Significance
Loss-of-function mutations in BRCA1 and its protein partner BARD1 lead to high risks of breast and ovarian cancer. Both BRCA1 and BARD1 proteins harbor RING domains, and missense mutations in the critical residues of the BRCA1 RING domain are among those known to predispose to cancer. The BRCA1 RING domain is a ubiquitin ligase, but the function of the BARD1 RING domain and the consequences of mutations in it are unknown. Evaluation of missense mutations at evolutionarily conserved zinc-binding residues of BARD1, each identified in a family severely affected with breast cancer, revealed that BARD1 is necessary for two core functions of the BRCA1/BARD1 complex: ubiquitylation of histone 2A on nucleosomes and transcriptional regulation of genes of estrogen metabolism.
Keywords: BARD1, BRCA1, breast cancer, ubiquitin, transcriptional repression
Abstract
Missense mutations that disrupt the RING domain of the tumor suppressor gene BRCA1 lead to increased risk of breast and ovarian cancer. The BRCA1 RING domain is a ubiquitin ligase, whose structure and function rely critically on forming a heterodimer with BARD1, which also harbors a RING domain. The function of the BARD1 RING domain is unknown. In families severely affected with breast cancer, we identified inherited BARD1 missense mutations Cys53Trp, Cys71Tyr, and Cys83Arg that alter three zinc-binding residues of the BARD1 RING domain. Each of these mutant BARD1 proteins retained the ability to form heterodimeric complexes with BRCA1 to make an active ubiquitin ligase, but the mutant BRCA1/BARD1 complexes were deficient in binding to nucleosomes and in ubiquitylating histone H2A. The BARD1 mutations also caused loss of transcriptional repression of BRCA1-regulated estrogen metabolism genes CYP1A1 and CYP3A4; breast epithelial cells edited to create heterozygous loss of BARD1 showed significantly higher expression of CYP1A1 and CYP3A4. Reintroduction of wild-type BARD1 into these cells restored CYP1A1 and CYP3A4 transcription to normal levels, but introduction of the cancer-predisposing BARD1 RING mutants failed to do so. These results indicate that an intact BARD1 RING domain is critical to BRCA1/BARD1 binding to nucleosomes and hence to ubiquitylation of histone H2A and also critical to transcriptional repression of BRCA1-regulated genes active in estrogen metabolism.
Among inherited mutations of BRCA1 that increase risk of breast and ovarian cancer are missense mutations that abrogate the function of the BRCA1 RING domain. The BRCA1 RING domain is a ubiquitin ligase (E3) enzyme, that catalyzes the covalent attachment of the signaling protein ubiquitin (Ub) onto protein substrates by binding to and activating E2 enzymes. As is true for most Ub ligases, BRCA1 modifies numerous proteins, including histone H2A, estrogen receptor α (ERα), and progesterone receptor (1–3). Although the biological consequences of protein ubiquitylation by BRCA1 are largely unknown, the BRCA1-dependent modification of histone H2A is critical for DNA damage repair and genome stability (4, 5). Cancer-predisposing missense mutations in the BRCA1 RING domain cause loss of the domain’s ligase activity, and hence a general defect in ubiquitylation. The mutations also lead to genomic instability (2, 6–8). Nevertheless, there are conflicting hypotheses regarding the role of BRCA1 Ub ligase activity in tumor suppression (4, 8–10). The alternative hypotheses have been difficult to resolve because BRCA1-mutant RING domains are inactive toward all ubiquitylation substrates, so the specific target(s) involved in tumor suppression activity of BRCA1 has not been identified.
Ub ligase activity of BRCA1 depends on BRCA1 forming a complex with BARD1, which also harbors a RING domain. The BRCA1/BARD1 heterodimer comprises the first ∼100 residues of each protein, bringing their two RING domains into close proximity. As part of the heterodimer, the BRCA1 RING domain interacts with an E2 enzyme to activate transfer of ubiquitin onto a substrate, but the role of the BARD1 RING domain has remained unknown. Mutations that result in truncation of the BARD1 protein are associated with increased risk of breast and ovarian cancer (11–15), but the functional and clinical consequences of missense mutations in the BARD1 RING domain have not been characterized. To address these questions, we evaluated three missense mutations in the BARD1 RING domain, newly discovered in families severely affected by breast cancer.
Results
Germline Mutations in the BARD1 RING Domain in Breast Cancer Families.
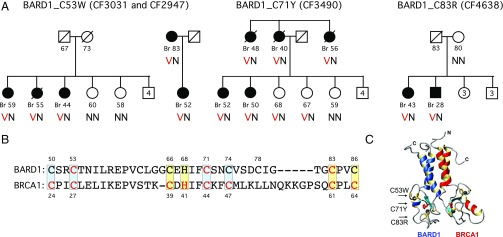
Truncating mutations of BARD1 are known to increase risk of breast and ovarian cancer (16, 17), but much less is known of the functional and clinical consequences of missense mutations in the BARD1 RING domain. In the course of genomic analysis of inherited predisposition to breast cancer, we identified three mutations in the RING domain of BARD1 in four families: BARD1 p.C53W (c.159T > G) at chr 2: 215,661,841 in families CF2947 and CF3031 (who are not related to each other); BARD1 p.C71Y (c.212G > A) at chr 2: 215,661,788 in family CF3490; and BARD1 p.C83R (c.247 A > G) at chr 2: 215,657,138 in family CF4638 (18) (Fig. 1A). Each of the three BARD1 mutations occurs at a residue completely conserved across all sequenced species. None of the three mutations appear in the Genome Aggregation Database (gnomAD) of 123,136 exome sequences (19) or among 10,000 cancer-free older female participants of the Women’s Health Initiative (whi.color.com/). For five of the 12 breast cancers in the four families, hormone receptor information was available: one tumor was triple negative and four were estrogen receptor and progesterone receptor positive. Most breast cancers from patients with BRCA1 or truncating BARD1 mutations are triple negative (17). Whether triple negative breast cancer will also predominate among patients with missense mutations in the BARD1 RING domain awaits additional data. The observation in family CF3490 of two unaffected women, ages 67 and 68, who carry BARD1 p.C71Y suggests that BARD1 RING domain mutations may not convey as high a risk as mutations in the comparable residues of the BRCA1 RING domain (Fig. 1B) (20).
