Significance
Cells must interact with their environment to survive. The lipids and proteins of the plasma membrane send and receive signals at the cell surface to respond to stimuli. When the lipid bilayer of the plasma membrane is damaged, cells release membrane-bound extracellular vesicles to repair the membrane. Cells also release signals on extracellular vesicles to communicate at a distance. Here, we identify proteins that regulate the formation of extracellular vesicles from the plasma membrane, providing additional tools to control their release that can be used to test potential functions of extracellular vesicles. Furthermore, we reveal that proteins regulating the asymmetric localization of the lipid phosphatidylethanolamine are critical for extracellular vesicle release, implicating this abundant but understudied lipid.
Keywords: extracellular vesicle, lipid asymmetry, retromer, microvesicle, flippase
Abstract
Cells release extracellular vesicles (EVs) that mediate intercellular communication and repair damaged membranes. Despite the pleiotropic functions of EVs in vitro, their in vivo function is debated, largely because it is unclear how to induce or inhibit their formation. In particular, the mechanisms of EV release by plasma membrane budding or ectocytosis are poorly understood. We previously showed that TAT-5 phospholipid flippase activity maintains the asymmetric localization of the lipid phosphatidylethanolamine (PE) in the plasma membrane and inhibits EV budding by ectocytosis in Caenorhabditis elegans. However, no proteins that inhibit ectocytosis upstream of TAT-5 were known. Here, we identify TAT-5 regulators associated with retrograde endosomal recycling: PI3Kinase VPS-34, Beclin1 homolog BEC-1, DnaJ protein RME-8, and the uncharacterized Dopey homolog PAD-1. PI3Kinase, RME-8, and semiredundant sorting nexins are required for the plasma membrane localization of TAT-5, which is important to maintain PE asymmetry and inhibit EV release. PAD-1 does not directly regulate TAT-5 localization, but is required for the lipid flipping activity of TAT-5. PAD-1 also has roles in endosomal trafficking with the GEF-like protein MON-2, which regulates PE asymmetry and EV release redundantly with sorting nexins independent of the core retromer. Thus, in addition to uncovering redundant intracellular trafficking pathways, our study identifies additional proteins that regulate EV release. This work pinpoints TAT-5 and PE as key regulators of plasma membrane budding, further supporting the model that PE externalization drives ectocytosis.
Cells release extracellular vesicles (EVs) that can mediate intercellular communication to influence development and disease (1, 2). EV release can also repair membranes to avoid cell death (3, 4). However, the in vivo functions of EVs are debated, because the molecules known to govern their formation are shared with other membrane trafficking pathways. For example, much is known about how multivesicular endosomes are formed by the recruitment of the endosomal sorting complex required for transport (ESCRT) to bud vesicles into the lumen of endosomes (5). The mechanisms of multivesicular endosome fusion with the plasma membrane to release exosomes by exocytosis are also characterized (1, 2). However, neither ESCRT nor exocytic proteins are specific to exosome release. The ESCRT machinery is one of the few membrane-sculpting factors known to bud vesicles away from the cytosol, and it regulates many membrane remodeling events (6), including plasma membrane budding away from the cytosol to release microvesicles by ectocytosis (1, 2). EV release by ectocytosis is poorly understood, even though microvesicles released from activated platelets improve coagulation and ciliary microvesicles are implicated in behavior (7, 8). Thus, to elucidate the in vivo functions of EVs and to better understand how membrane budding is regulated, it is necessary to identify the pathways that regulate EV release by ectocytosis.
We previously discovered that the conserved P4-ATPase TAT-5 maintains phosphatidylethanolamine (PE) asymmetry in the Caenorhabditis elegans plasma membrane and inhibits ectocytosis (9). When TAT-5 flippase activity is lost, the normally cytofacial lipid PE is externalized, and the plasma membrane overproduces EVs. EV production in tat-5 mutants depends on the plasma membrane recruitment of ESCRT (9), and ESCRT also acts during ectocytosis in mammalian cells and virus-infected cells (10, 11). Thus, proteins carrying out the final steps of budding have been identified, but it is unclear how cells regulate these pathways to control EV release and restrict ectocytosis to damaged membranes or selected cargos. In particular, no proteins were known to regulate TAT-5 activity during EV release. Unlike other P4-ATPases that require a β-subunit from the Cdc50 family of proteins to be chaperoned and active (12), the essential subclass of P4-ATPases, which includes worm TAT-5, yeast Neo1p, and mammalian ATP9A and ATP9B, act independently of Cdc50 proteins in yeast and mammals (13). Yeast Neo1p forms a complex with two large scaffolding proteins, the Dopey domain protein Dop1p and the GEF-like protein Mon2p (14). Together, they regulate clathrin-dependent retrograde trafficking from endosomes in yeast (15, 16). ATP9A also regulates recycling from endosomes (17), but it is unknown whether mammalian ATP9, Dopey, or Mon2 have a role in EV release. As cells regulate PE asymmetry and EV release during cell division, fusion, and death (18–20), it is important to determine how TAT-5 flippase activity is regulated to inhibit the outward budding of the plasma membrane and the release of EVs.
In this study, we identify proteins that regulate the trafficking and activity of TAT-5. We find that redundant retrograde recycling pathways traffic TAT-5 to the plasma membrane to inhibit EV release, suggesting that maintaining PE asymmetry in the plasma membrane is critical to maintain plasma membrane structure. We also discovered that the Dop1p ortholog PAD-1 is essential for TAT-5 to maintain PE asymmetry and inhibit EV release. We further reveal that TAT-5, PAD-1, and MON-2 regulate an intracellular trafficking pathway that is important for late endosome morphology. MON-2 and PAD-1 also play a redundant role in TAT-5 trafficking and EV release with proteins typically associated with retromer recycling. Our data demonstrate that relative increases in PE externalization consistently correlate with increased EV release, further supporting the model that PE exposure drives ESCRT-mediated ectocytosis. This work provides important insights into the mechanisms of EV release as well as intracellular trafficking.
Results
RME-8, PI3Kinase, and PAD-1 Inhibit EV Release.
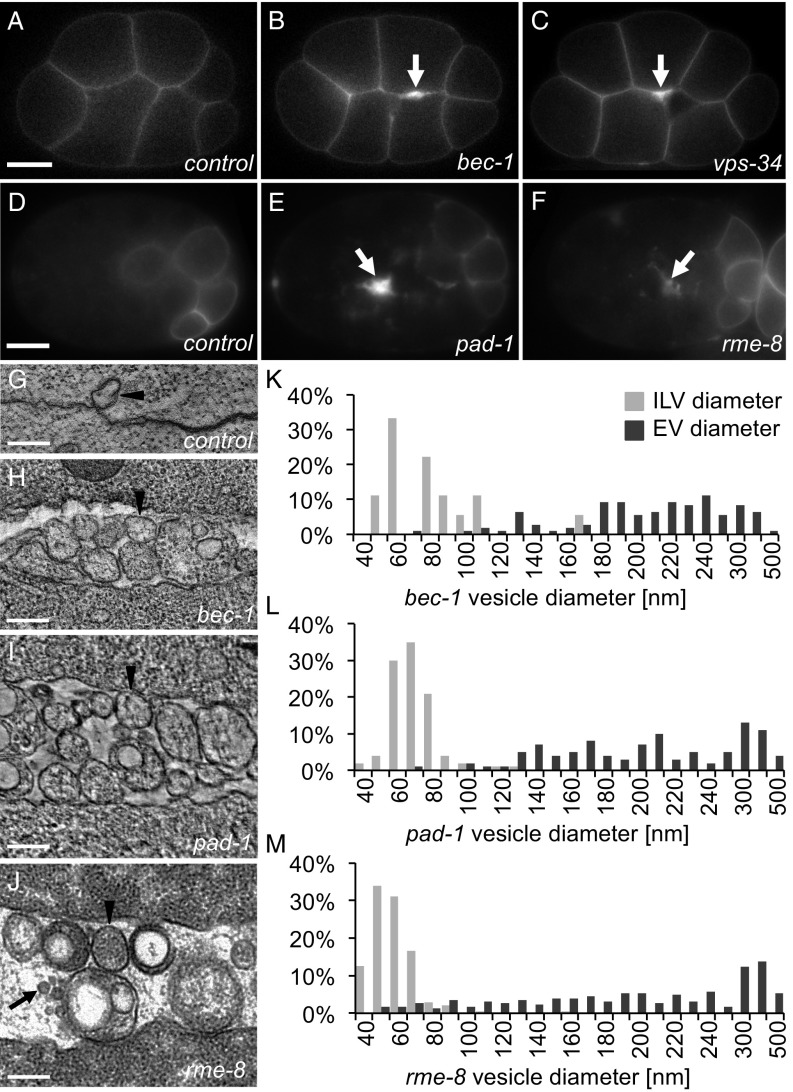
To understand how TAT-5 activity is regulated to inhibit EV budding, we performed an RNAi screen targeting 137 candidate interactors, many of which are involved in membrane trafficking and lipid biogenesis (Dataset S1). We screened for increased EV release using a plasma membrane reporter (PHPLC1∂1; Fig. 1 A–C), which labels plasma-membrane-derived EVs. We also used a degron-tagged plasma membrane reporter to specifically label EVs released before the onset of ZF1-mediated proteasomal degradation (ZF1::PHPLC1∂1; Fig. 1 D–F) (9, 21). We discovered four genes whose disruption results in increased membrane labeling at cell contacts (Table 1), similar to tat-5 mutants (9). These include the Beclin1 homolog BEC-1, the class III PI3Kinase VPS-34, the Dopey domain protein PAD-1, and the DnaJ domain protein RME-8 (Fig. 1 B, C, E, and F). We confirmed the presence of increased EVs using electron microscopy and tomography in bec-1, pad-1, and rme-8 mutants (Fig. 1 G–J). In bec-1 and pad-1 mutants, the EVs are larger than the intraluminal vesicles of multivesicular endosomes (Fig. 1 K and L), consistent with the >100-nm diameter of microvesicles formed by plasma membrane budding (2). In contrast, rme-8 knockdown resulted in EVs with diameters similar to both intraluminal vesicles and microvesicles (Fig. 1M), demonstrating that loss of RME-8 causes an increase in microvesicle release, but may also alter the size of microvesicles or result in exosome release. Thus, our screen identified four proteins that inhibit ectocytosis.
