Significance
The [PSI+] prion (infectious protein) is an amyloid (filamentous polymer) of the Sup35 protein, producing detrimental effects on yeast. We find that at their normal expression levels the core components of nonsense-mediated mRNA decay for mRNA quality control, Upf1p, Upf2p, and Upf3p, block the propagation of most new [PSI+] prion variants. The curing mechanism relies upon both Sup35p-binding by each Upf protein and by the trimeric Upf complex. Our results support the notion that normal protein–protein interactions prevent abnormal interactions.
Keywords: prion, antiprion system, [PSI+], Upf proteins, nonsense-mediated mRNA decay
Abstract
The yeast prion [PSI+] is a self-propagating amyloid of Sup35p with a folded in-register parallel β-sheet architecture. In a genetic screen for antiprion genes, using the yeast knockout collection, UPF1/NAM7 and UPF3, encoding nonsense-mediated mRNA decay (NMD) factors, were frequently detected. Almost all [PSI+] variants arising in the absence of Upf proteins were eliminated by restored normal levels of these proteins, and [PSI+] arises more frequently in upf mutants. Upf1p, complexed with Upf2p and Upf3p, is a multifunctional protein with helicase, ATP-binding, and RNA-binding activities promoting efficient translation termination and degradation of mRNAs with premature nonsense codons. We find that the curing ability of Upf proteins is uncorrelated with these previously reported functions but does depend on their interaction with Sup35p and formation of the Upf1p–Upf2p–Upf3p complex (i.e., the Upf complex). Indeed, Sup35p amyloid formation in vitro is inhibited by substoichiometric Upf1p. Inhibition of [PSI+] prion generation and propagation by Upf proteins may be due to the monomeric Upf proteins and the Upf complex competing with Sup35p amyloid fibers for available Sup35p monomers. Alternatively, the association of the Upf complex with amyloid filaments may block the addition of new monomers. Our results suggest that maintenance of normal protein–protein interactions prevents prion formation and can even reverse the process.
The yeast prion [PSI+] is a self-propagating amyloid of Sup35p (1–7). Sup35p normally functions as a soluble subunit of the translation termination factor (8, 9). However, its conversion to the prion amyloid aggregate sequesters Sup35p from the translation termination machinery, resulting in read-through of stop codons. [URE3] is similarly an amyloid prion of Ure2p (2, 10–14), a regulator of nitrogen catabolism (15), and [PIN+] is a prion of Rnq1p, an asparagine (N)- and glutamine (Q)-rich protein with unknown function (16–18).
A given prion protein sequence can become any of many distinct prion variants/strains, with different biological properties as a result of different amyloid structures, each quite stably propagating (19, 20). The yeast prion amyloids have a folded, parallel, in-register β-sheet architecture (21–24), a structure that can explain how prion proteins can template their conformation, resulting in the stable propagation of different prion variants (25, 26). These different prion variants act as alleles of a cytoplasmic protein-based gene.
The high frequency of toxic or even lethal variants of [PSI+] and [URE3] (27), and the rare occurrence in wild strains of even the mildest variants of these prions (28), indicates that these prions are detrimental to yeast and that the potential to develop or be infected by a prion is a risk with no reproducible compensating advantage other than the normal function of the prion domain (29, 30). Like the antipathogen systems against fungi, bacteria, and viruses, yeast should have evolved systems to prevent prion generation or to eliminate them or limit their pathogenic effects after they arise. Most of the known antiprion systems are closely related to molecular chaperones, well-characterized protein quality-control systems for dealing with protein aggregates (reviewed in ref. 26). Deletion of ribosome-associated Hsp70 chaperones Ssb1p and Ssb2p leads to elevated frequency of [PSI+] formation, either spontaneously or induced by Sup35p overproduction (31). Restored Ssbs did not eliminate [PSI+] variants arising in an ssb1Δ ssb2Δ strain, indicating that the normal level of Ssb1p and Ssb2p affects [PSI+] generation but not propagation (31). Sis1p is an Hsp40 that is necessary for cell growth (32) and for propagation of [PSI+], [URE3], and [PIN+] (33). Sis1p’s C-terminal domain is not necessary for cell growth in the absence of [PSI+] but becomes nearly essential in the presence of an otherwise mild [PSI+] (34). Thus, Sis1p prevents [PSI+] toxicity.
The disaggregating chaperone Hsp104 is both necessary for the propagation of [PSI+] and other amyloid-based yeast prions and cures [PSI+] on its overproduction (35, 36). Deletion or mutation (e.g., T160M) of the Hsp104 N-terminal domain eliminates the ability of overproduced Hsp104 to cure [PSI+] without affecting [PSI+] propagation, suggesting that these are different activities of Hsp104 (37). Indeed, the Hsp104 overproduction curing activity requires Sti1p and Hsp90, but neither affects [PSI+] propagation (38–40). This result was used to show that normal levels of WT Hsp104 cures many [PSI+] variants arising spontaneously in an hsp104T160M strain, and the frequency of spontaneous [PSI+] is >10-fold elevated in the hsp104T160M mutant (41). Like the curing by Hsp104 overproduction, the curing of [PSI+] by normal levels of Hsp104 is promoted by Sti1p and Hsp90.
Overproduction of either Btn2p or its paralogue Cur1p was found to cure the [URE3] prion (42). During the curing process, Btn2p colocalized with and collected Ure2p-GFP aggregates, suggesting that Btn2p sequesters prion aggregates, preventing their distribution to daughter cells (42, 43). Btn2p also partially colocalized with Sup35NM-GFP aggregates or huntingtin-like Q103-GFP aggregates, although [PSI+] was not cured by Btn2p or Cur1p overproduction (42), and Btn2p can collect other nonprion aggregates (44, 45). It was then found that almost all [URE3] variants isolated in a btn2Δcur1Δ strain were cured by restoring just normal levels of either Btn2p or Cur1p, or both (46). These results indicate that normal levels of Btn2p and Cur1p cure most [URE3] variants arising in the cell.
Most recently, Siw14p, a pyrophosphatase specific for 5PP-IP5 (5-diphosphoinositol pentakisphosphate) in the inositol polyphosphate synthesis pathway, was found to play a role as an antiprion factor (47). Restored normal levels of Siw14p eliminate about half of the [PSI+] variants generated in a siw14Δ strain by limiting the levels of some inositol poly-/pyrophosphates. Further study of this phenomenon revealed that inositol poly-/pyrophosphates are needed for nearly all [PSI+] variants (47). These studies indicate that yeast has antiprion systems that constantly act to block the formation of prions and to prevent the propagation of most prions that do arise. These systems are working in normal cells whose physiology is not distorted by overproduction or deficiency of any components.
Here, we report that normal levels of the Upf proteins (Upf1p, Upf2p, and Upf3p), components of the nonsense-mediated mRNA decay (NMD) apparatus (48), can cure [PSI+] variants arising in the absence of each of these proteins. Further, we present evidence that prion curing involves binding to Sup35p and formation of the Upf complex. Throughout this study, we were careful to distinguish the known effects of upf mutations on the translational effects of [PSI+] from the effect of upf mutations on the propagation of [PSI+] as a genetic element.
Results
Screening for [PSI+] Prion-Curing Factors.
We screened for [PSI+] prion-curing factors by a genetic method without protein overproduction (47). Pools of the MATa knockout collection were transformed with the centromeric plasmid p1520 (47), which carries LEU2 as a marker for yeast, the [PSI+]-suppressible nonsense allele ura3-14 (49), and the Sup35p-prion domain (NM) controlled by the GAL1 promoter. [PSI+] was induced by growth for 24 h in galactose to overproduce Sup35NM, and cells were plated on −Ura plates with dextrose to select Ura+ ([PSI+]) clones. These Ura+ clones were crossed with an isogenic WT strain, BY4742, to complement the knockout mutation in each clone. Ura− diploids were identified, and the corresponding original Ura+ clone was tested for curability by transient growth on guanidine, an inhibitor of Hsp104 that specifically cures prions. Guanidine-curable candidates were not simply recessive suppressors of the ura3-14 nonsense mutation complemented by mating with the WT strain but had to be [PSI+] clones. As shown below, this was confirmed by the transfer by cytoplasmic mixing (cytoduction) of the suppressor phenotype. From each Ura+ candidate that produced Ura− diploids and whose Ura+ phenotype was curable by growth on guanidine, DNA was extracted, and the deleted gene was identified by PCR of the bar-code region of KanMX. Among 30 candidates, upf1::kanMX was found eight times, and upf3::kanMX was found nine times. UPF1 and UPF3 encode components of the NMD apparatus.
A Normal Level of Upf1p Can Cure [PSI+] Variants Isolated in a upf1Δ Strain.
