Abstract
Recent comprehensive molecular analysis allowed the identification of unique molecular signatures in pheochromocytomas and paragangliomas. Here we summarize the main pathway clusters activated by pheochromocytoma and paraganglioma susceptibility genes: pseudohypoxic, kinase, and Wnt signaling. Molecular characterization and clustering of pheochromocytomas and paragangliomas may help in the application of principles of personalized medicine and in decision making for targeted therapy of these tumors.
Keywords: Pheochromocytoma, paraganglioma, genetics, signaling pathways
Oncogenic Signaling in Pheochromocytoma and Paraganglioma
Recent studies extended our knowledge on the genetic landscape of pheochromocytoma (PHEO) and paraganglioma (PGL), and described new clinical phenotypes and signaling pathways involved in the pathogenesis of these tumors [1–3]. PHEOs and PGLs, catecholamine-producing neuroendocrine tumors arising from adrenal and extra-adrenal chromaffin tissues, respectively, present a great therapeutic challenge especially when they become metastatic. PHEOs and PGLs are currently associated with germline and/or somatic mutations in more than 20 genes. These mutations are divided into three main clusters based on the activation of a particular signaling pathway (Figure 1) and each cluster is associated with unique clinical characteristics of patients with these tumors.
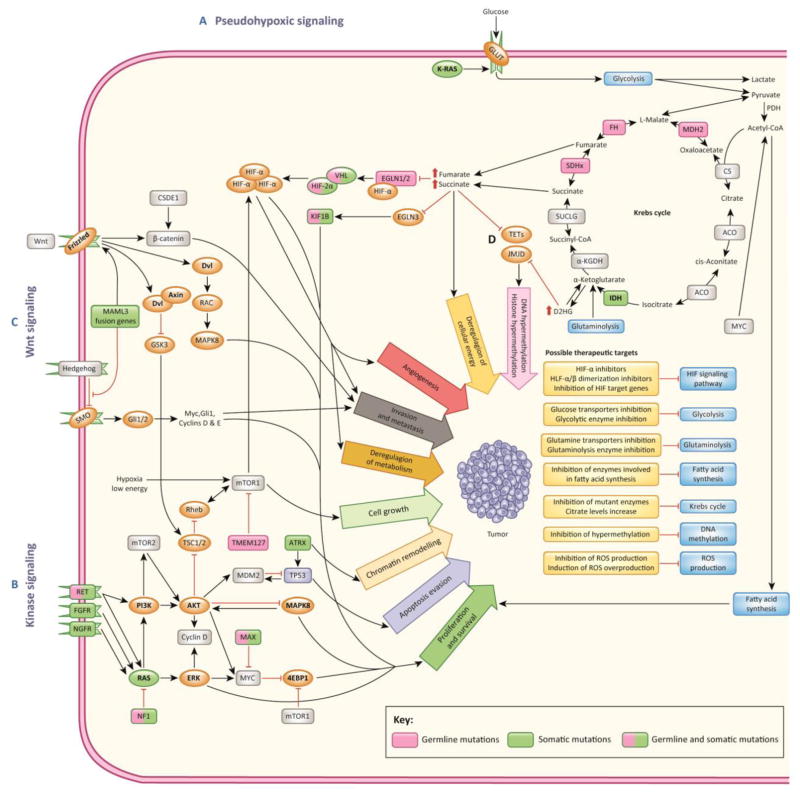
Figure 1. Main signaling clusters activated in PHEO and PGL.
