Significance
Plague is infamous as the cause of the Black Death (1347–1353) and later Second Pandemic (14th to 19th centuries CE), when devastating epidemics occurred throughout Europe, the Middle East, and North Africa. Despite the historical significance of the disease, the mechanisms underlying the spread of plague in Europe are poorly understood. While it is commonly assumed that rats and their fleas spread plague during the Second Pandemic, there is little historical and archaeological support for such a claim. Here, we show that human ectoparasites, like body lice and human fleas, might be more likely than rats to have caused the rapidly developing epidemics in pre-Industrial Europe. Such an alternative transmission route explains many of the notable epidemiological differences between historical and modern plague epidemics.
Keywords: Yersinia pestis, Black Death, SIR modeling, Bayesian analysis, Monte Carlo Markov chain
Abstract
Plague, caused by the bacterium Yersinia pestis, can spread through human populations by multiple transmission pathways. Today, most human plague cases are bubonic, caused by spillover of infected fleas from rodent epizootics, or pneumonic, caused by inhalation of infectious droplets. However, little is known about the historical spread of plague in Europe during the Second Pandemic (14–19th centuries), including the Black Death, which led to high mortality and recurrent epidemics for hundreds of years. Several studies have suggested that human ectoparasite vectors, such as human fleas (Pulex irritans) or body lice (Pediculus humanus humanus), caused the rapidly spreading epidemics. Here, we describe a compartmental model for plague transmission by a human ectoparasite vector. Using Bayesian inference, we found that this model fits mortality curves from nine outbreaks in Europe better than models for pneumonic or rodent transmission. Our results support that human ectoparasites were primary vectors for plague during the Second Pandemic, including the Black Death (1346–1353), ultimately challenging the assumption that plague in Europe was predominantly spread by rats.
Plague, caused by the bacterium Yersinia pestis, has been extensively studied due to its modern and historical significance. In the past, plague has famously caused at least three pandemics in human history: the First Pandemic beginning with the Justinianic Plague (6th to 8th centuries), the Second Pandemic beginning with the “Black Death” (14th to 19th centuries), and the Third Pandemic (beginning in the 19th century) (1). Today, plague persists primarily in rodent reservoirs in Asia, Africa, and the Americas, where it poses a recurrent threat to nearby human settlements (2).
The most common forms of plague infection are bubonic and pneumonic (2). Bubonic plague occurs when bacteria enter the skin, usually from the bite of an infected flea vector. The bacteria are then transported to the lymph nodes, causing characteristic swelling, or “buboes.” Bubonic plague is typically transmitted to humans from wild or commensal rodents (3), but transmission between people is also thought to occur by human ectoparasites, such as human fleas (Pulex irritans) or body lice (Pediculus humanus humanus) (4). Primary pneumonic plague occurs when aerosolized bacteria enter and infect the lungs. Pneumonic plague can also arise as a complication of bubonic or septicemic infections (2), known as secondary pneumonic plague. Individuals with pneumonic plague can transmit the disease through the respiratory route, although outbreaks of pneumonic plague are typically small because infected persons die rapidly without treatment (5). Septicemic plague occurs when bacteria infect the bloodstream, commonly from a primary pneumonic or bubonic infection (2).
A central focus of historical plague research has been to understand the spread and persistence of plague in Europe. Little is known about the transmission of plague in Europe, the Middle East, and North Africa during the Second Pandemic, including the Black Death, when the disease killed an estimated one-third of the population. Many studies (4, 6, 7) have suggested that human ectoparasites, like human fleas and body lice, were more likely than commensal rats to have caused the rapidly spreading epidemics. Proponents of the “human ectoparasite hypothesis” argue that plague epidemics during the Second Pandemic differ from the rat-associated epidemics that occurred later, during the Third Pandemic. Specifically, the geographic spread and total mortality of the Black Death far exceeds that of modern plague epidemics (8). While contemporaneous accounts of symptoms during the Second Pandemic are consistent with those of plague (7), there are no descriptions of rat epizootics, or “rat falls,” that often precede epidemics in the Third Pandemic (7–9). Some have noted that the climate of northern Europe could not have fostered the widespread distribution of Rattus rattus (10), a claim that is supported by a scarcity of rats in the archaeological record (6). Finally, epidemiological characteristics of plague in Europe, such as a high rate of household transmission (11), are suggestive of a more direct transmission route (12).
Despite support for human ectoparasite transmission, it has been difficult to assess their historical contribution because their role in modern plague epidemics appears to be relatively minor. Today, human ectoparasite diseases have declined in most developed countries, but they are still associated with poverty and unhygienic conditions (13). In the past, human ectoparasites have been efficient vectors for diseases such as epidemic typhus (14) and relapsing fever (15). In 1941, plague-infected body lice and human fleas were found on septicemic patients during an outbreak in Morocco (16), indicating that humans can transmit the disease to lice and human fleas. In addition, recent experimental studies have demonstrated that body lice can transmit the bacteria to naive rabbits (4, 17–19). However, the transmission from body lice and human fleas to humans has not yet been documented, and thus the importance of human ectoparasite transmission in current and historic settings remains an open question. Our theoretical analysis demonstrates that human ectoparasites may indeed play such a role.