We cured [PSI+] from one upf1Δ isolate, replaced [PIN+] by cytoduction (cytoplasmic transfer), and then induced [PSI+] formation by overproduction of Sup35NM. Ura+ isolates were mated with either the WT [psi−] strain BY4742 or a upf1Δ [psi−] strain. Almost all diploids formed with the WT were uniformly Ura−, but not the diploids with the upf1Δ [psi−] strain (Fig. 1). Because ura3-14 is expected to be a target of NMD, we took special care to determine whether [PSI+] was lost from the diploids formed in these tests or if its phenotype was merely unapparent.
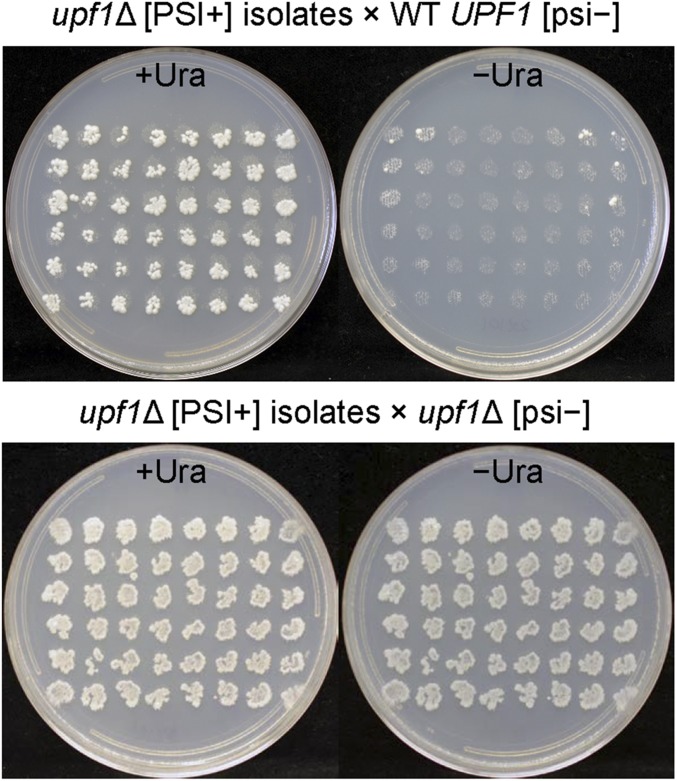
Fig. 1.
Most [PSI+] prion variants isolated in a upf1Δ strain are lost in the presence of the WT allele of UPF1. [PSI+] isolates in the upf1Δ strain MS114 were mated for 1 d on YPAD with either isogenic WT MS173 or the upf1Δ [psi−] strain MS177 and were replica-plated to minimal medium with and without uracil. The presence of p1520 (pCEN LEU2 ura3-14) in all strains enables scoring [PSI+]. (Upper) Diploids formed with WT are almost all Ura− as a result of the elimination of [PSI+]. Several isolates are not efficiently cured by mating with isogenic WT (Lower). Diploids formed with upf1Δ are Ura+, indicating stable maintenance of [PSI+].
Meiotic segregation of the upf1Δ/+ Ura− heterozygotes showed mostly 4 Ura−:0 Ura+ segregation. Only a few upf1Δ segregants showed a Ura+ or a very weak Ura+ phenotype; the majority (50–100%, average 76%) of the upf1Δ segregants were Ura−. Because upf1Δ segregants were largely Ura−, [PSI+] must have been lost in most of the heterozygous diploids before sporulation. In contrast, if, after mating, the diploids were immediately sporulated to prevent loss of [PSI+] in the diploids, the segregation was 2 Ura− G418sensitive:2 Ura+ G418resistant (37 tetrads). The combined meiotic analysis implies that the [PSI+] genetic element is eliminated by Upf1p, not that its phenotype is merely disguised. Based on their loss of [PSI+] on restoration of normal levels of Upf1p, these variants (all guanidine-curable and mitotically stable) are denoted “Upf1p-sensitive,” [PSI+u1s].
To confirm loss of [PSI+u1s] in WT strains, cytoduction (transfer of cytoplasm; see Methods) was performed using [PSI+u1s] upf1Δ strains as donors and WT or upf1Δ [psi−] strains as recipients. As expected, almost all cytoductants showed a Ura− phenotype in WT recipients and a Ura+ phenotype in upf1Δ recipients (Table 1). To verify that Ura− cytoductants from upf1Δ [PSI+u1s] strains to WT strains had indeed lost [PSI+u1s], reverse-cytoductions were conducted using Ura− cytoductants as donors. The majority of reverse-cytoductants were Ura− in the upf1Δ recipient, indicating that a large fraction of [PSI+u1s] variants was really lost in the WT recipients (Table 1). The appearance of a minority of Ura+ cytoductants on the return of cytoplasm to the upf1Δ host indicates that this [PSI+] variant was mostly, but not completely, lost in the WT host. This varies with prion isolate (see Tables S2 and S3, in which it can be seen that, of 24 [PSI+] isolates in a upf1∆ host, two showed retention of [PSI+] in a significant fraction of WT cells).
Table 1.
Confirmation of elimination of [PSI+] variants by normal level of Upf1p
| Donor | Recipient | Cytoductants | |
| Ura+ | Total | ||
| upf1Δ [PSI+u1s2] | WT ρo | 0 | 35 |
| upf1Δ [PSI+u1s5] | 0 | 40 | |
| upf1Δ [PSI+u1s6] | 0 | 36 | |
| upf1Δ [PSI+u1s2] | upf1Δ ρo | 37 | 38 |
| upf1Δ [PSI+u1s5] | 39 | 40 | |
| upf1Δ [PSI+u1s6] | 30 | 32 | |
| WT-2* Ura− | upf1Δ ρo | 7 | 146 |
| WT-5* Ura− | 33 | 150 | |
| WT-6* Ura− | 23 | 160 | |
| upf1Δ-2† Ura+ | upf1Δ ρo | 38 | 40 |
| upf1Δ-5† Ura+ | 37 | 40 | |
| upf1Δ-6† Ura+ | 39 | 40 | |
Upf1p-sensitive [PSI+] variants ([PSI+u1s]) were transferred by cytoduction (cytoplasmic mixing) from isolates 2, 5, and 6 into the WT strain MS173 and into the upf1Δ strain MS177. Cytoductants of each were used as reverse-cytoduction donors into upf1Δ strain MS114.
Ura− WT cytoductants from first cytoduction.
Ura+ upf1Δ cytoductants from first cytoduction.
It is well known that upf1 and upf3 mutations enhance the expression of genes, such as ura3-14, that have a premature termination codon, both by allowing longer survival of their mRNA (no NMD) and by decreasing the efficiency of termination, allowing more read-through of the premature termination codon (reviewed in ref. 48). However, the reverse cytoductants are Ura−, while the original [PSI+u1s] isolates are Ura+, both being upf1Δ, showing that [PSI+] can be scored in a upf1Δ strain and that [PSI+u1s] was lost in the WT strains rather than its phenotype simply being disguised. Additionally, the loss of [PSI+u1s] in WT strains was examined by investigating the formation of fluorescent structures. Sup35NM-GFP was expressed in Ura− WT cytoductants and Ura+ upf1Δ cytoductants to detect prion aggregates. Like the WT [psi−][PIN+] and upf1Δ [psi−][PIN+] strains, single or multiple dots rarely appeared in the Ura− WT cytoductants but were seen in many cells of normal [PSI+] strains and Ura+ upf1Δ cytoductants (Fig. S1 and Table S1). Taken together, these several lines of evidence prove that many [PSI+] variants are lost in the presence of UPF1 in WT strains, independent of the known effect of Upf1p on the phenotypic expression of the prion.
To confirm that Upf1p plays a role in eliminating [PSI+u1s], pRS313 (CEN vector) or pM25 (pRS313-UPF1), a single-copy plasmid with the UPF1 gene controlled by its native promoter, was transformed into [PSI+u1s] upf1Δ isolates. Transformants were selected in the presence of uracil and were replica-plated to plates lacking uracil. Almost none of the transformants carrying pM25 could grow on medium lacking uracil, indicating that [PSI+u1s] was eliminated by normal levels of Upf1p (Fig. S2). As shown in Table 2, three independent [PSI+u1s] variants examined were almost completely eliminated by the restoration of UPF1, while standard strong [PSI+] ([PSI+s]) or weak [PSI+] ([PSI+w]) variants were almost unaffected. More than 50 Ura− subclones that had lost pM25 were examined for uracil auxotrophy by replica-plating. Nearly all Ura− subclones remained Ura− after losing pM25, indicating that [PSI+u1s] variants are cured by pUPF1 and that the UPF1 gene does not merely affect the Ura+ phenotype (Table 2).
Table 2.