A) Mutations in PHEO/PGL susceptibility genes belonging to the pseudohypoxic signaling cluster result in direct or indirect activation of the hypoxia signaling (HIF) pathway even in the presence of sufficient amount of oxygen (pseudohypoxia). Inactivating mutations in SDHx, SDHAF2, FH, or MDH2 lead to accumulation of Krebs cycle metabolites, such as succinate, fumarate, pyruvate, or glutamine, which is followed by activation of pseudohypoxic signaling and transcription of HIF target genes. VHL, EGLN1/2, and HIF2A genes are directly involved in hypoxic signaling. Mutations in these genes are associated with activation of the HIF signaling pathway. Pseudohypoxic signaling drives cell proliferation, deregulation of cellular energy and metabolism, angiogenesis, invasion and metastasis. B) Kinase signaling cluster includes mutations in genes involved in abnormal activation of kinase signaling, particularly PI3K/AKT, mTOR, and RAS/MAPK pathways. Activation of these pathways is associated with cell proliferation and survival, evasion from apoptosis, and chromatin remodeling. Kinase signaling pathways in PHEO and PGL can be disrupted by mutations in NF1, KIF1B, MAX, RET, TMEM127, H-RAS, and ATRX genes. Rarely, mutations in K-RAS, FGFR genes and fusion genes involving NGFR, BRAF, or NF1 can be found in PHEOs/PGLs. C) Recently, the Wnt signaling pathway has been implied to play role in PHEO/PGL pathogenesis based on the identification of CSDE1 mutations and MAML3 fusion genes [3]. D) Mutations in SDHx and FH genes result in an accumulation of succinate and fumarate and mutated IDH1 activity that results in conversion of α-ketoglutarate to the oncometabolite D2HG. Succinate, fumarate, D2HG, and L2HG function as competitive inhibitors of α-ketoglutarate–dependent dioxygenases. Inhibition of reactions mediated by α-ketoglutarate-dependent dioxygenases leads to DNA and histone hypermethylation, activation of hypoxic responses, and inhibition of collagen maturation and folding.
Abbreviations: 4EBP1, eukaryotic translation initiation factor 4E binding protein 1; α-KGDH, alpha-ketoglutarate dehydrogenase; Acetyl-CoA, acetyl-coenzyme A; ACO, aconitase; AKT, AKT serine/threonine kinase; ATRX, ATRX, chromatin remodeler; BRAF, B-Raf proto-oncogene; CS, citrate synthase; CSDE1, cold shock domain containing E1; D2HG, D-2-hydroxyglutarate; Dvl, Dishevelled segment polarity protein; EGLN1/2/3, egl-9 prolyl hydroxylase 1, 2, 3; ERK, mitogen-activated protein kinase 1; FGFR, fibrablast growth factor receptor; FH, fumarate hydratase; Gli1/2, GLI family zinc finger 1, 2; GLUT, glucose transporter; GSK3, glycogen synthase kinase 3; HIF, hypoxia-inducible factor; HIF2A, hypoxia-inducible factor 2 alpha; H-RAS, HRas proto-oncogene, GTPase; IDH1/2, isocitrate dehydrogenase 1, 2; JMJD, Jumoni C domain-containing histone lysine demethylases; KIF1B, kinesin family member 1B; K-RAS, KRAS proto-oncogene, GTPase; L2HG, L-2-hydroxyglutarate; MAPK, mitogen activated kinase; MAML3, mastermind like transcriptional coactivator 3; MAX, MYC associated factor X; MDH2, malate dehydrogenase 2; MDM2, MDM2 proto-oncogene; mTOR, mechanistic target of rapamycin; MYC, MYC proto-oncogene; NF1, neurofibromin 1; NGFR, nerve growth factor receptor; PDH, pyruvate dehydrogenase; PHD1/2, egl-9 family hypoxia inducible factor 2, 1; PHEO/PGL, pheochromocytoma/paraganglioma; PI3K, phosphatidylinositol-3-kinase; RAC, Rho guanine nucleotide exchange factors; RAS, RAS type GTPase family; RET, RET proto-oncogene; Rheb, Ras homolog enriched in brain; ROS, reactive oxygen species; SDHx, succinate dehydrogenase subunits A, B, C, D, SDHAF2, succinate dehydrogenase complex assembly factor 2; SMO, Smoothened, frizzled class receptor; SUCLG, succinyl-coenzyme A ligase; TETs, tet methylcytosine dioxygenases; TMEM127, transmembrane protein 127; TP53, tumor protein 53; TSC1/2, TSC complex subunit 1, 2; VHL, von Hippel-Lindau tumor suppressor,