Mathematical modeling can provide strong insight into mechanisms of plague transmission for past epidemics. Previous epidemiological models of plague during the Second Pandemic are focused mainly on modeling the spread of the disease by commensal rats during a single outbreak (20, 21). In this study, we developed a susceptible–infectious–recovered (SIR) model for plague transmission with a human ectoparasite vector and compared it to models for pneumonic and rat–flea transmission. We applied these models to nine outbreaks during the Second Pandemic, to gain a broad understanding of the transmission dynamics of plague in European epidemics. We identified the best-fitting model for each outbreak and estimated the basic reproduction number, R0.
Methods
Historical Data.
We used data on the daily and weekly disease-induced mortality for nine plague outbreaks during the Second Pandemic (Table 1). These data were publicly available in secondary sources including published articles, books, and government reports. We digitized the epidemic data from printed tables and graphs, using the entire duration of each outbreak, apart from Eyam, which had two mortality peaks. The deterministic models we used cannot account for the stochasticity of infectious disease processes during the early phase of an epidemic; thus, for the outbreak in Eyam, we removed the first 279 data points and considered only the second, larger epidemic peak. To validate the models for pneumonic and rat-associated plague epidemics, we used three additional mortality curves from epidemics with known transmission routes during the Third Pandemic (Table S1).
Table 1.
Summary of the Second Pandemic mortality data
| Location | Date (MM/YYYY) | Population | Recorded mortality | Refs. |
| Givry, France | 07/1348–11/1348 | 1,500 | 636 | 22 |
| Florence, Italy | 05/1400–11/1400 | 60,000 | 10,215 | 23 |
| Barcelona, Spain | 04/1490–09/1490 | 25,000 | 3,576 | 24, 25 |
| London, England | 06/1563–01/1564 | 80,000 | 16,886 | 26 |
| Eyam, England | 06/1666–11/1666 | 350 | 197 | 26 |
| Gdansk, Poland | 03/1709–12/1709 | 50,000 | 23,496 | 27 |
| Stockholm, Sweden | 08/1710–02/1711 | 55,000 | 12,252 | 27 |
| Moscow, Russia | 07/1771–12/1771 | 300,000 | 53,642 | 28 |
| Island of Malta, Malta | 04/1813–11/1813 | 97,000 | 4,487 | 29 |
The present-day location, date (month/year), preplague population size, and recorded plague deaths, for nine plague outbreaks during the Second Pandemic.
Parameters.
The parameter values and initial conditions used in the models are shown in Table 2 and Table S2. Fixed values were taken from field, experimental, or epidemiological case studies when available. Unobservable parameters were estimated using Bayesian inference.
Table 2.
Parameters for three SIR models of plague transmission
| Parameter | Value | Definition | Refs. |
| Humans | |||
| U(0.001, 0.05) | Transmission rate for bubonic plague from mildly infectious humans to body lice | ||
| U(0.001, 1) | Transmission rate for bubonic plague from highly infectious humans to body lice | ||
| U(0.001, 1) | Transmission rate for pneumonic plague | ||
| U(0.001, 0.2) | Transmission rate for bubonic plague from rat fleas to humans | ||
| 8.0 (d) | Average low infectious period for bubonic plague | ||
| 2.0 (d) | Average high infectious period for bubonic plague | ||
| 2.5 (d) | Average infectious period for pneumonic plague | 5 | |
| 10.0 (d) | Average duration of infection for bubonic plague | 30 | |
| 0.4 | Probability of recovery from bubonic plague | 3 | |
| Lice (P. humanus humanus) | |||
| 0.11 (per d) | Natural lice growth rate | 31 | |
| 15.0 (per person) | Lice index at carrying capacity | 32, 33 | |
| 0.05 | Transmission rate for bubonic plague from body lice to humans | ||
| 3.0 (d) | Average infectious period for bubonic plague | 17 | |
| Rats (R. rattus) | |||
| U(0.001, 1) | Transmission rate for bubonic plague from rat fleas to rats | ||
| 5.2 (d) | Average infectious period for bubonic plague | 34 | |
| 0.1 | Probability of recovery from bubonic plague | 34 | |
| Fleas (X. cheopis) | |||
| 0.0084 (per d) | Natural flea growth rate | 35, 36 | |
| 6.0 | Average number of fleas at carrying capacity | 37, 38 | |
| 5.0 (d) | Death rate of fleas | 39 | |
| 3.0/ | Searching efficiency | 35, 36 | |
Single numbers are fixed values and distributions (U = uniform) are priors.
Human–Ectoparasite Model.
