Significance
Tissues are viscoelastic in nature and their physical properties play a fundamental role in development, tumorigenesis, and wound healing. Cell response to matrix elasticity is well understood through a “molecular clutch” which engages when stiffness is sufficiently high to expose binding sites in mechanosensitive proteins. Here we show that cell response to pure viscous surfaces (i.e., with no elastic component) can be explained through the same molecular clutch. Mechanisms used by cells to sense rigidity are more universal and can be used to unveil cell interaction with complex viscoelastic environments. The research presents a tool to understand cells within tissues and in turn opens new avenues to incorporate viscosity into the design of synthetic cellular microenvironments.
Keywords: matrix rigidity, molecular clutch, surface viscosity, mechanotransduction, cell differentiation
Abstract
Cell response to matrix rigidity has been explained by the mechanical properties of the actin-talin-integrin-fibronectin clutch. Here the molecular clutch model is extended to account for cell interactions with purely viscous surfaces (i.e., without an elastic component). Supported lipid bilayers present an idealized and controllable system through which to study this concept. Using lipids of different diffusion coefficients, the mobility (i.e., surface viscosity) of the presented ligands (in this case RGD) was altered by an order of magnitude. Cell size and cytoskeletal organization were proportional to viscosity. Furthermore, there was a higher number of focal adhesions and a higher phosphorylation of FAK on less-mobile (more-viscous) surfaces. Actin retrograde flow, an indicator of the force exerted on surfaces, was also seen to be faster on more mobile surfaces. This has consequential effects on downstream molecules; the mechanosensitive YAP protein localized to the nucleus more on less-mobile (more-viscous) surfaces and differentiation of myoblast cells was enhanced on higher viscosity. This behavior was explained within the framework of the molecular clutch model, with lower viscosity leading to a low force loading rate, preventing the exposure of mechanosensitive proteins, and with a higher viscosity causing a higher force loading rate exposing these sites, activating downstream pathways. Consequently, the understanding of how viscosity (regardless of matrix stiffness) influences cell response adds a further tool to engineer materials that control cell behavior.
Much research has been undertaken with respect to how matrix properties influence cellular behavior, including the use of synthetic materials. Indeed, controlling these properties has been shown to be key to defining principal cellular functions, driving cells toward growth or apoptotic pathways (1). Consequently, it is widely recognized that features such as stiffness (2), topography (3), or chemical modification (4) can define the cell response. This is of particular importance in stem cell lines, where these properties have the potential to either promote lineage commitment or self-renewal (3), with the promotion or restriction of cell spreading enough to drive osteogenic or adipogenic lineage commitment, respectively (5).
Viscosity defines the range of motion, or mobility, of the molecules on a surface and is a property conceptually distinct from stiffness but also illuminates a further surface property that can significantly affect cellular behavior. For example, mobility has an effect on cell adhesion (6), spreading (7), focal adhesion (FA) properties (8), and cell fate (9). It also has the potential to change the nature of the cell/material interface, in terms of protein conformation (10–12). The effect of viscoelastic substrates has also shown that a combination of viscosity and stiffness changes the cell response (13, 14), for example increased stress-relaxation compensates for lower elastic moduli (13), implying that increasing the magnitude of the viscous element affects cellular behavior. However, current evidence arises from studies on substrates with both viscous and elastic components. Previous work has begun to elucidate the role of viscosity, noting that the lateral mobility can alter the cell response (8). However, the reason for this influence of mobility (viscosity) on cell behavior has yet to be understood.
To respond to their environment cells must have a link between the cytoskeleton and the surrounding matrix. This is initiated by integrin proteins, leading to the recruitment of further proteins forming FAs, linking the cytoskeleton to the surface (15). The nature of this overall interaction, specifically its degree of activation, has a downstream effect on transcription factors, which, in turn, guide the cell toward a specific lineage. Changing the nature of the surface can affect one or all of these processes, with recruitment of various integrin types (16), associated proteins (17, 18), actin properties (19), and transcription factors/gene expression (20, 21), all varying based on the nature of the cell/matrix interaction.
Supported lipid bilayers (SLBs) provide a well-characterized and easily manipulated system through which to study how the mobility of the surface (i.e., viscosity) controls these various factors, without the contribution of elasticity. Previously it has been shown that, while normally nonfouling, SLBs functionalized with various proteins (14, 22, 23) or peptides (24, 25) can allow for cell adhesion and spreading, as well as control of differentiation (26). Furthermore, SLBs have been used to understand the role of traction forces in the endocytic turnover of integrins (27) and the formation of mature FAs (28, 29).