Pseudohypoxic Signaling Cluster
The hypoxia-inducible factor (HIF) signaling pathway is affected by mutations in genes encoding the hypoxia-inducible factor 2 alpha (HIF2A), succinate dehydrogenase subunits (SDHx [SDHA, SDHB, SDHC, SDHD]), succinate dehydrogenase complex assembly factor 2 (SDHAF2), von Hippel-Lindau tumor suppresor (VHL), egl-9 prolyl hydroxylase 1 and 2 (EGLN1/2), fumarate hydratase (FH), malate dehydrogenase 2 (MDH2), and isocitrate dehydrogenase (IDH). PHEOs and PGLs associated with mutations are pseudohypoxic, since the upregulation of HIF-α is not caused by hypoxia (i. e. insufficient oxygen levels) but by various other mechanisms (Figure 1). Gain-of-function mutations in HIF2A result in direct activation of HIF signaling and upregulation of HIF-α target genes [4]. Mutations in genes encoding Krebs cycle enzymes lead to deregulation of cellular energy, chromatin remodeling, changes in DNA methylation, and ROS production. These mutations lead to a metabolic shift that result in increased dependence on glycolysis, more or less impaired oxidative phosphorylation, and a substantial increase in some Krebs cycle oncometabolites, such as succinate, fumarate, or 2-hydroxyglutarate [5]. PHEOs and PGLs tumors in this cluster are aggressive and often metastatic. Furthermore, multiple and recurrent tumors are very common and the clinical outcome of patients in this cluster is the poorest compared to other susceptibility gene mutations.
Kinase Signaling Cluster
Dysregulation of the PI3K/mTOR pathway/receptor kinase signaling results from mutations in the RET proto-oncogene, neurofibromin 1 (NF1) tumor suppressor, H-RAS and K-RAS proto-oncogenes, transmembrane protein 127 (TMEM127), Myc-associated factor X (MAX), chromatin remodeler ATRX, and cold shock domain containing E1 (CSDE1) [3, 6]. The majority of PHEOs and PGLs in this cluster have overall good clinical outcome, except for those with ATRX mutations, where recurrence and metastases are more common [3]. PI3K/AKT and RAS/mitogen-activated protein kinases (MAPK) signaling pathways can also be altered by fusion genes involving NF1, B-Raf proto-oncogene (BRAF), and nerve growth factor receptor (NGFR), although these are very rare [3]. Activation of PI3K/AKT and RAS/MAPK signaling regulates mechanisms of cell growth, proliferation, apoptosis, and chromatin remodeling; and is also involved in the metabolic switch toward glycolysis and glutaminolysis in cancer cells [7]. Most PHEOs and PGLs in this cluster are adrenal and have a typical adrenergic phenotype compared to those in the HIF signaling cluster, where almost all tumors (except for VHL-associated) are extra-adrenal but all have the noradrenergic phenotype [8].
Wnt Signaling Cluster
PHEOs and PGLs overexpressing genes of the Wnt and Hedgehog pathways belong to so-called Wnt-altered subtype. These tumors are PHEOs and are related to somatic mutations in CSDE1 and the mastermind like transcriptional coactivator 3 (MAML3) fusion genes (upstream binding transcription factor, RNA polymerase I (UBTF)-MAML3 and transcription factor 4 (TCF4)-MAML3). MAML3 fusion-positive PHEOs display hypomethylating phenotype and increased Wnt and Hedgehog signaling. Wnt-altered tumors exhibit high expression of CHGA, a gene that encodes chromogranin A – a clinical marker of neuroendocrine tumors [3]. The Wnt pathway regulates a variety of developmental processes such as cell proliferation, adhesion and motility, and cell polarity and differentiation. Target genes of the Wnt signaling include for example the MYC proto-oncogene and cyclin D1 [9].
