Significance
Aminoglycosides remain a vital clinical asset. Gentamicin C complex in particular is remarkably potent in treating systemic Gram-negative infections, and semisynthetic gentamicins that combat pathogen resistance or show reduced toxicity remain attractive goals. We report here the roles of clustered genes and enzymes that define a methylation network in gentamicin biosynthesis and also identify a remote gene on the chromosome encoding the essential methyltransferase GenL, which is decisive for the proportions of the five major components present in the gentamicin C complex. This is an important step toward engineered fermentation to produce single components as valuable starting materials for semisynthesis of next-generation aminoglycoside antibiotics.
Keywords: aminoglycoside biosynthesis, methyltransferase, antibiotic, metabolic engineering
Abstract
Gentamicin C complex from Micromonospora echinospora remains a globally important antibiotic, and there is revived interest in the semisynthesis of analogs that might show improved therapeutic properties. The complex consists of five components differing in their methylation pattern at one or more sites in the molecule. We show here, using specific gene deletion and chemical complementation, that the gentamicin pathway up to the branch point is defined by the selectivity of the methyltransferases GenN, GenD1, and GenK. Unexpectedly, they comprise a methylation network in which early intermediates are ectopically modified. Using whole-genome sequence, we have also discovered the terminal 6′-N-methyltransfer required to produce gentamicin C2b from C1a or gentamicin C1 from C2, an example of an essential biosynthetic enzyme being located not in the biosynthetic gene cluster but far removed on the chromosome. These findings fully account for the methylation pattern in gentamicins and open the way to production of individual gentamicins by fermentation, as starting materials for semisynthesis.
Gentamicins are clinically valuable aminoglycoside antibiotics isolated as gentamicin C complex, a mixture of five components (Fig. 1), from the filamentous bacterium Micromonospora echinospora. Gentamicins are protein synthesis inhibitors used to combat Gram-negative bacterial infections. They are also being actively explored for use in other applications (1–3). However, gentamicins carry a serious risk of kidney damage and irreversible hearing loss (4, 5), which limits their utility. Encouragingly, individual components of the gentamicin mixture may have lower toxicity (6, 7). Individual components of gentamicin C can already be separated after manual chemical derivatization (8), but achieving this by low-cost direct fermentation would significantly advance interest in single components both as starting material for semisynthesis to novel gentamicins and for testing as therapeutics in their own right.
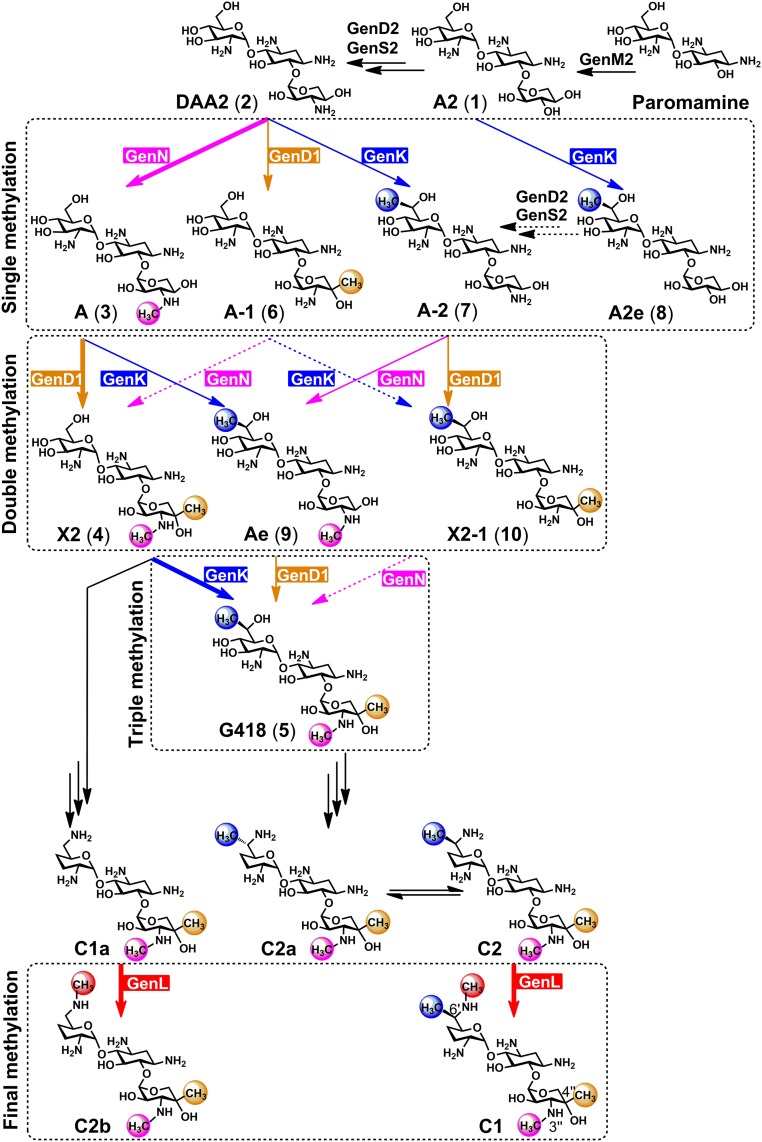
Fig. 1.
Methyltransfer reactions in gentamicin biosynthetic pathway. The determined major and minor alternative pathways in the methylation network are indicated by boldface type and solid arrows, respectively. The dashed arrows refer to conversions without substantial experimental support. Methyltransferases and their roles revealed in this study of GenN, GenD1, GenK, and GenL are highlighted in magenta, orange, blue, and red, respectively. The extents of methylation are shown in the dashed rectangle boxes. Carbon numbering is shown with the structure of 1.