Fig. 1.
Retrograde trafficking proteins inhibit microvesicle release. (A) PHPLC1∂1::mCh localizes primarily to the plasma membrane in control embryos at the eight-cell stage. (B and C) PH reporters localize to thickened membranes (arrow) in bec-1 (PHPLC1∂1::mCh) and vps-34 (PHPLC1∂1::GFP) maternal zygotic mutants. (D) In a 26-cell control embryo, GFP::ZF1::PHPLC1∂1 is degraded in most somatic cells, only persisting on the plasma membrane in a few posterior cells. (E and F) EVs labeled with GFP::ZF1::PHPLC1∂1 (arrow) accumulate between cells in pad-1 and rme-8 RNAi-treated embryos. (G) EVs are infrequently observed between wild-type N2 cells in a two-cell embryo tomogram. (H–J) Released EVs accumulate between cells in a tomogram from a three-cell bec-1 maternal-zygotic mutant, a three-cell embryo treated with pad-1 RNAi, and a 24-cell rme-8 RNAi embryo. Arrowheads point to microvesicle-sized EVs; arrows point to exosome-sized EVs. (K–M) Histograms of EV and intraluminal vesicle (ILV) diameters measured from TEM images of bec-1 maternal-zygotic mutant embryos and pad-1 RNAi embryos demonstrate that the majority of EVs are microvesicles, because they are larger than ILVs. In rme-8 RNAi, EVs could be both exosomes and microvesicles, because they are the same size as or larger than ILVs. [Scale bars: A, 10 µm (also applies to B and C); D, 10 µm (also applies to E and F); G–J, 200 nm.]
Table 1.
Summary of EV release and GFP::TAT-5 localization defects
| Genotype, RNAi | Increased EV release (thick membranes) | GFP::TAT-5 not in plasma membrane | ||
| % | n | % | n | |
| + | ||||
| − | 0 | 119 | 0 | 96 |
| tat-5 | 90 | 31 | 79 | 29 |
| pad-1 | 89 | 55 | 0 | 60 |
| mon-2 | 0 | 72 | 0 | 47 |
| vps-26 | 0 | 73 | 0 | 41 |
| vps-29 | 0 | 40 | 0 | 49 |
| vps-35 | 0 | 40 | 0 | 42 |
| snx-1 | 0 | 61 | 31 | 36 |
| snx-3 | 0 | 60 | 5 | 73 |
| snx-6 | 0 | 63 | 28 | 72 |
| lst-4 | 0 | 148 | 0 | 72 |
| rme-8 | 84 | 50 | 39 | 89 |
| rme-8(b1023) | ||||
| − | 28 | 101 | ||
| bec-1(ok691) | ||||
| − | 75 | 32 | 69 | 26 |
| vps-34(h510) | ||||
| − | 25 | 28 | 27 | 83 |
| pad-1(wur02) | ||||
| − | 100 | 11 | 0 | 3 |
| vps-26(tm1523) | ||||
| − | 0 | 41 | 0 | 52 |
| vps-29 | 0 | 45 | 0 | 46 |
| vps-35 | 0 | 43 | 0 | 35 |
| snx-1(tm847) | ||||
| − | 0 | 24 | 37 | 41 |
| pad-1 | 45* | 40 | 13 | 40 |
| snx-3 | 55* | 20 | 26 | 37 |
| snx-3(tm1595) | ||||
| − | 0 | 47 | 26 | 53 |
| mon-2 | 98 | 61 | ||
| pad-1 | 100 | 43 | ||
| snx-1 | 87 | 55 | ||
| snx-6 | 100 | 30 | 36 | 53 |
| chat-1(ok1681) | ||||
| F20C5.4, W03G11.2 | 0 | 18 | 0 | 18 |
| chat-1(ok1681); W03G11.2(ok2574) | ||||
| F20C5.4 | 0 | 36 | ||
| mon-2(xh22) | ||||
| − | 0 | 105 | 0 | 28 |
| mon-2 | 0 | 270 | 0 | 33 |
| pad-1 | 76 | 78 | 0 | 44 |
| vps-26 | 0 | 102 | 0 | 44 |
| vps-29 | 0 | 97 | 0 | 39 |
| vps-35 | 0 | 79 | 0 | 34 |
| snx-1 | 77 | 87 | 16 | 45 |
| snx-3 | 51 | 84 | 25 | 28 |
| snx-6 | 96 | 95 | 37 | 30 |
Mutant and control (+) worms expressing fluorescent reporters were treated with RNAi or untreated (−). The percentage of embryos showing the indicated phenotype is given. Embryos (n) from the late 4-cell to 102-cell stage were scored for thickened membrane labeling between cells, which is indicative of EV release. Embryos from the zygote to 15-cell stage were scored for whether GFP::TAT-5 was visible in the plasma membrane.
GFP::TAT-5 was used to score EV release in snx-1(tm847) pad-1 RNAi embryos and snx-1(tm847) snx-3 RNAi embryos, which is likely to underestimate EV release as GFP::TAT-5 localization to the plasma membrane was reduced.
PI3Kinase and RME-8 Are Required for TAT-5 Localization to the Plasma Membrane.
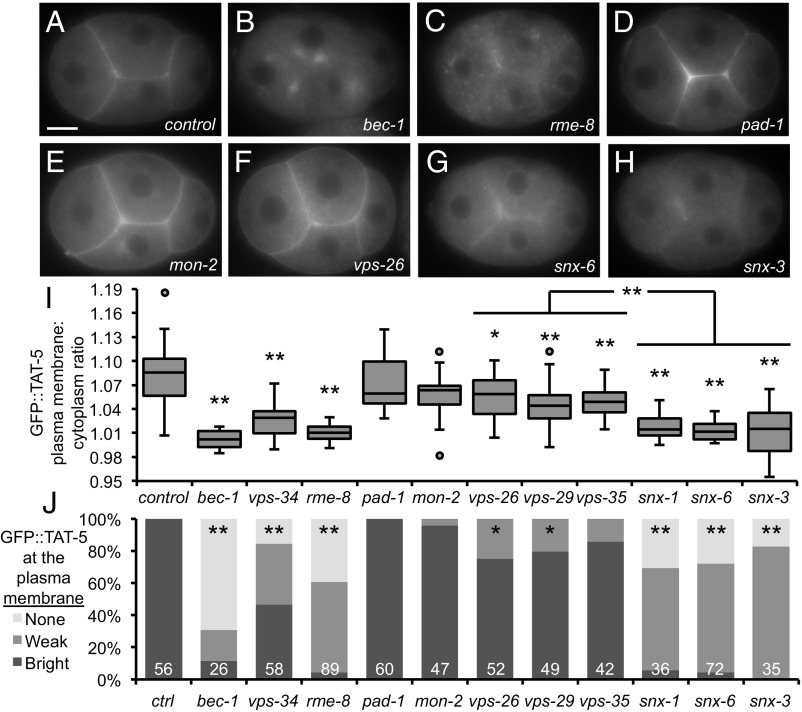
All four of the identified EV regulators are linked to retrograde endosomal trafficking. The class III PI3Kinase complex (PI3K), which includes the kinase VPS-34 and the Beclin1 ortholog BEC-1, phosphorylates phosphatidylinositol (PI) to create PI3P on autophagosomal and endosomal membranes (22). The lipid PI3P is important for the recruitment of trafficking factors to endosomes, including the DnaJ protein RME-8 involved in endosomal recycling (23). PAD-1 homologs in yeast and mammals have also been implicated in retrograde trafficking between endosomes and Golgi (15, 24). TAT-5 is a multipass transmembrane protein that localizes primarily to the plasma membrane (Fig. 2A). GFP::TAT-5 is also found on endocytic vesicles, colocalizing with the clathrin adaptor AP2 (Fig. S1 A–E), which shows that TAT-5 is endocytosed. TAT-5 is trafficked through various endosomes that are positive for markers of sorting or recycling endosomes, including RME-8 (Fig. S1 A, B, F, and G), as well as degradative endosomes or lysosomes (Fig. S1 A, B, H, and I). Although we did not observe significant colocalization with Golgi (Fig. S1 A, B, and J), we tested whether GFP::TAT-5 localization was changed in EV-releasing retrograde trafficking mutants. GFP::TAT-5 localized to prominent structures in the cytoplasm instead of the plasma membrane in bec-1, vps-34, or rme-8 mutants and after rme-8 RNAi treatment (Fig. 2 B, C, I, and J), potentially explaining the observed increase in EV release (Fig. 1 and Table 1). The overall levels of GFP::TAT-5 were also reduced after rme-8 knockdown (Fig. S2), suggesting that mislocalization could alter protein stability. In contrast, GFP::TAT-5 localized robustly to the plasma membrane in pad-1 knockdowns (Fig. 2 D, I, and J). In fact, GFP::TAT-5 was increased at the cell surface after pad-1 knockdown (Fig. S3A), likely due to the release of TAT-5 on EVs. To confirm that the normal localization of TAT-5 was not due to incomplete knockdown of PAD-1, we generated a deletion allele removing 99.5% of the pad-1 coding sequence and discovered that pad-1(wur02) mutants were >90% sterile, similar to tat-5 deletion mutants (9). We examined GFP::TAT-5 in rare maternal-zygotic pad-1 deletion mutant embryos and found that TAT-5 still localized to the plasma membrane after the complete loss of PAD-1 (Table 1 and Fig. S3B). Thus, PI3K and RME-8 are required for TAT-5 localization to the plasma membrane, while PAD-1 is not.