Restored normal level of Upf1p can eliminate most Upf1p-sensitive [PSI+] variants
| [PSI+] variant | Transformant clones | Phenotype of subclones from Ura− clones that lost pUPF1 | ||||
| Vector | pUPF1 | |||||
| Ura+ | Total | Ura+ | Total | Ura+ | Ura− | |
| [PSI+s] | 263 | 265 | 235 | 265 | 0 | 50 |
| [PSI+w] | 245 | 247 | 255 | 280 | 0 | 55 |
| [PSI+u1s2] | 271 | 276 | 15 | 217 | 1 | 54 |
| [PSI+u1s5] | 255 | 275 | 11 | 308 | 1 | 50 |
| [PSI+u1s6] | 209 | 218 | 9 | 235 | 2 | 54 |
upf1Δ strains MS277, MS281, MS2, MS5, and MS6 (top to bottom) carrying [PSI+s], [PSI+w], and [PSI+u1s] were transformed with the CEN plasmid pRS313 or with the same plasmid carrying UPF1 under its native promoter (pM25 = pUPF1). Transformants were selected in the presence of uracil and were replica-plated to a plate lacking uracil. More than 200 transformant clones were investigated in each case. Subclones of Ura− transformants that had lost pM25 were tested for uracil auxotrophy by replica-plating. The results, summed and shown in the rightmost column, show that [PSI+u1s] had been eliminated by pUPF1.
Overexpression of Hsp104 cures all [PSI+] variants (35), and overexpression of Btn2p or Cur1p cures all [URE3] variants (42). However, we found that overexpression of Upf1p or Upf3p from a GAL promoter did not cure [PSI+] from strain 779-6A carrying a conventional [PSI+s] variant.
Nonsense Suppression Phenotypes Produced by [PSI+u1s] or upf1Δ or Other [PSI+]s Are Distinguishable, Depending on the Assay Marker.
In [PSI+] cells, most of Sup35p is in filaments, producing inefficient translation termination and readthrough of termination codons, such as ade1-14, ade2-1, and ura3-14 (for this study). We compared the phenotypes of [PSI+u1s] variants with those of [PSI+s] and [PSI+w] using strains generated by cytoduction into the background of the bank strains and carrying p1520 (with ura3-14). In the white/red color assay on adenine-limited half-strength yeast extract/peptone/dextrose (YPD) medium, the upf1Δ [PSI+s] cells show essentially the same pink color as WT [PSI+s] cells, but the former grow dramatically better on −Ura plates than the latter (Fig. 2). The ade1-14 allele has a UGA terminator at codon 244 of the 306-residue protein (50), while ura3-14 was constructed to have the same terminator at codon 18 of the 267-residue Ura3 protein (49). NMD is far less effective if the codon is close to the 3′ end of the ORF than if it is near the beginning (51). This could explain the larger effect of upf1Δ on the Ura− phenotype than on the Ade− phenotype. When compared in a upf1Δ strain, the efficiency of ura3-14 nonsense suppression by [PSI+u1s] is slightly lower than that produced by [PSI+s] or [PSI+w] (Fig. 2). In addition, the efficiency of readthrough produced by [PSI+] alone is significantly higher than that resulting from upf1Δ in a [psi−] strain, and the strongest readthrough results from the combination in upf1Δ [PSI+] cells (Fig. 2). Although upf1Δ [psi−] cells barely grew after 3-d incubation on a plate lacking uracil, allowing them a further 7 d of growth made possible a distinction from WT [psi−] cells. Therefore, 3-d incubation was determined as a checkpoint for ura3-14 nonsense suppression by [PSI+] or [PSI+u1s] for this study.
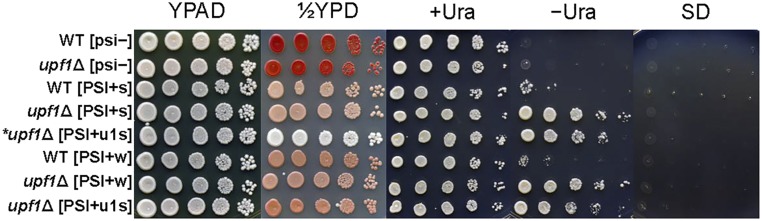
Fig. 2.
Comparison of nonsense-suppression phenotypes of [PSI+u1s] or other [PSI+]s in WT or upf1Δ hosts. Tenfold dilutions of cells of strains MS109, MS114, MS224, MS277, MS2, MS225, MS281, and MS286 (top to bottom) carrying p1520 with ura3-14 were plated on rich medium and minimal medium (SD) with and without uracil. [PSI+s] indicates a strong variant and [PSI+w] indicates a weak variant. In the fifth row, asterisked upf1Δ [PSI+u1s] is a bank-background original isolate without the ade1-14 allele. For photographs, cells on rich medium (YPAD and 1/2YPD) were grown at 30 °C for 2 d, and cells on minimal medium were grown at 30 °C for 3 d.
Upf1p-Sensitive [PSI+] Variants Have Seed Numbers Similar to Normal [PSI+].
The prion seed numbers (also called “propagons”) of [PSI+u1s] variants were investigated using a method developed previously (52). Low concentrations of guanidine specifically inhibit Hsp104’s filament-cleaving activity (53–55), preventing the generation of new seeds. Preexisting seeds segregate until there is one seed per cell. By measuring the number of prion-carrying cells in the colony, the number of preexisted seeds in the founder cell can be estimated (52). Using two different concentrations of guanidine, the seed number of [PSI+u1s] variants was counted. Both experiments showed that [PSI+u1s] variants have seed numbers not significantly different from those in normal [PSI+] variants (Table 3). This result contrasts with the Btn2p/Cur1p antiprion systems that cure [URE3]. Btn2p/Cur1p-hypersensitive [URE3] variants have substantially lower seed numbers (46), suggesting that these two different antiprion systems use distinct mechanisms to deal with yeast prions.
Table 3.
Seed number of [PSI+] variants
| Prion variant | Seed no. ± SD |
| Experiment 1 (4 mM guanidine HCl) | |
| upf1Δ [PSI+s] | 145.5 ± 59.3 |
| upf1Δ [PSI+w] | 71.8 ± 17.3 |
| upf1Δ [PSI+u1s2] | 45.5 ± 2.4 |
| upf1Δ [PSI+u1s5] | 106.0 ± 97.0 |
| upf1Δ [PSI+u1s6] | 59.0 ± 6.6 |
| Experiment 2 (5 mM guanidine HCl) | |
| upf1Δ [PSI+s] | 89.6 ± 15.4 |
| upf1Δ [PSI+w] | 47.1 ± 18.9 |
| upf1Δ [PSI+u1s2] | 40.6 ± 21.4 |
| upf1Δ [PSI+u1s5] | 66.2 ± 13.6 |
| upf1Δ [PSI+u1s6] | 63.4 ± 6.8 |
upf1Δ strain MS114 carrying the indicated [PSI+] variants was streaked for single colonies on YPAD medium containing 4 mM guanidine HCl (experiment 1) or 5 mM guanidine HCl (experiment 2). Single colonies were suspended in sterilized water and plated on −Ade plates. Ade+ colonies are a relative measure of the number of prion seeds in the cell founding the colony.
Spontaneous and Induced [PSI+] Generation Is Elevated in upf Mutants.
To test whether the normal level of Upf proteins can block many [PSI+] variants as they arise, the frequency of spontaneous and induced generation of [PSI+] was investigated in WT strains and in NMD-deficient upf1Δ, upf2Δ, and upf3Δ mutants. For these experiments, the same [PIN+], a prion of Rnq1, was transferred to all 5 mM guanidine-treated recipient strains. For spontaneous generation of [PSI+], the frequency of [PSI+] clones per 107 cells from upf1Δ or upf2Δ strains was elevated sevenfold and fivefold, respectively (Table 4). Interestingly, although all strains carried the same [PIN+], the upf3Δ mutant did not show elevated spontaneous [PSI+] generation. In [PSI+] generation induced by Sup35NM overproduction, upf1Δ, upf2Δ, and upf3Δ mutants produced 16-, 17-, and 13-fold more [PSI+] clones, respectively, than the WT strain (Table 4).
Table 4.
De novo generation of [PSI+] variants is enhanced in upf mutants
| Host | Spontaneous [PSI+], total Ura+ colonies per 107 cells | Induced [PSI+], total Ura+ colonies per 105 cells |
| WT | 3.7 ± 1.0 | 8.00 ± 3.4 |
| upf1Δ | 28.3 ± 7.3 | 130.3 ± 32.4 |
| upf2Δ | 19.0 ± 5.4 | 132.2 ± 22.5 |
| upf3Δ | 2.7 ± 1.5 | 105.7 ± 16.4 |
Strains MS327, MS330, MS333, and MS336 (top to bottom), used in this experiment, carry the same [PIN+]. For spontaneous [PSI+], cells were grown for 2 d in 2% glucose liquid culture at 30 °C, and 107 yeast cells were spread on standard synthetic complete medium (SC) plates without uracil. For induced [PSI+] formation, strains carrying p1520 with SUP35NM driven by a galactose-inducible promoter were grown for 2 d in 2% galactose/2% raffinose medium at 30 °C, and 105 yeast cells were spread on SC plates without uracil. The average number of colonies formed after 5 d of incubation at 30 °C is shown (the data from six independent experiments were combined). Numbers represent Ura+ colonies with [PSI+] that were confirmed by GuHCl curability. The number of Ura+ colonies ±SD is shown.