Gentamicins are modified sugars containing an unusual aminocyclitol ring (2-deoxystreptamine, 2-DOS) (9, 10). The enzymatic steps that lead to the 2-DOS scaffold, and thence to the pseudodisaccharide paromamine (11–13) and the pseudotrisaccharide gentamicin A2 (1) (14) (Fig. 1), have all been elucidated, and more recently rapid progress has also been made in delineating the later steps of gentamicin biosynthesis, greatly aided by the sequencing of the gentamicin biosynthetic gene cluster (15–17) and the related sisomicin cluster (18). We have previously used a combination of genetic and biochemical evidence to reveal that, starting from the intermediate 3″-dehydro-3″-oxo-gentamicin A2 (DAA2) (2) (Fig. 1), the S-adenosylmethionine (SAM)-dependent methyltransferase GenN catalyzes 3′′-N-methylation, followed by 4″-C-methylation catalyzed by a cobalamin- and radical SAM-dependent methyltransferase GenD1, to form gentamicin X2 (4) (19). In the same way, we (20) and others (21–24) have shown that a second cobalamin- and radical SAM-dependent methyltransferase, GenK, catalyzes 6′-C-methylation of gentamicin X2 (4) to initiate the branch of the gentamicin pathway that leads via G418 (5) to gentamicin components C2a, C2, and C1. The full mechanistic details of the subsequent transamination and dehydroxylation steps remain to be clarified, although the dehydrogenase GenQ (20), phosphotransferase GenP, and the pyridoxal-dependent enzymes GenB1, GenB2, GenB3, and GenB4 have all been implicated in this enigmatic process (20, 25, 26). Finally, the terminal step in both branches of the pathway involves the (partial) conversion of C1a into C2b and of C2 into C1, catalyzed by an unidentified 6′-N-methyltransferase (or possibly two different methyltransferases). Given the decisive role of methylation in shaping the pathway to the gentamicin C complex, we have used mutant strains of M. echinospora engineered to contain different complements of GenN, GenD1, and GenK to investigate the substrate selectivity of these known methyltransferases to determine the scope for altering the methylation pattern; and we have tracked down and identified, by combined biochemical and genetic approaches, the terminal methyltransferase (named GenL). GenL is located on its own 2.54 Mbp from the previously characterized gentamicin gene cluster. We show that GenL is necessary and sufficient to catalyze the 6′-N-methylation of both C2 and C1a. It follows that future judicious manipulation of the levels of (in particular) GenK and GenL activity in cells should permit targeted enrichment of individual components of the gentamicin C complex.
Results
Systematic Deletion of One or More of the Methyltransferase Genes genN, genD1, and genK Reveals Alternative Minor Pathways for Methylation.
Commercial samples of gentamicin C complex obtained by fermentation contain variable amounts of minor gentamicin metabolites as impurities. Careful analysis has detected as many as 21 such minor products in a single sample (27). For 6 of these, the structure could be determined by MS and MSn methods, and structures could also be proposed for 13 others (27). In previous work it has been shown that levels of certain metabolites with altered methylation patterns are enhanced upon deletion of either genN (19, 28) or genK (20). We therefore set out systematically to alter the complement of known methyltransferase genes and to study the metabolites produced.
First, the single mutants of genN and genD1, which were constructed in our previous studies (19, 20), were used to generate the series of mutants ΔgenD1ΔgenN, ΔgenD1ΔgenK, ΔgenNΔgenK, and ΔgenD1ΔgenKΔgenN. In these mutants, an internal fragment has been deleted in-frame, respectively, from genN, genD1, and genK. These mutants were confirmed by PCR and Southern blotting and then fermented (SI Appendix, Fig. S1 A–D). The extracts were analyzed by using liquid chromatography coupled with electrospray ionization high-resolution mass spectrometry (LC-ESI-HRMS) and tandem mass spectrometry (MS/MS).