Fig. 2.
TAT-5 is recycled by retromer-associated proteins. (A) GFP::TAT-5 localizes to the plasma membrane in a control four-cell embryo. (B) GFP::TAT-5 is mislocalized to large cytoplasmic structures in bec-1 maternal zygotic mutants. (C) GFP::TAT-5 shows a more dispersed localization to cytoplasmic structures in rme-8 RNAi. See Fig. S2 for reduced total levels of GFP::TAT-5. (D and E) GFP::TAT-5 localization is not altered after pad-1 or mon-2 RNAi. See Fig. S3 for GFP::TAT-5 localization to EVs. (F) GFP::TAT-5 still localizes to the plasma membrane in core retromer vps-26 mutants. (G and H) In contrast, GFP::TAT-5 plasma membrane localization is weak after snx-6 RNAi or in snx-3 deletion mutants. (I) Ratio of GFP::TAT-5 fluorescence intensity in the plasma membrane to the intensity of the cytoplasm. Disrupting PI3K subunits and RME-8 had highly significant effects on GFP::TAT-5 plasma membrane localization, as did SNX. More variability is seen in vps-34 mutants due to the presence of a mosaic rescuing transgene (Materials and Methods). There was no significant change in plasma membrane localization after pad-1 or mon-2 RNAi, but knocking down or deleting core retromer proteins resulted in mild, but significant, decreases. Student’s t test with Bonferroni correction was used for statistical analysis. *P < 0.05; **P < 0.001 (compared with control empty vector RNAi). (J) Embryos were categorized for the brightness of GFP::TAT-5 localization in the plasma membrane. PI3K subunits, RME-8, and sorting nexins prevented or decreased TAT-5 plasma membrane localization, while core retromer proteins resulted in weaker plasma membrane localization. *P < 0.01; **P < 0.0001 (Fisher’s exact test with Bonferroni correction). Number of embryos scored is indicated for each genotype. [Scale bar: A, 10 µm (also applies to B–H).]
PI3Kinase, RME-8, and Sorting Nexins Control TAT-5 Localization Independent of the Core Retromer.
PI3K and RME-8 have been specifically linked to retromer trafficking, which is involved in recycling transmembrane proteins to the plasma membrane in addition to recycling proteins from endosomes to Golgi (25). The retromer complex consists of a trimer of core subunits (VPS-26, -29, and -35) and lipid-binding sorting nexins (SNX). PI3K and RME-8 are essential for the retrograde trafficking of the Wntless homolog MIG-14 together with the core retromer and the SNX-PX protein SNX-3 (22, 26). RME-8 also binds to the SNX-BAR protein SNX-1 and its heterodimer partner SNX-6 to promote endosomal tubulation (23). Furthermore, PI3K and Snx3p have been shown to regulate endosomal trafficking of the TAT-5 ortholog Neo1p in yeast (16), although the known Snx3-binding domain is not conserved in animals (Fig. S4). Therefore, we tested whether GFP::TAT-5 localization was changed in retromer mutants. GFP::TAT-5 still localized to the plasma membrane in vps-26 deletion mutants (Fig. 2F) or after vps-26, -29, or -35 RNAi treatment (Table 1). However, there was a measurable drop in plasma membrane levels in vps-26 mutants, vps-29 RNAi, and vps-35 RNAi (Fig. 2 I and J), demonstrating that the core retromer had a mild effect on TAT-5 localization in contrast to PI3K and rme-8 knockdown. In contrast, GFP::TAT-5 appeared significantly more diffuse around the plasma membrane in snx-1 or -6 RNAi-treated embryos and in snx-3 deletion mutants (Fig. 2 G and H). Indeed, plasma membrane localization decreased similar to PI3K and rme-8 knockdown (Fig. 2 I and J and Table 1). These results indicate that sorting nexins, PI3K, and RME-8 play a more important role than the core retromer complex in TAT-5 localization.
To confirm that the core retromer proteins were not important for TAT-5 localization or EV release, we tested whether they could act redundantly. We performed RNAi on a series of deletion strains and obtained synthetic sterility and embryonic lethality when we targeted vps-29 or -35 in a vps-26 deletion strain (Table S1). However, these treatments did not significantly decrease GFP::TAT-5 localization to the plasma membrane or result in increased EV release (Table 1), demonstrating that the core retromer proteins are not acting redundantly during TAT-5 trafficking. These findings indicate that PI3K, RME-8, and sorting nexins control TAT-5 localization independent of the core retromer.
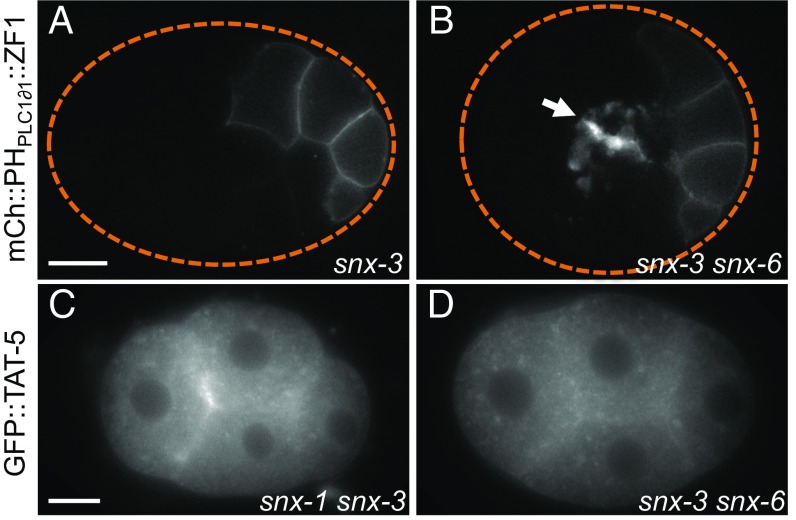
Given the significant role of sorting nexins on TAT-5 localization, we tested whether sorting nexins also inhibit EV release. Using the degron reporter to label EVs, we found that EV release was not increased after RNAi targeting snx-1, -3, or -6 or in snx-1 or -3 deletion mutants (Fig. 3A and Table 1), suggesting that TAT-5 was still able to maintain plasma membrane asymmetry despite its reduced levels at the plasma membrane. This could potentially be through its localization to cortical endosomes, as recent studies in yeast showed that the TAT-5 ortholog Neo1p maintains lipid asymmetry in the plasma membrane indirectly by regulating lipid asymmetry in endosomes (27). Given that sorting nexins acted independently of the core retromer, we tested whether other sorting nexins also regulated TAT-5 trafficking. Knocking down a SNX-BAR protein associated with endocytosis, the SNX9 ortholog LST-4 (28), did not disrupt GFP::TAT-5 localization to the plasma membrane or cause increased EV release (Table 1). We next tested whether retromer-associated sorting nexins could redundantly regulate EV release. We found significantly increased EV release in snx-1 mutants treated with snx-3 RNAi or in snx-3 mutants treated with snx-6 RNAi (Fig. 3B and Table 1). We next examined TAT-5 localization in these double mutants and saw TAT-5 vesicles dispersed throughout the cytoplasm (Fig. 3 C and D). However, TAT-5 was still visible at the plasma membrane, similar to single snx mutants (Table 1), although it was difficult to distinguish between weak GFP::TAT-5 fluorescence in released EVs or in the plasma membrane. Thus, the SNX-1–SNX-6 heterodimer is redundantly required with SNX-3 to traffic TAT-5 and inhibit EV release.
Fig. 3.
SNX-1/SNX-6 and SNX-3 redundantly inhibit EV release. (A) EV release was not increased in a 26-cell snx-3 mutant embryo expressing the mCherry::PHPLC1∂1::ZF1 plasma membrane reporter. (B) EVs labeled with mCherry::PHPLC1∂1::ZF1 (arrow) accumulate between cell contacts in a 26-cell snx-3 mutant treated with snx-6 RNAi. (C and D) GFP::TAT-5 is mislocalized to cytoplasmic compartments in snx-1 mutants treated with snx-3 RNAi, as well as in snx-3 mutants treated with snx-6 RNAi. [Scale bars: A, 10 μm (also applies to B); C, 10 μm (also applies to D).]
Cdc50 Family Proteins Do Not Regulate TAT-5 Trafficking or EV Release.
As Cdc50 family proteins act as functional β-subunits and chaperones for most P4-ATPases, including the PS flippase TAT-1 (12), we tested whether any of the three C. elegans Cdc50 homologs were needed for EV release or TAT-5 localization, specifically chat-1, F20C5.4, and W03G11.2. We did not observe increased EV release in triple-mutant embryos made with a combination of deletion alleles and RNAi (Table 1). Similarly, knocking down F20C5.4 and W03G11.2 in chat-1 deletion mutants did not disrupt GFP::TAT-5 exit from the endoplasmic reticulum or localization to the plasma membrane (Table 1). Thus, in contrast to TAT-1, which requires CHAT-1 for its plasma membrane localization and flippase activity (12), TAT-5 does not have a β-subunit from the Cdc50 family of proteins. This finding is similar to TAT-5 orthologs in other species (13), leaving open which proteins regulate TAT-5 activity.
