Significance
Recent advancements in computational protein design have enabled the generation of proteins and protein complexes that are not present in nature. A current major limitation of the research is in the subsequent structural characterization. The commonly used techniques—X-ray crystallography, NMR spectroscopy, and cryo-EM—are time- and resource-consuming and heavily rely on sample quantity and quality. These limitations are particularly problematic for protein complexes. In this work, we show the applicability of native MS ion mobility coupled with surface-induced dissociation as a structural biology screening tool for designed proteins. We show that information about stoichiometry, intersubunit connectivity, and complex topology is rapidly gained by this SID-IM-MS methodology.
Keywords: computational design, native mass spectrometry, heterododecamers, surface-induced dissociation, ion mobility
Abstract
Computational protein design provides the tools to expand the diversity of protein complexes beyond those found in nature. Understanding the rules that drive proteins to interact with each other enables the design of protein–protein interactions to generate specific protein assemblies. In this work, we designed protein–protein interfaces between dimers and trimers to generate dodecameric protein assemblies with dihedral point group symmetry. We subsequently analyzed the designed protein complexes by native MS. We show that the use of ion mobility MS in combination with surface-induced dissociation (SID) allows for the rapid determination of the stoichiometry and topology of designed complexes. The information collected along with the speed of data acquisition and processing make SID ion mobility MS well-suited to determine key structural features of designed protein complexes, thereby circumventing the requirement for more time- and sample-consuming structural biology approaches.
Protein–protein interactions are key contributors to biological complexity and processes. Protein complexes can accomplish tasks that cannot be performed by the isolated subunits, including metabolic channeling, signal transduction, and cell shape stabilization (1). Structures of homo- and heterooligomeric complexes solved by X-ray crystallography, NMR spectroscopy, and cryo-EM help to shed light on protein complex topologies and can provide atomic-resolution details on the intricacies underlying protein–protein interactions. Most of those solved structures are from naturally occurring protein assemblies. Inevitably, their diversity is limited, as their evolution is linked with the physiological context and cellular requirements (2, 3). The need for tailor-made, structurally defined nanomaterials to tackle a new generation of challenges in medicine and in industry makes it desirable to control the defined assembly of protein subunits into higher-order architectures not found in nature (4, 5). Several approaches have been utilized to drive the formation of higher-order protein structures, including the mediation of protein interactions by metals, disulfide bonds, genetic fusions, and ideal helix–helix interactions (6–10). More recently, a symmetric modeling approach was used in the Rosetta software suite for the design of cyclic oligomers (11), nanocages (12–15), and 2D layers (16). The extraordinary success of this method makes it interesting to further explore its potential to design additional heterooligomeric protein architectures.
Advances in the Rosetta design methodology and in gene synthesis enable the design and testing of many assemblies. However, there is still a great lack in methodologies to screen and characterize these assemblies with high confidence and efficiency on a structural level. Usually, high-resolution structural determination methods are used for structural analysis. However, the most commonly used technique, X-ray crystallography, is not generally applicable, as it contains time-consuming steps for sample preparation, measurement, and data analysis and bears the risk of introducing artifacts from structural rearrangements during the protein crystallization process (17). NMR is limited by the quantity and the size of the complex (18). Finally, cryo-EM is not yet universally available and is limited by sample preparation, size of the protein complex, measurement, data storage, and data analysis (19). Moreover, due to limitations in time and resources, atomic-resolution techniques are often not required for the initial screening and characterization of protein complexes. This is particularly true for a library of computationally designed protein complexes, where fast and high-throughput characterization methods are needed. The entire native MS work flow including data analysis for a sufficiently purified protein complex sample as shown in this work is accomplishable within 1 d.
Native MS has become an attractive structural biology tool to assess the key features of protein quaternary structures (20–23). In this technique, protein assemblies are ionized from a nondenaturing volatile salt solution, (partially) desolvated in vacuum, and detected by a mass analyzer to obtain their molecular masses. Additionally, ion mobility (IM) measurements can be used to also determine their collision cross-sections (CCSs) in a combined IM-MS approach. These IM-MS measurements provide information on the stoichiometry and shape of the intact protein complex in the gas phase (24–27). The protein complex ion of a particular charge state can be selected by a quadrupole and subjected to surface-induced dissociation (SID) before IM-MS. SID is a single-step, high-energy activation method, wherein noncovalent dissociation of the complex leads to generation of subcomplexes that are building blocks of the complex (24, 26). Consequently, a combination of native IM-MS and quadrupole SID-IM-MS can be used to obtain characteristics of protein assemblies, such as stoichiometry, intersubunit connectivity, and topology, using sample amounts typically as low as 5–50 pmol (26, 27).