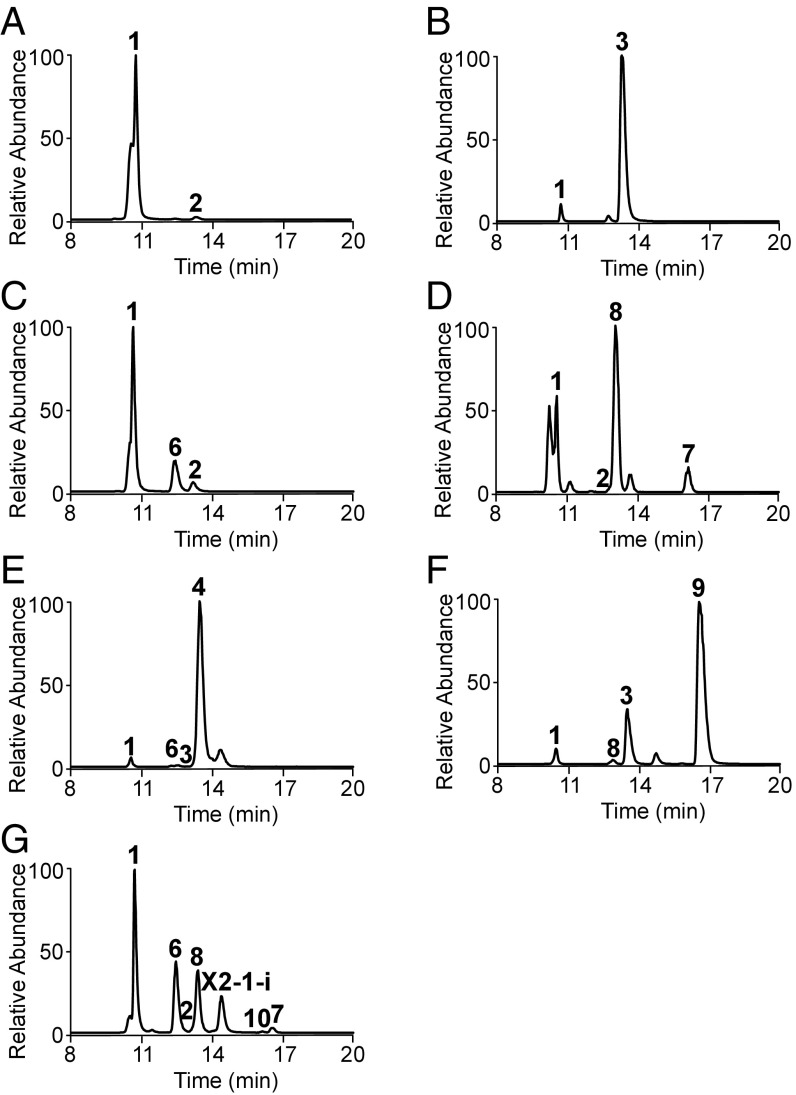
The triple-mutant ΔgenD1ΔgenKΔgenN produced small amounts of 2 (Fig. 1 and SI Appendix, Figs. S2A and S3A) (19), the upstream precursor of gentamicin A (3) (Figs. 1 and 2A and SI Appendix, Fig. S2B). NMR data confirmed the structure of 2 as a nonmethylated species as previously proposed (SI Appendix, Fig. S3A and Table S1). The intermediate 1 (Figs. 1 and 2A) was present as the major species, but no other gentamicin-related metabolite was present. The ΔgenD1ΔgenK mutant produced 1 and 3 (Figs. 1 and 2B), but no other methylated intermediates or other gentamicins. These results confirm that GenN is essential for the methylation on 3′′-N of 2. In extracts of the ΔgenNΔgenK mutant, intermediates 1 and 2 were detected at levels similar to those in the triple mutant, but an additional compound with the same molecular mass as 3 was observed, singly methylated on 3′′-N (Figs. 1 and 2C). Given the known regiospecificity of GenD1 (19), the site of modification is proposed to be at C-4′′ (SI Appendix, Fig. S2C). This metabolite, named here gentamicin A-1 (6) (Fig. 1 and SI Appendix, Fig. S2C), has been described as a trace component of a wild-type fermentation (27). In the extract of the ΔgenD1ΔgenN mutant, 2 was detected at levels similar to those in the triple mutant along with two 6′-C-methylated metabolites (Fig. 2D). One of these is formed as a result of the precocious action of GenK on 2 and is named here gentamicin A-2 (7) (Figs. 1 and 2D and SI Appendix, Figs. S2D and S3B and Table S1). It is assumed to have the same relative configuration at C-6′ as 5 (Fig. 1 and SI Appendix, Fig. S2E) and has also been previously described as a trace component of a wild-type fermentation (26). The second metabolite A2e (8) (Figs. 1 and 2D and SI Appendix, Fig. S2F) is formed by the known precocious action of GenK on 1 (19, 24).
Fig. 2.
LC-ESI-HRMS analysis of gentamicin-related intermediates from mutant strains. Extracted ion chromatogram of intermediates in (A) ΔgenD1ΔgenKΔgenN, (B) ΔgenD1ΔgenK, (C) ΔgenNΔgenK, (D) ΔgenD1ΔgenN, (E) ΔgenK, (F) ΔgenD1, and (G) ΔgenN.
We repeated our previous analysis of the ΔgenN, ΔgenD1 and ΔgenK mutants (19, 20) and scrutinized the extracts carefully for metabolites with an altered methylation pattern. The ΔgenK mutant produced 3 and 4 (SI Appendix, Fig. S2G) as expected but only a trace of the metabolite we have assigned as 6 (Figs. 1 and 2E), confirming that GenD1 fails to compete successfully with GenN for methylation of 2. The ΔgenD1 mutant in contrast produced a relatively large amount of the known doubly methylated metabolite gentamicin Ae (9) (Figs. 1 and 2F and SI Appendix, Fig. S2H) (19, 27), again a consequence of the precocious action of GenK. We confirmed the structure of 9 by NMR (SI Appendix, Fig. S3C and Table S1). The ΔgenN mutant produced low levels of 2, but mainly produced 1, as well as the singly methylated species 6, 7, 8, and an intermediate with the same molecular mass and MS/MS fragmentation as 9, which we name here gentamicin X2-1 (10) (Figs. 1 and 2G and SI Appendix, Fig. S2I). This compound, which has also been described as a trace component of wild-type gentamicin fermentation (27), is most likely methylated at both C-4″ and C-6′ positions. Another compound with the same molecular mass and MS/MS fragmentation as 10 was also observed, named here X2-1-i, which may be an isomer of it (Fig. 2G and SI Appendix, Fig. S2I). These species with various methylation patterns suggest a metabolic network in the pathway, in which methylations at the three sites occur relatively independently of each other rather than in a strictly fixed order.
Bioconversion in Vivo and in Vitro of Gentamicin Metabolites Confirms the Selectivity of Methylation by GenN, GenD1, and GenK.