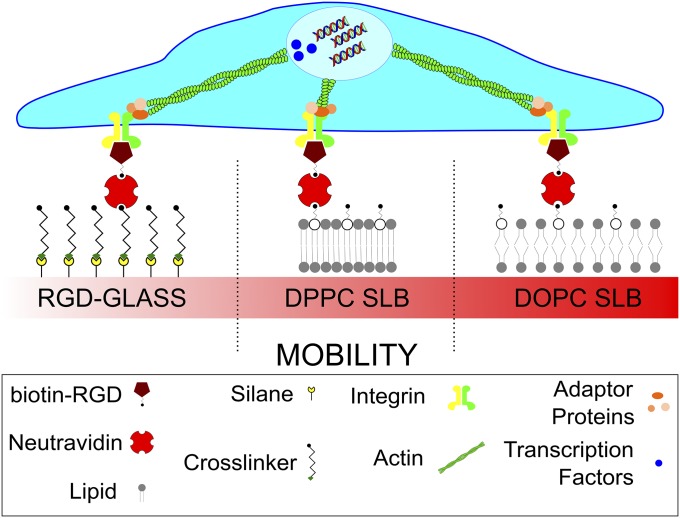
Here, the mobility/viscosity of RGD functionalized SLBs is manipulated. At a given temperature, defined by the intrinsic molecular properties of the constituent lipids, the acyl chains of the lipids melt and the system moves from a well-packed and immobile gel phase to a more disordered and mobile fluid phase. This viscosity changes the ligand mobility, as measured by the diffusion coefficient (D), and would consequently control the mobility of a ligand adsorbed to the surface. This study, summarized in Fig. 1, uses the fibronectin-derived RGD cell-binding tripeptide to understand the nature of and, more importantly, the processes dictating the cell response to mobility of ligands in these surfaces of varying viscosity. It was hypothesized that changing this mobility of the RGD ligands would change the nature of the cell/material interaction (Fig. 1), thus having a knock-on effect on further cell properties. It is shown here that in response to surface viscosity cell behavior is mediated by a dynamic clutch mechanism. Analogously to the role of elastic rigidity, viscosity triggers reduced actin flows, adhesion growth, YAP nuclear translocation, and myoblast differentiation. This work, especially on such a well-characterized model system, presents the opportunity to determine in depth the nature and processes that cells use to dictate their response to ligand mobility underpinned by viscosity. Understanding this can, in future, be utilized to provide further possible surfaces for controlling the cell response, as well as a means to understand how this understudied surface property defines cellular behavior.
Fig. 1.
Sketch of the systems used to control surface viscosity. DOPC and DPPC lipid bilayers were functionalized with same surface density of RGD. Functionalized glass was used as a control. The mobility of the ligands presented on the surface is driven by the viscosity of the bilayer. The proteins and processes cells use to detect this mobility, such as the nature of the FAs, actin flow, and protein translocation are determined. The consequence of these effects on cell differentiation is also evaluated.
Results and Discussion
RGD Functionalized Lipid Bilayers with Well-Controlled Surface Viscosity.
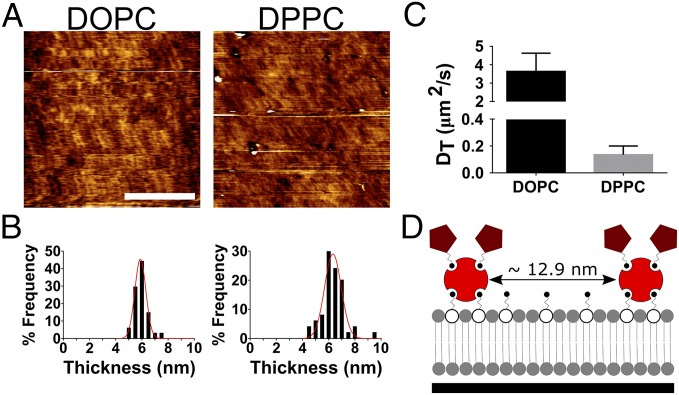
Fig. 2 shows the presence of SLBs of both the fluid- (1,2-dioleoyl-sn-glycero-3-phosphocholine, DOPC) and gel- (1,2-dipalmitoyl-sn-glycero-3-phosphocholine, DPPC) phase lipid on glass surfaces (Fig. 2A). The presence of both bilayers is confirmed via force mapping (Fig. 2B). The resulting force curves (representative curve in Fig. S1A), characteristic of an SLB (30, 31), was used to estimate the thickness of the DOPC and DPPC SLBs as 5.9 ± 0.4 nm and 6.3 nm ± 0.6 nm, respectively, a size range expected in single lipid bilayers (32–34). In conjunction with this, the lack of contrast seen in Fig. 2A demonstrates a predominantly homogenous surface with roughness (rms) values within the picometer range for all surfaces (DOPC = 0.4 ± 0.1 nm, DPPC = 0.5 ± 0.1 nm, and glass = 0.5 ± 0.1 nm). Further to this, fluorescence correlation spectroscopy (FCS) measurements (decay curves shown in Fig. S1 B and C) confirmed the difference in viscosity of the respective bilayers with differences between diffusion coefficients equaling an order of magnitude at 37 °C (Fig. 2C). These diffusion measurements are indicative of fluid-phase DOPC (D = 3.6 µm2/s) SLB (35) and a much less mobile gel-phase DPPC (D = 0.1 µm2/s) SLB. The original Saffman–Delbruck equation describes the relationship between the diffusion coefficient (D) and the viscosity (η) (36). However, this equation has been shown to have limitations across the wide range of conditions experienced in cell substrate interactions. There have been a number of attempts to further develop this equation including the Hughes–Pailthorpe–White model (37) so that its validity holds across a wider range of lengths [the original equation was noted to only accurately account for this relationship when the radius of the diffusing object was significantly smaller than the “Saffman–Delbruck length” (36)]. Eq. 1 (38) provides an alternative relationship between diffusion and the viscosity; shown to predict diffusion of membrane proteins where the Saffman–Delbruck cannot, it estimates a viscosity of 8.4 × 10−11 and 3.0 × 10−9 Pa⋅s ⋅m for DOPC and DPPC, respectively. Where D is the diffusion coefficient, kB is the Boltzmann constant, T the absolute temperature, ηm the membrane viscosity, R the radius (0.5 nm, the radius of a single lipid), and λ the characteristic length. This characteristic length is an indication of membrane perturbation and thus is assumed to be on the order of a single lipid (0.5 nm):
| [1] |
Upon functionalization of the bilayer, the interparticle distance of neutravidin/RGD on DOPC and DPPC was 12.9 nm in both cases with similar SD (0.4 and 0.2 nm, respectively) as detected by quantitative fluorescence methods described previously (Fig. 2D) (39). These results confirm that these SLB systems are in two wholly different phases, with a large difference in the mobility of their ligands, maximizing the cell’s response to these changes in viscosity. The effect of other physical properties, such as the stiffness, topography, and chemistry layer can also be disregarded. For the former of these it has been previously reported in the literature that the detected stiffness of an SLB is in the megapascal range (32), far above a cell’s detectable range (40). Topography was seen to be similar between the surfaces and contribution of chemistry is minimal due to the bilayers’ containing lipids of the same hydrophilic headgroups as well as being nonfouling (41). Furthermore, incorporation of ligands was seen not to be affected by the viscosity, thus ruling out any contribution of ligand density to the cell response.