We set out to computationally design two-component dihedral protein assemblies. These dihedral protein assemblies failed to form high-quality crystals required for X-ray crystallography. Hence, we used native MS coupled with IM and SID to characterize the designed protein complexes. We show that information about stoichiometry, intersubunit connectivity, and overall topology is rapidly gained by our methodology. The computationally designed protein complexes show the expected stoichiometry, intersubunit connectivity, and overall topology.
Results and Discussion
Computational Design.
We designed protein complexes with dihedral symmetry based on homodimeric and homotrimeric building blocks deposited in the Protein Data Bank (PDB). A total of 272 trimers and 1,054 dimers were selected as scaffolds; multidomain proteins, oligomers without distinct protein cores, and those without accessible structured surfaces were discarded. A set of D3 scaffolds that do not intercalate along the dihedral axis was also included as C3s. Additionally, only crystal structures with resolution better than 2.5 Å were accepted. Two trimeric building blocks were aligned in a D3 group, while three dimeric building blocks were aligned around the dihedral twofold axis, perpendicular to the trimer C3 axis, thereby also conforming to D3 symmetry (Fig. 1A). This resulted in an overall architecture that we labeled as “D32”: dihedral symmetry composed of cyclic homotrimers [C3, subunit A (Fig. 1A, green)] and cyclic homodimers [C2, subunit B (Fig. 1A, red)]. All 286,688 cross-pairs were subsequently docked, allowing freedom of movement and rotation along the rotational axes and relative to the center of the D32 architecture. The D32 architecture allowed us to utilize the library of dimers and trimers, which is significantly more numerous than that of higher cyclic symmetries. The architecture also results in unique connectivities, where the overall structure cannot assemble without both components being present. Where the trimeric and dimeric building blocks made favorable contacts, finer sampling was performed followed by RosettaDesign (28) to generate well-packed, hydrophobic protein interfaces. These designs were subsequently filtered by metrics including solvent-accessible surface area, buried unsatisfied hydrogen bonds, shape complementarity (29), and binding energy (13). All designs that passed these filters were subject to residue-by-residue reversion to the WT sequence and subsequently scored to minimize the amount of introduced mutations for each design. The RosettaScript and symmetry definition files for the dihedral designs can be found in Dataset S1.
Fig. 1.
Design of two-component dihedral complexes. (A) Dihedral symmetry contains a symmetrical cyclic axis perpendicular to a C2 axis. In our D32 symmetry, the C3 axis (green) is perpendicular to the C2 axes (red). Trimeric (green; A3) and dimeric (red; B2) scaffolds were aligned to their respective symmetrical axes. The two dfs sampled were radius from the center (r) and angle rotated along their symmetrical axes (ω). Where the building blocks meet, designed mutations were placed to generate an interface. (B) The designed complexes A6B6 (D32-01, D32-2, and D32-03) and their respective WT scaffold PDB ID codes are shown. The color scheme for subunit A (green) and subunit B (red) is shown. The expressed and purified designed complexes were confirmed through SEC (Superdex 200). Each SEC peak was further confirmed through SDS/PAGE separations to contain both of the components (Fig. S2).
Expression of Designs and Validation of Complex Formation.
A total of 48 designs were selected for experimental analysis, and the corresponding dimer–trimer pairs were coexpressed in Escherichia coli. Their ability to interact with each other was assessed based on the ability to copurify the untagged component with the hexahistidine-tagged component by immobilized metal ion affinity chromatography. Three complexes that displayed high expression levels and stability were subsequently verified to form higher-molecular mass complexes using analytical size exclusion chromatography (SEC), comparing the elution volume with the expected value (GE Healthcare) (Fig. 1B). Models for these three designs can be found in Dataset S2, and their amino acid sequences are reported in Fig. S1. Each design was also analyzed with SEC using buffers of different ionic concentrations (Fig. S2).
Stoichiometry and CCSs of Designed Protein Complexes.