Fig. 1.
Missense mutations in the BARD1 RING domain in breast cancer families. (A) Families with inherited mutations in critical residues of the BARD1 RING domain. Breast cancer (Br) is indicated by filled symbols; ages represent age at breast cancer diagnosis for affected individuals, or current age or age at death for individuals without breast cancer. Numbers inside symbols indicate additional siblings. For each family, V indicates the variant allele, and N indicates the reference allele. (B) RING domain sequences of BARD1 and BRCA1. Zinc-binding sites I and II are indicated in light blue and yellow, respectively. Residues with cancer-predisposing mutations in BARD1 (reported here) or in BRCA1 (reported elsewhere) are indicated in red. (C) The BRCA1/BARD1 heterodimer (based on protein database entry 1JM7) (52) indicates the positions of the missense mutations in the BARD1 RING domain (shown as blue subunit).
RING domain structures are stabilized by two zinc atoms that are bound by seven cysteine and one histidine residues. The three BARD1 RING domain mutations identified in breast cancer families alter cysteine residues that coordinate these zinc ions (Fig. 1 B and C). The amino acids of the resulting mutant proteins (tryptophan, tyrosine, and arginine) are not chemically capable of coordinating zinc in a manner similar to cysteine. In BRCA1, missense mutations in zinc-binding cysteine residues of the RING domain were among the first cancer-predisposing mutations identified (21). In BRCA1, these mutations cause structural changes that abrogate BRCA1 binding to ubiquitin-conjugating (E2) enzymes, thereby destroying the ubiquitin ligase activity of the BRCA1/BARD1 heterodimer (22). Because the BARD1 RING domain does not bind directly to E2 enzymes (22, 23), the consequences of comparable mutations in the BARD1 RING domain have remained undefined.
To better understand the effects of mutations in zinc-binding residues of the BARD1 RING domain, we evaluated BRCA1/BARD1 heterodimer complexes that contained each of the BARD1 missense mutations. For comparison, we similarly evaluated BARD1 p.C78S (c.233G > C) at chr 2: 215,657,152, which also lies in the BARD1 RING domain but alters a cysteine residue that is not conserved and does not coordinate zinc. BARD1 p.C78S has not been observed in any family in our study, but was reported as a “variant of unknown significance” in one individual of unknown phenotype in the ClinVar genetic database (NIH clinical variation database) (24).
Effect of BARD1 Mutations on Ubiquitylation of Nucleosomal Histone H2A.
The BARD1 RING domain binds to BRCA1 and stimulates BRCA1 ubiquitin ligase activity (6, 25, 26). To determine the effect of BARD1 mutations, we carried out in vitro assays for these activities using constructs of BRCA1 and BARD1 that have been shown to exhibit binding and ubiquitin ligase function similar to the full-length proteins (1, 2, 22). All wild-type and mutant BARD1 RING domains copurified with BRCA1, confirming that all BARD1 variants retained their ability to form heterodimers with BRCA1 (SI Appendix, Fig. S1).
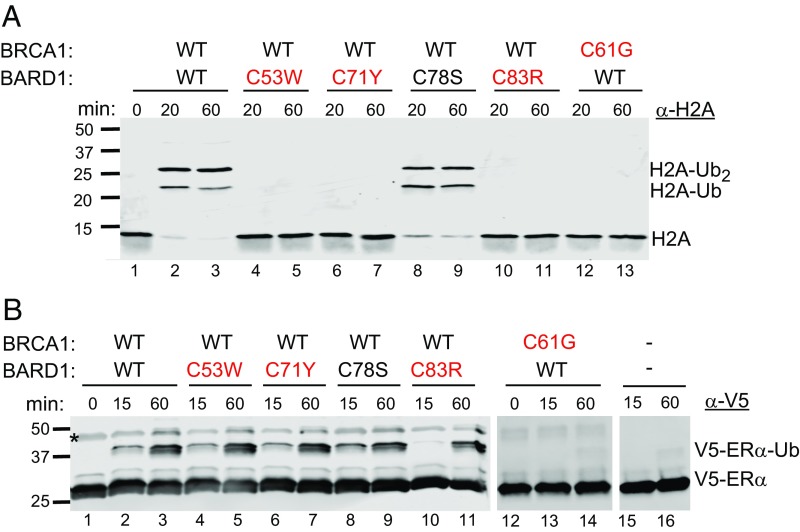
Ubiquitylation of lysine residues in the C-terminal tail of histone H2A by the BRCA1/BARD1 heterodimer plays an important role in DNA damage repair and gene silencing and is a possible explanation for the regulation of transcription of certain estrogen-metabolizing enzymes by BRCA1 (5, 27, 28). We therefore assessed the ability of BRCA1/BARD1 to modify H2A with ubiquitin in assays with recombinant mononucleosomes as the substrate. During reactions in which the wild-type BRCA1/BARD1 heterodimer served as the Ub ligase, ubiquitylated H2A products appeared as higher molecular weight bands, corresponding to H2A plus one or two ubiquitin molecules (Fig. 2A, bands H2A-Ub and H2A-Ub2), and the band for unmodified H2A disappeared (Fig. 2A, band H2A). As a positive control, the same reaction with BRCA1/BARD1 heterodimer containing the known loss-of-function mutation BRCA1 p.C61G had no detectable activity towards nucleosomes, consistent with its lack of ligase activity toward any substrate that has been measured to date. Reactions with the BRCA1/BARD1 heterodimer containing BARD1 p.C53W, BARD1 p.C71Y, or BARD1 p.C83R also yielded absence of ubiquitylated H2A product. In contrast, a BRCA1/BARD1 heterodimer containing BARD1 p.C78S yielded results similar to those of the wild-type heterodimer. These results indicate that the zinc-binding BRCA1 and BARD1 variants inherited in breast cancer families are defective in catalyzing ubiquitylation of H2A in nucleosomes.