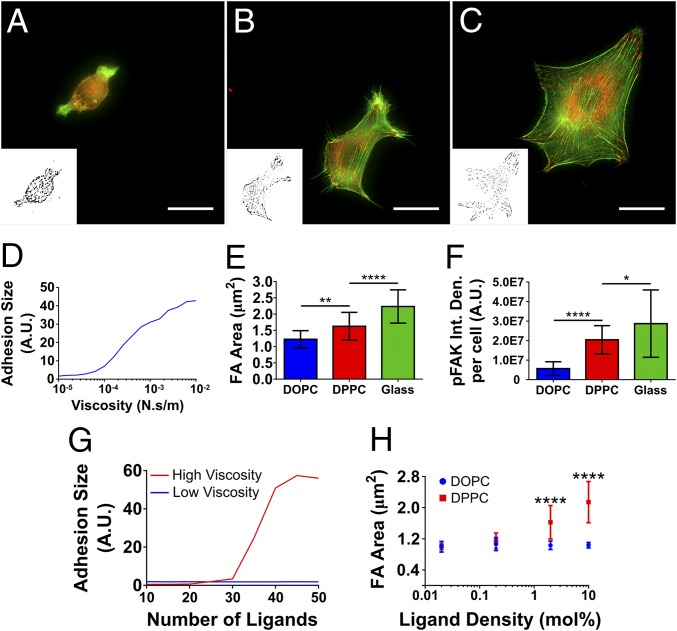
Fig. 2.
Characterization of SLBs. (A) The AFM (contact mode) images of both the DOPC (fluid phase) and DPPC (gel phase) bilayers. (Scale bar: 2 µm.) (B) The histograms of the thickness of both SLBs fitted to Gaussian distributions as measured via force mapping (DOPC n = 64, DPPC n = 50). (C) The diffusion coefficients of lipid bilayers as measured by fluorescence correlation spectroscopy (DOPC n = 10, DPPC n = 8). (D) A schematic representation of both the average interparticle distance (as calculated by 1/√n, where n is the particle density) and inferred average number of RGD groups per neutravidin (approximately two) molecule as determined by quantitative fluorescence microscopy.
Surface Viscosity Controls Cell Morphology and Actin Assembly.
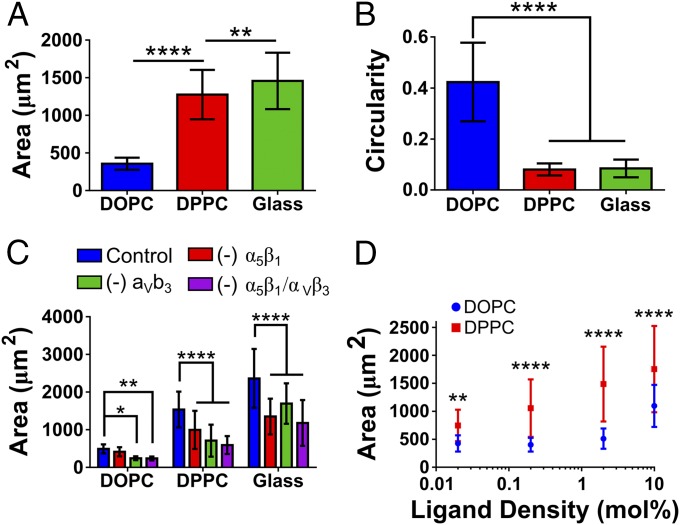
Previous work has established that the ligand mobility can have a significant effect on the cellular response in other systems (6, 7, 9, 42). Indeed, fluid (low-viscosity) SLBs lead to a cell phenotype resembling that observed on very soft elastic substrates, suggesting that force transmission is impaired (28, 29). The inclusion of a gel-phase bilayer in the current work has been used as an intermediate viscosity through which to bridge the gap between a mobile (DOPC) and immobile surface (glass), illuminating how cells respond to viscosity through the mobility of ligands on the surface. Fig. 3 demonstrates that the large-scale effects of a low-viscosity/high-ligand mobility surface on cells (i.e., DOPC) are as previously described in earlier work (27–29); cells had a smaller area (Fig. 3A) and a more circular morphology (Fig. 3B). This effect of the viscosity is limited as reducing it further (by the inclusion of 30% LysoPC) was not seen to reduce the cell area (Fig. S3). However, upon an increase in viscosity (i.e., DPPC) an increase in cell area and reduction in circularity is seen. A further increase in cell area is observed upon the removal of this viscosity altogether in the RGD-glass surface. Interestingly, this is in contrast to a biphasic cell spreading/viscosity relationship previously reported in viscoelastic substrates (8). However, we note that our results characterize a purely viscous rather than viscoelastic response and also a broader range of viscosities, leading to responses from an unstructured cytoskeleton in DOPC to well-formed stress fibers in DPPC and glass (Fig. S2).
Fig. 3.