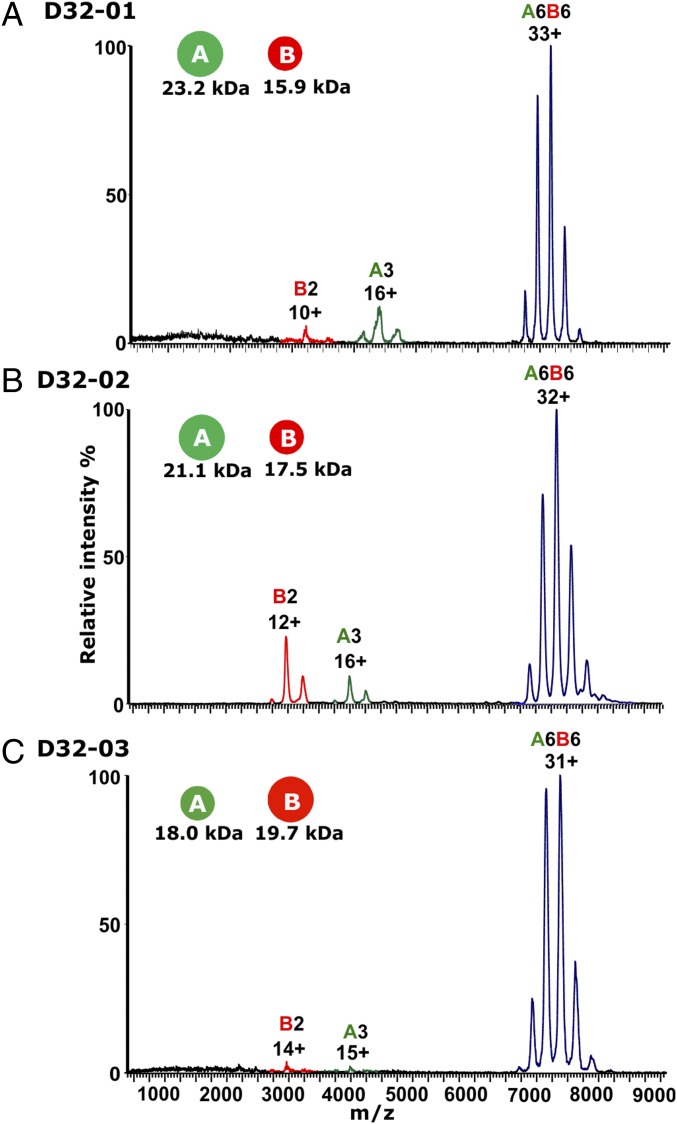
We used native MS to determine whether the designed protein interfaces resulted in the assembly of dimers and trimers to form the predicted heterododecamers. In a first step, we determined the stoichiometry and shape of the three complexes by IM-MS. We found differences in the effect of the ionic strength on the complex formation. Increasing ionic strength had no effect in the case of D32-01 but promoted complex formation in the cases of D32-02 and D32-03. (Fig. S3). At optimal ammonium acetate (AA) concentrations, a dominant species with a charge-state series around m/z 7,000–8,000 was observed for all three samples along with minor amounts of dimers and trimers (Fig. 2). The corresponding molecular masses of 235.9 ± 0.19 kDa for D32-01, 233.9 ± 0.49 kDa for D32-02, and 227.4 ± 0.12 kDa for D32-03 are in excellent agreement with those expected for the intended A6B6 heterododecamers (235.8 kDa for D32-01, 233.3 kDa for D32-02, and 226.3 kDa for D32-03) when taking into account the deviation from the theoretical mass that results from the additional mass of salt adducts as typically observed in native MS. Exact molecular mass determination under denaturing conditions further confirmed the integrity of the complex subunits with an absence of the N-terminal methionine in the case of the B subunits of D32-01 and D32-02 (Table S1). The CCSs of charge reduced complexes are 9,625 Å2 (CCS error 104 Å2) for the charge states 22–27+ for D32-01, 9,541 Å2 (CCS error 73 Å2) for the charge states 21–25+ for D32-02, and 8,964 Å2 (CCS error 42 Å2) for the charge states 20–24+ for D32-03 (Fig. S4). These can be compared with the CCS values calculated from the computational models for the designed complexes [10,277 Å2 (CCS error 92 Å2) for D32-01, 9,561 Å2 (CCS error 96 Å2) for D32-02, 10,055 Å2 (CCS error 83 Å2) for D32-03]. A deviation of the CCS by up to 11% most likely results from some extent of collapse in the gas phase as sometimes observed in native MS (30–32). Interestingly, the deviations of the CCS (7% for D32-01, 0.2% for D32-02, 11% for D32-03) do not correlate with the void volume of the designs (42,474 Å3 for D32-01, 7,389 Å3 for D32-02, 6,434 Å3 for D32-03). This suggests that the extent of gas-phase collapse is not exclusively determined by the void volume. In summary, our data show that the association of trimers and dimers results in well-defined heterododecamers with A6B6 stoichiometry and CCS values within 7, 0.2, and 11% , respectively, of those expected based on PSA-calculated CCS of the computational designs.
Fig. 2.
Mass spectra of D32 samples. Mass spectra are shown for (A) D32-01 in 100 mM AA, (B) D32-02 in 1,000 mM AA, and (C) D32-03 in 100 mM AA. Charge-state distributions for B dimer (B2), A trimer (A3), and heterododecamer (A6B6) are labeled in red, green, and blue, respectively, and the main charge state for each species is indicated.
