/SLC1A5 in promoting tumor progression. Individually, recurrence-free survival (RFS) in patients with high expression of YAP, TAZ, or EphA2 (Supplemental Fig. 7A–C) significantly correlated with decreased recurrence-free survival in all breast cancer subtypes. However, stratification for HER2-positive breast cancer patient data resulted in greater hazard ratios (HR) for high expression of YAP, TAZ, or EphA2 (Supplemental Fig. 7D–F), suggesting EphA2 and YAP/TAZ may play a more clinically relevant role in HER2-positive breast cancer compared to cases lacking HER2 overexpression.
Figure 6. EphA2 and YAP/TAZ expression correlates with increased glutaminolysis gene expression in human breast cancer patient data.
A,B) Log2(mRNA) expression from the Minn Breast 2 database (n=121). Pearson correlations (r) and T-distribution (p) coefficients are shown. C,D) Kaplan-Meier analysis of overall survival (OS) in HER2+ breast cancer patients (C: n=73; D: n=117) exhibiting low (black) or high (red) (C) EphA2, YAP, TAZ, and TEAD4 or (D) GLS and SLC1A5 expression. E–G) Immunohistochemistry of YAP in HER2-positive samples from a human breast carcinoma TMA. Representative images are shown in (E). Scale bar is 10 μm. F) Plot of YAP staining intensity per cell in HER2-positive TMA patient samples. ***p<0.005, Student’s t test. G) Samples from (F) further stratified according to levels of YAP nuclear localization, using the median percentage as the cutoff. Chi-square analysis (χ2) was calculated. H, I) BPTES sensitivity in (H) MMTV-Neu or (I) MCF10A-HER2 cells retrovirally transduced with pBABE (“Control”) or pBABE-EphA2 (“EphA2”) and treated with vehicle or BPTES (10 μM) for times indicated. Cell counts were calculated as fold change from vehicle controls from three independent experiments. *p<0.05, **p<0.01, ***p<0.005, (H) two-factor ANOVA or (I) one-way ANOVA, post-hoc Tukey’s.
Survival data suggests that enhanced expression of YAP and TAZ may confer an advantageous environment for tumor progression in HER2-positive breast cancer. To examine the role of YAP in tumor progression, YAP protein expression as well as YAP nuclear localization was examined in non-metastatic and metastatic HER2-positive breast cancer samples by performing IHC on a breast cancer tissue microarray (TMA) (Fig. 6E–G). Compared to localized, non-metastatic disease, YAP protein was significantly higher in tumors scored as metastatic (Fig. 6E–F). Likewise, nuclear YAP was found to be more strongly associated with metastatic progression (Fig. 6E, G). Our lab has shown that metastatic spread is also associated with enhanced EphA2 phosphorylation at Ser897, a measure of ligand-independent EphA2 activity, suggesting EphA2 and YAP/TAZ activity correlate in advanced disease (2). Together, these data are strongly supportive of EphA2 and YAP/TAZ coordinating to promote tumor progression in HER2-positive breast cancer, resulting in metastatic spread and reduced survival of patients.
Finally, we examined whether EphA2-overexpressing cells may be more sensitive to glutaminase inhibition. MMTV-Neu or MMTV-Neu-EphA2 cells were treated with vehicle or the glutaminase inhibitor BPTES, and cells were counted as a measure of cellular growth (Fig. 6H). Glutaminase inhibition reduced cellular growth, but this effect was significantly more dramatic in cells overexpressing EphA2 than control cells (Fig. 6H). A similar effect was observed in the MCF10A-HER2 (Fig. 6I), strongly supporting the clinical relevance of EphA2-mediated glutamine metabolism.
Discussion
The Hippo signaling pathway is an emerging growth control and tumor suppressor pathway that suppresses cell proliferation and stem cell function (27, 28). Genome-wide analyses identified increased expression and nuclear localization of the Hippo pathway downstream effectors and transcriptional co-activators YAP and TAZ in human cancer (16). While few mutations in components of the Hippo pathway have been discovered, amplification and overexpression of YAP and TAZ are observed in human breast cancer (12–15, 29). Although upregulation of YAP/TAZ is linked to proliferation, stem cell function, and drug resistance in breast cancer, how the Hippo pathway is deregulated is poorly understood because of lacking evidence of oncogenic driver mutations in this pathway. Here, we identify EphA2 as a receptor tyrosine kinase that stimulates YAP/TAZ activity. Overexpression of EphA2 induced YAP and TAZ nuclear accumulation in cell lines and MMTV-Neu tumors, and the expression of YAP/TAZ target genes (Cyr61, Ctgf, and Inhba), demonstrating that EphA2 functionally stimulates YAP/TAZ activity. Furthermore, the EphA2-YAP/TAZ signaling axis is clinically relevant, because there is a strong correlation between EphA2 and YAP/TAZ expression in a human breast cancer dataset, with hyperactivation of the pathway enhancing metastatic potential and reducing survival, particularly in HER2-positive patients.