The transmission of bubonic plague by a human ectoparasite vector, such as human fleas or body lice, is modeled by seven differential equations:
The five compartments for humans that are functions of time t: susceptible (), infectious with mild bacteremia , infectious with high bacteremia , recovered , and dead . The total living population is given by . The transmission of plague from vectors to humans occurs at rate . The model assumes that humans are mildly infectious for an average of 8 d (), and transmission is unlikely at rate . Humans with mild bacteremia may recover at rate , which is around 40% for untreated bubonic plague. Experimental studies have shown that fleas must feed on hosts with high levels of bacteremia for reliable transmission (40). Therefore, the model assumes that moribund humans transmit plague at a high rate to vectors for an average of 2 d (). Given the short duration of the outbreaks, we did not model natural births and deaths in the human population.
Human ectoparasite vectors are modeled in two compartments (, ). The susceptible vector population grows at the intrinsic growth rate . The growth of the vector population is limited by the carrying capacity , which is the product of the parasite index and the number of human hosts . Modern studies show that the rate of body louse infestation and abundance in affected human populations ranges from 10.5 to 67.7 lice on average per person (33, 41).
There are a limited number of studies that evaluate human fleas and body lice as vectors for plague (17–19). These studies have shown both vectors have similar transmission cycles for Y. pestis, and this makes it difficult to distinguish between the two species with either model structure or parameter values (17–19). Our model uses parameters specific to body lice; however, the ranges for the lice and flea parameters overlap. The duration of infection has been shown experimentally for both species, and is on average 4.5 d for human fleas and 3 d for body lice (17–19). The model assumes that infected human fleas and body lice do not recover. The transmission of plague by human fleas is hypothesized to occur through early phase transmission, an alternative to blocked transmission observed in rat fleas (Xenopsylla cheopis) that does not require a lengthy extrinsic incubation period (42).
Pneumonic Plague Model.
The direct human-to-human transmission of plague is modeled by three differential equations:
There are three compartments for humans (, ) and the total human population is . There is no compartment for recovered individuals because the case fatality rate of untreated pneumonic plague is close to 100% (43). The human-to-human transmission of pneumonic plague occurs at rate . The disease-induced mortality occurs at rate per day and is equal to the inverse of the infectious period, which is a mean of 2.5 d for pneumonic plague (5).
Rat–Flea Model.
Based on a metapopulation model for bubonic plague by Keeling and Gilligan (35, 36), the transmission of plague in a rodent epizootic, and the spillover to humans is modeled by 10 differential equations:
There are four compartments for rats (, , ) and the total rat population is . As epidemics within the rat population can only occur when a large proportion of the rats are susceptible to the disease, we assumed an initial black rat (Rattus rattus) population that was entirely susceptible. Although the expected ratio of urban rats to humans is about 1 rat to every 5 people (44), we allowed the prior in the model to have a maximum ratio of 1:1 rats to humans. Increasing the rat population in medieval cities allowed the simulated rat-borne plague outbreaks to more easily reach the mortality levels observed in humans during the Second Pandemic.
Rat fleas (X. cheopis) are modeled as the average number of fleas per rat, , and the number of free infectious fleas, . The flea population has a natural growth rate, , that is limited by the carrying capacity . We assumed that the flea population is limited by the number of rat hosts, because X. cheopis does not reproduce on humans (45). Plague is transmitted to rats at rate by free infectious fleas searching for a host with searching efficiency . We further assumed that fleas can transmit plague in the early phase (42). Rats die at a rate equal to the inverse of the infectious period , or recover with probability . When an infected rat dies, a number of free infectious fleas are released into the environment, depending on the average number of fleas per rat. Free infectious fleas die at rate . The model assumes that plague is a rodent disease and that human cases are a consequence of mortality in the rat population. Therefore, susceptible humans become infected by free infectious fleas at rate . Humans remain infected for an average of 10 d (), at which point they either recover at rate or die.
In the model by Keeling and Gilligan (35, 36), it is assumed that the force of infection from free infectious fleas is divided exclusively between rats and humans. However, the authors note that the true force of infection to humans is less because not every flea will find and infect a human (35). For our model, we sought to establish a range for that would accurately lower the force of infection to humans. To establish this range, we fitted the model to observed mortality for both rats and humans in Hong Kong in 1903 (Fig. S1) and found that the mean estimate for was 0.1 (Table S3). Using simulations, we found that should be less than 0.2 to preserve the characteristic delay and higher peak mortality of the rat epizootic compared with the human epidemic. Based on these observations, we constrained the prior for the transmission rate to humans to 0.0–0.2, which enabled us to use this model for outbreaks where only human mortality was available.
Bayesian Inference and Markov Chain Monte Carlo.