PAD-1 and TAT-5 Have Distinct Roles in EV Release from MON-2.
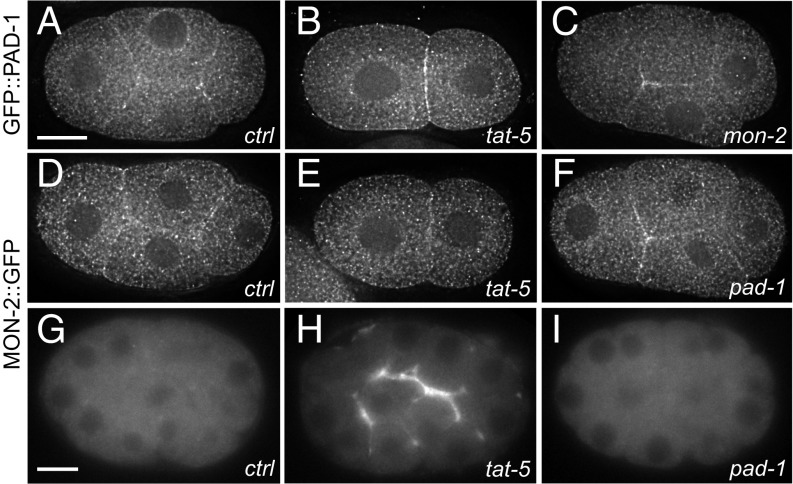
As the large Dopey domain protein PAD-1 inhibited EV release without disrupting TAT-5 localization, we characterized this protein more closely. Based on their yeast orthologs, TAT-5 is predicted to form a complex with PAD-1, as well as the large GEF-like protein MON-2 (14). We generated functional reporter strains for PAD-1 and MON-2 to observe their localization. GFP::PAD-1 and MON-2::GFP localize similarly to cytoplasmic puncta, but are also found at the plasma membrane (Fig. 4 A and D). Thus, TAT-5 localization is distinct but overlapping with the soluble proteins PAD-1 and MON-2, suggesting that their interaction from yeast could be conserved in C. elegans. However, yeast neo1 and dop1 are essential genes, while mon2 is not (29). Similarly, C. elegans tat-5 and pad-1 deletion mutants are sterile (9), in contrast to fertile mon-2 nonsense mutants (30), suggesting that TAT-5 and PAD-1 have more similar functions than TAT-5 and MON-2. We tested whether MON-2 also inhibits EV release using membrane reporters, but found that mon-2 nonsense mutants do not show increased EV release (Fig. 5A), even when additionally treated with mon-2 RNAi (Table 1). These data suggest that MON-2 does not directly inhibit EV release and indicate that TAT-5 and PAD-1 have separable roles in EV release from MON-2.
Fig. 4.
PAD-1 and MON-2 localize to the cell cortex and cytoplasm. (A) GFP::PAD-1 staining is found on cytoplasmic puncta as well as at the plasma membrane in a four-cell embryo. GFP was knocked into the endogenous pad-1 locus. (B) GFP::PAD-1 localization is not significantly altered after tat-5 RNAi treatment in a two-cell embryo. (C) GFP::PAD-1 localizes to the plasma membrane after mon-2 RNAi treatment in a four-cell embryo. (D) MON-2::GFP::3xFlag staining also localizes to cytoplasmic puncta as well as at the plasma membrane in a four-cell embryo. (E) MON-2::GFP::3xFlag localization is not significantly altered after tat-5 RNAi treatment in a two-cell embryo. (F) MON-2::GFP::3xFlag still localizes to the plasma membrane after pad-1 RNAi treatment in a four-cell embryo. (G) In live 26-cell embryos, MON-2::GFP::3xFlag puncta are barely visible. (H) MON-2::GFP::3xFlag is increased at the cell surface after tat-5 RNAi treatment, suggesting that cortical MON-2 is released outside cells in EVs. (I) MON-2::GFP::3xFlag is not released in EVs after pad-1 RNAi treatment, suggesting that PAD-1 is required for MON-2 localization in EVs. [Scale bars: A, 10 μm (also applies to B–F); G, 10 μm (also applies to H and I).]
Fig. 5.
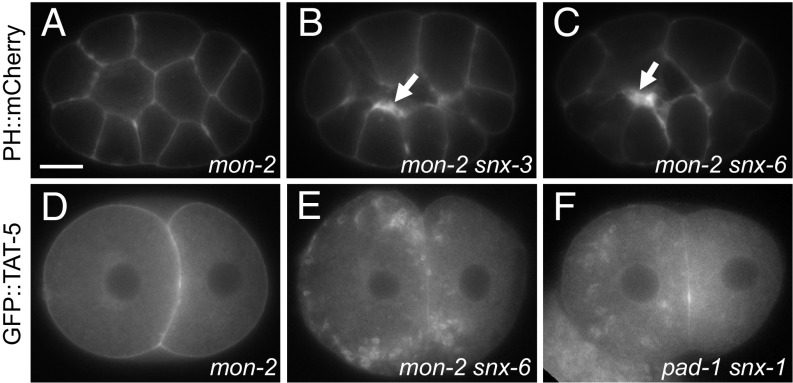
MON-2 inhibits EV release in snx-mutants. (A) The plasma membrane marker PHPLC1∂1::mCh appears normal in a 15-cell mon-2(xh22) mutant embryo. (B and C) Thickened patches of PHPLC1∂1::mCh (arrow) are observed in mon-2(xh22) mutants treated with snx-3 or -6 RNAi, indicating increased EV release. Similar phenotypes were also seen after snx-1 RNAi treatment (Table 1), demonstrating that sorting nexins and MON-2 redundantly inhibit EV release. (D) GFP::TAT-5 localizes to the plasma membrane in a two-cell mon-2(xh22) mutant embryo. (E) GFP::TAT-5 localizes to prominent cytoplasmic vesicles after snx-6 RNAi treatment in mon-2(xh22) mutants. Similar phenotypes were also seen after snx-1 or -3 RNAi treatment, suggesting that MON-2 acts redundantly with sorting nexins to control TAT-5 localization. (F) GFP::TAT-5 localizes to large cytoplasmic vesicles after pad-1 RNAi in snx-1(tm847) mutants, demonstrating that PAD-1 also acts redundantly with sorting nexins during TAT-5 trafficking. [Scale bar: A, 10 µm (also applies to B–F).]
TAT-5, PAD-1, and MON-2 Do Not Regulate Each Other’s Plasma Membrane Levels or Localization.
As the Neo1p–Dop1p–Mon2p complex maintains the levels of the individual proteins in yeast (14), we tested whether MON-2 and PAD-1 are required for TAT-5 levels or localization. GFP::TAT-5 localized robustly to the plasma membrane in mon-2 knockdowns (Fig. 2E), as well as in mon-2 mutant embryos (Fig. 5D). Additionally, overall GFP::TAT-5 levels were not significantly decreased after mon-2 or pad-1 knockdown (Fig. S2). We then examined whether MON-2 and PAD-1 could redundantly regulate TAT-5 localization. GFP::TAT-5 still localized to the plasma membrane after pad-1 was knocked down in a mon-2 mutant background (Fig. S3C). GFP::TAT-5 was increased at the cell surface of mon-2 mutants treated with pad-1 RNAi (Fig. S3C), similar to pad-1 RNAi on control strains (Fig. 2D), indicating that TAT-5 is released in EVs. As MON-2 is not required for EV release in pad-1 mutants, this suggests that MON-2 does not directly promote or inhibit EV release. In summary, these data demonstrate that MON-2 and PAD-1 are not required to maintain TAT-5 levels or to regulate TAT-5 localization to the plasma membrane.
We next tested whether TAT-5 regulates PAD-1 or MON-2 localization. In one- or two-cell stage embryos, there was no change in MON-2::GFP or GFP::PAD-1 localization after tat-5 knockdown (Fig. 4 B and E). In older embryos, increased localization was observed at the cell surface after tat-5 knockdown (Fig. S3 D and E), suggesting that MON-2 and PAD-1 can also be released in EVs. Thus, TAT-5 is not required to recruit MON-2 or PAD-1 to the plasma membrane or into EVs. We also tested whether MON-2 and PAD-1 are required for each other’s localization to the plasma membrane or into EVs. PAD-1 localized to the plasma membrane after mon-2 RNAi (Fig. 4C), and MON-2 localized to the plasma membrane after pad-1 RNAi (Fig. 4F). The normally dim cytosolic fluorescence of MON-2::GFP accumulates in EVs after tat-5 RNAi treatment (Fig. 4 G and H). In contrast, MON-2 did not accumulate in EVs after pad-1 knockdown (Fig. 4I), despite MON-2 localization to the plasma membrane (Fig. 4F). Together, these data suggest that MON-2, PAD-1, and TAT-5 do not regulate each other’s levels or localization in C. elegans, except that PAD-1 is needed for MON-2 sorting into EVs.
MON-2 and PAD-1 Regulate Endosomal Trafficking.