Cell-cell communication has been identified as the primary mode of YAP/TAZ regulation, and active Rho signaling is required for full activation (41). In fact, YAP/TAZ act as sensors of cell density to limit proliferation in response to excessive cell contacts (24). However, dysregulation of the upstream communicative pipeline hyperactivates YAP/TAZ, thus eliminating their anti-tumor role (27–30). The unique bidirectional nature of EphA2 signaling mirrors these intercellular communications that modulate YAP/TAZ activation. While interactions between ephrin-A1 and EphA2 located on neighboring cells suppresses cell proliferation, EphA2 ligand-independent signaling promotes tumor growth and cell migration (3, 4, 6–8). Indeed, we show that high EphA2 expression is associated with poorer survival in lymph-node positive breast cancer patients, especially those with HER2-positive tumors, confirming the role of EphA2 in tumor progression. In part, our data suggests that these tumor-promoting processes are initiated by EphA2 ligand-independent activation of Rho (2, 9, 48), although we cannot completely rule out how loss of EFNA1 may be contributing to this mechanism. Our data support that EphA2-mediated YAP/TAZ activation requires intact Rho signaling, as Rho/ROCK inhibition prevented nuclear accumulation through protein destabilization. While YAP and TAZ are commonly overexpressed and/or hyperactivated in human cancers, few mutations have been observed (27). Our data suggests that the RTK EphA2 may contribute to a previously unknown mechanism of YAP/TAZ dysregulation in human cancers.
Rho is known to promote glutaminase (GLS) activity, as the GLS1 inhibitor, 968, was first identified as an inhibitor for Rho-dependent transformation (40). However, how Rho activation affects glutaminase activity was poorly understood. Our data supports a role of TAZ in mediating Rho-dependent glutaminase activation. Prior studies show that Rho promotes YAP activity through actin dynamics and phosphorylation of LATS1/2 (64, 65), although Rho may also promote YAP/TAZ activity independently of Hippo signaling (41, 66). However, we were unable to detect changes of LATS1 phosphorylation in EphA2 overexpressing cells treated with Rho or ROCK inhibitors. It is possible that EphA2-induced YAP and TAZ activation through LATS2 or a LATS-independent mechanism, such as actin-binding angiomotin proteins which can bind to YAP directly (67). In addition to enhancement of YAP/TAZ nuclear translocation, EphA2 appears to also increase expression of YAP/TAZ in human breast cancer. Analyses of an Oncomine dataset revealed a positive correlation between EphA2 and YAP/TAZ mRNA expression. It is currently unclear how EphA2 may increase YAP/TAZ expression at the mRNA level.
While the focus of the present study remained on GLS and SLC1A5, the possibility remains that EphA2 and YAP/TAZ may promote expression of additional genes involved in glutamine metabolism. Numerous glutamine transporters, including SLC38A1 and LAT1, have been attributed to tumor promotion (68, 69), but strong evidence supports SLC1A5, also known as ASCT2, as an important contributor to tumor growth in breast cancer (70). Furthermore, two isozymes of glutaminase, GLS (kidney-type) and GLS2 (liver-type), have been identified in mammals. Whereas GLS overexpression is commonly observed in human cancer (38, 39, 71, 72), the role of GLS2 is more controversial, with both tumor-promoting and tumor-suppressive roles being described (73–77). Although we are unable to rule out GLS2 regulation, GLS was previously identified as a transcriptional target of YAP and TAZ in pulmonary hypertension (78), suggesting this more widely expressed form of glutaminase (37) may be targeted by YAP/TAZ in multiple disease contexts. Additional YAP targets in glutamine metabolism include glutamine synthetase (GLUL) and the glutamine transporter SLC38A1 in liver cancer (79, 80), implying the EphA2-YAP/TAZ signaling axis may enhance glutamine metabolism through many gene targets. However, in the context of breast cancer, transcriptional regulation of the more catalytically-active glutaminase isoform, GAC, by TAZ supports the tumor-promoting properties associated with EphA2 and YAP/TAZ overexpression. Indeed, EphA2 overexpression increased sensitivity to glutaminase inhibition, suggesting that EphA2 and YAP/TAZ may be potential biomarkers for a favorable patient response to GLS inhibitors (CB-839) currently in clinical trials (81).
In addition to YAP/TAZ transcription co-activators, the proto-oncogene MYC and the NF-kB pathway are known to increase glutamine metabolism in cancer (39, 40, 55, 56). While we cannot rule out a MYC contribution to the observed changes in glutamine metabolism, our data support a YAP/TAZ-TEAD interaction driven mechanism in HER2-positive breast cancer. In addition to YAP, we demonstrated that the TEAD4 transcription factor is bound to the GLS and SLC1A5 promoter regions, which was significantly reduced following EphA2 knockdown. Although these ChIP analyses largely correlate with observed trends in mRNA expression, our data indicates that YAP also associates with the GLS promoter, suggesting that YAP may indeed have a role in the regulation of GLS expression. Overexpression of MYC has been shown to only partially rescue the proliferation of YAP/TAZ knockdown in human breast cancer cells (16), possibly due to negative feedback on YAP/TAZ-TEAD4 activity (82). Furthermore, the TEAD family of transcription factors have been identified as the primary recruiters of YAP/TAZ to chromatin (16), supporting the TEAD family of transcription factors as a major contributor to EphA2-YAP/TAZ-dependent glutamine metabolism in HER2-positive breast cancer. Still, TEAD4 as well as other TEAD proteins, including TEAD1-3, have been reported to exhibit redundancy (83, 84). How the remaining TEAD family members contribute to EphA2-mediated glutamine metabolism needs further investigation.
Although evidence does not support a role of MYC in this model, the possibility remains that additional mechanism(s) may still increase glutamine metabolism in HER2-positive breast cancer. This is particularly true with GLS, since our data suggested GLS was more weakly associated with the EphA2-YAP/TAZ-TEAD4 pathway than SLC1A5. While indications of defined regulatory mechanisms contributing to SLC1A5 gene expression in cancer has been limited, previous evidence has demonstrated that GLS gene expression is enhanced by other transcription factors, most notably NF-κB in HER2-positive breast cancer cells (39, 40). It was also noted NF-κB-mediated GLS gene expression is dependent on Rho-GTPase activity (40), consistent with our TAZ-TEAD4 data. Taken together, we speculate that upon activation of Rho-GTPase, both NF-κB and TAZ-TEAD4 may promote glutamine metabolism in HER2-positive breast cancer by upregulating GLS expression. However, our data strongly support an EphA2- and YAP/TAZ-dependent up-regulation of SLC1A5, a novel and unique regulatory mechanism.