Physical characteristics of cells on differentially mobile SLBs. (A) The increase in the average area of C2C12 cells as viscosity increases. For all samples n = 64. (B) The concomitant decrease of circularity; from left to right n = 22, 22, and 25. In both cases statistical differences were determined by one-way ANOVA. (C) The reduction in the cell area upon incubation of cells with BMB5 and Gpen, inhibtiors of α5 and α3, respectively, both independently and simultaneously. For DOPC/DPPC/RGD-glass n (control) = 56/53/45; (- α5β1) = 60/59/62; (- αVβ3) = 45/61/60; (- α5β1 & αVβ3) = 58/60/65. (D) The changes in the cell area upon both DOPC and DPPC as the mole percent RGD-containing lipid is increases. For DOPC/DPPC in each ligand n = 21/23, 17/28, 19/27, and 19/29 on 0.02, 0.2, 2, and 10 mol % respectively. Statistical differences in C and D were determined via two-way ANOVA. In D the only the differences between DOPC and DPPC are shown. In DOPC statistical differences noted were only seen between 10 mol % on all other ligand densities. In DPPC a statistical difference between all ligand densities of at least P = 0.01 is seen. Representative images of both C and D are displayed in Figs. S3 and S5, respectively. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001.
Cell area decreases upon the blocking of α5β1 and αvβ3 integrins (Fig. 3C; representative images in Fig. S4), key mediators of cell binding to fibronectin. A significant increase in cell area was also noted on DPPC surfaces as the ligand density was increased over three orders of magnitude, in contrast to DOPC, where the cell area was only seen to increase at 10 mol % of RGD-lipid (Fig. 3D; representative images in Fig. S5). Drawing parallels to elastic stiffness, cells have been previously noted to respond to changes to surface ligand density to a greater extent at higher stiffness, a similar trend to that seen here (43). Regardless of ligand density, cells on DOPC are unable to exert force on the surface. This is in contrast to DPPC, where cells can spread to a greater extent as the ligand density is increased.
Cell shape has previously been noted to be dependent on the nature of the surface. For example, stiffer surfaces show greater cell spreading (2) as well as cell shape’s being a key regulator in cellular processes, such as lineage commitment (5), growth, and apoptosis (1). Here it is seen that increasing ligand mobility, by reducing viscosity, leads to a monophasic change in the cell shape to a smaller and more rounded morphology, with a concomitant reduction in cytoskeletal tension, which is mediated by α5β1 and αvβ3 integrins.
The Molecular Clutch Explains Actin Flow and FA Assembly in Response to Viscosity.
Cell morphology was smaller and more rounded on low-viscosity surfaces, with larger, more spread cells seen on high-viscosity surfaces, reminiscent of the cell response to stiffness on elastic substrates (2). The response of cells to increased stiffness can be interpreted through the “molecular clutch” model. First proposed by Mitchison and Kirschner (44) and framed computationally by Chan and Odde (45), this model links the ECM to the actin cytoskeleton, through integrins and FA complexes.
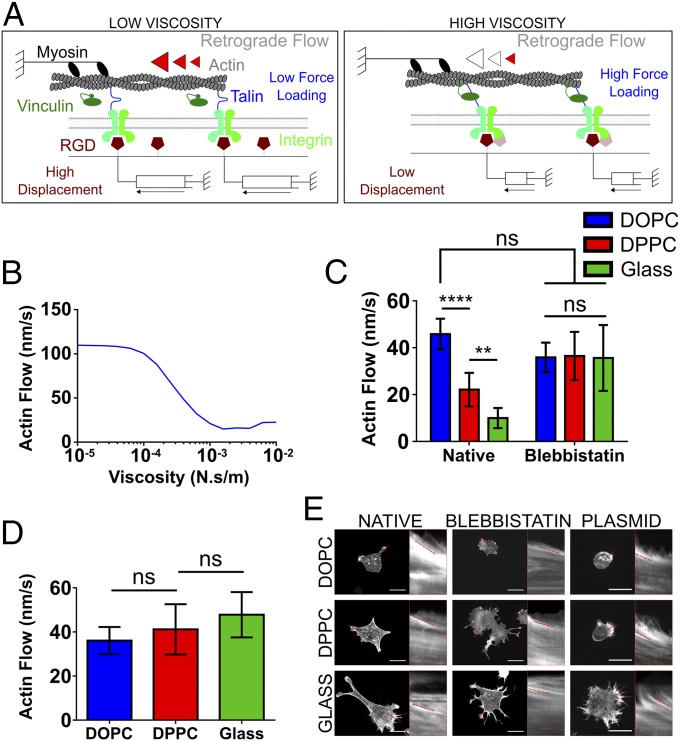
The model first considers myosin contractility, which powers a continuous flow of actin toward the cell center (retrograde flow). Once this actin flow is coupled to the ECM through FAs and integrins, myosin contractility is countered by the elastic resistance of the substrate to deformation, slowing the flow but increasing the rate of force loading on integrins and FA molecules. This force loading rate increases with increasing stiffness, enabling the sensing of stiffness. At a high stiffness, the high loading rate allows for force-sensitive proteins (talin) to unfold before the integrin–ECM connection breaks, leading to vinculin binding, the growth of adhesion sites, and downstream signaling (17). At lower stiffness the opposite is true, with slower force loading leading to integrin–ECM detachment before allowing force-sensitive talin unfolding. While this has been shown to be true on elastic stiffness, it is hypothesized here that the viscosity of the membrane is detected in a similar manner, since actin retrograde flow would also load forces faster when connected to a mobile substrate with high rather than low viscosity.