The [PSI+] variants arising at high frequency in upf2Δ or upf3Δ strains (Table 4) were further tested by cytoduction to determine whether normal levels of Upf2p or Upf3p can cure them (Table 5). As was found for [PSI+u1s] (Table 1), cytoduction to WT recipients resulted in Ura− cytoductants, but cytoduction into another strain with the same upf mutation allowed maintenance of [PSI+]. Returning cytoplasm from the WT Ura− cytoductants to the original upf2Δ or upf3Δ host produced mostly Ura− back-cytoductants, indicating that the Ura− phenotype in the WT strain was due to the loss of [PSI+], not to merely affecting the phenotype (Table 5). Thus, these are Upf2p-sensitive ([PSI+u2s]) and Upf3p-sensitive ([PSI+u3s]) [PSI+] prion variants. In addition, 12 [PSI+] clones each from those spontaneously generated and induced in the upf1Δ host (Table 4) were analyzed in the same way as our original isolates in Table 1. Cytoplasm was transferred to WT recipients, resulting in nearly all cytoductants being Ura− (Tables S2 and S3). Sample WT Ura− cytoductants were then used as donors to return cytoplasm to upf1Δ cells, with nearly all cytoductants again being Ura− (Tables S2 and S3), a pattern exactly similar to the previous cytoduction results using the original [PSI+u1s] isolates as a donor (Table 1) and indicating that both spontaneous and induced [PSI+] clones are indeed mostly Upf1p-sensitive [PSI+] prion variants.
Table 5.
[PSI+] variants isolated in upf2Δ and upf3Δ cells are cured by restoring Upf2p and Upf3p, respectively
| Donor | Recipient | Cytoductants | |
| Ura+ | Total | ||
| upf2Δ [PSI+u2s1] | WT ρo* | 0 | 43 |
| upf2Δ [PSI+u2s2] | 0 | 40 | |
| upf2Δ [PSI+u2s4] | 0 | 41 | |
| upf2Δ [PSI+u2s1] | upf2Δ ρo | 39 | 41 |
| upf2Δ [PSI+u2s2] | 35 | 38 | |
| upf2Δ [PSI+u2s4] | 36 | 39 | |
| upf3Δ [PSI+u3s1] | WT ρo† | 0 | 38 |
| upf3Δ [PSI+u3s2] | 0 | 40 | |
| upf3Δ [PSI+u3s3] | 1 | 35 | |
| upf3Δ [PSI+u3s1] | upf3Δ ρo | 39 | 40 |
| upf3Δ [PSI+u3s2] | 34 | 40 | |
| upf3Δ [PSI+u3s3] | 37 | 40 | |
| WT* | upf2Δ ρo | 2 | 40 |
| WT* | 1 | 40 | |
| WT* | 2 | 35 | |
| WT† | upf3Δ ρo | 1 | 40 |
| WT† | 1 | 40 | |
| WT† | 2 | 40 | |
[PSI+] variants isolated in upf2Δ strain MS62 and upf3Δ strain MS65 (Table 4) were transferred by cytoduction into the WT strain MS327, upf2Δ strain MS308, and upf3Δ strain MS68. Ura− cytoductants of each were used as reverse-cytoduction donors. The results show the loss of [PSI+] from the WT host in each case, meaning that these variants can be referred to as Upf2p-sensitive ([PSI+u2s]) or Upf3p-sensitive ([PSI+u3s]).
Ura− cytoductants from upf2Δ [PSI+u2s] to WT.
Ura− cytoductants from upf3Δ [PSI+u3s] to WT.
Does the elevated frequency of [PSI+] in upfΔ strains result from the nonsense-suppression effect of these mutations allowing more residual growth on –Ura plates and thus the selection of [PSI+] from a larger effective population? The suggested mechanism would predict that the [PSI+] variants arising in a upf mutant would be largely the same kind as variants arising in a WT cell, just more of them because there was a bigger population of cells from which they came. In fact, nearly all the variants that arise are cured by mating with a WT strain or by cytoduction into a WT strain. Thus, this mechanism cannot explain the observed results.
Upf1p-Sensitive [PSI+] Variants Are Stabilized in upf2Δ or upf3Δ Cells.
Upf1p acts with Upf2 and Upf3 in NMD, forming a trimeric Upf1p–Upf2p–Upf3p complex (i.e., the Upf complex) necessary for activity (56–58). We investigated whether [PSI+u1s] variants can propagate in upf2Δ or upf3Δ strains despite the presence of Upf1p. As is evident from Table 6, almost all cytoductants into upf2Δ and upf3Δ recipient strains showed a Ura+ phenotype, indicating that the curing of [PSI+u1s] variants requires all three Upf proteins. In each case, cytoduction from Ura+ cytoductants into WT recipients confirmed that [PSI+u1s] variants in the upf2Δ or upf3Δ strains had not changed and were eliminated by the presence of normal levels of the three Upf proteins. Thus, [PSI+u1s] variants can propagate if any one of the Upf proteins is absent (Table 6).
Table 6.
Upf1p-sensitive [PSI+] variants are stabilized in upf2Δ or upf3Δ cells
| Donor | Recipient | Cytoductants | |
| Ura+ | Total | ||
| upf1Δ [PSI+u1s2] | WT ρo | 0 | 35 |
| upf1Δ [PSI+u1s5] | 0 | 40 | |
| upf1Δ [PSI+u1s6] | 0 | 37 | |
| upf1Δ [PSI+u1s2] | upf1Δ ρo* | 45 | 45 |
| upf1Δ [PSI+u1s5] | 33 | 34 | |
| upf1Δ [PSI+u1s6] | 31 | 33 | |
| upf1Δ [PSI+u1s2] | upf2Δ ρo† | 31 | 31 |
| upf1Δ [PSI+u1s5] | 30 | 30 | |
| upf1Δ [PSI+u1s6] | 25 | 25 | |
| upf1Δ [PSI+u1s2] | upf3Δ ρo‡ | 30 | 30 |
| upf1Δ [PSI+u1s5] | 28 | 28 | |
| upf1Δ [PSI+u1s6] | 38 | 38 | |
| upf1Δ [PSI+u1s]* | WT ρo | 1 | 90 |
| upf2Δ [PSI+u1s]† | WT ρo | 0 | 90 |
| upf3Δ [PSI+u1s]‡ | WT ρo | 0 | 90 |
Upf1p-sensitive [PSI+] variants ([PSI+u1s]) were transferred by cytoduction from strain MS2, MS5, and MS6 into the [psi−] ρo WT strain MS173 and upf1Δ strain MS177, upf2Δ strain MS62, and upf3Δ strain MS65. Ura+ Cytoductants of each were used as reverse-cytoduction donors.
Ura+ cytoductants from upf1Δ [PSI+u1s] to upf1Δ [psi−].
Ura+ cytoductants from upf1Δ [PSI+u1s] to upf2Δ.
Ura+ cytoductants from upf1Δ [PSI+u1s] to upf3Δ.
Curing of [PSI+u1s] by Normal Levels of Upf1p Is Uncorrelated with Its Functions in Translation Termination and Nonsense-Mediated mRNA Degradation.
Upf1p is a multifunctional protein consisting of a cysteine- and histidine-rich zinc-finger domain (CH domain) at the N-terminal region and a C-terminal Helicase domain (59–61). These two domains engage in various functions, including ATP hydrolysis, ATP binding, RNA binding, and functional interaction with other factors required for activating NMD. To identify which function(s) of Upf1p is important for eliminating [PSI+u1s], plasmids expressing various mutants of Upf1p were generated using pM25 as a backbone (Fig. S3A and Table S4).
These plasmids were transformed into a upf1Δ strain carrying [PSI+u1s], and transformants were replica-plated to plates lacking uracil. Consistent with the results shown in Table 2 and Fig. S2, the WT allele of UPF1 could eliminate [PSI+u1s] from most cells, but the empty vector could not. Except for C72S and RR793KK, each of the mutant plasmids produced less efficient curing than pUPF1-WT, suggesting that the curing ability of Upf1p is impaired by amino acid substitutions of Upf1p (Table 7 and Fig. S3B); C72S and RR793KK mutations were previously reported to not affect the original functions of Upf1p (59, 61). Mutations in the CH domain (C62Y, C84S, and C125S) more severely affected curing ability than those in Helicase domain (K436Q, DE572AA, TR800AA, and RR793AA) (Table 7 and Fig. S3B).