Overall, these findings define a novel role of the RTK EphA2 in activation of the transcriptional co-activators YAP and TAZ to promote glutamine metabolism in HER2-positive breast cancer. Targeting the EphA2-YAP/TAZ signaling axis reveals great promise to reduce glutamine metabolism in HER2-positive breast cancer, possibly extending to other subtypes and additional forms of cancer. Therefore, our work describes the potential clinical benefit of using EphA2, YAP, and TAZ as future biomarkers in HER2-positive breast cancer to predict patient outlook and response to pharmacological agents that inhibit EphA2 or glutamine metabolism.
Materials and Methods
Cell Culture
MMTV-Neu tumor cells were isolated and cultured as reported previously (85, 86). MCF10A-HER2 cells were generated and cultured as previously described (85, 87, 88). All cells were maintained at low passages. Transient overexpression of EphA2 or Rho mutants was achieved by adenoviral infection with Ad-GFP, Ad-EphA2, Ad-Rho-T19N, or Ad-Rho-Q63L adenovirus for 72 hrs. EphA2 was stably overexpressed by infection with pBABE or pBABE-EphA2 lentivirus and selected with 2 μg/mL puromycin for 5 days. Flag-YAP was overexpressed using pBABE-YAP1 (a gift from Joan Brugge, Addgene plasmid #15682) (12). To achieve transient knockdown, cells were transfected with ON-TARGETplus Non-Targeting pool siRNA (D-001810-10), siGENOME YAP1 SMARTpool (M-012200-00) or individual siRNA’s (siYAP#1: M-012200-01, siYAP#2: M-012200-02, siYAP#3: M-012200-03) and siGENOME WWTR1 SMARTpool (M-016083-00) or individual siRNA’s (siTAZ#1: M-016083-01, siTAZ#2: M-016083-02, siTAZ#3: M-016083-04) (GE Healthcare) (all at 40 nM), as indicated, with Lipofectamine RNAiMAX reagent (Invitrogen) per manufacturer’s instructions for 72 hrs. To generate stable EphA2-knockdown cells, MCF10A-HER2 cells were transduced with lentiviral pLKO.1-blast (a gift from Keith Mostov; Addgene plasmid #26655) (89), pLKO-shEphA2#1 (sense: 5′-CGGACAGACATATAGGATATT-3′), or pLKO-shEphA2#2 (sense: 5′-GCGTATCTTCATTGAGCTCAA-3′) and selected with blasticidin (8 μg/mL) for 2 days. To inhibit Rho or ROCK activity, MMTV-Neu cells were treated with PBS control, Rho Inhibitor I (CT04, Cytoskeleton #CT04, 3 μg/mL) or the ROCK inhibitor Y-27632 (Tocris #1254, 10 μM) for 4–6 hrs as indicated in serum-free medium. Cells were incubated with EphA2-Fc (R&D Systems) or Fc (1 μg/mL) for 16 hrs.
Antibodies and Immunoblotting
The following antibodies (1:1000 except where indicated) were used against the corresponding proteins: EphA2 (Santa Cruz #sc-924), EphA2 (CST #6997), pEphA2-Tyr588 (CST #12677S), pEphA2-Ser897 (1:500, Abgent #AP3722a), EFNA1 (R&D Systems, #AF702), YAP/TAZ (CST #8418), pYAP-Ser397 (CST #13619), pYAP-Ser127 (CST #13008), pTAZ-Ser89 (1:500, CST #75275), MLC (CST #3672), pMLC2-Ser19 (CST #3671), LATS1 (CST #3477), pLATS1-Ser909 (CST #9157), ERBB2 (Neomarkers #MS-730-P0), β-actin (Santa Cruz #sc-47778), and β-tubulin (Sigma #T4026). IRDye 680LT goat anti-mouse (Licor #925-68020, 1:20,000), IRDye 800CW goat anti-rabbit (Licor #925-32211, 1:10,000), horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Promega #W401B, 1:5000), and HRP-conjugated goat anti-mouse (Promega #W402B, 1:5000) were used as secondary antibodies. For immunoblotting, pre-cleared lysates were electrophoresed by SDS-PAGE and transferred to nitrocellulose membranes, which were blocked for 1 hr in 5% nonfat dry milk or 5% bovine serum albumin (BSA). Membranes were incubated with primary antibodies overnight followed by incubation with secondary antibodies for 1 hr at room temperature and imaged using Licor Odyssey or enhance chemiluminescence (Clarity ECL kit, Bio-rad). Densitometry was performed using ImageJ software, and measured proteins were normalized using tubulin controls.
Immunohistochemistry
Tumors from EFNA1+/+ and EFNA1−/− MMTV-NeuT mice (2) were subjected to immunohistochemical staining (IHC) as described previously (2). Briefly, formalin-fixed paraffin-embedded (FFPE) tumor sections were rehydrated, and antigen retrieval was achieved using Retrievagen A (BD Biosciences), according to the manufacturer’s instructions. After blocking for endogenous peroxidases and several washes in PBS, tissue sections were probed with antibodies against EphA2 (Zymed Laboratories #34-7400, 10 μg/mL), YAP (CST #14074, 1:400), or TAZ (Abcam #ab84927, 1:400) overnight at 4°C. After washing, samples were incubated with biotinylated anti-rabbit IgG (Vector Laboratories #BA-1000) for 1 hr at room temperature and streptavidin peroxidase reagents (BD Pharmingen #51-75477E), liquid diaminobenzidine (DAB) (Invitrogen #00-2014), and hematoxylin were used for staining. Stained tissues were mounted with Cytoseal XYL and images of at least four fields of view were obtained using an Olympus inverted fluorescence microscope (40x).