Topology of Designs.
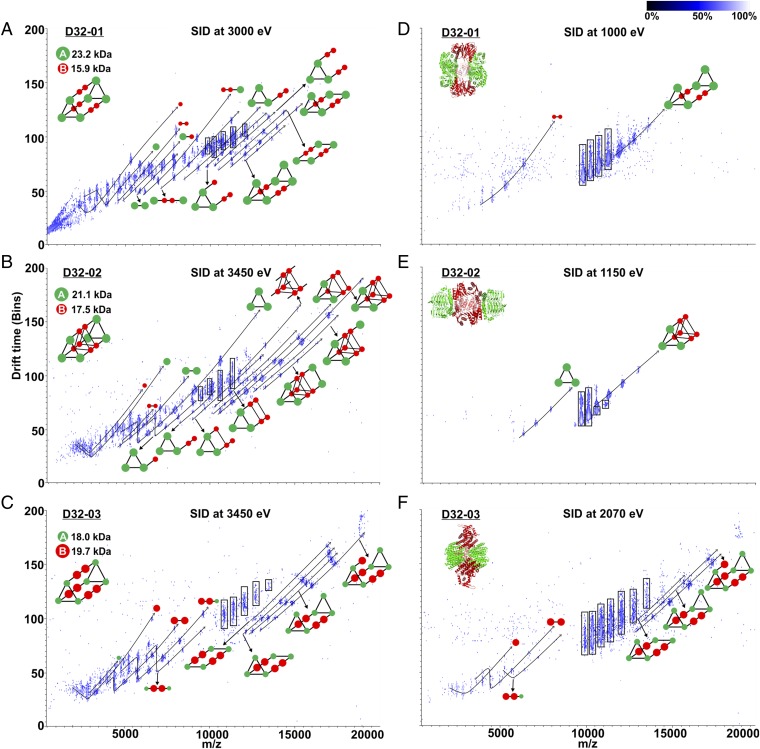
Next, we analyzed the complexes in the gas phase to obtain information on their topology and the underlying protein–protein interactions. For this purpose, we selected the heterododecamers at a single charge state in the mass spectrometer by a quadrupole and probed their composition by SID (25, 26, 33). In this technique, protein complexes are dissociated by collision with a surface. The SID products of a complex are often sufficient to gain insight into the complex architecture (26, 27). SID was performed by directing and accelerating the protein complex ions to an off-axis surface. The voltage settings are summarized in Table S2. All SID experiments were performed by using charge reduced protein complexes, as previous work [for example, by Quintyn et al. (25), Zhou and Wysocki (33), and Song et al. (26)] showed that SID of charge-reduced complexes reliably results in the generation of native-like subcomplexes that are reflective of the initial complex topology. The generated subcomplexes can be subsequently analyzed by IM TOF. We found that a sufficient variety of SID products is formed from the surface collision at a mid- to high-energy SID voltage to determine the topology of the protein designs from the charge-state distributions and corresponding masses of the generated subcomplexes (energies of [charge state × (100–180 V)] ∼ 3,200–5,800 eV). Based on the IM-MS data, we were able to unambiguously confirm the topology of all three designed A6B6 heterododecamers (Fig. 3 A–C). We observed subcomplexes collapse and assume that the subunits rearrange to stabilize charge/intramolecularly “solvate.” For example, SID of D32-01 generated AB2 with a CCS of 3,200 Å2 (clipped from model with no rearrangement: 3,714 Å2) and A5B4 with a CCS of 7,031 Å2 (clipped from model: 9,195 Å2). Consequently, we focused on using IM solely as an additional separation dimension to facilitate the identification and annotation of all m/z series. The corresponding masses of the SID-generated subcomplexes can be found in Table S3. All designs were thus shown to consist of two trimers connected by three dimers. Furthermore, only the SID of D32-02 results in generation of a B6 subcomplex, consistent with the fact that it is the only design where the three B dimers are in direct contact with each other.
Fig. 3.
IM-MS for midenergy (A–C) and low-energy (D–F) SID of A6B6 complexes. IM-MS (log scale) data are shown for 25+ D32-01 heterododecamer [(A) SID at 3,000 eV and (D) SID at 1,000 eV], for 23+ D32-02 heterododecamer D32-02 [(B) SID at 3,450 eV and (E) SID at 1,150 eV], and for 23+ D32-03 heterododecamer [(C) SID at 3,450 eV and (F) SID at 2,070 eV]. The charge-state series for SID-generated species were used to calculate the masses of subcomplexes and determine their identity (Table S3). Subcomplexes that could be unambiguously identified are shown. Selected (m/z 9,527, 10,296, and 10,482 for D32-01, D32-02, and D32-03, respectively) and charge-stripped complex ions are highlighted by rectangular boxes.