We fitted the deterministic models to the observed data using Bayesian inference and estimated unobservable parameters of interest. The models had a time-step of 1 d and were fitted to daily mortality or weekly mortality. Denoting the set of model parameters as , the probability of the observed data given is calculated as the product of a series of Poisson random variables with mean equal to the human mortality in the model at times :
The parameters that we fitted were the transmission rates for each model (, , , , ) and the size of the initial primary host population that was at risk [, ] or infected [, ]. We assumed uniformly distributed priors and obtained posterior distributions using Markov chain Monte Carlo (MCMC) simulations with an adaptive Metropolis–Hastings algorithm implemented in PyMC2 (46) (for examples of the implementation, see https://zenodo.org/record/1043924). We ran the MCMC simulations for 180,000 iterations with a burn-in of 80,000 iterations and a thinning of 10. We assessed convergence for each model by running three independent MCMC chains and verifying that the Gelman–Rubin statistic (47) was <1.05 for each parameter. We performed model comparison using the Bayesian information criterion (BIC) from the maximum-likelihood estimates of the model parameters (48). The model with the lowest BIC value was the unique preferred model if the second-best model had a BIC value of at least 10 larger (49).
Estimation of the Basic Reproduction Number.
We estimated the basic reproduction number in each model for the primary host using the next-generation matrix method (50).
Reporting Error.
We conducted the analysis again considering different levels of underreporting (10%, 25%, and 50%) for each outbreak. To do so, we incorporated a constant probability of reporting into the likelihood function.
Results
Model Fit and Selection.
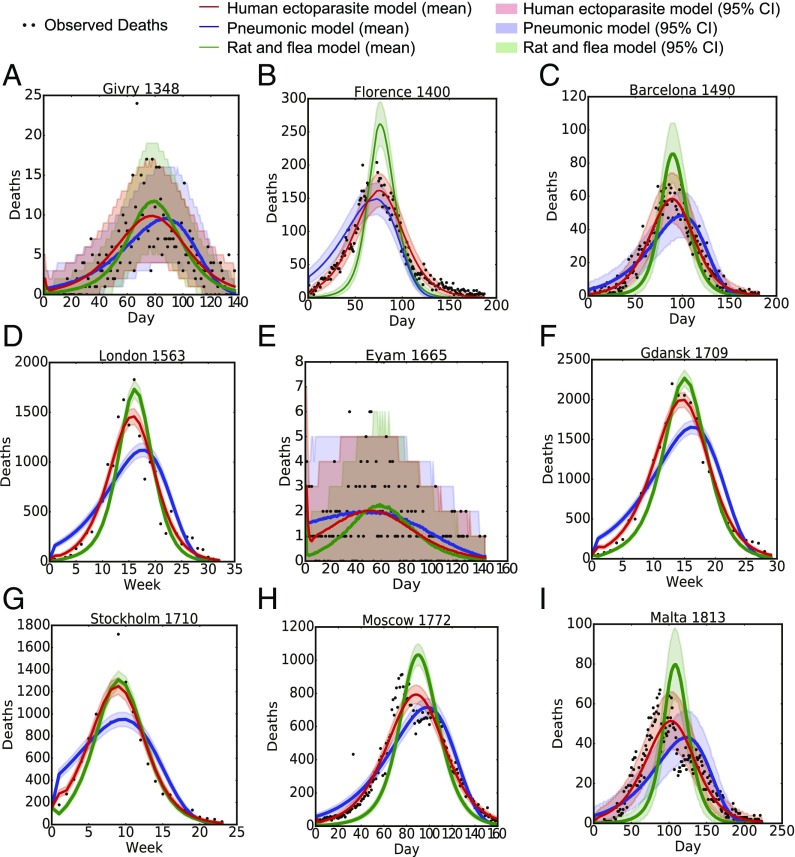
We used Bayesian MCMC and the mortality data to fit the three transmission models: human ectoparasite plague (EP), pneumonic plague (PP), and rat-borne plague (RP) (Fig. 1). The posterior means and 95% credible intervals for the fitted parameters in each model are given in Table S3. Fig. 1 shows the fit of each model to the observed mortality. For the smallest outbreaks, Eyam and Givry, it is difficult to visually distinguish between the models because the credible intervals are overlapping. In general, the human ectoparasite model fit the pattern of the observed data for the Second Pandemic outbreaks. However, the model could not account for irregularities in the observed mortality from Malta and Moscow, which have two peaks. For the pneumonic plague model, the mortality curve is right skewed compared with the observed mortality. Mortality in the rat model tended to grow slowly while plague spread through the rat population, and peaked higher than the observed mortality.
Fig. 1.
Fit of three models of plague transmission to mortality during Second Pandemic outbreaks. The observed human mortality data (black dots) and the fit (mean and 95% credible interval) of three models for plague transmission [human ectoparasite (red), pneumonic (blue), and rat–flea (green)] for nine plague outbreaks: (A) Givry, France (1348), (B) Florence, Italy (1400), (C) Barcelona, Spain (1490), (D) London, England (1563), (E) Eyam, England (1665), (F) Gdansk, Poland (1709), (G) Stockholm, Sweden (1710), (H) Moscow, Russia (1772), and (I) Island of Malta, Malta (1813).