Nuclear localization and staining intensity were calculated using CellProfiler software. Briefly, to quantitate nuclear localization, original images were dividing into red and blue channels, and nuclei were identified as the primary objects from the blue channel, as defined between 10 and 40 pixels in diameter. Thresholding was controlled using the Global Otzu method (three-class thresholding) with a threshold correction factor of 2, and lower and upper bounds of 0 and 1.0, respectively. Smoothing was automatically applied, and clumped objects were identified based on intensity. DAB stained nuclei were identified as secondary objects, using the propagation method with an automatic threshold and a regularization factor of 0.05. Positive nuclei were filtered based on mean intensity values, and classified as positive when the mean intensity value was greater than the threshold of 0.2. To detect EphA2 and YAP DAB staining intensity per cell, the original image was divided into red and blue channels, and overall DAB intensity was measured from the red channel image. Nuclei were identified as the primary objects as described above, except for use of two-class thresholding with a correction factor of 1. To calculate intensity per cell, total DAB intensity was divided by the total number of nuclei per image.
Immunofluorescence
MMTV-Neu or MCF10A-HER2 cells were infected with adenovirus as described above. Subsequently, cells were fixed with 2% paraformaldehyde for 15 min, permeabilized with 1% Triton X-100 in PBS for 5 min, and blocked with 3% bovine serum albumin (BSA) in PBS for 1 hr, all at room temperature. Cells were probed with anti-YAP (CST #14074, 1:100) or anti-TAZ (Abcam #ab84927, 1:50) overnight at 4°C, washed multiple times with 0.5% Tween 20 in PBS, followed by incubation with Alexa Fluor® 594 anti-rabbit secondary antibody (Invitrogen #A-11012, 1:500) for 1 hr at room temperature. Following several washes, coverslips were mounted using SlowFade® Diamond antifade reagent containing DAPI (Molecular Probes #S36963). Images of at least three fields of view per sample were obtained using an Olympus inverted fluorescence microscope (40x) or Deltavision OMX Super Resolution microscope (60x). ImageJ software was used to quantitate YAP or TAZ positive nuclei and generate z-stack videos.
Glutamate Assay
Intracellular glutamate concentrations were determined in duplicate using the Glutamate Assay Kit (Sigma), according to the manufacturer’s instructions and as previously described (2). MCF10A-HER2 or MCF10A-HER2-EphA2 cells were transfected with the designated siRNA’s as described above for 48 hrs and then cultured in serum-free and glutamine-free DMEM/F12 media for 24 hrs. Cells were stimulated with EGF (20 ng/mL) and L-glutamine (2.5 mM) with or without 5% FBS for 0, 5, 10, or 20 minutes, as indicated. Glutamate concentrations were calculated from known standards, and all data are corrected according to baseline values.
Growth Assays
MCF10A-HER2 cells were transfected to knockdown YAP or TAZ as described above. After 24 hrs, cells were supplemented with vehicle or dimethyl α-ketoglutarate (DKG) (dimethyl 2-oxoglutarate (Sigma), 4 mM), and cells were counted after 72 hrs. Cell number was normalized to non-targeting controls in each treatment group. To assess sensitivity to BPTES, MMTV-Neu, MMTV-Neu-EphA2, MCF10A-HER2, or MCF10A-HER2-EphA2 cells were treated with vehicle or BPTES (10 μM) for 1–2 days, as indicated, and cells were counted. Cell number was normalized to untreated controls for each cell line.
Quantitative Real-Time PCR
Cells were prepared as indicated and RNA was collected using the RNeasy Kit (Qiagen) and converted to cDNA by reverse transcription using iSCRIPT (Bio-Rad) according to the manufacturer’s instructions. Using Taqman Fast Advanced Master Mix, samples were amplified as previously described (2) in triplicate on the Step-One Plus (Applied Biosystems) the following Taqman probe sets: EPHA2 (Hs00171656_m1), YAP (Hs00902712_g1), TAZ (WWTR1, Hs00210007_m1), EphA2 (Mm00438726_m1), Cyr61 (Mm00487498_m1), Ctgf (Mm01192933_g1), Inhba (Mm00434339_m1), CYR61 (Hs00998500_g1), AXL (Hs01064444_m1), GLS (GAC splice variant, Hs01022166_m1, SLC1A5 (Hs01056542_m1), Actb (Mm02619580_g1), or ACTB (Hs01060665_g1). Quantitation was performed using the DDCt method.