As MON-2 and PAD-1 homologs in yeast and mammals have been implicated in retrograde trafficking between endosomes and Golgi (15, 24), we next tested whether they could interact genetically with the core retromer or with sorting nexins. We performed retromer RNAi in the mon-2 mutant background and indeed obtained synthetic developmental phenotypes, including sterility and embryonic lethality with both core retromers and sorting nexins (Table S1). However, EV release was only increased when we knocked down the SNX-BAR proteins SNX-1 and -6 or the SNX-PX protein SNX-3 in mon-2 mutants (Fig. 5 B and C), not when we targeted any of the core retromer proteins (Table 1), revealing that MON-2 redundantly inhibits EV release, similar to sorting nexins. We tested whether the increased EV release was due to changes in TAT-5 trafficking. Although GFP::TAT-5 was still visible in the plasma membrane (Table 1), GFP::TAT-5 also localized to prominent 0.5- to 2-µm cytoplasmic vesicles in mon-2 mutants treated with snx-1, -3, or -6 RNAi (Fig. 5E). Large GFP::TAT-5 vesicles were not seen when sorting nexins were knocked down in a wild-type background (Fig. 2 G and H), but were seen after pad-1 knockdown in a snx-1 mutant background (Fig. 5F). These data suggest that MON-2 and PAD-1 could redundantly regulate TAT-5 localization with sorting nexins or that MON-2 and PAD-1 could regulate another step of endolysosomal trafficking, which only impacts TAT-5 trafficking when sorting nexins are absent.
MON-2, PAD-1, and TAT-5 Regulate Multivesicular Endosome Size.
The large vesicles in mon-2 snx and pad-1 snx-1 double mutants appeared similar to the enlarged LMP-1-positive late endosomes or lysosomes we had previously observed in tat-5 mutants (9). As mammalian Mon2 has been implicated in recycling proteins from a degratory fate by trafficking them back from endosomes to the Golgi complex (24), we hypothesized that TAT-5, MON-2, and PAD-1 could also regulate intracellular trafficking events together. Therefore, we tested whether mon-2 and pad-1 mutants also showed enlarged LMP-1 vesicles. Indeed, both mon-2 and pad-1 RNAi-treated embryos, as well as mon-2 mutants, have enlarged clusters of LMP-1–positive late endosomes or lysosomes (Fig. S5 B–D). These data confirm that mon-2 RNAi and the mon-2 nonsense mutation result in a loss of MON-2 function. In contrast to mon-2 and pad-1 mutants, LMP-1–positive vesicles were not enlarged after snx-6 knockdown in control embryos (Fig. S5F), suggesting that the enlarged vesicles in double mutants are due to loss of MON-2 or PAD-1. To determine whether the enlarged LMP-1–positive vesicles were late endosomes or lysosomes, we measured the diameter of multivesicular endosomes and electron-dense lysosomes from electron micrographs. Multivesicular endosomes were significantly larger in pad-1 and tat-5 RNAi-treated embryos in comparison with wild-type embryos (Fig. S5 H and I; 320 ± 140 nm in control, 470 ± 290 nm in pad-1, and 410 ± 170 nm in tat-5). In contrast, electron-dense lysosomes had a similar diameter (Fig. S5J; control, 490 ± 150 nm; pad-1, 500 ± 200 nm; and tat-5, 470 ± 120 nm). Intriguingly, despite the increased size of multivesicular endosomes, the number of intraluminal vesicles was not significantly changed (control, 8 ± 9; pad-1, 8 ± 10; and tat-5, 10 ± 7), suggesting that PAD-1 and TAT-5 are not required for the formation of intraluminal vesicles. Given that intraluminal vesicles have the same budding topology as microvesicles and that there is no increase in intraluminal vesicle number, PAD-1 and TAT-5 are likely to have different functions at the plasma membrane and on endosomal membranes. Therefore, we predict that MON-2, PAD-1, and TAT-5 regulate an intracellular trafficking pathway that impacts late endosome size.
RME-8, MON-2, and SNX Prevent the Missorting of TAT-5 into Late Endosomes.
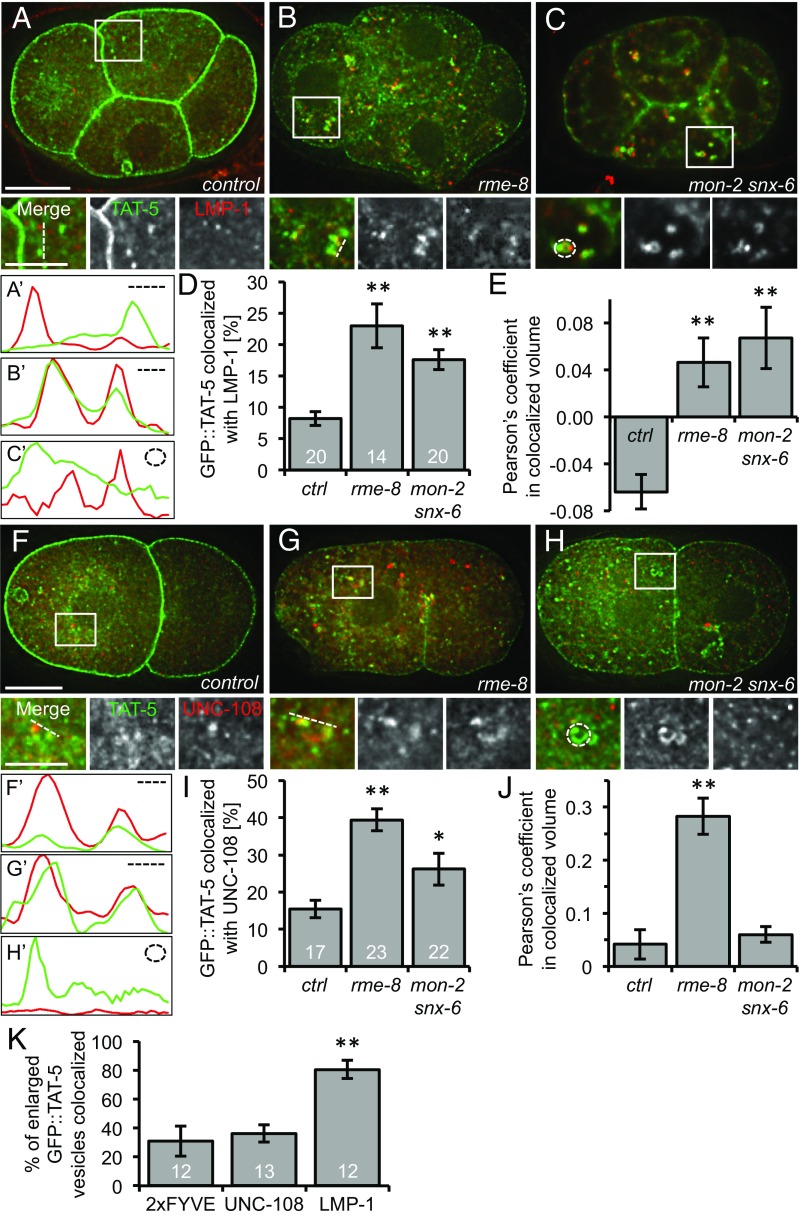
We next asked whether TAT-5 is missorted into late endosomes or lysosomes in mutants that cause increased EV release. GFP::TAT-5 rarely colocalizes with LMP-1 staining in control embryos (Fig. 6 A and D), and the Pearson’s coefficient showed a negative correlation (Fig. 6E). In contrast, GFP::TAT-5 showed a significant twofold to threefold increase in colocalization with LMP-1 in rme-8 RNAi-treated embryos and in mon-2 mutants treated with snx-6 RNAi (Fig. 6 B–D). Indeed, the Pearson’s coefficient showed a positive correlation in these mutants (Fig. 6E), indicating that more TAT-5 localized to late endosomes or lysosomes. The large GFP::TAT-5–containing vesicles found in mon-2 mutants treated with snx RNAi frequently have LMP-1–positive subdomains (Fig. 6 C′ and K), which in combination with the TEM data (Fig. S5 G–J), suggests that TAT-5 is mislocalized to late endosomes in mon-2 snx-6 mutants. Thus, TAT-5 localizes to degradative endosomes in these EV-releasing mutants.
Fig. 6.
TAT-5 localization to late endosomes increases in EV-releasing mutants. (A) GFP::TAT-5 (green) staining localizes primarily to the plasma membrane in control four-cell embryos. TAT-5 is also found in cytoplasmic vesicles, but they do not often colocalize with LMP-1 (red) staining in late endosomes or lysosomes, as shown in Insets. (B) Mislocalization of GFP::TAT-5 in rme-8 RNAi causes increased colocalization with LMP-1. (C) Large GFP::TAT-5-positive cytoplasmic vesicles colocalize with LMP-1 in a mon-2(xh22) mutant embryo treated with snx-6 RNAi. (A′–C′) Line scans of GFP::TAT-5 and LMP-1 intensity next to the dotted lines or circle in Insets. (D) The percentage of TAT-5 colocalized with LMP-1 is significantly increased in 2- to 12-cell embryos after rme-8 RNAi and in mon-2(xh22) mutants treated with snx-6 RNAi compared with control embryos. Number of embryos scored is indicated for each genotype. (E) The Pearson’s coefficient of GFP::TAT-5 colocalization with LMP-1 increases significantly from a negative correlation in control embryos to a positive correlation in rme-8 RNAi as well as in mon-2(xh22) mutants treated with snx-6 RNAi. (F) Cytoplasmic GFP::TAT-5 vesicles (green) rarely colocalize with mCherry::UNC-108 staining in Golgi and endosomes (red) in two-cell embryos, as shown in Insets. (G) TAT-5 colocalization with the Rab2 homolog UNC-108 increases after rme-8 RNAi. (H) TAT-5 colocalization with UNC-108 increased slightly in mon-2 mutants treated with snx-6 RNAi. (F′–H′) Line scans of GFP::TAT-5 and UNC-108 intensity next to the dotted lines or circle in Insets. (I) The percentage of TAT-5 colocalized with UNC-108 is significantly increased in 2- to 12-cell embryos after rme-8 RNAi and in mon-2(xh22) mutants treated with snx-6 RNAi compared with control embryos. Number of embryos scored is indicated for each genotype. (J) The Pearson’s coefficient of GFP::TAT-5 colocalization with LMP-1 increases significantly in rme-8 RNAi, but not in mon-2(xh22) mutants treated with snx-6 RNAi. (K) Enlarged GFP::TAT-5 vesicles colocalize significantly more often with LMP-1 (n = 54 vesicles) than the PI3P reporter 2xFYVE (n = 85) or UNC-108 (n = 89). Percentages are expressed in relation to the total number of large vesicles. Number of embryos scored is indicated for each genotype. Student’s t test with Bonferroni correction was used for statistical analysis. *P < 0.05; **P < 0.001. [Scale bars: A and F, 10 µm (also apply to B and C and G and H, respectively); Insets in A and F, 5 µm (also apply to Insets in B and C and G and H, respectively).]