We compared the three competing models using the BIC. Our results (Table 3) show that the human ectoparasite model had the lowest BIC value for all outbreaks, except Eyam and Givry. For the remaining outbreaks, the difference in BIC for the human ectoparasite model and the other candidate models was greater than 10, which provides strong evidence against the pneumonic and rat–flea models (50). For Eyam and Givry, the difference between the human ectoparasite model and another model was less than 10; therefore, neither model could be excluded.
Table 3.
Comparison of transmission models and posterior estimates for the basic reproduction number for different plague models and outbreaks
| Location | Model | BIC | ΔBIC | R0 |
| Givry (1348) | EP | 1,287 | 0 | 1.82 [1.82, 1.82] |
| PP | 1,333 | 46 | 1.10 [1.10, 1.10] | |
| RP | 1,287 | 0 | 1.61 [1.61, 1.61] | |
| Florence (1400) | EP | 2,662 | 0 | 1.76 [1.76, 1.76] |
| PP | 4,569 | 1,907 | 1.09 [1.09, 1.09] | |
| RP | 10,157 | 7,495 | 2.03 [2.03, 2.03] | |
| Barcelona (1490) | EP | 1,942 | 0 | 1.91 [1.91, 1.91] |
| PP | 2,410 | 468 | 1.09 [1.09, 1.09] | |
| RP | 3,392 | 1,450 | 2.04 [2.04, 2.04] | |
| London (1563) | EP | 1,585 | 0 | 1.64 [1.64, 1.64] |
| PP | 4,647 | 3,062 | 1.06 [1.06, 1.06] | |
| RP | 3,882 | 2,297 | 1.52 [1.52, 1.52] | |
| Eyam (1666) | EP | 1,171 | 0 | 1.48 [1.48, 1.49] |
| PP | 1,174 | 3 | 1.04 [1.04, 1.04] | |
| RP | 1,205 | 34 | 1.24 [1.24, 1.24] | |
| Gdansk (1709) | EP | 797 | 0 | 1.64 [1.64, 1.64] |
| PP | 3,841 | 3,044 | 1.06 [1.06, 1.06] | |
| RP | 2,212 | 1,415 | 1.46 [1.46, 1.46] | |
| Stockholm (1710) | EP | 726 | 0 | 1.75 [1.75, 1.75] |
| PP | 2,118 | 1,392 | 1.06 [1.06, 1.06] | |
| RP | 1,062 | 336 | 1.30 [1.30, 1.30] | |
| Moscow (1771) | EP | 3,912 | 0 | 1.79 [1.79, 1.79] |
| PP | 6,789 | 2,877 | 1.09 [1.09, 1.09] | |
| RP | 15,946 | 12,034 | 1.76 [1.76, 1.76] | |
| Malta (1813) | EP | 2,761 | 0 | 1.57 [1.57, 1.57] |
| PP | 3,580 | 819 | 1.06 [1.06, 1.06] | |
| RP | 6,445 | 3,684 | 1.79 [1.79, 1.79] |
The models are designated as human ectoparasite (EP), primary pneumonic plague (PP), and rat–flea (RP). Values in bold represent the best-fitting models that were within 10 points of the lowest BIC. The R0 (mean [95% confidence interval]) was estimated using the next-generation matrix method.
To verify our model comparison method, we fitted the models to three additional Third Pandemic outbreaks with known transmission routes (Fig. S2). We found that the model with the lowest BIC matched the known modes of transmission for the outbreaks in Hong Kong (rats) and Harbin (pneumonic) (Table S5). However, we could not distinguish between two of the models for a small outbreak of rat-associated plague in Sydney, suggesting together with the results from Eyam and Givry, that our model comparison method is better suited for sufficiently large outbreaks (>750 deaths).
Basic Reproduction Number R0.
By definition, the basic reproduction number, R0, is the average number of secondary cases produced by a primary case, given an entirely susceptible population. In practice, R0 is an important threshold for disease invasion. For each of the three models, we calculated R0 from the posterior estimates of the fitted parameters (Table 3). For all of the models, R0 was greater than 1, which is above the threshold for disease invasion. Using the human ectoparasite model, the estimated R0 was 1.48–1.91 for all pre-Industrial outbreaks.
Reporting Error.
We considered the impact of different levels of constant underreporting of deaths throughout the epidemics on model selection (Table S6). We found that underreporting of 10% and 25% did not change the results of the model selection; under these conditions, the human ectoparasite model was the best fit for all outbreaks in Europe except Eyam and Givry. Underreporting of 50% changed the best-fitting models of Gdansk and Givry to pneumonic plague. For these cities, 50% underreporting resulted in the death of more than 90% of the population, giving preference to a pneumonic plague model where all infected individuals die from plague.