Chromatin Immunoprecipitation
MCF10A-HER2/Flag-YAP and stable EphA2 knockdown cells were generated as described above and subjected to chromatin immunoprecipitation as previously described (90), with modification. Briefly, cells were fixed in 1% formaldehyde solution (50 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% formaldehyde) for 10 min at room temperature, followed by quenching using 125 mM glycine. Crosslinked cells were washed and collected in PBS, and nuclear enrichment was achieved by resuspension in LB1 (50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100) for 10 min at 4°C and LB2 (10 mM Tris pH 8, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) for 5 min at 4°C, both with rotation. The collected cell pellets were lysed in LB3 (10 mM Tris pH 8, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine), and samples were sonicated using a Biorupter XL to generate DNA fragments of 500–1000 bp, with Triton X-100 (1% final) and protease inhibitor cocktail (Roche) added afterward. Diluted nuclear extracts were precleared with salmon sperm DNA/Protein A Agarose slurry (EMD Millipore) for 30 min at 4°C, with rotation, and 50 μL was saved as the input control. The beads were pelleted, and the supernatant was immunoprecipitated with 3 μg of normal rabbit IgG (Santa Cruz #sc-3888), TEAD4 (Abcam #ab58310), or FLAG M2 (Sigma #F2426) at 4°C overnight with rotation. Immunocomplexes were collected with salmon sperm DNA/Protein A Agarose slurry for 1 hr at 4°C with rotation, followed by multiple rounds of washing and elution twice in 200 μL of elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at room temperature with agitation. Crosslinks were reversed by addition of 16 μL 5 M NaCl and 1 μg RNase A at 65°C overnight, followed by treatment with proteinase K (20 μg) for 1 hr at 50°C. Immunoprecipitated DNA was collected by phenol-collected by phenol-chloroform extraction followed by ethanol precipitation with 20 μg glycogen and resuspended in 50 μL 1x TE buffer (10 mM Tris pH 8, 1 mM EDTA). For quantitation, 1/50 of the DNA, 1 mM forward primer, 1 mM reverse primer (see Supplemental Table 1 for ChIP primer sequences), and SYBR Green PCR Master Mix (Applied Biosystems) were combined and qRT-PCR was performed in triplicate using the Step-One Plus (Applied Biosystems) system.
Human Breast Cancer Tissue Microarray and Gene Expression Dataset Analysis
YAP immunohistochemical staining was performed on a commercially available human breast cancer tissue microarray (Cybrdi, #CC08-10-001). Using two fields of view per sample, YAP staining intensity and nuclear localization were determined using CellProfiler software. HER2-overexpressing tumors were further stratified for metastases based on the TNM classification and staging system, with patients staged with lymph node (N>0) and/or distant metastases (M=1) being classified as having metastatic disease. For YAP activation, the percentage of cells with YAP nuclear localization was calculated using the CellProfiler software, as described above, and samples were classified as having low or high activation when below or above the median percentage of YAP-positive nuclei (27.5%), respectively.
To assess TEAD4 binding sites, TEAD4 ChIP-seq data acquired from multiple cell lines by the Myers Laboratory was examined using the ENCODE database (http://genome.ucsc.edu) (57). H3K4Me3 (GEO GSM733680 and GSM733720) and H3K27Ac (GEO GSM733656 and GSM733674) data submitted by the Bernstein Laboratory were collected to indicate promoter regions and active enhancer elements, respectively.
To determine if gene expression is correlative in human breast tissue, the Minn Breast 2 database (62) was searched on ONCOMINE (http://www.oncomine.org) (62, 91)for mRNA expression of EPHA2 (Gene ID 203499_at), YAP (Gene ID 213342_at), WWTR1 (TAZ, Gene ID 202133_at), TEAD4 (Gene ID 204281_at), GLS (GAC isoform specific, Gene ID 221510_s_at), and SLC1A5 (Gene ID 208916). The reported data was collected and reported on a log2 scale.
EPHA2 (203499_at), YAP (213342_at, 224894_at), WWTR1 (TAZ, 202133_at), TEAD4 (41037_at), GLS (GAC isoform specific, 221510_s_at), and SLC1A5 (208916_at) overall and recurrence-free survival was determined using Kaplan-Meier Plotter online (http://www.kmplot.com) (63), using automatic cutoff. Data was analyzed from 1764 breast cancer patients and 73–156 patients exhibiting tumors with the HER2-positive subtype. EPHA2 RFS was examined in lymph-node positive patients. Patient samples were classified according to gene expression as high or low expressers of each designated gene or gene set.
Statistics
Correlation was determined using Pearson’s Correlation coefficient and tested using Student’s T-distribution. For statistical analysis of Kaplan-Meier plots, hazard ratio (HR) with 95% confidence intervals and logrank p-values were calculated from stratified patient data. Categorical data was analyzed by Chi-Square analysis, and for comparisons between two groups, unpaired Student’s t-test was performed. Statistical analysis for multiple comparisons was performed using one- or two-way ANOVA for one or more characteristics, respectively. To identify where differences existed among groups, post hoc analyses consisting of Dunnett’s or Tukey’s methods were completed, as indicated. All statistical tests performed were two-tailed, and p<0.05 was considered statistically significant.
Supplementary Material
EphA2 overexpression leads to intranuclear accumulation of YAP.
Nuclear exclusion of YAP upon Rho inhibition.
Supplemental Figure 1. EphA2 activates YAP/TAZ in in MCF10A-HER2 cells
Supplemental Figure 2. EphA2-EphrinA1 interactions do not significantly contribute to YAP/TAZ activation.
Supplemental Figure 3. Rho and ROCK inhibition do not enhance LATS1 phosphorylation in MMTV-Neu cells.
Supplemental Figure 4. EphA2-mediated YAP/TAZ activation requires Rho catalytic activity.
Supplemental Figure 5. YAP and TAZ promote glutamine metabolism in MCF10A-HER2-EphA2 cells.
Supplemental Figure 6. YAP and TEAD4 are associated with GLS and SLC1A5 promoters.
Supplemental Figure 7. YAP/TAZ and EphA2 expression strongly correlated with decreased patient survival in HER2+ breast cancer.