To test this hypothesis, a modified version of the previously described clutch model (16, 17) was implemented, in which each RGD was considered to be bound to a viscous dashpot instead of an elastic spring (Fig. 4A). As expected, the model predicted that the effects of increasing viscosity were analogous to those of increasing stiffness. That is, talin unfolding occurs only above a threshold in viscosity, leading to a reduction in actin flow (Fig. 4B). To test this model, first actin retrograde flows were measured in the different conditions by using live cells transfected with fluorescent actin. As predicted, actin flows decreased as the viscosity increased (Fig. 4C), demonstrating the mechanical nature of the differences observed. These were abolished upon inhibition of the contractile machinery with blebbistatin (Fig. 4C); by blocking the activity of myosin II, the actin flow rate is therefore controlled only by the rate of F-actin polymerization at the cell edge. Indeed, the rate of actin flow was observed to be similar in all cases to that of the most mobile surface, DOPC. Second, it was verified that the response was mediated as predicted by talin unfolding. To this end, cells were transfected with the head domain of vinculin (VD1), containing the talin binding domain. Previous work has shown that VD1 acts as a dominant negative for the binding to talin of endogenous vinculin, preventing the stiffness response mediated by talin unfolding (17). As predicted, despite no significant differences between FAs in native and VD1+ cells (Fig. S6), VD1 transfection prevented the reduction in actin flows induced by increased viscosity (Fig. 4D).
Fig. 4.
Viscosity-dependent actin flow. (A) Schematic representation of the inferred impact of viscosity of the molecular clutch. Myosin contractility pulls on actin filaments, leading to retrograde flow and movement of RGD ligands bound to lipids. On low-viscosity substrates (Left), ligand movement generates low forces, which don’t significantly slow actin flow. On high-viscosity substrates (Right), ligand movement generates high forces, slowing retrograde flow and triggering talin-mediated reinforcement and adhesion growth. (B) The clutch model prediction concerning the actin flow substituting stiffness for viscosity; this shows that as the viscosity of the surface increases there is a concomitant decrease in actin flow. The retrograde flow of actin in LifeAct-RFP transfected C2C12 cells is shown in C with and without blebbistatin, an inhibitor of mechanotransduction. In the native samples from left to right n = 11, 12, and 17, and in the blebbistatin-containing samples n = 9, 9, and 18. (D) The average actin flow after transfection with the VD1 plasmid, which produces the vinculin head domain capable of dominantly binding talin over endogenous vinculin. From left to right n = 10, 12, and 13. (E) Representative images and kymographs for all surfaces. (Scale bars: 25 µm.) ns, P > 0.05, **P ≤ 0.01, ****P ≤ 0.0001.
Third, the prediction that increased viscosity should also lead to the formation of FAs was verified. To this end, the recruitment of two FA markers was measured: vinculin (representative images shown in Fig. 5 A–C) and phosphorylated FAK (pFAK). Further to the decrease in actin flow, noted above, the model also predicts that talin unfolding occurring above the threshold will also lead to progressive adhesion growth (Fig. 5D); this proved true with the FA area’s increasing with viscosity (Fig. 5E). Furthermore, FAs on DPPC and RGD-glass looked better defined in contrast to the more heterogeneous clustering seen on DOPC. This is not only supported by model predictions but also by related work on ligand mobility (8). In conjunction with this, by measuring the activity of pFAK (Fig. 5F; representative images Fig. S7) within these FAs it was seen that the degree of FAK phosphorylation was also increased in line with viscosity.
Fig. 5.
Properties and activity of FAs. (A–C) The presence of FAs in cells on DOPC, DPPC, and RGD-glass, respectively (red, vinculin; green, actin). Insets show the binary images used to quantify FAs. (Scale bars: 25 µm.) (D) The model prediction of the increase in adhesion size as the viscosity increases. (E) The FAs size in the cells on each of the surfaces. From left to right n = 19, 20, and 20. (F) The activity of the FAs on each surface, a represented by the amount of pFAK. From left to right n = 26, 26, and 15. In both cases (E and F) one-way ANOVA was used to determine statistical differences, which are given as P values, indicated as *P < 0.05, **P < 0.01, and ****P < 0.0001. (G) Model predictions regarding ligand density at high and low viscosity, demonstrating that when viscosity is high (e.g., DPPC) the adhesion size will decrease as the number of ligands, or clutches, decreases. This is in contrast to low viscosity (e.g., DOPC), where no difference is seen. (H) The change in FA size as the ligand density on the fluid-phase (DOPC) and gel-phase (DPPC) SLBs is increased as mole percent of functionalized lipid over three orders of magnitude. For DOPC/DPPC in each ligand n = 21/23, 17/28, 19/27, and 19/29 on 0.02, 0.2, 2, and 10 mol %, respectively. The numbers below each point show the estimated interligand distance between RGD molecules at each ligand density, with the asterisk at 12.9 nm indicating that this is has been measured (as shown in Fig. 2D) and has been used to estimate the remaining distances. Statistical differences were determined via two-way ANOVA, with P values indicated as previous stated. Only the statistical differences between DOPC and DPPC are shown. On DOPC there was no statistical difference between ligand densities. On DPPC 0.02 mol % and 0.2 mol % showed no statistical difference, with differences noted between all other surfaces. Figs. S4 and S5 show representative images of E and F, respectively.
Finally, the effect of ligand density was assessed. The clutch model predicts that reduced ligand density should have no effect on very mobile substrates, as talin unfolding does not occur. However, as viscosity (or stiffness) increases in less-dense substrates force is distributed among fewer clutches, leading to a higher force per clutch, increased reinforcement, and increased adhesion growth (46). If viscosity or stiffness increases sufficiently, adhesions can no longer grow and eventually collapse (Fig. 5G and ref. 46). This leads to smaller adhesions in less-dense substrates. Whereas this system did not have enough resolution to examine the intermediate regime, it was verified that, as predicted (Fig. 5G), adhesion size was not affected by ligand density at very low viscosities (DOPC) but increased with ligand density at very high viscosities (DPPC) (Fig. 5H; representative images in Fig. S5).