Fig. 2.
Nucleosome and ERα ubiquitylation activities of BARD1 mutant proteins. (A) Wild-type BRCA1 in complex with BARD1 p.C53W, BARD1 p.C71Y, or BARD1 p.C83R is deficient at in vitro ubiquitylation of nucleosomal histone H2A, as is BRCA1 p.C61G in complex with wild-type BARD1. In contrast, wild-type BRCA1 in complex with BARD1 p.C78S has normal ubiquitylation activity. (B) In vitro ubiquitylation of ERα is not altered by any mutant BARD1 proteins but is affected by BRCA1 p.C61G. ERα ubiquitylation was carried out for the specified amount of time with heterodimers incorporating the BRCA1 or BARD1 variant indicated and then Western blot probed for anti-V5 tag on the ligand-binding domain of ERα. The asterisk indicates nonspecific antibody binding to BRCA1. Mutations identified in breast cancer families are labeled in red.
To evaluate the generality of loss of ubiquitylating activity by BARD1 mutants, we tested ligase activity toward another known substrate, the ligand-binding domain of ERα. BRCA1/BARD1 activity toward ERα is important for regulation of estrogen signaling in cells, and this activity is defective in BRCA1 RING domain mutations (2, 29). In vitro ubiquitylation assays were carried out and the substrate and products were visualized by Western blot analysis for the V5 tag on the ERα ligand-binding domain (Fig. 2B). For wild-type BARD1 and all mutant BARD1 proteins, the BRCA1/BARD1 complex produced a product that corresponds to attachment of one ubiquitin to the ERα ligand-binding domain (Fig. 2B, band ERα-Ub), whereas the BRCA1/BARD1 complex including BRCA1 p.C61G was inactive. Similarly, when BRCA1/BARD1 complexes were assayed for autoubiquitylation activity, in which BRCA1 serves as a proxy substrate (6), heterodimers incorporating BARD1 mutant proteins produced products, whereas the heterodimer incorporating BRCA1 p.C61G did not (SI Appendix, Fig. S2). Together, the results of in vitro ubiquitylation assays indicate that while BRCA1 p.C61G causes a complete loss of Ub ligase function (due to its inability to bind an E2 ubiquitin species that brings ubiquitin to the substrate), the newly discovered BARD1 RING mutations specifically fail to ubiquitylate nucleosomal H2A.
BRCA1/BARD1 has been reported to ubiquitylate free H2A; that is, outside the context of a nucleosome (1, 30, 31). In assays containing free H2A (at much higher concentration than in the nucleosome reactions), ubiquitylated product is generated by heterodimers incorporating each of the BARD1 variants (SI Appendix, Fig. S3, band H2A-Ub). In contrast, heterodimers incorporating to BRCA1 p.C61G and wild-type BARD1 do not ubiquitylate free H2A. Thus, while the BRCA1 p.C61G protein is inactive in both contexts, the BARD1 RING mutations only fail to ubiquitylate H2A on nucleosomes. Ubiquitylation of free H2A is far less efficient than ubiquitylation of H2A on nucleosomes; higher concentrations of free H2A must be used and most free H2A remains unmodified. It is possible that efficient histone 2A ubiquitylation by the BRCA1/BARD1 complex is orchestrated by recruitment of nucleosomes by BARD1 to target histone H2A as a substrate.