Table 7.
Mutations in the CH domain or the helicase region of Upf1p can impair elimination of Upf1p-sensitive [PSI+] variants
| UPF1 alleles in plasmid | Average % Ura+ transformant clones, ± SD | Phenotype of subclones from Ura− clones that lost plasmid | |
| Ura+ | Ura− | ||
| Vector | 99.9 ± 0.1a | ||
| pUPF1-WT | 4.7 ± 1.4b | 2 | 114 |
| pUPF1-C62Y | 92.4 ± 3.5c | 1 | 84 |
| pUPF1-C72S | 2.5 ± 2.2b | 0 | 109 |
| pUPF1-C84S | 90.7 ± 5.7c | 1 | 97 |
| pUPF1-C125S | 86.1 ± 3.3c | 0 | 88 |
| pUPF1-K436Q | 70.4 ± 7.1d | 1 | 121 |
| pUPF1-DE572AA | 71.8 ± 2.9d | 1 | 94 |
| pUPF1-TR800AA | 68.2 ± 4.5d | 1 | 113 |
| pUPF1-RR793AA | 51.9 ± 2.0e | 0 | 78 |
| pUPF1-RR793KK | 4.4 ± 1.0b | 0 | 81 |
Primary [PSI+u1s] upf1Δ isolates (MS2, MS5, and MS6) and three other [PSI+u1s] isolated in the upf1Δ strain MS114 (MS318, MS319, and MS320) were transformed with the CEN vector pRS313 or the vector carrying UPF1 (pM25) or UPF1 mutated by amino acid substitution(s) under its native promoter. Substituted amino acid(s) and their positions are shown. Transformants were selected in the presence of uracil and were replica-plated to a plate lacking uracil. More than 400 transformant clones were investigated in each case. The average percent of Ura+ transformants, calculated using six independent experiments, ± SD, is shown. Numbers with different letters are significantly different at a P value of <0.05 based on the Tukey test. Ura− subclones that had lost each plasmid were tested for uracil auxotrophy by replica-plating. Their failure to become Ura+ again shows that the prion was cured.
Both C62Y and C84S mutations dramatically reduced [PSI+] curing ability by ∼90%, but the former stabilizes nonsense-containing mRNA (like upf1Δ), and the latter does not (like WT UPF1) (60). The K436Q mutation produces strong nonsense suppression, while DE572AA resembles the WT (59), but these mutants have similar curing ability, reduced by 70.4% and 71.8%, respectively (Table 7). In this sense, there is no clear correlation of [PSI+]-curing activity with these or other known activities of Upf1p (Table S4).
Upf1p Colocalizes with [PSI+] Prion Aggregates in Vivo.
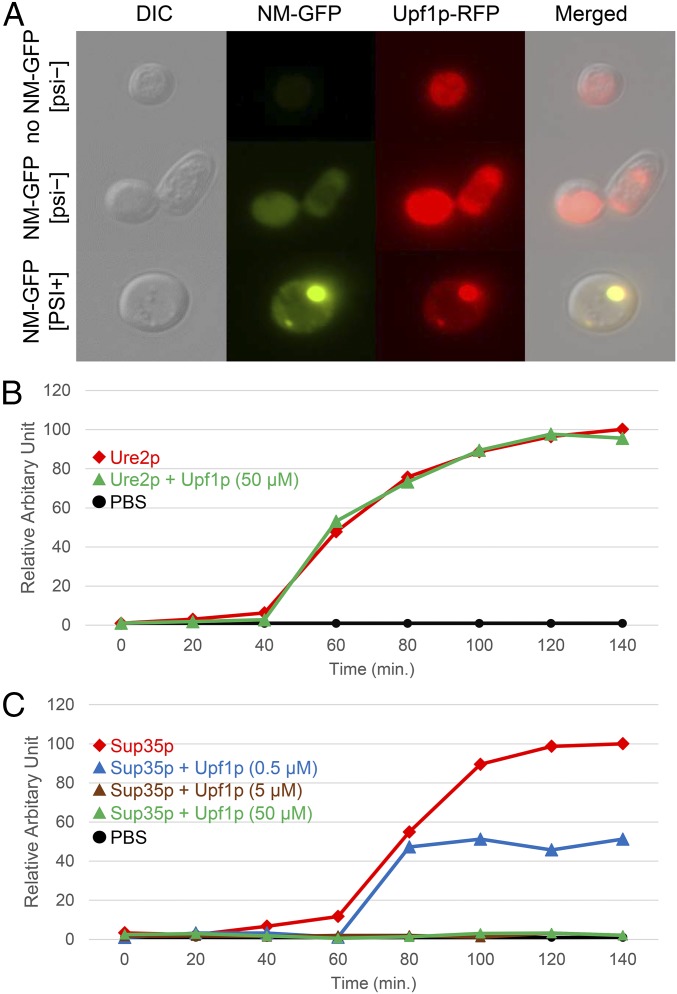
Direct binding of Upf1p and soluble Sup35p or Sup35p aggregates has been previously identified in extracts (62). To examine the interaction between Upf1p and both forms of Sup35p in vivo, strain MS307, which has RFP-tagged Upf1p, was transformed with pSL1066, a centromeric plasmid with CUP1-promoted Sup35NM-GFP, to decorate [PSI+] aggregates (63). After 48 h growth in medium containing 50 µM CuSO4, the localization of each protein was investigated using confocal microscopy. As expected, Upf1p-RFP showed cytoplasmic localization without Sup35NM-GFP aggregates (Fig. 3A). In cells with Sup35NM-aggregates, Upf1p-RFP colocalized with Sup35NM-GFP aggregates and was not detected free in the cytoplasm (Fig. 3A, Bottom), indicating that Upf1p colocalizes with [PSI+] prion aggregates in vivo. Since deficiency of Upf1p is known to produce nonsense suppression [both by allowing mRNAs to survive longer and by other mechanisms (64)], it is possible that the association of Upf1p with Sup35p aggregates in [PSI+] cells contributes to the nonsense-suppression phenotype of this prion.
Fig. 3.
Upf1p colocalizes with Sup35p prion aggregates in vivo and has an inhibitory effect on the assembly of Sup35p amyloid in vitro. (A) Colocalization of Upf1p-RFP and Sup35NM-GFP aggregates in strain MS307 was observed after 48 h of Sup35NM-GFP induction from pSL1066 and visualized by fluorescence confocal microscopy. (Top) Sup35NM-GFP not expressed in [psi−] cells. (Middle) Sup35NM-GFP expressed in [psi−] cells. (Bottom) Sup35NM-GFP expressed in [PSI+] cells. (Magnification: 1,500×.) (B) Kinetics of Ure2p amyloid formation with and without Upf1p. Thioflavin-T was used for monitoring amyloid formation. Purified Ure2p (5 µM) was incubated with 50 µM FLAG peptide (red line) or 50 µM Upf1p (green line), with constant shaking at 37 °C. (C) Kinetics of Sup35p amyloid formation with and without Upf1p. Purified Sup35p (5 µM) was incubated with 50 µM FLAG peptide (red line) or Upf1p (0.5 µM, blue line; 5 µM, brown line; or 50 µM, green line). The brown, green, and black lines are densely packed near the x axis. B and C show representative individual experiments; data were normalized to the maximum fluorescence level of Ure2p or Sup35p incubated alone, which was set as 100%.
Upf1p Inhibits Sup35p Amyloid Formation but Not Ure2p Amyloid Formation in Vitro.
The effect of Upf1p on the assembly of Sup35p into amyloid was tested in vitro to understand how normal levels of Upf1p cure [PSI+]. For this experiment, full-length recombinant Sup35p and Ure2p were expressed with His6 tags in Escherichia coli, and FLAG-tagged Upf1p was purified in yeast using the published method (62). Amyloid formation of Sup35p or Ure2p was monitored using thioflavin T binding. As expected, even a 10-fold excess of Upf1p did not affect Ure2p amyloid formation (Fig. 3B, green line). However, a decinormal amount of Upf1p was sufficient to slow the growth of Sup35p amyloid by half (Fig. 3C, blue line), and Sup35p amyloid did not assemble at all in presence of an equal amount or a 10-fold excess of Upf1p (Fig. 3C, brown line and green line, respectively). As internal control, we tested whether FLAG peptide has an effect on amyloid formation or whether Upf1p can form amyloid by itself. However, we did not detect a substantial difference between Sup35p alone and Sup35p mixing with FLAG peptide, and there was no fluorescent signal from Upf1p alone. This indicates that Upf1p has an inhibitory effect on Sup35p amyloid formation in vitro.
Both Sup35p-Binding Activity and Upf Complex Formation Are Required for Efficient Elimination of [PSI+] Prion Variants.