It is important to note that collision-induced dissociation (CID), which is the most commonly used gas-phase dissociation technique in MS/MS, typically results in the ejection of an unfolded monomer and generation of the complementary (n − 1)-mer subcomplex (34–36) and is thus—in contrast to SID—not generally applicable for the analysis of complex topologies. Supercharging can in some cases promote the generation of structure-informative fragments by CID, but the retention of structural information seems to heavily rely on protein complex properties, like subunit flexibility and charge density (37). Alternatively, it has been shown that protein–protein interactions can be disrupted by varying the ionic strength or percentage of organic solvent to generate subcomplexes for MS analysis (26, 38), and those are sometimes sufficient to determine a complex topology. In the case of D32 complexes, solution disruption showed the presence of B2 dimers and A3 trimers but did not show the other subcomplexes seen in SID (Fig. 3 A–C). SID alleviates the need to screen multiple in-solution disruption conditions that require extensive sample handling and large sample amounts. Furthermore, SID also provides more extensive connectivity information than solution disruption.
Protein–Protein Interactions Within the Designs.
We also varied the SID acceleration voltage to determine which subcomplexes are readily generated at low-energy surface collision. By plotting the abundance of (sub-)complexes against the SID collision energy, we found that D32-01 and D32-02 start to dissociate at around 750 and 500 eV, respectively (Fig. S5). Whereas D32-01 predominantly dissociates into B2 and complementary A6B4, D32-02 predominantly dissociates into A3 and complementary A3B6 at low SID energy (Fig. 3 D and E). This is consistent with the expectation of the small (designed) interface area between dimers and trimers being preferentially dissociated at low-energy SID. The dissociation of D32-01 into B2 and A6B4 requires only the dissociation of two relatively small A–B interactions (2 × 662 Å2). For D32-02, the additional interactions between the dimers favor the dissociation into A3 and A3B6, as this only require the dissociation of three relatively small A–B interactions (3 × 619 Å2) (Table S4).
Interestingly, a relatively high collision energy of around 1,800 eV is necessary to initiate dissociation of D32-03 (Fig. S5), and a significant amount of B subunit is generated above this SID energy threshold (Fig. 3F). This is particularly surprising, as the B dimer in the D32-03 complex has a large interface area between the subunits (B–B interaction: 3,131 Å2) and is expected to be far more stable than the designed interfaces between dimers and trimers. Solely based on the protein–protein interactions within the proposed D32-03 design, we would have expected B2 and complementary A6B4 as predominant SID products, as this only requires the dissociation of two relatively small A–B interactions (2 × 614 Å2) (Table S4).
Examination of D32-03 B Dimer.
Because D32-03 required higher energies to dissociate and because it did not produce dominant B2 dimers, as we predicted, we compared the properties of the D32-03 B subunit with the WT scaffold dimer (PDB ID code 3L9F) to test whether the designed substitutions altered the homooligomerization interface. We confirmed that both WT and D32-03 B subunit are dimers with almost identical CCSs (Fig. S6). CD spectroscopy measurements further confirmed that the dimers do not differ significantly in their secondary structure content (Fig. S7). Gas-phase CID of the two dimers showed a similar energy-dependent unfolding as monitored by IM, and the dissociation of the dimers to monomers by SID also displayed an almost identical energy dependency (Fig. S8). Both dimers start to unfold by CID at an energy of 440 eV as indicated by an increase in their drift time. Based on our SID data, dimers start to dissociate at around 600 eV, and the intensity of monomers equals that of the dimer at 900 eV. In summary, our data show that the introduction of mutations to design a protein–protein interaction surface does not interfere with the homooligomerization interface and the structure of the dimer.
In Vitro Assembly of D32-03.
To exclude that the association of dimers and trimers in vivo had influenced the ability to form the complex with the expected structure, we analyzed the mixture of separately expressed, hexahistidine-tagged dimers and trimers as well. We confirmed by MS that the D32-03 complex is formed in vitro, and using SID-IM-MS, we were further able to show that the in vitro- and in vivo-formed D32-03 complexes show the same gas-phase dissociation properties (Fig. S9). Whereas we can exclude that the D32-03 B subunit differs significantly from the WT scaffold dimer, it is not obvious whether the unexpected behavior of D32-03 is a result of the gas-phase collapse as indicated by IM, a rearrangement at high SID energies, or a combination thereof. At this point, we do not fully understand the dissociation behavior of D32-03.