Supplemental Table 1. ChIP Primers
Acknowledgments
We would like to thank Drs. Victoria Youngblood and Wenqiang Song (Vanderbilt University, Nashville, TN) for providing MMTV-Neu tumor samples and pBABE-EphA2 vector, respectively. Additionally, we would like to express our gratitude to the ENCODE Consortium, as well as the Bradley Bernstein laboratory (Broad Institute) and the Richard Myers laboratory (Hudson Alpha Institute for Biotechnology) for providing a platform to examine ChIP-seq data. Funding: Super resolution microscopy experiments were performed in part through the use of the Vanderbilt Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, DK59637 and EY08126). Sample preparation was supported by the Vanderbilt Translational Pathology Shared Resource (NCI/NIH Grant 2P30 CA068485-14). This work was supported by NIH grants R01 CA177681 (J.C.), R01 CA95004 (J.C.), T32 CA009592 (D.N.E. and V.M.N.), F30 CA216891-01 (E.S.), F31 CA220804-01 (L.C.K.), a VA Merit Award 5101BX000134 (J.C.), a pilot project from the Vanderbilt Breast SPORE grant CA098131 (J.C.), and a Susan G. Komen Award PDF#17480733 (D.N.E.).
Footnotes
Author Contributions: D.N.E., A.B.R., and J.C. conceptualized the project; D.N.E., V.M.N., S.W., E.S., D.B.S., and L.C.K. performed the experiments and analyzed the data; and D.N.E., and J.C. wrote the manuscript.
Competing Interests: The authors declare that they have no competing interests.
Data and Materials Availability: All unique plasmids, cell lines, and mouse models not commercially available require a Materials Transfer Agreement from Vanderbilt University Medical Center.
References and Notes
- 1.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youngblood VM, Kim LC, Edwards DN, Hwang Y, Santapuram PR, Stirdivant SM, Lu P, Ye F, Brantley-Sieders DM, Chen J. The Ephrin-A1/EPHA2 Signaling Axis Regulates Glutamine Metabolism in HER2-Positive Breast Cancer. Cancer research. 2016;76:1825–1836. doi: 10.1158/0008-5472.CAN-15-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nature reviews Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brantley-Sieders DM, Jiang A, Sarma K, Badu-Nkansah A, Walter DL, Shyr Y, Chen J. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PloS one. 2011;6:e24426. doi: 10.1371/journal.pone.0024426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantley-Sieders DM, Zhuang G, Hicks D, Fang WB, Hwang Y, Cates JM, Coffman K, Jackson D, Bruckheimer E, Muraoka-Cook RS, Chen J. The receptor tyrosine kinase EphA2 promotes mammary adenocarcinoma tumorigenesis and metastatic progression in mice by amplifying ErbB2 signaling. The Journal of clinical investigation. 2008;118:64–78. doi: 10.1172/JCI33154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, Basilion JP, Sedor J, Wu J, Danielpour D, Sloan AE, Cohen ML, Wang B. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaught D, Brantley-Sieders DM, Chen J. Eph receptors in breast cancer: roles in tumor promotion and tumor suppression. Breast cancer research : BCR. 2008;10:217. doi: 10.1186/bcr2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J. Regulation of tumor initiation and metastatic progression by Eph receptor tyrosine kinases. Advances in cancer research. 2012;114:1–20. doi: 10.1016/B978-0-12-386503-8.00001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang WB, Ireton RC, Zhuang G, Takahashi T, Reynolds A, Chen J. Overexpression of EPHA2 receptor destabilizes adherens junctions via a RhoA-dependent mechanism. Journal of cell science. 2008;121:358–368. doi: 10.1242/jcs.017145. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang G, Brantley-Sieders DM, Vaught D, Yu J, Xie L, Wells S, Jackson D, Muraoka-Cook R, Arteaga C, Chen J. Elevation of receptor tyrosine kinase EphA2 mediates resistance to trastuzumab therapy. Cancer research. 2010;70:299–308. doi: 10.1158/0008-5472.CAN-09-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macrae M, Neve RM, Rodriguez-Viciana P, Haqq C, Yeh J, Chen C, Gray JW, McCormick F. A conditional feedback loop regulates Ras activity through EphA2. Cancer cell. 2005;8:111–118. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Su L, Ou Q. Yes-associated protein promotes tumour development in luminal epithelial derived breast cancer. European journal of cancer. 2012;48:1227–1234. doi: 10.1016/j.ejca.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhao D, Zhi X, Zhou Z, Chen C. TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis. 2012;33:59–67. doi: 10.1093/carcin/bgr242. [DOI] [PubMed] [Google Scholar]
- 15.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer research. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 16.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nature cell biology. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes & development. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes & development. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong X, Nguyen HT, Chen Q, Zhang R, Hagman Z, Voorhoeve PM, Cohen SM. Opposing activities of the Ras and Hippo pathways converge on regulation of YAP protein turnover. EMBO J. 2014;33:2447–2457. doi: 10.15252/embj.201489385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes & development. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, Zhao S, Xiong Y, Lei QY, Guan KL. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 28.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews Drug discovery. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 30.Fan R, Kim NG, Gumbiner BM. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2569–2574. doi: 10.1073/pnas.1216462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends in biochemical sciences. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. The Journal of clinical investigation. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson NE, Tryndyak VP, Beland FA, Pogribny IP. An in vitro investigation of metabolically sensitive biomarkers in breast cancer progression. Breast cancer research and treatment. 2012;133:959–968. doi: 10.1007/s10549-011-1871-x. [DOI] [PubMed] [Google Scholar]