For the full function of NMD, both Upf complex formation and binding between each of the Upf proteins and Sup35p are necessary (58, 61, 64). The CH domain of Upf1p plays roles in both Sup35p binding and Upf complex formation by interaction with Upf2p (62, 64, 65). Upf2p, another core NMD component, has a highly acidic domain (AC, amino acids 886–938) for Sup35p binding and a Upf1p-interacting domain (U1I, amino acids 939–1,089) for Upf complex formation (58, 64, 66). To distinguish which of these functions is important for [PSI+] prion curing, a series of UPF2 plasmids lacking the AC domain, the U1I domain, or both, were generated (Fig. S4A) and transformed into upf2Δ strains carrying [PSI+] variants. Transformants were then replica-plated on medium with and without uracil (Fig. S4B).
Loss of Sup35p-binding activity by deletion of the AC domain decreased the ability to cure [PSI+u1s] or [PSI+u2s] to half that of WT Upf2p, and blocking Upf complex formation by deletion of the U1I domain had a similar effect (Table 8). Deletion of both domains completely eliminated the curing ability for [PSI+u1s] or [PSI+u2s], similar to the empty vector (Table 8 and Fig. S4B). This result suggests that both Sup35p binding of Upf2p and formation of the Upf complex are required for efficient curing of the [PSI+] variants and also supports our previous result that each of the Upf proteins is needed for efficient curing of [PSI+] variants that arise in the absence of one Upfp.
Table 8.
Both the Sup35p-binding activity of Upf2p and Upf complex formation are required for efficient curing of [PSI+] prion variants in a upf2Δ strain
| Prion isolate | UPF2 alleles in plasmid | Average % of Ura+ transformant clones | Phenotype of subclones from Ura− clones that lost plasmid | |
| Ura+ | Ura− | |||
| [PSI+u1s] | Vector | 96.0 ± 3.0a | 0 | 20 |
| pUPF2-WT | 4.6 ± 1.9b | 2 | 52 | |
| pUPF2-ΔAC | 55.5 ± 2.5c | 2 | 48 | |
| pUPF2-ΔU1I | 55.2 ± 1.8c | 0 | 41 | |
| pUPF2-ΔACU1I | 89.4 ± 2.7a | 0 | 45 | |
| [PSI+u2s] | Vector | 95.4 ± 4.3a | 0 | 20 |
| pUPF2-WT | 5.1 ± 1.0b | 0 | 36 | |
| pUPF2-ΔAC | 56.6 ± 1.6c | 1 | 44 | |
| pUPF2-ΔU1I | 48.5 ± 4.5c | 1 | 50 | |
| pUPF2-ΔACU1I | 93.9 ± 3.8a | 0 | 41 | |
upf2Δ strains (MS321, MS322, MS323) carrying [PSI+u1s] (cytoductants from Table 6) and upf2Δ strains carrying three [PSI+u2s] (primary isolates in upf2Δ strain MS62 from Table 5) (MS299, MS300, and MS302) were transformed with the CEN vector pRS313 or with the vector carrying the UPF2 or UPF2 mutants under its native promoter. Deletion mutants of pUPF2 were denoted as ΔAC lacking the highly acidic domain (amino acids 886–938), ΔU1I lacking the Upf1p-interacting domain (amino acids 939–1,089), and ΔACU1I for the double mutant. Transformants were selected in the presence of uracil and were replica-plated to a plate lacking uracil. More than 200 transformant clones were investigated in each case. The average percent of Ura+ transformants, calculated using six independent experiments, ±SD, is shown. Numbers with different letters are significantly different at a P value of <0.05 based on the Tukey test. Ura− subclones that had lost each plasmid were tested for uracil auxotrophy by replica-plating to confirm that the Ura− transformants had lost [PSI+].
Discussion
Using a screen for antiprion factors active against [PSI+] without overproduction, we found that some variants are cured by Upf proteins. By isolating many [PSI+] variants in a upf1Δ strain and then restoring the normal level of Upf1p (by mating with an isogenic WT strain, by cytoduction into a UPF1 strain, or by transformation with the UPF1 gene on a single-copy plasmid), we found that most [PSI+] variants (Upf1p-sensitive, u1s) are eliminated by restoration of normal levels of Upf1p. Although the known effects of Upf proteins on suppression necessarily complicated the situation, our genetic analysis shows that the loss of the [PSI+] prion is occurring, rather than simply effects on the detection system. In each case, following exposure to Upf1p, the cytoplasm was returned to the upf1∆ state (by meiosis, reverse cytoduction, or loss of the UPF1 plasmid), and the phenotype of [PSI+] was gone, showing that the prion had been lost when the cells had Upf1p. This curing occurs in a WT strain, indicating that Upf1p has an antiprion action. We find that [PSI+] arises at greater frequency in upf mutants. This is not simply the detection of weaker [PSI+] variants, as replacing the corresponding UPF gene cures nearly all the [PSI+] variants. Ssb1p and Ssb2p, Hsp70 family chaperones central to the ribosome-associated chaperone system, lower the frequency of [PSI+] generation, but their restoration to normal levels does not cure the [PSI+] variants arising in their absence, suggesting that the Upf proteins are not acting by affecting Ssb protein activity.
From yeasts to humans, NMD is a conserved surveillance mechanism targeting for degradation cytoplasmic mRNA containing premature termination codons (PTCs) (48, 61). Efficient activation of NMD requires the conserved core components Upf1p, Upf2p, and Upf3p. The upf mutants also led to inefficient translation termination (readthrough of PTCs), in part by allowing the PTC-containing mRNAs to survive longer (59, 60). Yeast and human Upf proteins interact with each other and with both the eukaryotic translation termination factors eRF1 (Sup45p) and eRF3 (Sup35p) (48, 61, 62, 64, 67). Upf1p, the central actor in NMD, is a multifunctional protein classified as a superfamily I RNA helicase with ATPase, ATP-binding, RNA-binding, and Upf2p-binding activities defined by a series of point mutants (59, 60, 65, 68). We used these mutations to examine which functions of Upf1p can affect [PSI+] curing ability. In general, all substitutions led to a decrease in curing ability to some degree (Table 8 and Fig. S3), but there was no close correlation between [PSI+] curing ability and nonsense suppression or PTC-containing mRNA decay.
Upf1p was detected in Sup35p aggregates in extracts of [PSI+] cells (62), and we observed colocalization of Upf1p and [PSI+] aggregates in vivo (Fig. 3A), suggesting that Upf1p can interact with both the soluble and amyloid forms of Sup35p in yeast cells. Although there could be substantial difference(s) between the test tube situation and living cells, our in vitro Sup35p amyloid formation assay indicates that Upf1p has an inhibitory effect on the assembly of Sup35p amyloid (Fig. 3C).
What is the mechanism of the curing of [PSI+] by the Upf proteins? The frequency with which [PSI+] arises is generally increased by the mutation of any of the UPF genes (Table 4), and all three are needed for curing the sensitive [PSI+]s (Table 6). Point mutations in UPF1 show no clear correlation of [PSI+]-curing ability with NMD efficiency or nonsense-suppression activity or with the helicase, ATPase, ATP-binding, or RNA-binding activities of Upf1p. However, there is some weak correlation with Upf2p-binding activity (Table S4). These lines of evidence all point to the formation of the complex of Upf proteins with the Sup35p-Sup45p termination factor as key to the curing effect. More striking are the effects of Upf2p mutants defective in binding to Sup35p or to Upf1p: Each lowers curing activity by half, and the combined double mutant completely eliminates [PSI+] curing. Moreover, Upf1p alone at a 1/10 molar concentration blocks amyloid formation by Sup35p in vitro. Accordingly, both the formation of the Upf1-2–1-3/Sup35–45 complex and monomeric Upf proteins may compete with the amyloid filaments for Sup35p monomers, inhibiting fiber growth enough to allow other cellular systems to destroy the prion. Alternatively, the association of the Upf complex and Upf proteins with the filaments may block growing points (presumably the filament ends) sufficiently to prevent fiber growth. Our proposed mechanisms are reminiscent of a negative effect of another Sup35p-binding protein, Sup45p, on [PSI+], namely, that overproduction of Sup45p inhibits [PSI+] induction by overproduction of Sup35p but does not affect prion propagation (69).
The association of Upf proteins with Sup35p amyloid filaments in extracts of [PSI+] cells and the colocalization of [PSI+] filaments with Upf1p in vivo (Fig. 3) suggest that the Sup35p amyloid depletes Upf1p (and probably other Upfs) from the available pool. It is likely that this Upf protein deficiency effect contributes to the nonsense-suppression phenotype of [PSI+] strains.