Despite the good agreement between model predictions and experimental trends it is noted that the computational clutch model predicts an initiation of reinforcement and adhesion growth at values above 10−4 N⋅s/m, greater than those detected in freely diffusing SLBs shown in Fig. 2, and calculated through Eq. 1 (38). However, a characteristic length (λ) of 0.5 nm is likely not to be accurate here, due to the much larger impact of the cell on the lipid bilayer (as oppose to that on a single lipid molecule). λ in Eq. 1 describes the uncertainty in the relationship between the diffusion coefficient and the viscosity and has been reported to vary by three orders of magnitude for protein inclusions in lipid bilayers (38). To this end, λ was calculated to account for a length scale similar to that of cells on top of the bilayer (10 µm for DOPC and 20 µm for DPPC, derived from the equivalent average cell radius). This produces an estimated viscosity of 1 × 10−6 Pa⋅s⋅m for DOPC and 1 × 10−4 for DPPC, bringing the latter into the range that leads to engagement of the clutch. While calculating specific corrected values for viscosity is challenging due to the complexity of the system, these estimated values likely give a good approximation of the scale of change of the viscosity within the cell area.
The Molecular Clutch Explains Force Transduction in Response to Viscosity.
One means through which cells sense external mechanical cues is through translation into biochemical signals (e.g., transcription factors), thus having an effect on gene expression. For example, cellular response to stiffness leads to downstream up-regulation of specific proteins such as RhoA, YAP, and TAZ (20, 47). In turn, up-regulation of these proteins in mesenchymal stem cells (MSCs) promotes lineage commitment to the more contractile osteogenic lineage (5, 20).
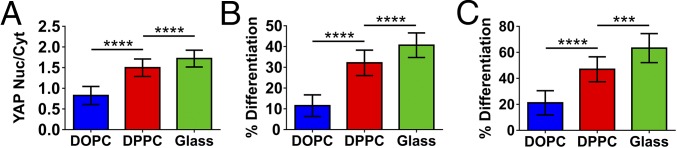
In light of this, the initial downstream effect of viscosity on the cell was determined using YAP localization, as shown in Fig. 6A (representative images in Fig. S8). YAP was seen to translocate to the nucleus to a greater extent on less-mobile surfaces, providing further support for the hypothesis of viscosity being interpreted via the molecular clutch model (17). It has been previously shown that YAP activity is regulated by the formation of stress fibers, cytoskeletal tension (20), and traction force (17), which shows a link between the extent of activity and both the morphological characteristics (Fig. 2) and the retrograde actin flow (Fig. 4). This regulation by YAP induced by response of the cell to varying viscosity has the ability to control further cell properties such as cell differentiation. Indeed, it has previously been shown that MSC differentiation can be prevented through YAP inhibition (20).
Fig. 6.
Downstream effects of viscosity. The cell response is controlled by the force exerted on the surface by the cell, which is in turn defined by the surface’s physical properties; further to this, the signals are then transduced via transcription factors such as YAP and myogenin, driving further cell behaviors, such as differentiation. (A) Shows the increased ratio of the mechnanosensitive YAP in the nucleus as the viscosity is increased (from left to right n = 21, 30, and 55). (B) The further downstream effects of viscosity by increased in the number of nuclei expressing myogenin, as transcription factor involved in the early stages of differentiation of C2C12 cells (from left to right n = 16, 27, and 27). (C) Terminal differentiation of C2C12 cells, through the expression of sarcomeric myosin (from left to right n = 18, 15, and 12). In all cases statistical significance was determined by one-way ANOVA. ***P ≤ 0.001, ****P ≤ 0.0001.
As a determination of the applicability of this system the response of cells was characterized by their lineage commitment as shown in Fig. 6 B and C (representative images in Fig. S8). It was observed that a greater degree of differentiation, as indicated initially by the up-regulation of the transcription factor myogenin, and subsequently by more cells staining positive for sarcomeric myosin, was seen on more viscous surfaces. This is in contrast to previously reported work using viscosity to differentiate MSCs (48), where higher amounts of differentiation markers as well as cell spreading were seen on more mobile surfaces; this is despite the current work and previous work demonstrating that cells do not spread (27–29) or differentiate on mobile surfaces.
The stiffness of a substrate is important in myogenic differentiation (49); due to the similar nature of cell response to the viscosity of the surface, it could be assumed that analogous pathways are activated and promoted here. One such candidate for viscosity-induced differentiation in this system is FAK. The activity of FAK has been shown to be of significant importance in the myogenic differentiation pathway of C2C12 murine myoblast cells, with overexpression of FAK rescuing differentiation on nonconducive substrates (50). Further work has also noted the role of FAK phosphorylation in the differentiation pathways of skeletal muscle (51). One hypothesis may therefore be that the increased FAK phosphorylation, induced by changes in viscosity, activates the differentiation pathways. These results may therefore indicate that the sensing of the viscosity of a surface, through mechanosensitive pathways, has a significant effect on cell differentiation.
This work has elucidated the process involved in cell sensing of viscosity, from its detection through FAs through to its overall effect on cell fate/differentiation. We have engineered RGD functionalized lipid bilayers with controlled ligand density and significantly different viscosity (i.e., diffusion coefficient or mobility). We show that viscosity has a significant effect on mechanotransduction proteins, downstream transcriptional regulators, and differentiation. Overall, the effect of pure viscous flow in cell mechanotransduction is explained by the molecular clutch model. It is shown that at higher viscosities (DPPC) upon exertion of force by the cell the ligand will oppose higher resistance to movement through the tightly packed lipid membrane. This builds force quickly, allowing stable integrin binding, talin unfolding, vinculin bridging to the actin cytoskeleton, and FA assembly, thereby slowing down actin flow. Conversely, at lower viscosity (DOPC) the force loading rate will be slower, as the ligand will exert less resistance to movement through the membrane upon force exertion by the cell. This prevents protein unfolding and increases actin flow.