Conclusion
We designed dihedral complexes based on dimers and trimers derived from nature; 3 of 48 designs were found to form highly stable complexes, but they failed to form crystals of sufficient quality for high-resolution X-ray structure determination. Thus, we used native MS as an alternative structural biology technique to assess the success of our computational approach. IM-MS in combination with SID of the three designs, D32-01, D32-02, and D32-03, confirmed that they are indeed A6B6 heterododecamers consisting of two trimers, with each monomer of the trimer connected by the dimers, further implying the dihedral architecture. Low-energy dissociation products of D32-01 and D32-02 by SID are consistent with the designed heterooligomerization interfaces, and midenergy dissociation products by SID suggest that the two designed complexes have the intended structure. In the case of D32-03, low-energy SID products deviate from the expected dominant products based on the protein–protein interface analysis, although each of the SID products formed is consistent with the overall structure. To exclude the possibility of formation of unexpected protein–protein interactions in the designed B dimer and within the complex, we confirmed that the designed B dimer does not deviate significantly from its scaffold dimer and that the B dimer associates both in vivo and in vitro with the corresponding trimer to form a heterododecamer. Although overall SID results show that the complex has the intended design, we cannot exclude at this point that the unexpected dissociation of this complex might be due to structural rearrangement in the gas phase because of the high-onset energy for dissociation. Consistent with some level of restructuring even before high-energy surface activation, D32-03 dodecamer and separately expressed dimer are collapsed in the gas phase as indicated by the significant deviations (11 and 13%, respectively) from the expected CCS values.
In summary, native MS-SID-IM is a beneficial structural biology tool as shown in this work. Its high speed and low sample consumption also make it perfectly suited for the rapid screening of (designed) proteins to determine their quaternary structures and facilitate the decision on which ones should be investigated by high-resolution structural biology methods. Native MS-SID-IM is highly suitable for rapidly assessing proposed quaternary structure models, and this technology has reached a point where even ab initio quaternary structure determination is possible (26, 27).
Materials and Methods
Protein Expression and Purification.
To produce the heterododecameric dihedral complexes, genes were synthesized and cloned into pET29b+ vectors (Gen9) via NdeI and XhoI, with an additional ribosome binding site in front of the gene encoding the second component to allow for the coexpression of both components in E. coli BL21 (DE3) cells. By construct design, a hexahistadine tag is only present on one of two components (Fig. S1). After induction with isopropyl β-d-1-thiogalactopyranoside (1 mM) at OD600 of 0.8, cell growth was continued at 18 °C for 16 h. After centrifugation, cells were resuspended in Tris-buffered saline (TBS; 50 mM Tris, 500 mM NaCl, pH 8.0) supplemented with lysozyme and DNase at 0.25 mg/mL (Sigma-Aldrich) and disrupted by sonication (15 W for 5 min total). Protein complexes were purified from the clarified lysate by immobilized metal ion affinity chromatography (binding: TBS, 30 mM imidazole; wash: TBS, 60 mM imidazole; elution: TBS, 500 mM imidazole) followed by SEC.
Analytical SEC.
Protein oligomers were analyzed by analytical SEC. A total amount of 1–5 mg protein was applied on a Superdex 200 Increase 10/300 GL size exclusion column (GE Healthcare). TBS or AA was used as mobile phase, and eluting proteins were detected by UV absorption at a wavelength of 280 nm.
CD Spectroscopy.
CD measurements were performed with a J-815 spectrophotometer (Jasco). Data were recorded at a scanning rate of 100 nm/min from 190 to 300 nm (data pitch, 1 nm; bandwidth, 1 nm). Proteins in TBS were measured at 25 °C with a concentration of 10 µM.
Native MS.
Proteins were buffer-exchanged into AA using Micro Bio-Spin P-6 Gel Columns (Bio-Rad), and protein concentrations were determined by UV absorbance using a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific). Proteins were diluted to a final concentration of 5 μM. For experiments requiring charge reduction (SID and CCS analyses) during nanoelectrospray ionization (nESI), triethylammonium acetate (TEAA; Sigma-Aldrich) was added at a ratio of 5:1 AA:TEAA. Borosilicate glass capillaries were prepared in house with a P-97 micropipette capillary puller (Sutter Instruments) and were filled with 5–10 µL of the protein solution. A platinum wire was inserted into the solution, and 1.2–1.5 kV were applied for nESI into a SYNAPT G2 HDMS mass spectrometer (Waters Corporation) with an SID device incorporated between a truncated trap traveling wave ion guide and the IM cell as described previously. The following instrument parameters were used: sampling cone, 20 V; extraction cone, 2 V; source temperature, 20 °C; trap gas flow, 4–5 mL/min; helium cell gas flow, 120 mL/min; IM nitrogen gas flow, 60 mL/min; trap wave velocity, 150 m/s; trap wave height, 4 V; ion mobility spectrometry (IMS) wave velocity, 300 m/s; IMS wave height, 20 V; transfer wave velocity, 65 m/s; transfer wave height, 2 V; and backing pressure, 4–6 mbar. The trap direct current bias was varied from 45 (for SID at 0 V accelerating voltage) to 240 (for SID 180 V accelerating voltage). Laboratory frame collision energy is obtained by multiplying the precursor ion charge state by the accelerating voltage. Details on the SID settings can be found in Table S2. Mass spectra were recorded for 5–10 min and smoothed (smoothing method: mean; smooth window: 50; number of smooths: 10). Tandem MS SID spectra were recorded for 20–30 min for low-energy SID and 60–80 min for high-energy SID.