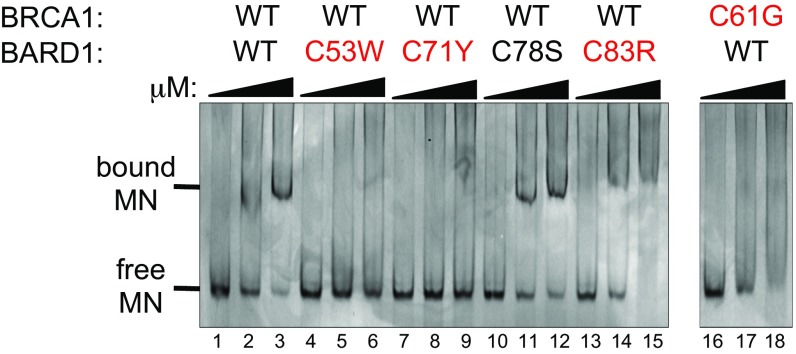
The simplest explanation for why mutant BARD1 RING domains are only inactive toward H2A in nucleosomes is that the mutant BRCA1/BARD1 complex is unable to recognize and bind nucleosomes. To test this hypothesis, we assessed the ability of BRCA1/BARD1 complexes to bind to nucleosomes in electrophoretic mobility shift assays, using mononucleosomes reconstituted from purified recombinant histones and DNA as substrates (Fig. 3). With increasing amounts of wild-type BRCA1/BARD1 complex, the band corresponding to unbound mononucleosomes decreased in intensity and a new band corresponding to a higher molecular weight species appeared (Fig. 3, lanes 1–3). Results similar to wild type were observed for BARD1 p.C78S (Fig. 3, lanes 10–12). In contrast, complexes incorporating BARD1 p.C53W (Fig. 3, lanes 4–6) or BARD1 p.C71Y (Fig. 3, lanes 7–9) did not appear to bind nucleosomes, as evidenced by no change in the band corresponding to unbound mononucleosomes. Complexes incorporating BARD1 p.C83R (Fig. 3, lanes 13–15) or BRCA1 p.C61G (Fig. 3, lanes 16–18) appeared to bind mononucleosomes, as evidenced by the disappearance of the free mononucleosome band, but the apparent binding does not result in a distinct bound species at the same molecular weight as seen with wild-type BRCA1/BARD1. Instead, binding by these complexes produced a diffuse staining that likely corresponds to a distribution of dynamically associated species. Notably, the mutated site in these two proteins, Cys83 of BARD1 and Cys61 of BRCA1, are structurally homologous within their corresponding RING domains (Fig. 1B), which may underlie their similar behavior on electrophoretic shift assays. Together with the mononucleosome ubiquitylation assay (Fig. 2A), these results indicate that both BRCA1 and BARD1 RING domains contain nucleosome-binding sites and that sites of both RING domains must be intact for formation of the nucleosomal complex required for ubiquitylation of H2A in chromatin.
Fig. 3.
Mononucleosome binding activities of BARD1 mutant proteins. Mononucleosome binding by wild-type BRCA1/BARD1 and by complex with neutral BARD1 p.C78S is observed as the appearance of a distinct band (marked as “bound MN”) at increasing concentration of added protein. In contrast, BARD1 and BRCA1 mutations identified in breast cancer families are deficient at binding mononucleosomes in vitro. Electromobility gel shift assays were carried out at increasing concentrations (10, 20, and 30 μM) of the indicated BRCA1/BARD1 complexes and visualized by ethidium bromide staining of DNA incorporated into the mononucleosomes (MN). Mutations identified in breast cancer families are labeled in red.
Effect of BARD1 Mutations on Transcriptional Regulation of Estrogen Metabolizing Genes.
Ubiquitylation of the C-terminal tail of histone H2A is associated with gene silencing (4, 32, 33). We therefore hypothesized a link between regulation of gene expression by BRCA1/BARD1 and loss of H2A ubiquitylation activity due to mutation of BARD1. Appropriate genes to test this hypothesis are CYP1A1 and CYP3A4, two members of the cytochrome P450 family that are transcriptionally regulated by BRCA1, and that catalyze transformation of estrogen into metabolites that generate DNA damage-inducing free radicals (28, 34, 35). Depletion of BRCA1 results in increased CYP1A1 and CYP3A4 transcription and increased estrogen metabolite-mediated DNA damage (28).
To determine the consequences of loss of function of BARD1 on CYP1A1 and CYP3A4 transcription, we created BARD1 knockout alleles in MCF10A breast epithelial cells by CRISPR/Cas9 gene editing. Guide RNAs were developed that efficiently deleted BARD1 exon 1 in cell pools (SI Appendix, Fig. S4A and Table S1). Of 182 clones screened, 64 were heterozygous, but none were homozygous for the BARD1 deletion. We hypothesize that homozygous loss of BARD1 is lethal. We therefore isolated several BARD1+/− heterozygous clones (SI Appendix, Fig. S4B). As expected, BARD1+/− cells have decreased levels of BARD1 RNA and protein (SI Appendix, Fig. S4 C and E). Furthermore, BARD1+/− cells have higher levels of BRCA1 RNA but lower levels of BRCA1 protein (SI Appendix, Fig. S4 D and E), presumably due to the reliance of BRCA1 on BARD1 for stability (30).