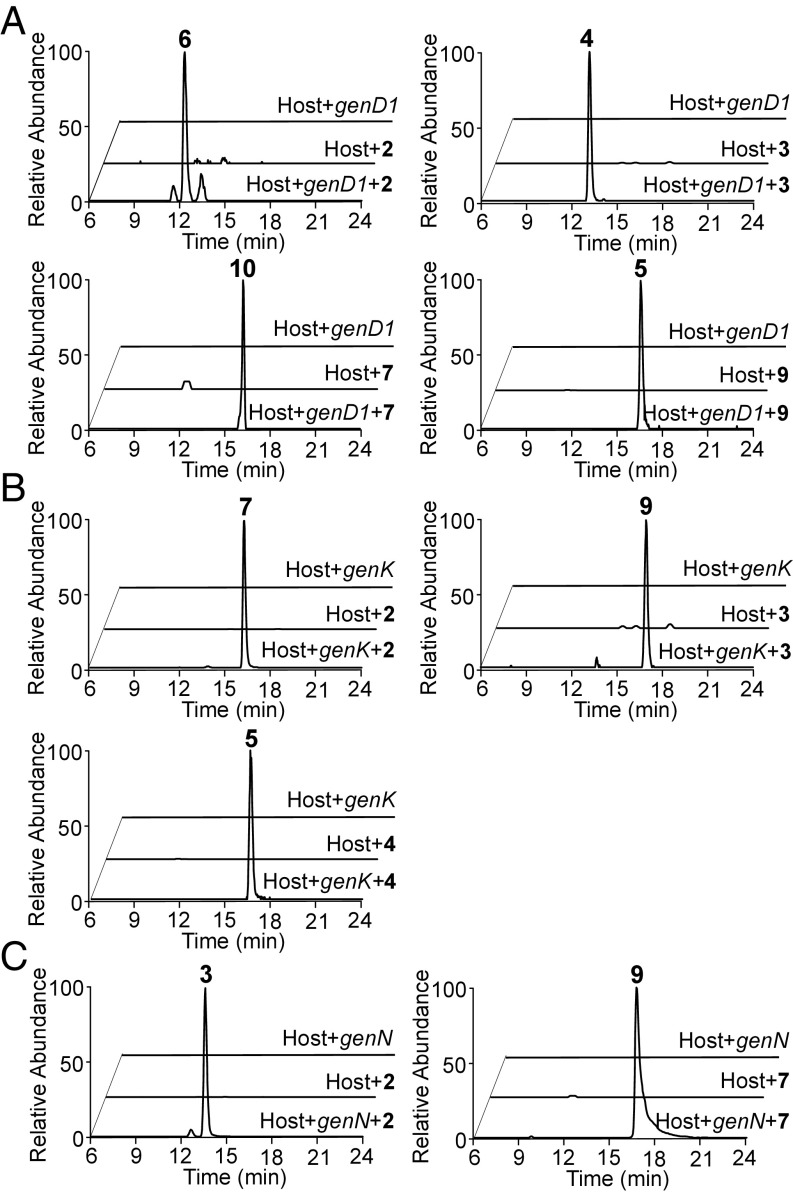
Established gentamicin intermediates 2, 3, and 4 as well as gentamicin metabolites with altered methylation patterns 6, 7, 9, and 10 were isolated from the mutants described above, as described in Materials and Methods (SI Appendix). We aimed to interrogate directly the ability of individual methyltransferases to use these compounds as substrates and verify which of the possible conversions in the methylation network actually occur. To do this, the gentamicin biosynthetic gene cluster (GenBank accession no. KY971520) from grmB to genN was knocked out by deletion of a 40,524-bp fragment to generate the ΔBN mutant, which was confirmed by PCR and Southern blotting analysis (SI Appendix, Fig. S1E). Complementation of the ΔBN mutant by using plasmid pWHU91 (SI Appendix, Table S2) containing genN and gmrA under the control of the PermE* promoter, respectively, generated ΔBN::gmrA+genN (SI Appendix, Fig. S5B and Table S3). In parallel, ΔBN::gmrA+genD1 and ΔBN::gmrA+genK were constructed in the same way with pWHU92 and pWHU93 (SI Appendix, Figs. S5 C and D, and Table S3). The gene gmrA that confers gentamicin self-resistance was present to avoid toxicity of gentamicin metabolites in the host strain. The strain ΔBN::gmrA was constructed as a negative control by using pWHU90 (SI Appendix, Table S2), which contains gmrA only, under the control of the constitutive PermE* promoter (SI Appendix, Fig. S5A).
The results from LC-ESI-HRMS and MS/MS showed that feeding either 2, 3 (the normal substrate), 7, or 9 to ΔBN::gmrA+genD1 led to detectable methylation, which is presumed to be at C-4″ (Fig. 3A). Likewise, feeding 2, 3, or 4 (the normal substrate) to ΔBN::gmrA+genK in each case led to detectable methylation (presumed to be at C-6′) (Fig. 3B). In contrast, feeding of 6 to this strain did not produce 10. Meanwhile, feeding of 2 (the normal substrate) to ΔBN::gmrA+genN generated 3, and feeding of 7 to this strain generated 9 (Fig. 3C). However, neither 6 nor 10 were methylated in this strain.
Fig. 3.
Production of gentamicin intermediates in feeding experiments. Extracted ion chromatograms are shown from LC-ESI-HRMS of feeding experiments: (A) ΔBN::gmrA+genD1 fed with 2, 3, 7, and 9, respectively. (B) ΔBN::gmrA+genK fed with 2, 3, and 4, respectively. (C) ΔBN::gmrA+genN fed with 2 and 7, respectively. Host refers to ΔBN::gmrA.
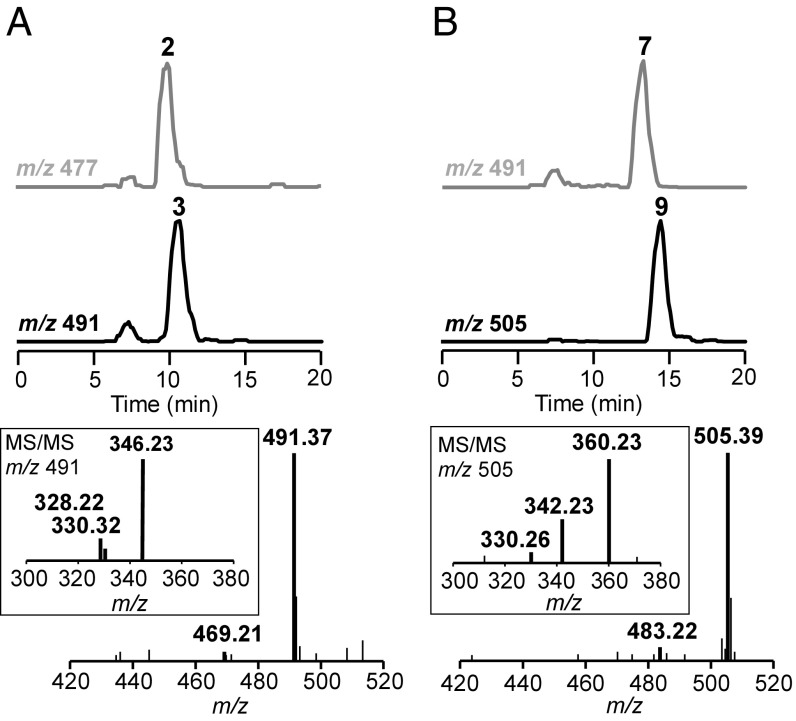
Consistent with the in vivo biotransformation results, purified His-tagged recombinant GenN catalyzed methylation of 2 and 7 into 3 and 9, respectively, with SAM as a methyl donor (Fig. 4). No methylated product was observed when 6 and 10 were used as substrates under the same assay conditions.