CCS Determination.
Beta-lactoglobulin, transthyretin, and avidin were obtained from Sigma-Aldrich, and serum amyloid P component was obtained from EMD Millipore. Published protocols were used for the determination of experimental CCS (39, 40). Beta-lactoglobulin (charge states 7–8+ for monomer and 11–13+ for dimer), transthyretin (charge states 14–16+ for tetramer), avidin (charge states 15–17+ for tetramer), and serum amyloid P component (charge states 22–26+ for pentamer and 31–34+ for decamer) were used as CCS calibrants. N2-CCS values for the calibrants were derived from the work by Bush et al. (39). For dodecamers, the scan range was kept at m/z 400–20,000 (IM pusher frequency: 0.2729 ms per bin). For D32-03 WT and designed dimers, the scan range was kept at m/z 400–14,000 (IM pusher frequency: 0.1813 ms per bin). CCS calibrations were performed in three independent repeats. Theoretical CCS values were calculated based on the projection superposition approximation method (41). The following calculation parameters were used: temperature, 300 K; buffer gas, N2; orientation averaging accuracy, 0.01; number of orientations, 50–10,000; shape factor surface density, 2.0; mesh factor, 1.0; and surface factor accuracy, 0.01.
Supplementary Material
Acknowledgments
We thank Jacob Bale for his initial developments of the interface design protocol and Stephen Rettie for liquid chromatography/MS analysis. This work was partially supported by National Science Foundation Grant CHE-1332907 (to N.P.K. and D.B.) and NIH Grant R01 GM113658 (to V.H.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713646115/-/DCSupplemental.
References
- 1.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 2.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh JA, et al. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell. 2013;153:461–470. doi: 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rother M, Nussbaumer MG, Renggli K, Bruns N. Protein cages and synthetic polymers: A fruitful symbiosis for drug delivery applications, bionanotechnology and materials science. Chem Soc Rev. 2016;45:6213–6249. doi: 10.1039/c6cs00177g. [DOI] [PubMed] [Google Scholar]
- 5.Abe S, Maity B, Ueno T. Design of a confined environment using protein cages and crystals for the development of biohybrid materials. Chem Commun (Camb) 2016;52:6496–6512. doi: 10.1039/c6cc01355d. [DOI] [PubMed] [Google Scholar]
- 6.Salgado EN, Faraone-Mennella J, Tezcan FA. Controlling protein-protein interactions through metal coordination: Assembly of a 16-helix bundle protein. J Am Chem Soc. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Churchfield LA, Medina-Morales A, Brodin JD, Perez A, Tezcan FA. De novo design of an allosteric metalloprotein assembly with strained disulfide bonds. J Am Chem Soc. 2016;138:13163–13166. doi: 10.1021/jacs.6b08458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salgado EN, et al. Metal templated design of protein interfaces. Proc Natl Acad Sci USA. 2010;107:1827–1832. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi N, et al. Self-assembling nano-architectures created from a protein nano-building block using an intermolecularly folded dimeric de novo protein. J Am Chem Soc. 2015;137:11285–11293. doi: 10.1021/jacs.5b03593. [DOI] [PubMed] [Google Scholar]
- 10.Kaltofen S, et al. Computational de novo design of a self-assembling peptide with predefined structure. J Mol Biol. 2015;427:550–562. doi: 10.1016/j.jmb.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Fallas JA, et al. Computational design of self-assembling cyclic protein homo-oligomers. Nat Chem. 2017;9:353–360. doi: 10.1038/nchem.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bale JB, et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389–394. doi: 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsia Y, et al. Design of a hyperstable 60-subunit protein dodecahedron. [corrected] Nature. 2016;535:136–139. doi: 10.1038/nature18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King NP, et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King NP, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonen S, DiMaio F, Gonen T, Baker D. Design of ordered two-dimensional arrays mediated by noncovalent protein-protein interfaces. Science. 2015;348:1365–1368. doi: 10.1126/science.aaa9897. [DOI] [PubMed] [Google Scholar]