To confirm that TAT-5 is mislocalized to late endosomes, we tested whether TAT-5 colocalizes with the Rab2 homolog UNC-108, which is found on the Golgi and recruited to endosomes after early endosome markers, such as RAB-5 and PI3P, and acts before late endosome markers, such as RAB-7 (31, 32). GFP::TAT-5 colocalized with a subset of mCherry-tagged UNC-108 in control embryos, showing a positive Pearson’s coefficient (Fig. 6 F, I, and J). TAT-5 colocalization with UNC-108 increased a significant twofold to threefold after rme-8 knockdown (Fig. 6 G, I, and J). UNC-108 colocalization also increased in mon-2 mutants treated with snx-6 RNAi (Fig. 6 H and I), although the Pearson’s coefficient was only significantly increased after rme-8 RNAi (Fig. 6J). In contrast to LMP-1, the large TAT-5 vesicles were not often positive for UNC-108 (Fig. 6 H and K), suggesting that the enlarged endosomes had already progressed past the Rab2-positive stage. These data confirm that GFP::TAT-5 is mislocalized to late endosomes in mutants that cause increased EV release, suggesting that TAT-5 cannot function from late endosomes.
We also tested whether TAT-5 is mislocalized to early endosomes in EV-releasing mutants. We used the PI3P binding domain of EEA-1 (2xFYVE) to label early endosomes (33). GFP::TAT-5 showed minor colocalization with mCherry-tagged 2xFYVE in control embryos (Fig. S6 A and C), which was not increased in mon-2 mutants treated with snx-6 RNAi (Fig. S6 B and C), confirming that TAT-5 is not mislocalized to early endosomes. Furthermore, the large TAT-5 vesicles in mon-2 snx-6 mutants were not often PI3P-positive (Fig. 6K and Fig. S6B), suggesting that they are not early endosomes. Together, these data suggest that TAT-5 is mislocalized to degradative compartments in trafficking mutants that cause increased EV release.
TAT-5 Localization Is Necessary to Maintain PE Asymmetry.
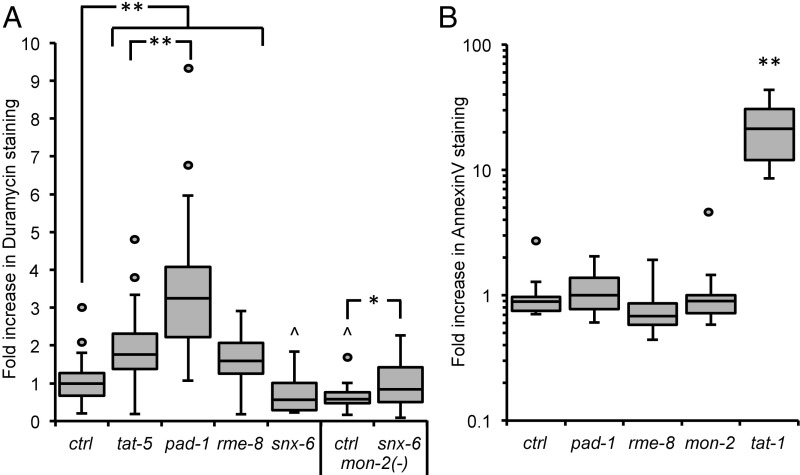
As the TAT-5 ortholog Neo1p maintains plasma membrane asymmetry from endosomes (27), we tested whether PE is externalized when TAT-5 is not localized to the plasma membrane. We stained live gonads with the PE-binding lantibiotic duramycin (34) and saw increased duramycin staining in rme-8 knockdown animals (Fig. 7A), indicating that PE is externalized on the surface of cells. In contrast, PE externalization was not increased after snx-6 RNAi (Fig. 7A), despite the reduction in TAT-5 localization to the plasma membrane and consistent with the observation that there was no increase in EV release (Table 1). In fact, snx-6 knockdowns displayed significantly less PE externalization than control embryos. Similar to rme-8 knockdown, knocking down snx-6 in a mon-2 mutant background significantly increased duramycin staining in comparison with untreated mon-2 mutants (Fig. 7A), demonstrating that PE is externalized when TAT-5 is mislocalized to late endosomes and EVs are released. PE externalization was significantly decreased in untreated mon-2 mutants, which could be through increased TAT-5 activity internalizing PE or through decreased activity of an unidentified protein responsible for externalizing PE. Thus, TAT-5 is not able to maintain PE asymmetry in the plasma membrane when mislocalized to late endosomes.
Fig. 7.
PAD-1 and RME-8 are required to maintain PE asymmetry. (A) Duramycin staining is significantly increased on dissected gonads after pad-1, rme-8, or tat-5 RNAi treatment compared to control embryos (ctrl), indicating that PAD-1 and RME-8 are required for TAT-5 to maintain PE asymmetry. The increase after pad-1 RNAi is also significantly more than tat-5 knockdown, suggesting that PAD-1 may influence PE asymmetry through more than TAT-5 activity. Duramycin staining is significantly decreased after snx-6 RNAi treatment and in untreated mon-2(xh22) mutants. Duramycin staining is increased on dissected gonads after snx-6 RNAi treatment in mon-2(xh22) nonsense mutants, in comparison with untreated mon-2(xh22) mutants, demonstrating that MON-2 and SNX-6 are required redundantly for PE asymmetry. (B) Annexin V staining is not increased on dissected gonads after mon-2, pad-1, or rme-8 RNAi treatment (P > 0.05), indicating that PS is not externalized. Annexin V staining is strongly increased in tat-1(kr15) mutants, together indicating that MON-2, PAD-1, and RME-8 are not required for TAT-1 PS flippase activity. Student’s t test with Bonferroni correction was used for statistical analysis. Asterisks indicate significantly increased values, and carets indicate significantly decreased values. *P < 0.05; **P < 0.001; ^P < 0.05.
We next wanted to test whether RME-8 was generally required for lipid asymmetry or whether it specifically regulated TAT-5. Knocking down RME-8 did not result in an increase in Annexin V staining (Fig. 7B), indicating that the lipid phosphatidylserine (PS) remains cytofacial and is not externalized in rme-8 mutants. This suggests that RME-8 is not required to localize the related P4-ATPase TAT-1 that maintains PS asymmetry in the plasma membrane (35). Thus, RME-8 is not likely to regulate the trafficking of all P4-ATPases, but is specifically required for TAT-5 to localize to the plasma membrane.
PAD-1 Is Necessary to Maintain PE Asymmetry.
PAD-1 is not directly required for TAT-5 localization, but does inhibit EV release. As TAT-5 was released in EVs in pad-1 mutants similar to an ATPase-dead tat-5 mutant (9), we tested whether PAD-1 is specifically required for TAT-5 flippase activity to maintain PE asymmetry. In pad-1 RNAi-treated worms, duramycin staining was increased on the surface of cells (Fig. 7A), indicating that PE is externalized and suggesting that PAD-1 is required for TAT-5 flippase activity. Moreover, duramycin staining was significantly increased after pad-1 RNAi in comparison with tat-5 RNAi (P < 0.001), implying that PAD-1 could also regulate PE externalization through another protein in addition to TAT-5. To determine whether PAD-1 is a specific cofactor for TAT-5, we tested whether PAD-1 was needed for the flippase activity of TAT-1 to maintain PS asymmetry. Annexin V staining was not increased when PAD-1 was depleted, indicating that PS is not externalized (Fig. 7B). This shows that PAD-1 does not regulate the activity of the PS flippase TAT-1, suggesting that PAD-1 specifically controls TAT-5 flippase activity to maintain PE asymmetry in the plasma membrane and inhibit EV release by ectocytosis.