- 34.Asiago VM, Alvarado LZ, Shanaiah N, Gowda GA, Owusu-Sarfo K, Ballas RA, Raftery D. Early detection of recurrent breast cancer using metabolite profiling. Cancer research. 2010;70:8309–8318. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch. 2004;447:469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- 36.Kekuda R, Prasad PD, Fei YJ, Torres-Zamorano V, Sinha S, Yang-Feng TL, Leibach FH, Ganapathy V. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J Biol Chem. 1996;271:18657–18661. doi: 10.1074/jbc.271.31.18657. [DOI] [PubMed] [Google Scholar]
- 37.Aledo JC, Gomez-Fabre PM, Olalla L, Marquez J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mammalian genome : official journal of the International Mammalian Genome Society. 2000;11:1107–1110. doi: 10.1007/s003350010190. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Kim DH, Jung WH, Koo JS. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr Relat Cancer. 2013;20:339–348. doi: 10.1530/ERC-12-0398. [DOI] [PubMed] [Google Scholar]
- 39.Qie S, Chu C, Li W, Wang C, Sang N. ErbB2 activation upregulates glutaminase 1 expression which promotes breast cancer cell proliferation. J Cell Biochem. 2014;115:498–509. doi: 10.1002/jcb.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 42.Brantley DM, Cheng N, Thompson EJ, Lin Q, Brekken RA, Thorpe PE, Muraoka RS, Cerretti DP, Pozzi A, Jackson D, Lin C, Chen J. Soluble Eph A receptors inhibit tumor angiogenesis and progression in vivo. Oncogene. 2002;21:7011–7026. doi: 10.1038/sj.onc.1205679. [DOI] [PubMed] [Google Scholar]
- 43.Xu MZ, Chan SW, Liu AM, Wong KF, Fan ST, Chen J, Poon RT, Zender L, Lowe SW, Hong W, Luk JM. AXL receptor kinase is a mediator of YAP-dependent oncogenic functions in hepatocellular carcinoma. Oncogene. 2011;30:1229–1240. doi: 10.1038/onc.2010.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer research. 2011;71:2728–2738. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 45.Lehmann W, Mossmann D, Kleemann J, Mock K, Meisinger C, Brummer T, Herr R, Brabletz S, Stemmler MP, Brabletz T. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun. 2016;7:10498. doi: 10.1038/ncomms10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gundry C, Marco S, Rainero E, Miller B, Dornier E, Mitchell L, Caswell PT, Campbell AD, Hogeweg A, Sansom OJ, Morton JP, Norman JC. Phosphorylation of Rab-coupling protein by LMTK3 controls Rab14-dependent EphA2 trafficking to promote cell:cell repulsion. Nat Commun. 2017;8:14646. doi: 10.1038/ncomms14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miura K, Wakayama Y, Tanino M, Orba Y, Sawa H, Hatakeyama M, Tanaka S, Sabe H, Mochizuki N. Involvement of EphA2-mediated tyrosine phosphorylation of Shp2 in Shp2-regulated activation of extracellular signal-regulated kinase. Oncogene. 2013;32:5292–5301. doi: 10.1038/onc.2012.571. [DOI] [PubMed] [Google Scholar]
- 48.Parri M, Taddei ML, Bianchini F, Calorini L, Chiarugi P. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer research. 2009;69:2072–2081. doi: 10.1158/0008-5472.CAN-08-1845. [DOI] [PubMed] [Google Scholar]
- 49.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 50.Herraiz C, Calvo F, Pandya P, Cantelli G, Rodriguez-Hernandez I, Orgaz JL, Kang N, Chu T, Sahai E, Sanz-Moreno V. Reactivation of p53 by a Cytoskeletal Sensor to Control the Balance Between DNA Damage and Tumor Dissemination. Journal of the National Cancer Institute. 2016;108 doi: 10.1093/jnci/djv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willenborg M, Panten U, Rustenbeck I. Triggering and amplification of insulin secretion by dimethyl alpha-ketoglutarate, a membrane permeable alpha-ketoglutarate analogue. Eur J Pharmacol. 2009;607:41–46. doi: 10.1016/j.ejphar.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Elgadi KM, Meguid RA, Qian M, Souba WW, Abcouwer SF. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol Genomics. 1999;1:51–62. doi: 10.1152/physiolgenomics.1999.1.2.51. [DOI] [PubMed] [Google Scholar]
- 53.Cassago A, Ferreira AP, Ferreira IM, Fornezari C, Gomes ER, Greene KS, Pereira HM, Garratt RC, Dias SM, Ambrosio AL. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1092–1097. doi: 10.1073/pnas.1112495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, Mackinnon AL, Parlati F, Rodriguez ML, Shwonek PJ, Sjogren EB, Stanton TF, Wang T, Yang J, Zhao F, Bennett MK. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 55.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schubeler D, Bauer A. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015;11:e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 64.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes & development. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 67.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes & development. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang K, Cao F, Fang W, Hu Y, Chen Y, Ding H, Yu G. Activation of SNAT1/SLC38A1 in human breast cancer: correlation with p-Akt overexpression. BMC Cancer. 2013;13:343. doi: 10.1186/1471-2407-13-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang Z, Cho HT, Williams L, Zhu A, Liang K, Huang K, Wu H, Jiang C, Hong S, Crowe R, Goodman MM, Shim H. Potential Biomarker of L-type Amino Acid Transporter 1 in Breast Cancer Progression. Nucl Med Mol Imaging. 2011;45:93–102. doi: 10.1007/s13139-010-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N, Harvey K, Beith JM, Selinger CI, O’Toole SA, Rasko JE, Holst J. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35:3201–3208. doi: 10.1038/onc.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, Hsieh AL, Gouw AM, Thomas AG, Gao P, Sun L, Song L, Yan B, Slusher BS, Zhuo J, Ooi LL, Lee CG, Mancuso A, McCallion AS, Le A, Milone MC, Rayport S, Felsher DW, Dang CV. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. The Journal of clinical investigation. 2015;125:2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang F, Zhang Q, Ma H, Lv Q, Zhang T. Expression of glutaminase is upregulated in colorectal cancer and of clinical significance. Int J Clin Exp Pathol. 2014;7:1093–1100. [PMC free article] [PubMed] [Google Scholar]