- 17.Krissinel E. Macromolecular complexes in crystals and solutions. Acta Crystallogr D Biol Crystallogr. 2011;67:376–385. doi: 10.1107/S0907444911007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marion D. An introduction to biological NMR spectroscopy. Mol Cell Proteomics. 2013;12:3006–3025. doi: 10.1074/mcp.O113.030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Leiro R, Scheres SH. Unravelling biological macromolecules with cryo-electron microscopy. Nature. 2016;537:339–346. doi: 10.1038/nature19948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenzen K, van Duijn E. Native mass spectrometry as a tool in structural biology. Curr Protoc Protein Sci. 2010;17:17.12. doi: 10.1002/0471140864.ps1712s62. [DOI] [PubMed] [Google Scholar]
- 21.Kirshenbaum N, Michaelevski I, Sharon M. Analyzing large protein complexes by structural mass spectrometry. J Vis Exp. 2010;40:1954. doi: 10.3791/1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lössl P, van de Waterbeemd M, Heck AJ. The diverse and expanding role of mass spectrometry in structural and molecular biology. EMBO J. 2016;35:2634–2657. doi: 10.15252/embj.201694818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liko I, Allison TM, Hopper JT, Robinson CV. Mass spectrometry guided structural biology. Curr Opin Struct Biol. 2016;40:136–144. doi: 10.1016/j.sbi.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Huang C, Wysocki VH. Surface-induced dissociation of ion mobility-separated noncovalent complexes in a quadrupole/time-of-flight mass spectrometer. Anal Chem. 2012;84:6016–6023. doi: 10.1021/ac300810u. [DOI] [PubMed] [Google Scholar]
- 25.Quintyn RS, Yan J, Wysocki VH. Surface-induced dissociation of homotetramers with D2 symmetry yields their assembly pathways and characterizes the effect of ligand binding. Chem Biol. 2015;22:583–592. doi: 10.1016/j.chembiol.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Song Y, Nelp MT, Bandarian V, Wysocki VH. Refining the structural model of a heterohexameric protein complex: Surface induced dissociation and ion mobility provide key connectivity and topology information. ACS Cent Sci. 2015;1:477–487. doi: 10.1021/acscentsci.5b00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano CA, et al. Biogenic manganese oxide nanoparticle formation by a multimeric multicopper oxidase Mnx. Nat Commun. 2017;8:746. doi: 10.1038/s41467-017-00896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leaver-Fay A, et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 30.Hogan CJ, Jr, Ruotolo BT, Robinson CV, Fernandez de la Mora J. Tandem differential mobility analysis-mass spectrometry reveals partial gas-phase collapse of the GroEL complex. J Phys Chem B. 2011;115:3614–3621. doi: 10.1021/jp109172k. [DOI] [PubMed] [Google Scholar]
- 31.Hall Z, Politis A, Robinson CV. Structural modeling of heteromeric protein complexes from disassembly pathways and ion mobility-mass spectrometry. Structure. 2012;20:1596–1609. doi: 10.1016/j.str.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Devine PWA, et al. Investigating the structural compaction of biomolecules upon transition to the gas-phase using ESI-TWIMS-MS. J Am Soc Mass Spectrom. 2017;28:1855–1862. doi: 10.1007/s13361-017-1689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M, Wysocki VH. Surface induced dissociation: Dissecting noncovalent protein complexes in the gas phase. Acc Chem Res. 2014;47:1010–1018. doi: 10.1021/ar400223t. [DOI] [PubMed] [Google Scholar]
- 34.Zhou M, Dagan S, Wysocki VH. Impact of charge state on gas-phase behaviors of noncovalent protein complexes in collision induced dissociation and surface induced dissociation. Analyst (Lond) 2013;138:1353–1362. doi: 10.1039/c2an36525a. [DOI] [PubMed] [Google Scholar]
- 35.Hall Z, Hernández H, Marsh JA, Teichmann SA, Robinson CV. The role of salt bridges, charge density, and subunit flexibility in determining disassembly routes of protein complexes. Structure. 2013;21:1325–1337. doi: 10.1016/j.str.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai YT, et al. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat Chem. 2014;6:1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. Charge-state dependent compaction and dissociation of protein complexes: Insights from ion mobility and molecular dynamics. J Am Chem Soc. 2012;134:3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- 38.Zhou M, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bush MF, et al. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 40.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 41.Bleiholder C, Johnson NR, Contreras S, Wyttenbach T, Bowers MT. Molecular structures and ion mobility cross sections: Analysis of the effects of He and N2 buffer gas. Anal Chem. 2015;87:7196–7203. doi: 10.1021/acs.analchem.5b01429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.