Alterations in these three aforementioned major signaling pathways lead to the dysregulation of many other cellular processes, including numerous metabolic (e. g. activation of MAPK signaling or switch to aerobic glycolysis) and epigenetic changes (such as modifications in DNA methylation, histones, chromatin remodeling, or nucleosome positioning) [10]. For instance, in SDHx- or FH-mutated tumors the accumulation of succinate or fumarate occurs, respectively, and mutated IDH1 demonstrates neomorphic activity leading to conversion of α-ketoglutarate to oncometabolite D-2-hydroxyglutarate (D2HG). Succinate, fumarate, and D2HG act as competitive inhibitors of α-ketoglutarate-dependent dioxygenases [tet methylcytosine dioxygneases (TET), Jumoni C domain-containing histone lysine demethylases (JMJD), EGLNs, lysyl hydroxylase (LHD)] (Figure 1). L-2-hydroxyglutarate (L2HD), a product of lactate dehydrogenase A (LDHA) and MDH1/2 metabolism in hypoxic cells, is another α-ketoglutarate-dependent dioxygenase inhibitor. Inhibiton of reactions mediated by α-ketoglutarate-dependent dioxygenases results in dysregulation of HIF degradation, DNA and histone hypermethylation, and inhibition of collagen maturation and folding (reviewed in [5]).
Metabolic profiling of PHEOs and PGLs demonstrated accumulation of certain metabolites (e. g. succinate, fumarate, L2HD, pyruvate) or a decrease of others (e. g. citrate) [11]. Some of these metabolic changes are specific for tumors with certain genetic background, for example different metabolic profile of SDHx-mutated tumors compared to sporadic PHEOs and PGLs (reviewed in [5]). Based on this, the tumor metabolic profile can serve as a ground for decision making in personalized and targeted therapy (Figure 1). In addition, new data on genetic, metabolic, and biochemical alterations of PHEO and PGL are allowing to look for new tumor-specific targets. Metabolic profiling should become an integral part of the diagnostic process preceding genetic testing, because known genotype–metabolic phenotype correlation could help in genetic testing decision making in a particular patient. Some promising therapeutic strategies include modifying or inhibiting the metabolic processes and enzymes that participate in metabolic reprogramming; replenishing depleted substrates for Krebs cycle; or using demethylating agents, as summarized in the Figure 1 (reviewed in [5]). Further understanding of the metabolic and genetic basis of PHEO and PGL will undoubtedly lead to the development of effective forms of therapy for these tumors.
Acknowledgments
This research was supported, in part, by the Intramural Research Program of the NIH, NICHD.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gimenez-Roqueplo AP, et al. An Update on the Genetics of Paraganglioma, Pheochromocytoma, and Associated Hereditary Syndromes. Horm Metab Res. 2012;44:328–333. doi: 10.1055/s-0031-1301302. [DOI] [PubMed] [Google Scholar]
- 2.Favier J, et al. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat Rev Endocrinol. 2015;11:101–111. doi: 10.1038/nrendo.2014.188. [DOI] [PubMed] [Google Scholar]
- 3.Fishbein L, et al. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell. 2017;31:181–193. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhuang Z, et al. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367:922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jochmanova I, Pacak K. Pheochromoctyoma: The First Metabolic Endocrine Cancer. Clin Cancer Res. 2016;22:5001–5011. doi: 10.1158/1078-0432.CCR-16-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comino-Méndez I, et al. ATRX driver mutation in a composite malignant pheochromocytoma. Cancer Genet. 2016;209:272–277. doi: 10.1016/j.cancergen.2016.04.058. [DOI] [PubMed] [Google Scholar]
- 7.Icard P, Lincet H. A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochim Biophys Acta. 2012;1826:423–433. doi: 10.1016/j.bbcan.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Eisenhofer G, et al. Measurements of plasma methoxytyramine, normetanephrine, and metanephrine as discriminators of different hereditary forms of pheochromocytoma. Clin Chem. 2011;57:411–420. doi: 10.1373/clinchem.2010.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielinska M, et al. Review paper: origin and molecular pathology of adrenocortical neoplasms. Vet Pathol. 2009;46:194–210. doi: 10.1354/vp.46-2-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo RA, et al. Recurrent Mutations of Chromatin-Remodeling Genes and Kinase Receptors in Pheochromocytomas and Paragangliomas. Clin Cancer Res. 2016;22:2301–2310. doi: 10.1158/1078-0432.CCR-15-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao JU, et al. Genotype-specific differences in the tumor metabolite profile of pheochromocytoma and paraganglioma using untargeted and targeted metabolomics. J Clin Endocrinol Metab. 2015;100:E214–222. doi: 10.1210/jc.2014-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]