Prion variants can be cured without protein overproduction by Btn2p or Cur1p for [URE3-1] (46) and by Hsp104 or Siw14p for [PSI+] (41, 47). Normal levels of the Ssb chaperones largely block [PSI+] generation (31). These previously reported antiprion systems and the current study indicate that the yeast cell has an exquisite antiprion defense system, specialized for different prions, to repress the generation of new prion variants and block their propagation once they have arisen. While the Upf system is primarily directed at NMD, it certainly also serves as a strong and natural barrier to [PSI+] prion variants.
Although we think of these systems as somewhat analogous to antiviral or antibacterial systems, there is a fundamental difference, in that viruses and bacteria represent an outside invader, whereas prions are an inside-the-cell risk. This is illustrated by the Upf proteins blocking prion formation or curing prions by their normal interactions with Sup35p.
It is remarkable that the Ssb1 system lowers [PSI+] generation frequency by ∼10-fold (31), the Hsp104 antiprion system apparently lowers [PSI+] frequency by ∼13-fold (41), and the Upf antiprion effect described here has a 5- to 10-fold or greater effect on [PSI+]. If these systems work independently, then the true frequency of formation of the [PSI+] prion may be quite high. Our studies of Btn2p, Cur1p, Hsp104, Siw14p, and the Upf proteins have revealed classes of prions that are not able to propagate in normal strains because of the respective antiprion systems but are able to arise when those systems are not active. It is an assumption that these curable variants arise in wild strains, justified by the ease with which infectious yeast prions arise from recombinant proteins in the absence of other factors (e.g., refs. 6, 7, 14, 70, and 71). Not included in these variants are the known frankly lethal variants of [PSI+] and the near-lethal variants of [PSI+] and [URE3], which are rarely studied (27). It remains likely that there are other lethal variants of yeast prions for which a permissive condition has not yet been discovered, making their observation impossible so far.
The existence of lethal yeast prion variants (27), the rarity of even the mildest yeast prion variants in wild strains (28), and evidence for the existence of several antiprion systems, including the present study, strongly suggest that yeast prions are not generally advantageous to the cell. The present data suggest that a potentially prion-forming protein will generally be stabilized by maintenance of its normal protein–protein interactions. This phenomenon is somewhat similar to the stabilization of transthyretin against amyloid formation by analogs of thyroid hormone that bind transthyretin even better than the hormone itself, an effect now being used in therapy of transthyretin amyloidosis (72). Enhancing normal interactions may prove to be a general route to deterring the abnormal interactions that constitute amyloidoses of humans (and yeast).
Methods
Nomenclature.
Yeast prions are shown in brackets to indicate they are nonchromosomal genes, e.g., [PSI+] or [URE3]. Specific types of prion variants are indicated within the brackets, e.g., “[PSI+u1s]” for Upf1-sensitive [PSI+].
Strains and Media.
Strains used in this study are listed in Table 9. Gene-disruption mutants were generated by PCR-amplifying yeast genomic DNA of the corresponding strains from the Saccharomyces cerevisiae knockout collection (73). Media used were as described by Sherman (74). Induction of GAL1-promoted genes was conducted using galactose/raffinose-containing medium as previously described (46). The ade1-14 allele from strain 74-D694 (31) was amplified by PCR, transformed into strain 5335 (derived from BY4741) selecting for cotransformation by pRS313. Mixed transformants were plated on 1/2 YPD (0.5% yeast extract, 2% peptone, 2% dextrose, 2% agar), and red clones were isolated and confirmed by PCR and sequencing.
Table 9.
Strains used in this study
| Strain | Genotype | Source |
| BY4741/MS157 | MATa ura3 leu2 his3 met15 [psi−][PIN+] | (77) |
| BY4742/MS317 | MATα ura3 leu2 his3 lys2 [psi−][PIN+] | (77) |
| MS2,5,6 | MATa ura3 leu2 his3 met15 upf1::kanMX [PSI+u1s (2, 5, 6)] primary [PSI+] isolates | This study |
| 5335 | MATα ura3 leu2 his3 lys2 kar1Δ15 [psi−][PIN+] | (47) |
| MS173 | MATα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 [psi−][PIN+] | This study |
| MS177 | MATα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 upf1::kanMX [psi−][PIN+] | This study |
| MS109, MS327 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 [psi−][PIN+] | This study |
| MS114, MS330 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf1::kanMX [psi−][PIN+] | This study |
| MS224 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 [PSI+s] | This study |
| MS225 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 [PSI+w] | This study |
| MS277 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf1::kanMX [PSI+s] | This study |
| MS281 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf1::kanMX [PSI+w] | This study |
| MS286 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf1::kanMX [PSI+u1s] | This study |
| MS333 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf2::kanMX [psi−][PIN+] | This study |
| MS336 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf3::kanMX [psi−][PIN+] | This study |
| MS299, 300, 302 | MATα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 upf2::kanMX [PSI+u2s] | This study |
| MS303, 304, 305 | MATα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 upf3::kanMX [PSI+u3s] | This study |
| MS308 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf2::kanMX [psi−][PIN+] | This study |
| MS68 | MATa ura3 leu2 his3 met15 ade1-14 kar1Δ15 upf3::kanMX [psi−][PIN+] | This study |
| MS62 | MATα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 upf2::kanMX [psi−][PIN+] | This study |
| MS65 | MATα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 upf3::kanMX [psi−][PIN+] | This study |
| MS318, 319, 320 | MATa ura3 leu2 his3 met15 ade1-14 upf1::kanMX [PSI+u1s] | This study |
| MS321 | MATa ura3 leu2 his3 met15 ade1-14 upf2::kanMX [PSI+u1s] | This study |
| MS307 | Matα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 UPF1-mCherry [psi−][PIN+] | This study |
| MS339 | MATα ura3 leu2 his3 lys2 ade1-14 kar1Δ15 UPF1-mCherry [PSI+][PIN+] | This study |
| BJ2168 | MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407 gal2 | Gift of A. Jacobson, University of Massachusetts Medical School, Worcester, MA |
Except for strain BJ2168, all strains were isogenic to BY4741, the knockout bank parent.
Plasmids.
Plasmids used in this study are listed in Table 10. UPF1, with 500 bp upstream of the ORF, was amplified and ligated into pRS313 (CEN HIS3) cut with BamHI and XhoI forming pM25. Plasmids expressing Upf1p with amino acid replacements were generated from pM25 using the Q5 Site-Directed Mutagenesis Kit (New England BioLabs). UPF2 and truncated UPF2s, with 500 bp upstream of the ORF, were amplified and ligated into pRS313 using Gibson Assembly Master mix (New England BioLabs). SUP35NM-GFP was excised from pH1329 and ligated into pH770 (2 µ HIS3 PGAL1) cut with BamHI and XhoI, forming pM18.
Table 10.
Plasmids used in this study
| Name | Description | Source |
| p1520 | pCEN LEU2 URA3-14 PGAL1:SUP35NM | (47) |
| pRS313 | CEN HIS3 | (78) |
| pM25 | pRS313 PUPF1:UPF1 | This study |
| pH770 | pRS423 PGAL1 | Gift of H. Edskes, National Institute of Diabetes and Digestive and Kidney Diseases, NIH, Bethesda |
| pM18 | pRS423 PGAL1:SUP35NM-GFP | This study |
| pM28 | pM25-UPF1-K486Q | This study |
| pM29 | pM25-UPF1-DE572AA | This study |
| pM30 | pM25-UPF1-TR800AA | This study |
| pM31 | pM25-UPF1-RR793AA | This study |
| pM32 | pM25-UPF1-RR793KK | This study |
| pM33 | pM25-UPF1-C62Y | This study |
| pM34 | pM25-UPF1-C72S | This study |
| pM35 | pM25-UPF1-C84S | This study |
| pM36 | pM25-UPF1-C125S | This study |
| pSL1066 | pCEN URA3 PCUP1:SUP35NM-GFP | (63) |
| pM27 | pFA6a-link-yomCherry-HIS5 | Addgene 44841 |
| pKT-41 | pET17b-URE2 Full-length | (11, 13) |
| pM41 | pET13b-SUP35 Full-length | Gift of F. Shewmaker, Uniformed Services University of the Health Sciences, Bethesda, MD |
| pM46 | pG1-FLAG-UPF1 | A. Jacobson, University of Massachusetts Medical Center, Worcester, MA (62) |
| pM54 | pRS313 PUPF2:UPF2 | This study |
| pM55 | pM54 UPF2-ΔAC | This study |
| pM56 | pM54 UPF2-ΔU1I | This study |
| pM57 | pM54 UPF2-ΔACU1I | This study |
Cytoduction.