Fig. 4.
LC-ESI-MS analysis of in vitro activities of GenN on gentamicin intermediates 2 and 7. In the presence of SAM, GenN catalyzed methylation of (A) 2 to 3 and (B) 7 to 9, respectively. The extracted ion traces of substrates are in gray and those of the products are in black.
The Enzyme GenL, Required for the Terminal 6′-N-Methylation Step in Both Branches of the Gentamicin Biosynthetic Pathway, Has Been Identified.
Preliminary experiments showed that C2 was readily bioconverted into C1 when fed to M. echinospora ΔBN::gmrA, which lacks the entire gentamicin gene cluster except the resistance gene gmrA. It appeared therefore that the enzyme catalyzing the terminal 6′-N-methylation step must be encoded by a gene outside the limits of the known cluster. Also, cell-free lysate of M. echinospora ΔBN was shown to carry out this conversion in the presence of SAM, and the enzymatic activity proved stable to initial fractionation using selective ammonium sulfate precipitation. We therefore contemplated searching for the elusive terminal methyltransferase by two parallel routes, one by conventional enzyme purification from M. echinospora and the other by bioinformatic analysis of genome sequence followed by the cloning and testing of plausible candidates. In the end, the genome-assisted approach proved quicker.
The whole-genome sequence of M. echinospora ATCC15835 was determined and assembled as described in Materials and Methods. To identify candidate genes for the methylation at 6′-N of C1a and C2, we chose the protein sequences of two authentic aminoglycoside N-methyltransferases as probes. One of these was IstU, which is proposed to catalyze an analogous 6′-N-methylation step during biosynthesis of istamycin in Streptomyces tenjimariensis (12, 16). Four candidate genes—orf0086, orf1678, orf12159, and orf3626 (GenBank accession nos. MF036116, MF036117, MF036132, and MF036133) with 40, 36, 38, and 27% protein sequence identities, respectively, to IstU (SI Appendix, Table S4)—were chosen, and each was inactivated in M. echinospora ATCC15835 through in-frame deletion. However, none of these mutants was found to abolish the methylation at 6′-N.
The second probe sequence chosen was GenN. In total, 19 candidate genes (GenBank accession nos. MF036118–MF036131 and MF036134–MF036138) were identified using the BLASTP program (28), with percentage sequence identities to GenN ranging from 26.67 to 40.35% (SI Appendix, Table S4). Each candidate gene was cloned into expression vector pET28a(+) in Escherichia coli, and cell extracts were assayed for their ability to catalyze SAM-dependent conversion of C2 into C1, monitored by LC-ESI-HRMS (SI Appendix, Fig. S6A). The results showed that only the cell-free lysate of BL21(DE3)/pET28a(+)-genL(orf5365) transformed C2 to C1. No other candidate, including boiled BL21(DE3)/pET28a(+)-genL, catalyzed this conversion. The cell-free lysate of BL21(DE3)/pET28a(+)-genL also catalyzed the conversion of C1a to C2b significantly (SI Appendix, Fig. S6B). These results strongly suggested that this enzyme, named GenL, is the elusive terminal methyltransferase catalyzing methylation on 6′-N of C1a and C2. The genL (GenBank accession no. MF036122) is separated from the known gentamicin biosynthetic cluster by 2.54 Mbp.
To confirm the function of genL, the gene was removed through in-frame deletion from the genome of M. echinospora ATCC15835. This mutant was verified through Southern blotting (SI Appendix, Fig. S4 A and B) and then fermented, followed by LC-ESI-HRMS analysis of the extract. The results showed selective loss of the C1 and C2b components of the gentamicin C complex (Fig. 1 and SI Appendix, Fig. S4C), confirming that genL is the unique and essential gene catalyzing 6′-N-methylation on C2 and C1a in gentamicin biosynthesis. Complementation in trans with the wild-type genL gene (SI Appendix, Fig. S5E) substantially restored C1 production (60% of wild type) and gave C2b levels 240% of wild type (SI Appendix, Fig. S4C).
GenL Is Highly Specific for Terminal 6′-N-Methylation of C1a and C2.
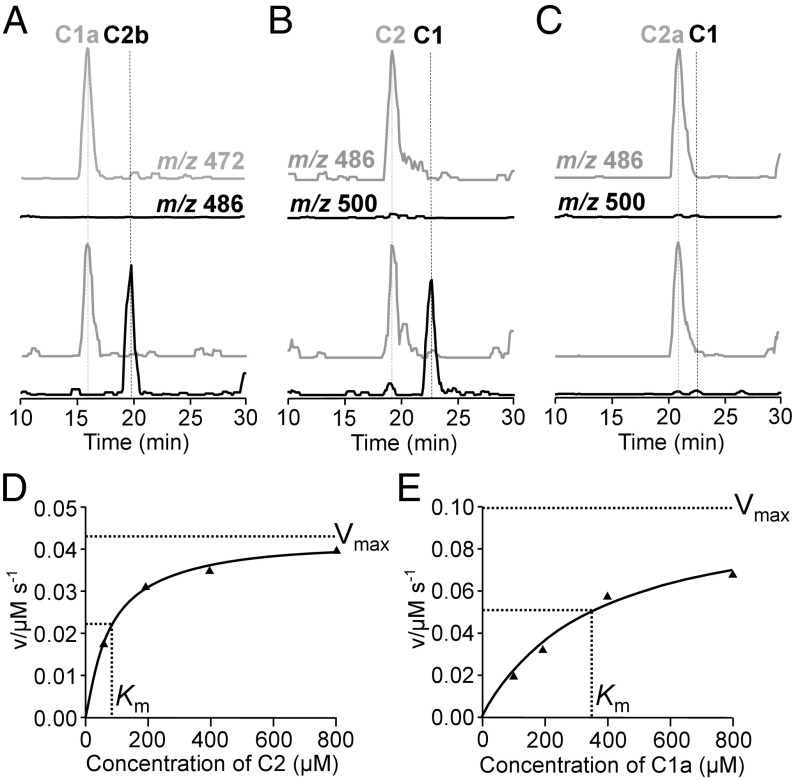
Recombinant GenL was expressed as an N-terminally His6-tagged protein and purified from E. coli BLR cells as described in Materials and Methods. After affinity purification on a Co2+-NTA column and gel-filtration chromatography on Sephadex S200, GenL was essentially homogenous as judged by SDS/PAGE and eluted as a single peak of protein with an estimated Mr of 43,000, roughly consistent with its being a dimer in solution (calculated Mr of the subunit 29,000). This purified enzyme preparation was active in the conversion of both C2 to C1 and of C1a to C2b (Fig. 5 A and B), as monitored by HPLC-MS, but was inactive against C2a, the C-6′ epimer of C2 (Fig. 5C).