Discussion
Our study supports human ectoparasite transmission of plague during the Second Pandemic, including the Black Death. Using recent experimental data on human fleas and body lice as plague vectors, we have developed a compartmental model that captures the dynamics of human ectoparasite transmission. We have shown that, in seven out of nine localities, the human ectoparasite model was the preferred model to explain the pattern of plague mortality during an outbreak, rather than models of pneumonic and rat–flea plague transmission (Table 3). The small size of the plague outbreaks in Eyam and Givry made it difficult to distinguish transmission routes based on mortality data. For Eyam, both the human ectoparasite model and the pneumonic model produced a similar quality fit for the observed mortality. This agrees with a previous modeling study of Eyam (1665), which found that the dominant mode of transmission was an unspecified route of human-to-human transmission, rather than rodent transmission (11). Overall, our results suggest that plague transmission in European epidemics occurred predominantly through human ectoparasites, rather than commensal rat or pneumonic transmission.
The strength of our study is that we compared three plague transmission models, each representing a known or hypothetical mode of plague transmission, for nine plague outbreaks across the spatial and temporal extent of the Second Pandemic in Europe. Our study thus provides a more general understanding of plague epidemics in Europe than previous modeling studies that focus on single outbreaks, or single transmission routes (11, 20, 35, 36, 51). However, since we considered nine outbreaks over several centuries, we were limited to using simple models that could be applied systematically. Consequently, these models did not account for local conditions that can affect disease transmission, like war, famine, immunity, and public health interventions. Additionally, we did not model mixed transmission routes, and this makes it difficult to fully assess the contribution of pneumonic plague, which commonly occurs during bubonic outbreaks (52). Secondary pneumonic plague develops in an estimated 20% of bubonic cases, and this creates potential for primary pneumonic spread, even if it is not the dominant transmission route (52). Finally, we do not consider events leading up to the introduction of the disease and our results cannot be extended to plague transmission between localities, which may have involved different transmission mechanisms.
Recent studies have found human ectoparasites during plague outbreaks in the Democratic Republic of Congo (41), Tanzania (53), and Madagascar (54), but their role in these outbreaks is not clear. In the absence of modern studies on human ectoparasites as vectors for plague, our results yield inferences about the conditions necessary to produce outbreaks driven by human ectoparasite transmission. Our estimated values for R0 using the human ectoparasite model were consistently between 1.5 and 1.9 for all nine cities. The main components of R0 in the human ectoparasite model are the ectoparasite index and the transmission rates (). From the fitted models, we obtained estimates for the transmission rates () from humans to ectoparasites during the early and late stages of plague infection. We found that the majority of ectoparasite infections occurred during the period of high infectivity in humans, consistent with experimental evidence (40). Inferences like these not only improve our understanding of human ectoparasites as plague vectors in the past but also have important implications for limiting the size of plague outbreaks today.
Many studies have sought to clarify the mechanisms underlying the spread and maintenance of plague during the Second Pandemic. Mathematical modeling is an important tool to examine the role of different transmission mechanisms, particularly in the absence of definitive experimental, historical, and archaeological information. Here, we demonstrate that human ectoparasites appear to have been the dominant transmission mode for plague during the Second Pandemic. This alternative mode of transmission could account for many of the epidemiological differences between the Second Pandemic and those caused by rats during the Third Pandemic. Plague is undeniably a disease of significant scientific, historic, and public interest, and is still present in many parts of the world today. It is therefore crucial that we understand the full spectrum of capabilities that this versatile, pandemic disease has exhibited in the past.
Supplementary Material
Acknowledgments
We thank W. Ryan Easterday and Jukka Corander for their valuable comments. We acknowledge funding from the European Research Council under the FP7-IDEAS-ERC Program (Grant 324249). We also acknowledge funding from the Centre for Ecological and Evolutionary Synthesis.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715640115/-/DCSupplemental.