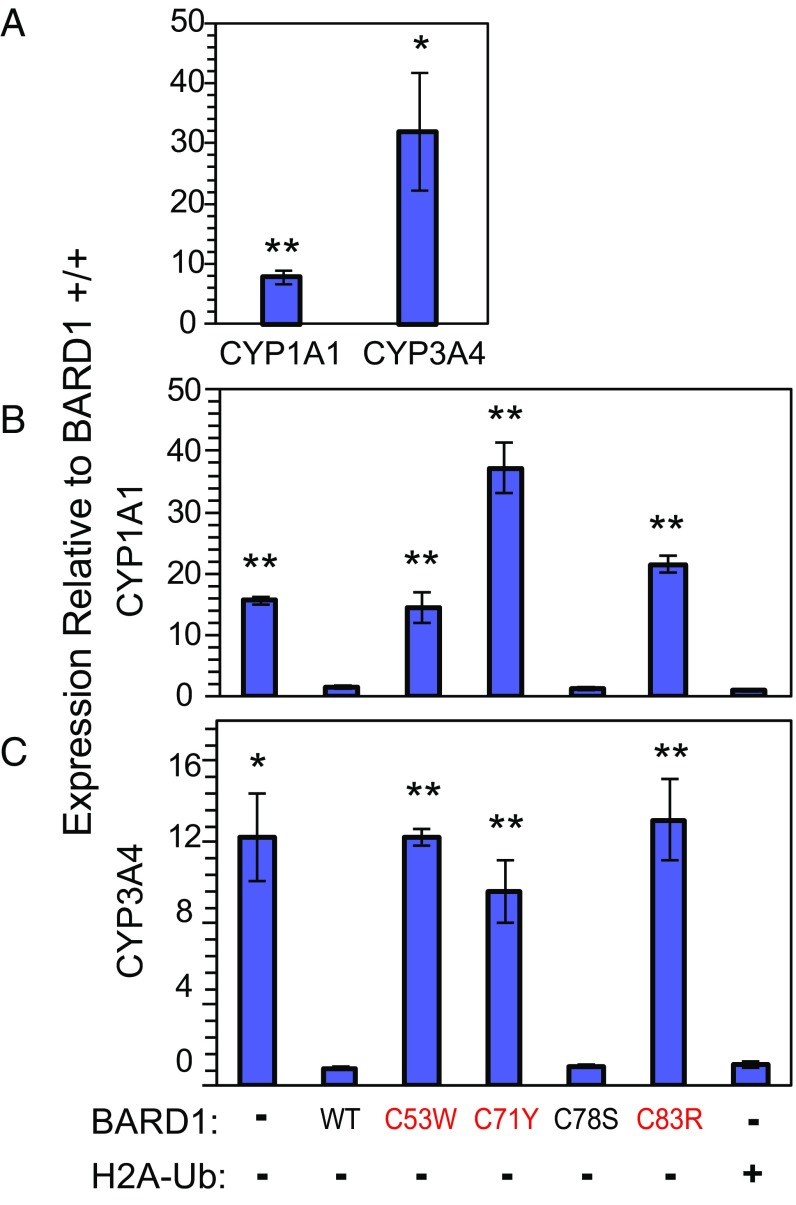
We compared expression of CYP1A1 and CYP3A4 in BARD1+/+ versus BARD1+/− MCF10A cells by quantitative real-time PCR (qRT-PCR). Expression levels of both CYP1A1 and CYP3A4 were significantly higher in BARD1+/− cells than in BARD1+/+ cells (Fig. 4A); increases were 7.8-fold (P = 0.008) for CYP1A1 and 32-fold (P = 0.017) for CYP3A4. By comparison, CYP1A1 and CYP3A4 expression increased 3-fold and 2-fold in similarly edited BRCA1+/− MCF10A cells (28). These data suggest that BARD1 plays a role in the repression of some genes associated with estrogen metabolism, and that haploinsufficiency of BARD1 leads to a substantial increase in the transcription of these genes.
Fig. 4.
Expression of estrogen metabolism genes in BARD1+/− MCF10A breast epithelial cells. (A) Expression of CYP1A1 and CYP3A4 in BARD1+/− cells relative to BARD1+/+ cells was measured by qRT-PCR. Transcriptional repression by the BRCA1/BARD1 complex is relaxed in BARD1 haploinsufficient cells, leading to increased expression of CYP1A1 and CYP3A4. For each gene, bar heights and error bars are means and SEs of relative expression levels derived from ΔΔCt measurements, with GAPDH standardization, from three replicate experiments. Comparisons were made by t tests for paired data. *P < 0.05; **P < 0.01. Details are shown in SI Appendix, Table S2. (B) Effects on CYP1A1 gene expression of introducing wild-type BARD1, one of the BARD1 mutants, or H2A-Ub fusion by transient transfection into BARD1+/− cells, accompanied by cotransfection with wild-type BRCA1. BARD1 mutants that fail to ubiquitylate nucleosomal H2A also fail to restore gene silencing of CYP1A1. Species introduced are noted below C. Mutations identified in breast cancer families are labeled in red. (C) Effects on CYP3A4 gene expression of introducing wild-type BARD1, one of the BARD1 mutants, or H2A-Ub fusion by transient transfection into BRCA1+/− cells, accompanied by cotransfection with wild-type BRCA1. BARD1 mutants that fail to ubiquitylate nucleosomal H2A also fail to restore gene silencing of CYP3A4.
To test whether mutant BARD1 proteins could rescue loss of BARD1 transcriptional regulatory function, we transiently transfected BARD1+/− cells with plasmids carrying either wild-type BARD1 or one of the mutant BARD1 genes, with simultaneous coexpression of wild-type BRCA1 to ensure formation of BRCA1/BARD1 heterodimers, and again measured CYP1A1 and CYP3A4 expression by qRT-PCR (Fig. 4 B and C and SI Appendix, Fig. S5). Introduction of either wild-type BARD1 or BARD1 p.C78S resulted in CYP1A1 and CYP3A4 expression returning to the low levels measured in BARD1+/+ cells. In contrast, the BARD1 mutations with defects in H2A ubiquitylation failed to restore gene silencing of either CYP1A1 or CYP3A4. These results indicate that BARD1 proteins that retain capacity to ubiquitylate H2A in nucleosomes in vitro are also able to restore repression of CYP1A1 and CYP3A4 in cells, whereas BARD1 proteins with a defect in nucleosome binding and ubiquitylation are unable to functionally restore CYP1A1 or CYP3A4 repression.