Discussion
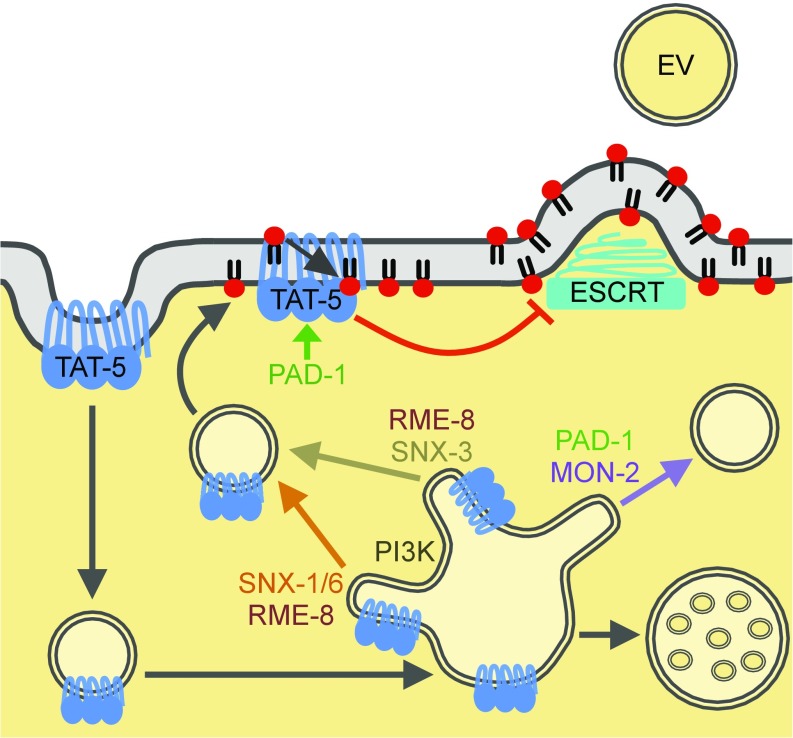
In summary, this study identifies proteins that regulate microvesicle release through TAT-5 and lipid asymmetry, shedding light onto the mechanisms of ectocytosis (Fig. 8). We show that TAT-5 is endocytosed and needs to be recycled from endosomes to maintain PE asymmetry and prevent ESCRT-mediated microvesicle budding. TAT-5 recycling depends on retrograde trafficking by the class III PI3K, RME-8, and redundant sorting nexin pathways. Our data also revealed two separable roles for the complex of TAT-5, PAD-1, and MON-2. TAT-5 and PAD-1 are required at the plasma membrane to maintain PE asymmetry, while the complex including MON-2 regulates endosomal trafficking. Thus, our study also reveals insights into redundant intracellular trafficking pathways.
Fig. 8.
Model of TAT-5 trafficking to inhibit EV release. TAT-5 maintains PE asymmetry in the plasma membrane to inhibit recruitment of the ESCRT machinery to release EVs by plasma membrane budding. PAD-1 is required for TAT-5 flippase activity, which inhibits the externalization of PE and EV release. TAT-5 is endocytosed and needs to be recycled from sorting endosomes to the plasma membrane through both SNX-1–SNX-6–mediated and SNX-3–mediated tubulation and vesicle formation. RME-8 and PI3K mediate TAT-5 recycling through multiple pathways. MON-2 and PAD-1 regulate an unknown step of endosomal trafficking, here drawn as a third recycling pathway preventing membrane cargos from being delivered to multivesicular endosomes for degradation. When both SNX-mediated recycling and MON-2/PAD-1–mediated trafficking are lost, TAT-5 is mislocalized to late endosomes where it can no longer maintain plasma membrane asymmetry.
The retromer complex and assorted sorting nexins help localize TAT-5 to the plasma membrane, but TAT-5 is primarily trafficked independently of the core retromer subunits in C. elegans. This is in contrast to yeast, where Neo1p reporters were mislocalized to the vacuole in core retromer mutants (36). As snx-1, -3, or -6 knockdown led to stronger trafficking defects than core retromer deletion mutants, these proteins act independently of the core retromer complex. Retromer-independent roles have been shown for SNX1 and SNX6 during GPCR and E-cadherin trafficking (37, 38), and SNX5/6 binds directly to the mannose-6-phosphate receptor to regulate its trafficking independent of the core retromer (39, 40). Snx3p also binds directly to Neo1p in yeast (16), but this domain is not conserved in TAT-5. Thus, although we predict that these sorting nexins will bind TAT-5, how they recognize TAT-5 as cargo is unclear. The SNX-BAR heterodimer SNX1–SNX6 is thought to traffic distinct cargos from the SNX-PX protein SNX3 (41), so it was unusual that we discovered similar effects on TAT-5 localization after disrupting either SNX-1–SNX-6 or SNX-3. It will be interesting to determine whether they recognize TAT-5 through the same domain or distinct sequences, especially considering that EV release was only increased when both snx-3 and -6 were disrupted. Thus, studying TAT-5 trafficking has identified two SNX-dependent, retromer-independent trafficking pathways that redundantly inhibit EV release.
RME-8 also plays redundant roles with the core retromer during Notch signaling (42), confirming that it also has retromer-independent functions. Furthermore, although RME-8 is required for both SNX1–SNX6- and SNX3-dependent cargos (23, 26), RME-8 establishes endosomal microdomains independent of the core retromer and SNX-3 to prevent cargo from being degraded (43). This is consistent with our observation that increased TAT-5 was observed in late endosomal compartments after depleting RME-8. Interestingly, RME-8–dependent trafficking may be specific to the TAT-5 subclass of P4-ATPases, because PS asymmetry in the plasma membrane was not disrupted when RME-8 was depleted. Most P4-ATPases, including the PS flippase TAT-1, have β-subunits from the Cdc50 family of proteins that act as chaperones important for their localization (12). Only TAT-5 orthologs localize without Cdc50 proteins (13), which our data suggest is due to the role of PI3K, RME-8, and sorting nexins in localizing TAT-5 to the plasma membrane.
Despite similar losses of GFP::TAT-5 from the plasma membrane after disrupting PI3K, RME-8, and SNX, only PI3K and RME-8 mutants showed increased EV release. Our data imply that TAT-5 is able to maintain PE asymmetry and inhibit EV release from some endosomes, but not from late endosomes. Increased EV release disrupts gastrulation movements (9) and may explain why rme-8 mutants are lethal (44), while individual snx deletions are viable (23, 45). Our data suggest that RME-8 is likely to regulate EV release through SNX-1– and SNX-3–dependent mechanisms, as EV release is increased in double SNX mutants, but we cannot rule out a role for other RME-8 interactors, such as Hsc70 family chaperones (44). Similarly, as part of the PI3K complex, BEC-1 and VPS-34 are likely to regulate EV release through SNX-, RME-8-, and retromer-independent roles, due to their pleiotropic effects on membrane trafficking (46). Thus, it appears that TAT-5 trafficking is essential for survival, and animals have developed redundant trafficking pathways to maintain TAT-5 localization.
MON-2 and PAD-1 represent another endosomal trafficking pathway, whose precise role remains to be determined. MON-2 and PAD-1 are not normally required for the plasma membrane localization of TAT-5, but MON-2 redundantly regulates EV release with both SNX-1 and -3 with similarly enlarged TAT-5 endosomes, demonstrating that MON-2 acts in a separate pathway from the sorting nexins. Therefore, MON-2 and PAD-1 could traffic another EV regulator, control another endosomal trafficking step, or provide a third redundant recycling pathway alongside SNX-1–SNX-6 and SNX-3. Similarly, the enlarged late endosomes observed in mon-2, pad-1, and tat-5 mutants could be caused by increased endosome–endosome fusion, decreased intraluminal vesicle budding, or decreased endosomal tubulation. As pad-1 and tat-5 mutants did not disrupt intraluminal vesicle formation (this study and ref. 9), despite its topological similarity to microvesicle budding and similar ESCRT dependence, TAT-5, MON-2, and PAD-1 are likely to regulate membrane trafficking into or out of endosomes. As this complex is required for the recycling of retrograde cargos in yeast and mammals (16, 17, 24), we favor the hypothesis that TAT-5, MON-2, and PAD-1 regulate a third redundant endosomal recycling pathway (Fig. 8), which normally does not traffic significant amounts of TAT-5 as cargo.
MON-2 and PAD-1 are also likely to interact with proteins beyond TAT-5 to regulate lipid asymmetry and inhibit EV release. PAD-1 is needed to maintain PE asymmetry, suggesting that it could activate TAT-5 flippase activity; however, the duramycin staining data also suggest that PAD-1 may inhibit an unidentified PE scramblase that causes PE externalization. Similarly, the decrease in PE externalization after disrupting MON-2 or SNX-6 could suggest that MON-2 and SNX-6 are required for scramblase activity. Identifying the proteins opposing TAT-5 flippase activity will be key to testing these hypotheses. Furthermore, PAD-1 is required for the release of MON-2 in EVs, suggesting that PAD-1 could regulate where MON-2 localizes on the plasma membrane or act as a scaffold for sorting EV cargo. Given that human Mon2 is required for HIV budding (47), which uses similar release mechanisms to EVs (11), the role of MON-2 in regulating EV release requires further investigation. Thus, MON-2 and PAD-1 may coordinate multiple regulators of PE localization and EV cargo to preserve plasma membrane morphology.
It remains to be determined whether PAD-1 and TAT-5 orthologs in other species have a role in EV release, but it is clear that TAT-5 needs to be recycled to maintain PE asymmetry in the plasma membrane. Furthermore, the correlation of increased EV release and PE externalization observed across several C. elegans mutants supports the hypothesis that PE asymmetry regulates plasma membrane budding. Whether PE externalization directly causes EV budding through its conical shape and effects on membrane curvature is still unclear. PE externalization also indirectly alters the lipid content of the cytosolic face of the plasma membrane and could thereby recruit membrane-sculpting proteins such as ESCRT (Fig. 8), as previously observed (9). Provocatively, PE asymmetry is lost during cytokinesis and needs to be reestablished for abscission (48), a step that also requires the ESCRT machinery (6). During abscission, EV release is seen at the intercellular bridge in both C. elegans embryos and mammalian cells (49, 50). Intriguingly, tat-5 mutant embryos occasionally have multinuclear cells due to sporadic retraction of the cleavage furrow (9), which suggests that successful abscission requires that EV budding is restricted to the intercellular bridge. Thinning membrane bridges may therefore be a crucial function of EV budding regulated by PE asymmetry. Thus, TAT-5 localization and activity are critical to maintain PE asymmetry in the plasma membrane and inhibit EV release by ectocytosis, which may have functional roles during cell division.