- 73.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nature reviews Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiang L, Xie G, Liu C, Zhou J, Chen J, Yu S, Li J, Pang X, Shi H, Liang H. Knock-down of glutaminase 2 expression decreases glutathione, NADH, and sensitizes cervical cancer to ionizing radiation. Biochim Biophys Acta. 2013;1833:2996–3005. doi: 10.1016/j.bbamcr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J, Wang C, Chen M, Cao J, Zhong Y, Chen L, Shen HM, Xia D. Epigenetic silencing of glutaminase 2 in human liver and colon cancers. BMC Cancer. 2013;13:601. doi: 10.1186/1471-2407-13-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J, Zhang C, Lin M, Zhu W, Liang Y, Hong X, Zhao Y, Young KH, Hu W, Feng Z. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget. 2014;5:2635–2647. doi: 10.18632/oncotarget.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang C, Liu J, Zhao Y, Yue X, Zhu Y, Wang X, Wu H, Blanco F, Li S, Bhanot G, Haffty BG, Hu W, Feng Z. Glutaminase 2 is a novel negative regulator of small GTPase Rac1 and mediates p53 function in suppressing metastasis. Elife. 2016;5:e10727. doi: 10.7554/eLife.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertero T, Oldham WM, Cottrill KA, Pisano S, Vanderpool RR, Yu Q, Zhao J, Tai Y, Tang Y, Zhang YY, Rehman S, Sugahara M, Qi Z, Gorcsan J, 3rd, Vargas SO, Saggar R, Saggar R, Wallace WD, Ross DJ, Haley KJ, Waxman AB, Parikh VN, De Marco T, Hsue PY, Morris A, Simon MA, Norris KA, Gaggioli C, Loscalzo J, Fessel J, Chan SY. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. The Journal of clinical investigation. 2016;126:3313–3335. doi: 10.1172/JCI86387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cox AG, Hwang KL, Brown KK, Evason KJ, Beltz S, Tsomides A, O’Connor K, Galli GG, Yimlamai D, Chhangawala S, Yuan M, Lien EC, Wucherpfennig J, Nissim S, Minami A, Cohen DE, Camargo FD, Asara JM, Houvras Y, Stainier DY, Goessling W. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nature cell biology. 2016;18:886–896. doi: 10.1038/ncb3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park YY, Sohn BH, Johnson RL, Kang MH, Kim SB, Shim JJ, Mangala LS, Kim JH, Yoo JE, Rodriguez-Aguayo C, Pradeep S, Hwang JE, Jang HJ, Lee HS, Rupaimoole R, Lopez-Berestein G, Jeong W, Park IS, Park YN, Sood AK, Mills GB, Lee JS. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology. 2016;63:159–172. doi: 10.1002/hep.28223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garber K. Cancer anabolic metabolism inhibitors move into clinic. Nat Biotechnol. 2016;34:794–795. doi: 10.1038/nbt0816-794. [DOI] [PubMed] [Google Scholar]
- 82.von Eyss B, Jaenicke LA, Kortlever RM, Royla N, Wiese KE, Letschert S, McDuffus LA, Sauer M, Rosenwald A, Evan GI, Kempa S, Eilers M. A MYC-Driven Change in Mitochondrial Dynamics Limits YAP/TAZ Function in Mammary Epithelial Cells and Breast Cancer. Cancer cell. 2015;28:743–757. doi: 10.1016/j.ccell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 83.Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS, Davis RJ, Mao J. Tead and AP1 Coordinate Transcription and Motility. Cell Rep. 2016;14:1169–1180. doi: 10.1016/j.celrep.2015.12.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rao M, Song W, Jiang A, Shyr Y, Lev S, Greenstein D, Brantley-Sieders D, Chen J. VAMP-associated protein B (VAPB) promotes breast tumor growth by modulation of Akt activity. PloS one. 2012;7:e46281. doi: 10.1371/journal.pone.0046281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muraoka RS, Koh Y, Roebuck LR, Sanders ME, Brantley-Sieders D, Gorska AE, Moses HL, Arteaga CL. Increased malignancy of Neu-induced mammary tumors overexpressing active transforming growth factor beta1. Mol Cell Biol. 2003;23:8691–8703. doi: 10.1128/MCB.23.23.8691-8703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 88.Ueda Y, Wang S, Dumont N, Yi JY, Koh Y, Arteaga CL. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J Biol Chem. 2004;279:24505–24513. doi: 10.1074/jbc.M400081200. [DOI] [PubMed] [Google Scholar]
- 89.Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nature cell biology. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EphA2 overexpression leads to intranuclear accumulation of YAP.
Nuclear exclusion of YAP upon Rho inhibition.
Supplemental Figure 1. EphA2 activates YAP/TAZ in in MCF10A-HER2 cells
Supplemental Figure 2. EphA2-EphrinA1 interactions do not significantly contribute to YAP/TAZ activation.
Supplemental Figure 3. Rho and ROCK inhibition do not enhance LATS1 phosphorylation in MMTV-Neu cells.
Supplemental Figure 4. EphA2-mediated YAP/TAZ activation requires Rho catalytic activity.
Supplemental Figure 5. YAP and TAZ promote glutamine metabolism in MCF10A-HER2-EphA2 cells.
Supplemental Figure 6. YAP and TEAD4 are associated with GLS and SLC1A5 promoters.
Supplemental Figure 7. YAP/TAZ and EphA2 expression strongly correlated with decreased patient survival in HER2+ breast cancer.
Supplemental Table 1. ChIP Primers