Mutants in kar1 can mate, but nuclear fusion does not occur. Subsequent cell divisions result in separation of the two parental nuclei, but the cytoplasm of each daughter cell is a mixture of the cytoplasm of the parents. Although this process is fundamentally symmetric, it is treated as the transfer of cytoplasmic genes from one strain (the donor) to another (the recipient). The recipient’s mitochondrial DNA is eliminated by growth in the presence of ethidium bromide, and the transfer of ρ+ (as measured by ability to grow on glycerol) from the ρ+ donor to the ρo recipient shows the transfer of cytoplasm from donor to recipient. Donor and recipient strains were isogenic to the knockout bank strains BY4741 and BY4742. Donors of induced [PSI+] variants and recipients carried the plasmid p1520 (CEN LEU2 ura3-14) bearing PGAL1-SUP35NM (47). Recipients were made ρ0 by growing on medium containing 25 µg/mL ethidium bromide. Donor and recipient cells were mixed in distilled water with an ∼3× excess of donor cells and were spotted on a YPAD (74) plate to allow mating. After 7 h incubation at 30 °C, the cell mixture was streaked for single colonies on medium selecting against the donor. After 3 d incubation at 30 °C, single colonies were replica-plated to yeast extract, peptone, glycerol (YPG) medium, to medium selective for diploids, or to medium lacking uracil. Clones growing on YPG medium, but not those growing on diploid-selection medium, were cytoductants. Clones that grew on the −Ura plate propagated [PSI+].
Measuring [PSI+] Prion Seed Number.
Following Cox et al. (52), freshly grown [PSI+] cells were streaked for single colonies on YPAD medium containing 4 mM or 5 mM guanidine HCl. Single colonies with the underlying agar block were suspended in distilled water and plated on medium lacking uracil. Colonies showing the Ura+ phenotype were counted and were assumed to represent [PSI+] prion seeds of the cell founding that colony. For each strain, at least 10 individual colonies were tested. A sample of Ura+ colonies was checked for guanidine curability.
Amyloid Assembly and Expression of Recombinant Proteins.
Fluorescence of thioflavin-T (Sigma-Aldrich) was used to measure Sup35p and Ure2p amyloid formation as described (42, 75). To form amyloid, Sup35p and Ure2p were diluted from purified stock solutions to 5 µM in 300 µL of 1× PBS and were shaken at 200 rpm at 37 °C. Aliquots were sampled at the specific time points, further mixed with thioflavin-T (final concentration 50 µM), and incubated at room temperature for 10 min without shaking. Fluorescence signal measurement was performed using a SupraMax M5 (Molecular Devices) with an excitation of 420 nm and emission of 495–500 nm.
Proteins used in the assay were expressed in E. coli strain BL21-CodonPlus (DE3)-RIPL (Agilent Technologies). Full-length Sup35p and Ure2p were prepared from pM41 and pKT-41, respectively. Cells were grown at 37 °C in LB medium to an optical density of 0.6 at 600 nm and were induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside. The collected cells were lysed by sonication in buffer containing 20 mM NaH2PO4·H2O, 500 mM NaCl, and 20 mM imidazole (His-tag column-binding buffer). After cell debris was removed by centrifugation, proteins in the supernatant were purified using His GraviTrap (GE Healthcare Life Sciences). The eluted proteins were desalted using a PD-10 desalting column (GE Healthcare Life Sciences). The final proteins were eluted in 1× PBS buffer. FLAG-tagged full-length Upf1p was expressed in yeast strain BJ2168 using pG1-FLAG-Upf1p, as described previously (76). Total yeast protein was extracted using CelLytic Y (Sigma-Aldrich), and Upf1p was further purified using ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich), following the manufacturer’s manual. Purified protein was eluted in 3×FLAG peptides (Sigma-Aldrich) containing 1× PBS buffer. Protein concentrations were determined by Bradford assay or absorbance at 280 nm. In all cases, protein aliquots were frozen at −80 °C before use.
Supplementary Material
Acknowledgments
We thank Sue Liebman (University of Nevada), Allan Jacobson (University of Massachusetts), and Frank Shewmaker (Uniformed Services University for Health Sciences) for strains and plasmids. This work was supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717495115/-/DCSupplemental.
References
- 1.Cox BS. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 2.Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 3.King C-Y, et al. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 5.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 6.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 8.Stansfield I, et al. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolova L, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 10.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masison DC, Maddelein M-L, Wickner RB. The prion model for [URE3] of yeast: Spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 14.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 18.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 19.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 21.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baxa U, et al. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 23.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci USA. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorkovskiy A, Thurber KR, Tycko R, Wickner RB. Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc Natl Acad Sci USA. 2014;111:E4615–E4622. doi: 10.1073/pnas.1417974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickner RB, et al. Yeast prions: Proteins templating conformation and an anti-prion system. PLoS Pathog. 2015;11:e1004584. doi: 10.1371/journal.ppat.1004584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci USA. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly AC, Shewmaker FP, Kryndushkin D, Wickner RB. Sex, prions, and plasmids in yeast. Proc Natl Acad Sci USA. 2012;109:E2683–E2690. doi: 10.1073/pnas.1213449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickner RB, et al. Yeast prions: Structure, biology, and prion-handling systems. Microbiol Mol Biol Rev. 2015;79:1–17. doi: 10.1128/MMBR.00041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci USA. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirkland PA, Reidy M, Masison DC. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics. 2011;188:565–577. doi: 10.1534/genetics.111.129460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 36.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moosavi B, Wongwigkarn J, Tuite MF. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI(+)] prion but not for prion propagation. Yeast. 2010;27:167–179. doi: 10.1002/yea.1742. [DOI] [PubMed] [Google Scholar]
- 39.Reidy M, Masison DC. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol Cell Biol. 2010;30:3542–3552. doi: 10.1128/MCB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorkovskiy A, Reidy M, Masison DC, Wickner RB. Hsp104 disaggregase at normal levels cures many [PSI+] prion variants in a process promoted by Sti1p, Hsp90, and Sis1p. Proc Natl Acad Sci USA. 2017;114:E4193–E4202. doi: 10.1073/pnas.1704016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kryndushkin DS, Shewmaker F, Wickner RB. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008;27:2725–2735. doi: 10.1038/emboj.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanneganti V, Kama R, Gerst JE. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol Biol Cell. 2011;22:1648–1663. doi: 10.1091/mbc.E10-11-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kryndushkin D, Ihrke G, Piermartiri TC, Shewmaker F. A yeast model of optineurin proteinopathy reveals a unique aggregation pattern associated with cellular toxicity. Mol Microbiol. 2012;86:1531–1547. doi: 10.1111/mmi.12075. [DOI] [PubMed] [Google Scholar]
- 46.Wickner RB, Bezsonov E, Bateman DA. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc Natl Acad Sci USA. 2014;111:E2711–E2720. doi: 10.1073/pnas.1409582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickner RB, Kelly AC, Bezsonov EE, Edskes HK. [PSI+] prion propagation is controlled by inositol polyphosphates. Proc Natl Acad Sci USA. 2017;114:E8402–E8410. doi: 10.1073/pnas.1714361114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He F, Jacobson A. Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu Rev Genet. 2015;49:339–366. doi: 10.1146/annurev-genet-112414-054639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manogaran AL, Kirkland KT, Liebman SW. An engineered nonsense URA3 allele provides a versatile system to detect the presence, absence and appearance of the [PSI+] prion in Saccharomyces cerevisiae. Yeast. 2006;23:141–147. doi: 10.1002/yea.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell. 2001;7:1121–1130. doi: 10.1016/s1097-2765(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 51.Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 52.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: A possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- 54.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung G, Jones G, Masison DC. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc Natl Acad Sci USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 57.He F, Brown AH, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–170. [PMC free article] [PubMed] [Google Scholar]
- 58.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kervestin S, Jacobson A. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Czaplinski K, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
- 64.Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He F, Ganesan R, Jacobson A. Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast. Mol Cell Biol. 2013;33:4672–4684. doi: 10.1128/MCB.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 67.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serdar LD, Whiteside DL, Baker KE. ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat Commun. 2016;7:14021. doi: 10.1038/ncomms14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derkatch IL, Bradley ME, Liebman SW. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc Natl Acad Sci USA. 1998;95:2400–2405. doi: 10.1073/pnas.95.5.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel BK, Liebman SW. “Prion-proof” for [PIN+]: Infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson SM, Connelly S, Fearns C, Powers ET, Kelly JW. The transthyretin amyloidoses: From delineating the molecular mechanism of aggregation linked to pathology to a regulatory-agency-approved drug. J Mol Biol. 2012;421:185–203. doi: 10.1016/j.jmb.2011.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 74.Sherman F. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink GR, editors. Academic; San Diego: 1991. pp. 3–21. [Google Scholar]
- 75.Reidy M, Sharma R, Roberts BL, Masison DC. Human J-protein DnaJB6b cures a subset of Saccharomyces cerevisiae prions and selectively blocks assembly of structurally related amyloids. J Biol Chem. 2016;291:4035–4047. doi: 10.1074/jbc.M115.700393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 77.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 78.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.