To test the relationship between BARD1 H2A ubiquitylation activity observed in vitro and gene expression changes in BARD1 haploinsufficient cells, we carried out a complementation experiment with ubiquitylated H2A. The BRCA1/BARD1 heterodimer attaches ubiquitin to the most C-terminal lysine residues on H2A, including the C-terminal residue, Lys129, of the histone tail (30). An H2A-Ub fusion expressed exogenously rescues heterochromatin silencing in BRCA1-negative cells (4) and homologous recombination in BARD1-silenced cells (5). Similarly, transfection of H2A-Ub fusion into BARD1+/− cells rescued CYP1A1 and CYP3A4 gene silencing, with their expression returning to levels similar to BARD1+/+ cells (Fig. 4 B and C and SI Appendix, Fig. S5). The ability of a mimic of BRCA1/BARD1-ubiquitylated H2A to rescue the increase in CYP1A1 and CYP3A4 expression observed in the BARD1+/− cells is strong, if indirect, evidence that ubiquitylation of H2A is the BARD1 function responsible for transcriptional regulation.
Discussion
We identified three missense mutations in highly conserved zinc-coordinating cysteine residues of the BARD1 RING domain in families severely affected with breast cancer. BRCA1/BARD1 heterodimers incorporating any of these mutations fail to ubiquitylate nucleosomal histone H2A. The same mutations are defective in mononucleosome binding, indicating that the BARD1 RING domain is instrumental in anchoring the BRCA1/BARD1 complex to chromatin. Engineered BARD1 haploinsufficiency in MCF10A cells led to increased expression of BRCA1-regulated estrogen metabolism genes CYP1A1 and CYP3A4 (28, 36).
A potential role of BRCA1 ubiquitylation of H2A in transcriptional regulation is based on the observation that expression of H2A with ubiquitin fused at its C terminus rescues heterochromatin silencing in a BRCA1-deficient cell line (4). Here we report that in BARD1 haploinsufficient cells, loss of transcriptional regulation of certain cytochrome P450 genes is rescued by reintroduction of wild-type BARD1, or of a BARD1 mutant protein with no effect on H2A ubiquitylation, or of H2A-Ub fusion, but is not rescued by BARD1 mutant proteins that fail to ubiquitylate H2A. Altogether our results suggest that the BARD1 RING domain binds a substrate required for nucleosome ubiquitylation, and that this function is linked to transcriptional regulation by BRCA1/BARD1 in breast epithelial cells.
Our results also indicate that BARD1 plays a major role in formation of a BRCA1/BARD1/nucleosome complex. BRCA1 ubiquitylates H2A on its C-terminal tail at Lys127 and Lys129 both in vitro and in vivo (1, 31). Another heterodimeric RING Ub ligase, RING1B/BMI1 of the polycomb repressive complex 1, also ubiquitylates H2A on the C-terminal tail but at a different position, Lys119 (32). While both RING1B/BMI1 and BRCA1/BARD1 activities lead to gene silencing, the two enzymes are not redundant (27) and the different sites of ubiquitylation play different roles in the cell (37). Binding of the RING1B/BMI1 complex to a nucleosome involves contact between the nucleosome and BMI1, the subunit of the heterodimer analogous to BARD1 (38). The contact is made by an extended loop on BMI1 that does not have a structural homolog in BARD1. But in contrast to the BARD1 mutants tested here, mutation of BMI1 at residues in the nucleosome interface did not affect nucleosome binding or activity (38), even though mutation of a conserved positively charged patch in RING1B or in BRCA1 resulted in similar losses of activity (38). Because the binding region on the ligase-active subunits (i.e., RING1B and BRCA1) is conserved, while the nucleosome binding sites on BMI1 and BARD1 are not, we propose that BARD1 and BMI1 function to position their ligase-active partners in specific orientations on the nucleosome to produce different ubiquitylated H2A products.
The observation that BARD1 mutants that show loss of activity toward H2A in vitro are the same BARD1 mutants that are unable to rescue gene repression lends further support to the hypothesis that BRCA1/BARD1 acts as a transcriptional repressor through its activity toward H2A. H2A with ubiquitin attached to a lysine on its C-terminal tail is a known repressive marker found in compact chromatin (4, 32, 33). BRCA1 recruitment to promoter regions of progesterone-responsive genes is correlated with appearance of ubiquitylated H2A in the promoter region and subsequent termination of progesterone signaling (3). BRCA1 activity toward H2A has also been associated with heterochromatin silencing in neuronal tissue culture (4, 27), suggesting that the role of BRCA1 as a transcriptional repressor via H2A ubiquitylation may not be limited to epithelial cells. The demonstration that BARD1 plays an active role in transcriptional repression, and the demonstration of a link between Ub ligase activity of BRCA1/BARD1 and transcriptional repression, add to the substantial body of evidence that BRCA1 acts as a transcriptional repressor.