References
- 1.Bramanti B, Stenseth NC, Walløe L, Lei X. Plague: A disease which changed the path of human civilization. Adv Exp Med Biol. 2016;918:1–26. doi: 10.1007/978-94-024-0890-4_1. [DOI] [PubMed] [Google Scholar]
- 2.Dennis DT, et al. Plague Manual: Epidemiology, Distribution, Surveillance and Control. WHO; Geneva: 1999. [PubMed] [Google Scholar]
- 3.Kugeler KJ, Staples JE, Hinckley AF, Gage KL, Mead PS. Epidemiology of human plague in the United States, 1900–2012. Emerg Infect Dis. 2015;21:16–22. doi: 10.3201/eid2101.140564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drancourt M, Houhamdi L, Raoult D. Yersinia pestis as a telluric, human ectoparasite-borne organism. Lancet Infect Dis. 2006;6:234–241. doi: 10.1016/S1473-3099(06)70438-8. [DOI] [PubMed] [Google Scholar]
- 5.Gani R, Leach S. Epidemiologic determinants for modeling pneumonic plague outbreaks. Emerg Infect Dis. 2004;10:608–614. doi: 10.3201/eid1004.030509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hufthammer AK, Walløe L. Rats cannot have been intermediate hosts for Yersinia pestis during medieval plague epidemics in Northern Europe. J Archaeol Sci. 2013;40:1752–1759. [Google Scholar]
- 7.Walløe L. Medieval and modern bubonic plague: Some clinical continuities. Med Hist Suppl. 2008;27:59–73. [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn SK. The Black Death Transformed: Disease and Culture in Early Renaissance Europe. Oxford Univ Press; London: 2002. [Google Scholar]
- 9.Ell SR. Some evidence for interhuman transmission of medieval plague. Rev Infect Dis. 1979;1:563–566. doi: 10.1093/clinids/1.3.563. [DOI] [PubMed] [Google Scholar]
- 10.Davis DE. The scarcity of rats and the Black Death: An ecological history. J Interdiscip Hist. 1986;16:455–470. [Google Scholar]
- 11.Whittles LK, Didelot X. Epidemiological analysis of the Eyam plague outbreak of 1665–1666. Proc Biol Sci. 2016;283:20160618. doi: 10.1098/rspb.2016.0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Advisory Committee appointed by the Secretary of State for India Reports on the plague investigations in India. XXIV. General considerations regarding the spread of infection, infectivity of houses etc. in Bombay City and Island. J Hyg (Lond) 1907;7:874–894. doi: 10.1017/s0022172400033702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brouqui P. Arthropod-borne diseases associated with political and social disorder. Annu Rev Entomol. 2011;56:357–374. doi: 10.1146/annurev-ento-120709-144739. [DOI] [PubMed] [Google Scholar]
- 14.Smallman-Raynor M, Cliff AD. War Epidemics: An Historical Geography of Infectious Diseases in Military Conflict and Civil Strife, 1850–2000. Oxford Univ Press; Oxford: 2004. [Google Scholar]
- 15.Bryceson AD, et al. Louse-borne relapsing fever. Q J Med. 1970;39:129–170. [PubMed] [Google Scholar]
- 16.Blanc G, Baltazard M. Rôle des ectoparasites humains dans la transmission de la peste. Bull Acad Natl Med. 1942;126:446. [Google Scholar]
- 17.Houhamdi L, Lepidi H, Drancourt M, Raoult D. Experimental model to evaluate the human body louse as a vector of plague. J Infect Dis. 2006;194:1589–1596. doi: 10.1086/508995. [DOI] [PubMed] [Google Scholar]
- 18.Zhao WH, Guo M, Duan B, Su LQ. Study on carrier time in Pulex irritans after infection of Yersinia pestis. China Trop Med. 2016;16:28–30. [Google Scholar]
- 19.Ayyadurai S, Sebbane F, Raoult D, Drancourt M. Body lice, Yersinia pestis orientalis, and Black Death. Emerg Infect Dis. 2010;16:892–893. doi: 10.3201/eid1605.091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massad E, Coutinho FA, Burattini MN, Lopez LF. The Eyam plague revisited: Did the village isolation change transmission from fleas to pulmonary? Med Hypotheses. 2004;63:911–915. doi: 10.1016/j.mehy.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 21.Monecke S, Monecke H, Monecke J. Modelling the black death. A historical case study and implications for the epidemiology of bubonic plague. Int J Med Microbiol. 2009;299:582–593. doi: 10.1016/j.ijmm.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Biraben JN. 1975. Les Hommes et la Peste en France et dans les Pays Européens et Méditerranéens. Tome I. La Peste dans l'Histoire (Mouton, Paris). French.
- 23.Carmichael AG. Plague and the Poor in Renaissance Florence. Cambridge Univ Press; Cambridge, UK: 1986. [Google Scholar]
- 24.Smith RS. Barcelona “Bills of Mortality” and population, 1457–1590. J Polit Econ. 1936;44:84–93. [Google Scholar]
- 25.Ferrán y Clua J, Viñas y Cusí F, Grau Rd. 1907. La Peste Bubónica: Memoria sobre la Epidemia Ocurrida en Porto en 1899 (Tipografia Sucesor de Sánchez, Barcelona). Spanish.