Fig. 5.
In vitro methyltransfer activities of GenL. In the presence of SAM, GenL converted gentamicin C1a (A) and C2 (B) to gentamicin C2b (A) and C1 (B), respectively, but had no effect on gentamicin C2a (C). The extracted ion traces on LC-MS of substrates are in gray and those of the products are in black. The Michaelis–Menten saturation curves of GenL-catalyzed methylations of C2 (D) and C1a (E) are also determined.
Kinetic analysis of the SAM-dependent methyltransferase activity of GenL was carried out with either C2 or C1a as substrate, using a continuous colorimetric assay based on enzymatically coupling the production of S-adenosylhomocysteine (SAH) to a change in absorbance at 510 nm (29, 30). The data were fitted to the integrated form of the Michaelis–Menten equation. For C2, kcat was 0.043·s−1, and Km was 77.8 ± 8 µM; while for C1a, kcat was 0.040·s−1, and Km was 346.8 ± 86 µM (Fig. 5 D and E). The same assay was also used to screen several other pathway intermediates (C2a, G418, JI-20A, and JI-20B) (20), and other gentamicin-related aminoglycosides, including sisomicin, kanamycin A, kanamycin B, and tobramycin. All of these were found to be essentially inactive as substrates for GenL, except for a slow reaction with sisomicin.
Discussion
Methylation is an important reaction in the biosynthesis of specialized metabolites in both plants and microorganisms, where it contributes significantly to the chemical diversity of the products (31, 32). The results presented here significantly advance our understanding of the way in which methylation shapes the branched biosynthetic pathway to the gentamicin C complex, an established drug used to combat serious infections. These insights are key to the success of efforts to redirect the course of biosynthesis toward specific components of the complex and give an appreciation of the scope for altering the methylation pattern on the gentamicin pseudotrisaccharide scaffold to generate various metabolites for testing as antibiotics or for other therapeutic indications (3). Methyltransferases involved in the biosynthesis of natural products have considerable potential for the engineering of new derivatives (33–35). There is renewed interest in aminoglycoside antibiotics (3, 36) driven in part by the increasing incidence of pathogenic strains resistant to many other antibiotics but also by encouraging evidence that toxicity and vulnerability to resistance mechanisms can be ameliorated by semisynthesis (37). For example, etimicin is prepared from gentamicin C1a (38) and plazomicin (currently in phase III trials) from the gentamicin-like sisomicin (39). If the single components of the gentamicin C complex can be efficiently prepared by low-cost direct fermentation, it will widen the scope for such efforts.
In previous work, three consecutive methylation steps in the later stages of gentamicin biosynthesis have been firmly established as being catalyzed, respectively, by the 3′′-N-methyltransferase GenN, the 4″-C-methyltransferase GenD1, and the 6′-C-methyltransferase GenK (19, 20, 28). However, here we have shown that the three methylations may take place in an alternative order, giving rise to a form of network. A commercial batch of gentamicin C has been shown to contain as trace contaminants gentamicins lacking one or more of these methyl groups (27), and specific deletion of genN has been shown, for example, to increase the yield of several 3′′-N-desmethylgentamicins (19, 28). To explore this further, we therefore constructed the complete combinatorial set of seven mutants in which one, two, or all three of methyltransferase genes had been independently deleted in-frame. LC-ESI-HRMS analysis of these mutants gave indications of the substrate flexibility of these enzymes.
First, the ΔgenD1ΔgenKΔgenN mutant produced no intermediate more advanced than 2, which confirms that no other methyltransferase present in M. echinospora can methylate this molecule. Also as expected, in ΔgenD1ΔgenK, ΔgenNΔgenK, and ΔgenD1ΔgenN, 3, 6, and 7, respectively, were seen. In the ΔGenK mutant, GenN clearly out-competes GenD1 because the products are those of the normal pathway: 3 and 4. In the ΔgenD1 mutant, GenK and GenN collaborate to produce doubly methylated 9 via either 3 or 7, while in the ΔgenN mutant, GenK and GenD1 collaborate to produce doubly labeled 10 via either 6 or 7.
Purification of the intermediates accumulated in these mutants also provided a panel of gentamicin metabolites. Further insight into methyltransferase substrate specificity was gained by feeding these individual metabolites to strains of M. echinospora stripped of the resident gentamicin biosynthetic gene cluster, except for a resistance gene and genN, genD1, or genK. The results showed that GenK and GenD1 are somewhat more versatile than GenN in accepting alternative substrates (Fig. 3). Even so, 6 is not a substrate for either GenK or GenN, so 10 must be formed via 7, and since 10 is not a substrate for GenN, neither 6 nor 10 are competent intermediates in biosynthesis of the gentamicin C complex. Thus, the only minor alternative pathways that can contribute to production of the gentamicin C complex lead from 3 to 9 to 5, inverting the normal order in which GenD1 and GenK act, or from 2 to 7 to 9 to 5, if GenK acts before GenN and GenD1 (Fig. 1). Agar diffusion assays against Bacillus pumilis showed that, with the important exception of the natural intermediate X2, derivatives tested that lacked any of the core methylations of the gentamicin C complex were essentially inactive (SI Appendix, Fig. S7), emphasizing the importance of both the pattern and the extent of this modification for antibiotic activity. The relatively tight selectivity of the three methyltransferases rationalizes why the conventional pathway dominates in the wild type. Although these results provide proof of concept for directing biosynthesis toward gentamicin-related metabolites with altered methylation patterns, enzyme engineering to modify substrate specificity would be required to make this an efficient process, and further modification of those templates would be required to restore antibiotic activity.