By understanding the nature of the cell response to viscosity, coupled with the knowledge of the stiffness response, it is reasonable to conclude that this may lead to further, combinatorial approaches to changing the physical properties of surfaces. The fine tuning of viscoelastic properties of surfaces will allow better, more defined control of the desired cellular response. It, therefore, has the potential to provide new and as yet not fully realized methods of manipulating cellular response. In addition, it will enhance our understanding of cell behavior in tissues which are viscoelastic by nature.
Materials and Methods
Further elaboration on the methods can be found in Supporting Information.
Production and Functionalization of SLBs.
Vesicle solutions were made by drying lipid solutions in rehydration (R) buffer above the relevant transition temperature. To make the SLBs the vesicles were diluted in fusion (F) buffer immediately before use and incubated on cleaned glass surfaces for 20 min at room temperature (RT) for DOPC and at 70 °C for DPPC and washed.
All samples were functionalized with 0.1 mg/mL neutravidin (Fisher) and 2 µL/mL cyclic-RGD (Peptides International) for 15 min each, washing in between. SLBs were characterized by atomic force microscopy (AFM) (Nanowizard 3 Bioscience AFM; JPK). Quantitative fluorescence microscopy was carried out as previously described (44) and used to quantify the amount of neutravidin on the surface. FCS was used to determine the diffusion coefficients and thus the viscosity of the membrane.
Cell Culture and Transfection.
C2C12 mouse myoblasts were used in all experiments and transfected with conditions as stated on the website. The pEGFPC1/GgVcl 1-258 (aka VD1) plasmid (Addgene plasmid no. 46270) was a gift from Susan Craig, The Johns Hopkins School of Medicine, Baltimore. Transformed cells were cultured for 24 h and used.
Statistical Analysis.
In all figures values are given as the mean ± the SD. One-way or two-way ANOVA tests were carried out as appropriate. Significance was taken as the P values, which are given as follows: not significant (ns), <0.05, *≤ 0.05, **≤ 0.01, ***≤ 0.001, and ****≤ 0.0001.
Supplementary Material
Acknowledgments
This work was supported by European Research Council HealInSynergy Grant 306990 and Engineering and Physical Sciences Research Council Grants EP/P001114/1 and EP/F500424/1, the Spanish Ministry of Economy and Competitiveness (Grant BFU2016-79916-P), the European Commission (Grant Agreement SEP-210342844), the Generalitat de Catalunya (Grant 2014-SGR-927), and Obra Social “La Caixa.”.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All the original data related to this article and the MATLAB code used to generate the clutch model are within the depository of the University of Glasgow (doi: 10.5525/GLA.RESEARCHDATA.567).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710653115/-/DCSupplemental.
References
- 1.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 2.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Dalby MJ, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 4.Benoit DS, Schwartz MP, Durney AR, Anseth KS. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat Mater. 2008;7:816–823. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 6.Seo JH, Yui N. The effect of molecular mobility of supramolecular polymer surfaces on fibroblast adhesion. Biomaterials. 2013;34:55–63. doi: 10.1016/j.biomaterials.2012.09.063. [DOI] [PubMed] [Google Scholar]
- 7.Seo JH, et al. The significance of hydrated surface molecular mobility in the control of the morphology of adhering fibroblasts. Biomaterials. 2013;34:3206–3214. doi: 10.1016/j.biomaterials.2013.01.080. [DOI] [PubMed] [Google Scholar]
- 8.Kourouklis AP, Lerum RV, Bermudez H. Cell adhesion mechanisms on laterally mobile polymer films. Biomaterials. 2014;35:4827–4834. doi: 10.1016/j.biomaterials.2014.02.052. [DOI] [PubMed] [Google Scholar]
- 9.González-García C, Moratal D, Oreffo ROC, Dalby MJ, Salmerón-Sánchez M. Surface mobility regulates skeletal stem cell differentiation. Integr Biol. 2012;4:531–539. doi: 10.1039/c2ib00139j. [DOI] [PubMed] [Google Scholar]
- 10.Guerra NB, et al. Subtle variations in polymer chemistry modulate substrate stiffness and fibronectin activity. Soft Matter. 2010;6:4748–4755. [Google Scholar]
- 11.Salmerón-Sánchez M, et al. Role of material-driven fibronectin fibrillogenesis in cell differentiation. Biomaterials. 2011;32:2099–2105. doi: 10.1016/j.biomaterials.2010.11.057. [DOI] [PubMed] [Google Scholar]
- 12.Llopis-Hernández V, Rico P, Ballester-Beltrán J, Moratal D, Salmerón-Sánchez M. Role of surface chemistry in protein remodeling at the cell-material interface. PLoS One. 2011;6:e19610. doi: 10.1371/journal.pone.0019610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri O, et al. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lautscham LA, et al. Biomembrane-mimicking lipid bilayer system as a mechanically tunable cell substrate. Biomaterials. 2014;35:3198–3207. doi: 10.1016/j.biomaterials.2013.12.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elosegui-Artola A, et al. Rigidity sensing and adaptation through regulation of integrin types. Nat Mater. 2014;13:631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elosegui-Artola A, et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. 2016;18:540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- 18.Teo BK, et al. Nanotopography modulates mechanotransduction of stem cells and induces differentiation through focal adhesion kinase. ACS Nano. 2013;7:4785–4798. doi: 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]
- 19.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, et al. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J. 2011;100:1902–1909. doi: 10.1016/j.bpj.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C-J, et al. Type I collagen-functionalized supported lipid bilayer as a cell culture platform. Biomacromolecules. 2010;11:1231–1240. doi: 10.1021/bm901445r. [DOI] [PubMed] [Google Scholar]