- 26.Creighton C. A History of Epidemics in Britain: From A.D. 664 to the Extinction of Plague. Cambridge Univ Press; Cambridge, UK: 1891. [Google Scholar]
- 27.Frandsen KE. The Last Plague in the Baltic Region 1709–1713. Museum Tusculanum Press; Copenhagen: 2010. [Google Scholar]
- 28.Alexander JT. Bubonic Plague in Early Modern Russia: Public Health and Urban Disaster. Oxford Univ Press; Oxford: 2003. [Google Scholar]
- 29.Burrell WH. Appendix V. To the Second Report on Quarantine: Report of Dr. W. H. Burrell on the Plague of Malta 1813. George E. Eyre and William Spottiswoode for Her Majesty’s Sationery Office; London: 1854. [Google Scholar]
- 30.The Advisory Committee appointed by the Secretary of State for India Reports on the plague investigations in India XXVIII. Additional experiments on the septicaemia in human plague, with an account of experiments on the infectivity of the excreta. J Hyg (Lond) 1908;8:221–235. doi: 10.1017/s0022172400003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans FC, Smith FE. The intrinsic rate of natural increase for the human louse, Pediculus humanus L. Am Nat. 1952;86:299–310. [Google Scholar]
- 32.Peacock AD. The louse problem at the Western Front. BMJ. 1916;1:784–788. doi: 10.1136/bmj.1.2892.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foucault C, et al. Oral ivermectin in the treatment of body lice. J Infect Dis. 2006;193:474–476. doi: 10.1086/499279. [DOI] [PubMed] [Google Scholar]
- 34.Tollenaere C, et al. Susceptibility to Yersinia pestis experimental infection in wild Rattus rattus, reservoir of plague in Madagascar. EcoHealth. 2010;7:242–247. doi: 10.1007/s10393-010-0312-3. [DOI] [PubMed] [Google Scholar]
- 35.Keeling MJ, Gilligan CA. Bubonic plague: A metapopulation model of a zoonosis. Proc Biol Sci. 2000;267:2219–2230. doi: 10.1098/rspb.2000.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keeling MJ, Gilligan CA. Metapopulation dynamics of bubonic plague. Nature. 2000;407:903–906. doi: 10.1038/35038073. [DOI] [PubMed] [Google Scholar]
- 37.Guernier V, et al. Fleas of small mammals on Reunion Island: Diversity, distribution and epidemiological consequences. PLoS Negl Trop Dis. 2014;8:e3129. doi: 10.1371/journal.pntd.0003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrion AL. Final report on a rat-flea survey of the city of San Juan, Porto Rico. Public Health Rep. 1932;47:193–201. [Google Scholar]
- 39.Bacot AW, Martin CJ. The respective influences of temperature and moisture upon the survival of the rat flea (Xenopsylla cheopis) away from its host. J Hyg (Lond) 1924;23:98–105. doi: 10.1017/s0022172400008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boegler KA, Graham CB, Johnson TL, Montenieri JA, Eisen RJ. Infection prevalence, bacterial loads, and transmission efficiency in Oropsylla montana (Siphonaptera: Ceratophyllidae) one day after exposure to varying concentrations of Yersinia pestis in blood. J Med Entomol. 2016;53:674–680. doi: 10.1093/jme/tjw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piarroux R, et al. Plague epidemics and lice, Democratic Republic of the Congo. Emerg Infect Dis. 2013;19:505–506. doi: 10.3201/eid1903.121542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisen RJ, et al. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kool JL. Risk of person-to-person transmission of pneumonic plague. Clin Infect Dis. 2005;40:1166–1172. doi: 10.1086/428617. [DOI] [PubMed] [Google Scholar]
- 44.Davis DE, Fales WT. The distribution of rats in Baltimore, Maryland. Am J Hyg. 1949;49:247–254. doi: 10.1093/oxfordjournals.aje.a119275. [DOI] [PubMed] [Google Scholar]
- 45.Seal SC, Bhatacharji LM. Epidemiological studies on plague in Calcutta. I. Bionomics of two species of ratfleas and distribution, densities and resistance of rodents in relation to the epidemiology of plague in Calcutta. Indian J Med Res. 1961;49:974–1007. [PubMed] [Google Scholar]
- 46.Patil A, Huard D, Fonnesbeck CJ. PyMC: Bayesian stochastic modelling in python. J Stat Softw. 2010;35:1–81. [PMC free article] [PubMed] [Google Scholar]
- 47.Gelman A, Rubin DR. A single series from the Gibbs sampler provides a false sense of security. In: Bernardo JM, et al., editors. Bayesian Statistics 4. Oxford Univ Press; Oxford: 1992. pp. 625–631. [Google Scholar]
- 48.Schwartz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 49.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 50.Diekmann O, Heesterbeek JA, Roberts MG. The construction of next-generation matrices for compartmental epidemic models. J R Soc Interface. 2010;7:873–885. doi: 10.1098/rsif.2009.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Didelot X, Whittles LK, Hall I. Model-based analysis of an outbreak of bubonic plague in Cairo in 1801. J R Soc Interface. 2017;14:20170160. doi: 10.1098/rsif.2017.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alsofrom DJ, Mettler FA, Jr, Mann JM. Radiographic manifestations of plaque in New Mexico, 1975–1980. A review of 42 proved cases. Radiology. 1981;139:561–565. doi: 10.1148/radiology.139.3.7232721. [DOI] [PubMed] [Google Scholar]
- 53.Laudisoit A, et al. Plague and the human flea, Tanzania. Emerg Infect Dis. 2007;13:687–693. doi: 10.3201/eid1305.061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratovonjato J, Rajerison M, Rahelinirina S, Boyer S. Yersinia pestis in Pulex irritans fleas during plague outbreak, Madagascar. Emerg Infect Dis. 2014;20:1414–1415. doi: 10.3201/eid2008.130629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.