The biosynthetic genes for all aminoglycoside antibiotics so far studied are found clustered together on the genome, and this has greatly facilitated their analysis (13, 17). A surprising feature of the gentamicin biosynthesis in M. echinospora is that the methyltransferase gene genL, which we have shown here is essential to catalyze the terminal methylations, is located 2.54 Mbp away from the known biosynthetic gene cluster. This was previously annotated as an orphan methyltransferase, along with many other similar genes in M. echinospora, and it is not located near any recognizable genes of specialized metabolism. The dependence of a biosynthetic gene cluster on the collaboration of a remotely located gene is not unprecedented (40), and its discovery here demonstrates the power of a genome-driven approach to uncover such “missing” elements of a biosynthetic pathway. Whether GenL also catalyzes a different reaction in primary or specialized metabolism is unknown. We have characterized recombinant GenL as a homodimeric SAM-dependent enzyme with exactly the expected substrate specificity. GenL showed no activity against earlier intermediates in the pathway. GenL bears very limited sequence similarity to the 3′′-N-methyltransferase GenN, and it would be of considerable interest to compare the crystal structures, substrate ranges, and reaction mechanisms of these two enzymes.
The separation of gentamicin C components has previously been at least partially achieved by chemical derivatization, column chromatography, and deprotection (8). The present work shows the feasibility, with the identity and role of every methyltransferase established, of achieving the same goal more conveniently by direct fermentation of an appropriately engineered mutant strain. Given the growing global threat of antibiotic resistance, it is of pressing importance to maximize the potential of the existing arsenal of antibacterial compounds, as well as to seek new targets. The results that we have obtained should encourage the use of individual congeners as starting points for semisynthesis to create novel aminoglycosides, Also, the reportedly lower toxicity of certain components of the gentamicin C complex (6, 7) deserves to be fully and critically evaluated, and the present work brings this prospect closer.
Materials and Methods
Methods describing gene cloning, gene deletion and complementation, fermentation of strains and feeding experiments, LC-ESI-HRMS analyses, isolation and characterization of compounds, genome sequencing, cell-free assay, protein overexpression and purification, enzymatic assay, and kinetic characterization are described in detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Ian Garrard and Svetlana Ignatova (Brunel University) for expert help with purification of gentamicin C components. This work was supported by National Natural Science Foundation of China Grant 31470186; by the 973 Program Grant 2012CB721005 from the Ministry of Science and Technology of China; by Open Project Grant MMLKF15-12 from the State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University (to Y.S.); by Medical Research Council (MRC) Grants G1001687 and MR/M019020/1 (to P.F.L.); and by an MRC postgraduate studentship (1343325) (to A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY971520 and MF036116–MF036138).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711603115/-/DCSupplemental.
References
- 1.Hainrichson M, Nudelman I, Baasov T. Designer aminoglycosides: The race to develop improved antibiotics and compounds for the treatment of human genetic diseases. Org Biomol Chem. 2008;6:227–239. doi: 10.1039/b712690p. [DOI] [PubMed] [Google Scholar]
- 2.Chandrika NT, Garneau-Tsodikova S. A review of patents (2011-2015) towards combating resistance to and toxicity of aminoglycosides. MedChemComm. 2016;7:50–68. doi: 10.1039/C5MD00453E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidou L, Bugaud O, Belakhov V, Baasov T, Namy O. Characterization of new-generation aminoglycoside promoting premature termination codon readthrough in cancer cells. RNA Biol. 2017;14:378–388. doi: 10.1080/15476286.2017.1285480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bockenhauer D, Hug MJ, Kleta R. Cystic fibrosis, aminoglycoside treatment and acute renal failure: The not so gentle micin. Pediatr Nephrol. 2009;24:925–928. doi: 10.1007/s00467-008-1036-2. [DOI] [PubMed] [Google Scholar]
- 5.McWilliam DJ, Antoine DJ, Smyth RL, Pirmohamed M. Aminoglycoside-induced nephrotoxicity in children. Pediatr Nephrol. 2017;32:2015–2025. doi: 10.1007/s00467-016-3533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandoval RM, et al. A non-nephrotoxic gentamicin congener that retains antimicrobial efficacy. J Am Soc Nephrol. 2006;17:2697–2705. doi: 10.1681/ASN.2005101124. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi M, et al. Comparisons of cochleotoxicity among three gentamicin compounds following intratympanic application. Acta Otolaryngol. 2008;128:245–249. doi: 10.1080/00016480701558948. [DOI] [PubMed] [Google Scholar]
- 8.Crote J, Himmelsbach R, Johnson D. Methodology for the rapid separation of gentamicin components and regiospecific synthesis of gentamicin conjugates. Tetrahedron Lett. 2012;53:6751–6754. [Google Scholar]
- 9.Houghton JL, Green KD, Chen W, Garneau-Tsodikova S. The future of aminoglycosides: The end or renaissance? ChemBioChem. 2010;11:880–902. doi: 10.1002/cbic.200900779. [DOI] [PubMed] [Google Scholar]
- 10.Park SR, Park JW, Ban YH, Sohng JK, Yoon YJ. 2-Deoxystreptamine-containing aminoglycoside antibiotics: Recent advances in the characterization and manipulation of their biosynthetic pathways. Nat Prod Rep. 2013;30:11–20. doi: 10.1039/c2np20092a. [DOI] [PubMed] [Google Scholar]