Materials and Methods
C. elegans worms were maintained and crossed according to standard protocols (51). Strains were maintained at room temperature, except that the temperature-sensitive rme-8(b1023) mutant was maintained at 15 °C. See Table S2 for a list of strains used. Additional methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Anne Haberberger, Theresa Henninger, Sarah Leone, Hanaa Ghanawi, Francisca Alagboso, Martin Boos, Jiapei Chen, Teresa Kee, Maïté Carre-Pierrat at the University of Lyon, Sebastian Markert and the EM facility staff at the Skirball Institute, New York Structural Biology Center, and Biocenter Würzburg for technical assistance. The imaging facility of the Rudolf Virchow Center provided Huygens deconvolution software and assistance with confocal and colocalization studies. We thank Jaime Lisack, Birgit Singer-Krüger, Anne Norris, and Barth Grant for critical reading of this manuscript. Strains and reagents were obtained from Julie Ahringer, Michael Ailion, Bruce Bowerman, Michael Glotzer, Barth Grant, Takao Inoue, Renaud Legouis, Philip Thorpe, Zheng Zhou, the National Bioresource Project for the Nematode C. elegans (Japan), the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation, the C. elegans Reverse Genetics Core Facility at the University of British Columbia, and the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs Grant P40 OD010440. This work was primarily supported by the Rudolf Virchow Center; and A.M.W. was initially funded by NIH Grant R03 AI099555 (to J.F.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714085115/-/DCSupplemental.
References
- 1.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 2.Beer KB, Wehman AM. Mechanisms and functions of extracellular vesicle release in vivo–What we can learn from flies and worms. Cell Adhes Migr. 2017;11:135–150. doi: 10.1080/19336918.2016.1236899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jimenez AJ, et al. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 4.Gong Y-N, et al. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell. 2017;169:286–300.e16. doi: 10.1016/j.cell.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5:a016766. doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, et al. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melki I, Tessandier N, Zufferey A, Boilard E. Platelet microvesicles in health and disease. Platelets. 2017;28:214–221. doi: 10.1080/09537104.2016.1265924. [DOI] [PubMed] [Google Scholar]
- 9.Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–1959. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolte-’t Hoen E, Cremer T, Gallo RC, Margolis LB. Extracellular vesicles and viruses: Are they close relatives? Proc Natl Acad Sci USA. 2016;113:9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B, et al. Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLoS Genet. 2010;6:e1001235. doi: 10.1371/journal.pgen.1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen JP, et al. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front Physiol. 2016;7:275. doi: 10.3389/fphys.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbosa S, Pratte D, Schwarz H, Pipkorn R, Singer-Krüger B. Oligomeric Dop1p is part of the endosomal Neo1p-Ysl2p-Arl1p membrane remodeling complex. Traffic. 2010;11:1092–1106. doi: 10.1111/j.1600-0854.2010.01079.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu K, Surendhran K, Nothwehr SF, Graham TR. P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol Biol Cell. 2008;19:3526–3535. doi: 10.1091/mbc.E08-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalton LE, Bean BDM, Davey M, Conibear E. Quantitative high-content imaging identifies novel regulators of Neo1 trafficking at endosomes. Mol Biol Cell. 2017;28:1539–1550. doi: 10.1091/mbc.E16-11-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka Y, et al. The phospholipid flippase ATP9A is required for recycling pathway from endosomes to the plasma membrane. Mol Biol Cell. 2016;27:3883–3893. doi: 10.1091/mbc.E16-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emoto K, et al. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc Natl Acad Sci USA. 1996;93:12867–12872. doi: 10.1073/pnas.93.23.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emoto K, Toyama-Sorimachi N, Karasuyama H, Inoue K, Umeda M. Exposure of phosphatidylethanolamine on the surface of apoptotic cells. Exp Cell Res. 1997;232:430–434. doi: 10.1006/excr.1997.3521. [DOI] [PubMed] [Google Scholar]
- 20.Irie A, Yamamoto K, Miki Y, Murakami M. Phosphatidylethanolamine dynamics are required for osteoclast fusion. Sci Rep. 2017;7:46715. doi: 10.1038/srep46715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fazeli G, Trinkwalder M, Irmisch L, Wehman AM. C. elegans midbodies are released, phagocytosed and undergo LC3-dependent degradation independent of macroautophagy. J Cell Sci. 2016;129:3721–3731. doi: 10.1242/jcs.190223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruck A, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400. doi: 10.4161/auto.7.4.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi A, et al. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 2009;28:3290–3302. doi: 10.1038/emboj.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahajan D, et al. Mammalian Mon2/Ysl2 regulates endosome-to-Golgi trafficking but possesses no guanine nucleotide exchange activity toward Arl1 GTPase. Sci Rep. 2013;3:3362. doi: 10.1038/srep03362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallon M, Cullen PJ. Retromer and sorting nexins in endosomal sorting. Biochem Soc Trans. 2015;43:33–47. doi: 10.1042/BST20140290. [DOI] [PubMed] [Google Scholar]
- 26.Harterink M, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takar M, Wu Y, Graham TR. The essential Neo1 protein from budding yeast plays a role in establishing aminophospholipid asymmetry of the plasma membrane. J Biol Chem. 2016;291:15727–15739. doi: 10.1074/jbc.M115.686253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almendinger J, et al. A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS One. 2011;6:e18325. doi: 10.1371/journal.pone.0018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wicky S, Schwarz H, Singer-Krüger B. Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol Cell Biol. 2004;24:7402–7418. doi: 10.1128/MCB.24.17.7402-7418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanamori T, et al. β-catenin asymmetry is regulated by PLA1 and retrograde traffic in C. elegans stem cell divisions. EMBO J. 2008;27:1647–1657. doi: 10.1038/emboj.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Q, et al. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008;135:1069–1080. doi: 10.1242/dev.016063. [DOI] [PubMed] [Google Scholar]
- 32.Cattin-Ortolá J, Topalidou I, Dosey A, Merz AJ, Ailion M. The dense-core vesicle maturation protein CCCP-1 binds RAB-2 and membranes through its C-terminal domain. Traffic. 2017;18:720–732. doi: 10.1111/tra.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews R, Ahringer J. Asymmetry of early endosome distribution in C. elegans embryos. PLoS One. 2007;2:e493. doi: 10.1371/journal.pone.0000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stafford JH, Thorpe PE. Increased exposure of phosphatidylethanolamine on the surface of tumor vascular endothelium. Neoplasia. 2011;13:299–308. doi: 10.1593/neo.101366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruaud A-F, et al. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic. 2009;10:88–100. doi: 10.1111/j.1600-0854.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, Takar M, Cuentas-Condori AA, Graham TR. Neo1 and phosphatidylethanolamine contribute to vacuole membrane fusion in Saccharomyces cerevisiae. Cell Logist. 2016;6:e1228791. doi: 10.1080/21592799.2016.1228791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nisar S, Kelly E, Cullen PJ, Mundell SJ. Regulation of P2Y1 receptor traffic by sorting Nexin 1 is retromer independent. Traffic. 2010;11:508–519. doi: 10.1111/j.1600-0854.2010.01035.x. [DOI] [PubMed] [Google Scholar]
- 38.Schill NJ, Hedman AC, Choi S, Anderson RA. Isoform 5 of PIPKIγ regulates the endosomal trafficking and degradation of E-cadherin. J Cell Sci. 2014;127:2189–2203. doi: 10.1242/jcs.132423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonetti B, Danson CM, Heesom KJ, Cullen PJ. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J Cell Biol. 2017;216:3695–3712. doi: 10.1083/jcb.201703015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvainickas A, et al. Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J Cell Biol. 2017;216:3677–3693. doi: 10.1083/jcb.201702137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas M, Hierro A. Retromer. Curr Biol. 2017;27:R687–R689. doi: 10.1016/j.cub.2017.05.072. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Lamarca MJ, Snowdon LA, Seib E, Klein T, Bray SJ. Rme-8 depletion perturbs Notch recycling and predisposes to pathogenic signaling. J Cell Biol. 2015;210:303–318. doi: 10.1083/jcb.201411001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norris A, et al. SNX-1 and RME-8 oppose the assembly of HGRS-1/ESCRT-0 degradative microdomains on endosomes. Proc Natl Acad Sci USA. 2017;114:E307–E316. doi: 10.1073/pnas.1612730114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Grant B, Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol Biol Cell. 2001;12:2011–2021. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D, et al. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261–1264. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- 46.Schink KO, Tan K-W, Stenmark H. Phosphoinositides in control of membrane dynamics. Annu Rev Cell Dev Biol. 2016;32:143–171. doi: 10.1146/annurev-cellbio-111315-125349. [DOI] [PubMed] [Google Scholar]
- 47.Tomita Y, et al. The cellular factors Vps18 and Mon2 are required for efficient production of infectious HIV-1 particles. J Virol. 2011;85:5618–5627. doi: 10.1128/JVI.00846-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emoto K, Inadome H, Kanaho Y, Narumiya S, Umeda M. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J Biol Chem. 2005;280:37901–37907. doi: 10.1074/jbc.M504282200. [DOI] [PubMed] [Google Scholar]
- 49.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.König J, Frankel EB, Audhya A, Müller-Reichert T. Membrane remodeling during embryonic abscission in Caenorhabditis elegans. J Cell Biol. 2017;216:1277–1286. doi: 10.1083/jcb.201607030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.