- 23.Andreasson-Ochsner M, et al. Single cell 3-D platform to study ligand mobility in cell-cell contact. Lab Chip. 2011;11:2876–2883. doi: 10.1039/c1lc20067d. [DOI] [PubMed] [Google Scholar]
- 24.Svedhem S, et al. In situ peptide-modified supported lipid bilayers for controlled cell attachment. Langmuir. 2003;19:6730–6736. [Google Scholar]
- 25.Ananthanarayanan B, Little L, Schaffer DV, Healy KE, Tirrell M. Neural stem cell adhesion and proliferation on phospholipid bilayers functionalized with RGD peptides. Biomaterials. 2010;31:8706–8715. doi: 10.1016/j.biomaterials.2010.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans SF, et al. Solid-supported lipid bilayers to drive stem cell fate and tissue architecture using periosteum derived progenitor cells. Biomaterials. 2013;34:1878–1887. doi: 10.1016/j.biomaterials.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 27.Yu CH, et al. Integrin-beta3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat Commun. 2015;6:8672. doi: 10.1038/ncomms9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu CH, Law JBK, Suryana M, Low HY, Sheetz MP. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc Natl Acad Sci USA. 2011;108:20585–20590. doi: 10.1073/pnas.1109485108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu CH, et al. Integrin-matrix clusters form podosome-like adhesions in the absence of traction forces. Cell Rep. 2013;5:1456–1468. doi: 10.1016/j.celrep.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pera I, Stark R, Kappl M, Butt H-J, Benfenati F. Using the atomic force microscope to study the interaction between two solid supported lipid bilayers and the influence of synapsin I. Biophys J. 2004;87:2446–2455. doi: 10.1529/biophysj.104.044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdulreda MH, Moy VT. Atomic force microscope studies of the fusion of floating lipid bilayers. Biophys J. 2007;92:4369–4378. doi: 10.1529/biophysj.106.096495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picas L, Rico F, Scheuring S. Direct measurement of the mechanical properties of lipid phases in supported bilayers. Biophys J. 2012;102:L01–L03. doi: 10.1016/j.bpj.2011.11.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alessandrini A, Facci P. Phase transitions in supported lipid bilayers studied by AFM. Soft Matter. 2014;10:7145–7164. doi: 10.1039/c4sm01104j. [DOI] [PubMed] [Google Scholar]
- 34.Attwood SJ, Choi Y, Leonenko Z. Preparation of DOPC and DPPC supported planar lipid bilayers for atomic force microscopy and atomic force spectroscopy. Int J Mol Sci. 2013;14:3514–3539. doi: 10.3390/ijms14023514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Przybylo M, et al. Lipid diffusion in giant unilamellar vesicles is more than 2 times faster than in supported phospholipid bilayers under identical conditions. Langmuir. 2006;22:9096–9099. doi: 10.1021/la061934p. [DOI] [PubMed] [Google Scholar]
- 36.Saffman PG, Delbrück M. Brownian motion in biological membranes. Proc Natl Acad Sci USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes BD, Pailthorpe BA, White LR. The translational and rotational drag on a cylinder moving in a membrane. J Fluid Mech. 2006;110:349–372. [Google Scholar]
- 38.Gambin Y, et al. Lateral mobility of proteins in liquid membranes revisited. Proc Natl Acad Sci USA. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair PM, Salaita K, Petit RS, Groves JT. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat Protoc. 2011;6:523–539. doi: 10.1038/nprot.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balaban NQ, et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 41.Andersson AS, Glasmästar K, Sutherland D, Lidberg U, Kasemo B. Cell adhesion on supported lipid bilayers. J Biomed Mater Res A. 2003;64:622–629. doi: 10.1002/jbm.a.10442. [DOI] [PubMed] [Google Scholar]
- 42.Bathawab F, et al. Lateral chain length in polyalkyl acrylates determines the mobility of fibronectin at the cell/material interface. Langmuir. 2016;32:800–809. doi: 10.1021/acs.langmuir.5b03259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler A, et al. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- 45.Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 46.Oria R, et al. Force loading explains spatial sensing of ligands by cells. Nature. 2017;552:219–224. doi: 10.1038/nature24662. [DOI] [PubMed] [Google Scholar]
- 47.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 48.Koçer G, Jonkheijm P. Guiding hMSC adhesion and differentiation on supported lipid bilayers. Adv Healthc Mater. 2017;6:1600862. doi: 10.1002/adhm.201600862. [DOI] [PubMed] [Google Scholar]
- 49.Engler AJ, et al. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han J-W, Lee H-J, Bae G-U, Kang J-S. Promyogenic function of integrin/FAK signaling is mediated by Cdo, Cdc42 and MyoD. Cell Signal. 2011;23:1162–1169. doi: 10.1016/j.cellsig.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Graham ZA, Gallagher PM, Cardozo CP. Focal adhesion kinase and its role in skeletal muscle. J Muscle Res Cell Motil. 2015;36:305–315. doi: 10.1007/s10974-015-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bazellières E, et al. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat Cell Biol. 2015;17:409–420. doi: 10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DC. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 54.Litvinov RI, et al. Resolving two-dimensional kinetics of the integrin αIIbβ3-fibrinogen interactions using binding-unbinding correlation spectroscopy. J Biol Chem. 2012;287:35275–35285. doi: 10.1074/jbc.M112.404848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roca-Cusachs P, Iskratsch T, Sheetz MP. Finding the weakest link: Exploring integrin-mediated mechanical molecular pathways. J Cell Sci. 2012;125:3025–3038. doi: 10.1242/jcs.095794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.