The three familial BARD1 variants presented here exhibit loss of nucleosomal H2A ubiquitylation, but maintain activity toward other substrates such as ERα and BRCA1 autoubiquitylation (Fig. 2 and SI Appendix, Figs. S2 and S3). This contrasts with RING missense mutations in BRCA1 that are inactive against all known substrates. That BARD1 variants in complex with BRCA1 are active toward nonnucleosomal substrates is consistent with the established role of the BRCA1 RING domain as the site to which E2 Ub-conjugating enzymes bind (22). While we cannot exclude additional undiscovered substrates responsible for BRCA1 and BARD1 tissue-specific tumor suppression, nucleosomal H2A was the only substrate tested that shows a loss of activity for both BARD1 and BRCA1 variants, strongly implicating H2A as an important BRCA1/BARD1 substrate for tumor suppression activity. We note that the importance of BRCA1 ligase function for tumor suppression remains a topic of debate (4, 8–10, 39). The BARD1 mutations presented here provide a powerful opportunity to better define the functions and substrates of BRCA1 involved in tumor suppression through identification of similar dysfunction in vivo for mutations of BRCA1 and BARD1.
We propose that the BRCA1/BARD1 complex acts as a tumor suppressor in estrogen-dependent tissues through H2A ubiquitylation, by suppressing the expression of genes that encode estrogen-metabolizing enzymes. Loss-of-function mutations of BRCA1 and BARD1 lead to increased risk of breast, ovarian, and fallopian tube cancer (11–15, 40), tissues that are exposed to high levels of estrogen and estrogen metabolites over a woman’s lifetime. Prophylactic removal of the ovaries and fallopian tubes well before menopause (by about age 40) significantly reduces risk of breast cancer in BRCA1 mutation carriers (41–43), reflecting the strong influence of estrogen level in tumor formation in these women. Cytochrome P450 enzymes transform estrogen into metabolites that produce free radicals that induce double-stranded DNA breaks (28, 34). BRCA1 binds to promoters of a subset of P450 genes, and BRCA1 haploinsufficiency results in increased transcription of the P450 genes CYP1A1 and CYP3A4 (28). In addition, carriers of BRCA1 mutations have higher concentrations of DNA-damaging estrogen metabolites in their serum and urine (44, 45). Our results indicate that BARD1 haploinsufficiency also results in increased transcription of CYP1A1 and CYP3A4, and that introduction of cancer-predisposing BARD1 mutations cannot restore P450 gene repression in BARD1 haploinsufficent cells. Together with comparable findings for BRCA1 (28), our results indicate that two functional copies of both BRCA1 and BARD1 are required for proper transcriptional control of estrogen-metabolizing enzymes.
A role for BRCA1/BARD1 in DNA damage repair is well established. Ubiquitylated H2A is a DNA damage repair signal, and ubiquitylation of H2A by BRCA1/BARD1 is essential for efficient homologous recombination (5), reflected in the observation that two of the BARD1 mutations identified in severely affected families, BARD1 p.C53W and BARD1 p.C71Y, are defective in homologous recombination in vitro (46). This link suggests that BRCA1 or BARD1 mutations deficient in H2A modification are also likely to be defective in repair of double-stranded breaks induced by estrogen metabolites. In consequence, carriers of these mutations would accumulate more estrogen-induced DNA damage over time than would other women. The convergence of damaging effects of BRCA1 and BARD1 mutants both on transcriptional repression activity and on DNA damage repair activity, mediated by a shared H2A ubiquitylation mechanism, begins to reveal the molecular basis of the high risks of breast and ovarian cancer among women who carry these mutations.
Materials and Methods
A delineation of methods used, including genomics (47), protein constructs and purification (2, 23, 48, 49), ubiquitylation assays, nucleosome binding assays, gene editing (50), transfection assays (5), and RNA assays (51), is available in SI Appendix, SI Methods. This project was approved by the Human Subjects Division of the University of Washington (study 1583). Written informed consent was obtained from all participants.
Supplementary Material
Acknowledgments
We thank Peter Brzovic, Sarah Pierce, Dana Miller, Tobias Ritterhoff, Katherine Reiter, and Pearl Magala for thoughtful discussion and critical reading of the manuscript and Prof. Jo Morris (University of Birmingham) for sharing the H2A-Ub fusion construct. This work was supported by NIH Grants R01GM088055 (to R.E.K.), R35CA197458 (to M.-C.K.), R01CA175716 (to M.-C.K. and T.W.), R01GM110430 (to C.C.), and T32CA080416 (to M.D.S.); the Breast Cancer Research Foundation (M.-C.K.); Komen Foundation for the Cure (M.-C.K.); the Li Ka Shing Foundation (J.E.C.); and the Heritage Medical Research Foundation (J.E.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715467115/-/DCSupplemental.