- 11.Llewellyn NM, Spencer JB. Biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics. Nat Prod Rep. 2006;23:864–874. doi: 10.1039/b604709m. [DOI] [PubMed] [Google Scholar]
- 12.Thibodeaux CJ, Melançon CE, III, Liu HW. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed Engl. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudo F, Eguchi T. Biosynthetic genes for aminoglycoside antibiotics. J Antibiot (Tokyo) 2009;62:471–481. doi: 10.1038/ja.2009.76. [DOI] [PubMed] [Google Scholar]
- 14.Park JW, et al. Genetic dissection of the biosynthetic route to gentamicin A2 by heterologous expression of its minimal gene set. Proc Natl Acad Sci USA. 2008;105:8399–8404. doi: 10.1073/pnas.0803164105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unwin J, et al. Gene cluster in Micromonospora echinospora ATCC15835 for the biosynthesis of the gentamicin C complex. J Antibiot (Tokyo) 2004;57:436–445. doi: 10.7164/antibiotics.57.436. [DOI] [PubMed] [Google Scholar]
- 16.Kharel MK, et al. A gene cluster for biosynthesis of kanamycin from Streptomyces kanamyceticus: Comparison with gentamicin biosynthetic gene cluster. Arch Biochem Biophys. 2004;429:204–214. doi: 10.1016/j.abb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Aboshanab KMA. 2005. Genetic studies on the biosynthesis of the major aminoglycoside antibiotics: Isolation, analysis and comparison of the biosynthetic gene clusters for 12 aminoglycoside antibiotics. PhD thesis (Bergische Universität Wuppertal, Wuppertal, Germany)
- 18.Hong WR, et al. Molecular cloning and sequence analysis of the sisomicin biosynthetic gene cluster from Micromonospora inyoensis. Biotechnol Lett. 2009;31:449–455, and erratum (2009) 31:1115. doi: 10.1007/s10529-008-9887-y. [DOI] [PubMed] [Google Scholar]
- 19.Huang C, et al. Delineating the biosynthesis of gentamicin x2, the common precursor of the gentamicin C antibiotic complex. Chem Biol. 2015;22:251–261. doi: 10.1016/j.chembiol.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo J, et al. Specificity and promiscuity at the branch point in gentamicin biosynthesis. Chem Biol. 2014;21:608–618. doi: 10.1016/j.chembiol.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karki S, Kim JY, Park SH, Kwon HJ. Gene inactivation study on gntK, a putative C-methyltransferase gene in gentamicin biosynthesis from Micromonospora echinospora. J Korean Soc Appl Biol Chem. 2012;55:439–442. [Google Scholar]
- 22.Hong W, Yan L. Identification of gntK, a gene required for the methylation of purpurosamine C-6′ in gentamicin biosynthesis. J Gen Appl Microbiol. 2012;58:349–356. doi: 10.2323/jgam.58.349. [DOI] [PubMed] [Google Scholar]
- 23.Li D, Li H, Ni X, Zhang H, Xia H. Construction of a gentamicin C1a-overproducing strain of Micromonospora purpurea by inactivation of the gacD gene. Microbiol Res. 2013;168:263–267. doi: 10.1016/j.micres.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, et al. GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: Isolation and characterization of a cobalamin-dependent radical SAM enzyme. J Am Chem Soc. 2013;135:8093–8096. doi: 10.1021/ja312641f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao L, et al. Characterization of a key aminoglycoside phosphotransferase in gentamicin biosynthesis. Bioorg Med Chem Lett. 2013;23:1438–1441. doi: 10.1016/j.bmcl.2012.12.064. [DOI] [PubMed] [Google Scholar]
- 26.Gu Y, et al. Biosynthesis of epimers C2 and C2a in the gentamicin C complex. ChemBioChem. 2015;16:1933–1942. doi: 10.1002/cbic.201500258. [DOI] [PubMed] [Google Scholar]
- 27.Li B, Van Schepdael A, Hoogmartens J, Adams E. Mass spectrometric characterization of gentamicin components separated by the new European Pharmacopoeia method. J Pharm Biomed Anal. 2011;55:78–84. doi: 10.1016/j.jpba.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Ni X, et al. Biosynthesis of 3″-demethyl-gentamicin C components by genN disruption strain of Micromonospora echinospora and test their antimicrobial activities in vitro. Microbiol Res. 2016;185:36–44. doi: 10.1016/j.micres.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Dorgan KM, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 32.Tsodikov OV, Hou C, Walsh CT, Garneau-Tsodikova S. Crystal structure of O-methyltransferase CalO6 from the calicheamicin biosynthetic pathway: A case of challenging structure determination at low resolution. BMC Struct Biol. 2015;15:13. doi: 10.1186/s12900-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi R, et al. Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics. Chem Biol. 2009;16:401–410. doi: 10.1016/j.chembiol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q, van der Donk WA. Catalytic promiscuity of a bacterial α-N-methyltransferase. FEBS Lett. 2012;586:3391–3397. doi: 10.1016/j.febslet.2012.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett MR, Shepherd SA, Cronin VA, Micklefield J. Recent advances in methyltransferase biocatalysis. Curr Opin Chem Biol. 2017;37:97–106. doi: 10.1016/j.cbpa.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Becker B, Cooper MA. Aminoglycoside antibiotics in the 21st century. ACS Chem Biol. 2013;8:105–115. doi: 10.1021/cb3005116. [DOI] [PubMed] [Google Scholar]
- 37.Huth ME, et al. Designer aminoglycosides prevent cochlear hair cell loss and hearing loss. J Clin Invest. 2015;125:583–592. doi: 10.1172/JCI77424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao M, Fan J. A new semisynthetic antibiotic: Etimicin. Bull Med Rec. 2002;31:24–33. [Google Scholar]
- 39.Aggen JB, et al. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob Agents Chemother. 2010;54:4636–4642. doi: 10.1128/AAC.00572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazos O, et al. Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. Chem Biol. 2010;17:160–173. doi: 10.1016/j.chembiol.2010.01.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.