References
- 1.Kalb R, Mallery DL, Larkin C, Huang JT, Hiom K. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 2014;8:999–1005. doi: 10.1016/j.celrep.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eakin CM, Maccoss MJ, Finney GL, Klevit RE. Estrogen receptor α is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci USA. 2007;104:5794–5799. doi: 10.1073/pnas.0610887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo V, Beato M. BRCA1 counteracts progesterone action by ubiquitination leading to progesterone receptor degradation and epigenetic silencing of target promoters. Cancer Res. 2011;71:3422–3431. doi: 10.1158/0008-5472.CAN-10-3670. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Densham RM, et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat Struct Mol Biol. 2016;23:647–655. doi: 10.1038/nsmb.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashizume R, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 7.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci USA. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drost R, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Shakya R, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334:525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato K, et al. A DNA-damage selective role for BRCA1 E3 ligase in claspin ubiquitylation, CHK1 activation, and DNA repair. Curr Biol. 2012;22:1659–1666. doi: 10.1016/j.cub.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 11.Thai TH, et al. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum Mol Genet. 1998;7:195–202. doi: 10.1093/hmg/7.2.195. [DOI] [PubMed] [Google Scholar]
- 12.Sauer MK, Andrulis IL. Identification and characterization of missense alterations in the BRCA1 associated RING domain (BARD1) gene in breast and ovarian cancer. J Med Genet. 2005;42:633–638. doi: 10.1136/jmg.2004.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh T, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratajska M, et al. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res Treat. 2012;131:89–97. doi: 10.1007/s10549-011-1403-8. [DOI] [PubMed] [Google Scholar]
- 15.DeLeonardis K, et al. Challenges in interpreting germline mutations in BARD1 and ATM in breast and ovarian cancer patients. Breast J. 2017;23:461–464. doi: 10.1111/tbj.12764. [DOI] [PubMed] [Google Scholar]
- 16.Norquist BM, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couch FJ, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33:304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lolas Hamameh S, et al. Genomic analysis of inherited breast cancer among Palestinian women: Genetic heterogeneity and a founder mutation in TP53. Int J Cancer. 2017;141:750–756. doi: 10.1002/ijc.30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris JR, et al. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum Mol Genet. 2006;15:599–606. doi: 10.1093/hmg/ddi476. [DOI] [PubMed] [Google Scholar]
- 21.Friedman LS, et al. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 22.Brzovic PS, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 24.Landrum MJ, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meza JE, Brzovic PS, King MC, Klevit RE. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J Biol Chem. 1999;274:5659–5665. doi: 10.1074/jbc.274.9.5659. [DOI] [PubMed] [Google Scholar]
- 26.Wu LC, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 27.Abdouh M, Hanna R, El Hajjar J, Flamier A, Bernier G. The polycomb repressive complex 1 protein BMI1 is required for constitutive heterochromatin formation and silencing in mammalian somatic cells. J Biol Chem. 2016;291:182–197. doi: 10.1074/jbc.M115.662403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savage KI, et al. BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 2014;74:2773–2784. doi: 10.1158/0008-5472.CAN-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, et al. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-α. Mol Endocrinol. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia Y, Pao GM, Chen HW, Verma IM, Hunter T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem. 2003;278:5255–5263. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 31.Thakar A, Parvin J, Zlatanova J. BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. J Biomol Struct Dyn. 2010;27:399–406. doi: 10.1080/07391102.2010.10507326. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 33.Zhou W, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizutani A, et al. Extensive chromosomal breaks are induced by tamoxifen and estrogen in DNA repair-deficient cells. Cancer Res. 2004;64:3144–3147. doi: 10.1158/0008-5472.can-03-3489. [DOI] [PubMed] [Google Scholar]
- 35.Cavalieri E, et al. Catechol estrogen quinones as initiators of breast and other human cancers: Implications for biomarkers of susceptibility and cancer prevention. Biochim Biophys Acta. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Harte MT, et al. BRD7, a subunit of SWI/SNF complexes, binds directly to BRCA1 and regulates BRCA1-dependent transcription. Cancer Res. 2010;70:2538–2547. doi: 10.1158/0008-5472.CAN-09-2089. [DOI] [PubMed] [Google Scholar]
- 37.Kim B-J, et al. The histone variant macroH2A1 is a BRCA1 ubiquitin ligase substrate. Cell Rep. 2017;19:1758–1766. doi: 10.1016/j.celrep.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finch AP, et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation. J Clin Oncol. 2014;32:1547–1553. doi: 10.1200/JCO.2013.53.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabai-Kapara E, et al. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc Natl Acad Sci USA. 2014;111:14205–14210. doi: 10.1073/pnas.1415979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kauff ND, et al. Risk-reducing salpingo-oophorectomy in women with a BRCA1 or BRCA2 mutation. N Engl J Med. 2002;346:1609–1615. doi: 10.1056/NEJMoa020119. [DOI] [PubMed] [Google Scholar]
- 42.Domchek SM, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304:967–975. doi: 10.1001/jama.2010.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finkelman BS, et al. Breast and ovarian cancer risk and risk reduction in Jewish BRCA1/2 mutation carriers. J Clin Oncol. 2012;30:1321–1328. doi: 10.1200/JCO.2011.37.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaikwad NW, et al. The molecular etiology of breast cancer: Evidence from biomarkers of risk. Int J Cancer. 2008;122:1949–1957. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaikwad NW, et al. Urine biomarkers of risk in the molecular etiology of breast cancer. Breast Cancer (Auckl) 2009;3:1–8. doi: 10.4137/bcbcr.s2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee C, et al. Functional analysis of BARD1 missense variants in homology-directed repair of DNA double strand breaks. Hum Mutat. 2015;36:1205–1214. doi: 10.1002/humu.22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh T, et al. Genetic predisposition to breast cancer due to mutations other than BRCA1 and BRCA2 founder alleles among Ashkenazi Jewish women. JAMA Oncol. 2017;3:1647–1653. doi: 10.1001/jamaoncol.2017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 49.Dhall A, et al. Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J Biol Chem. 2014;289:33827–33837. doi: 10.1074/jbc.M114.591644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.