Significance
Microbial communities living on and within plants and animals contribute to host function. How host evolution shapes associated microbial communities, and in turn, how these microbes affect the ecology of their hosts is relatively unknown. Here, we demonstrate that evolution occurring across plant species affects root microbial diversity and composition. Greater similarity in root microbiota among host plant species leads to reduced plant performance through negative soil feedbacks. Additionally, drought shifts the composition of root microbiomes, where changes in the relative abundance of specific bacterial taxa are associated with increased drought tolerance of plants. Our work highlights the potential role of host-associated microbial communities in mediating interactions between hosts and their biotic and abiotic environment.
Keywords: root microbiome, plant–soil feedback, plant drought, plant microbiome, host microbial ecology
Abstract
Across plants and animals, host-associated microbial communities play fundamental roles in host nutrition, development, and immunity. The factors that shape host–microbiome interactions are poorly understood, yet essential for understanding the evolution and ecology of these symbioses. Plant roots assemble two distinct microbial compartments from surrounding soil: the rhizosphere (microbes surrounding roots) and the endosphere (microbes within roots). Root-associated microbes were key for the evolution of land plants and underlie fundamental ecosystem processes. However, it is largely unknown how plant evolution has shaped root microbial communities, and in turn, how these microbes affect plant ecology, such as the ability to mitigate biotic and abiotic stressors. Here we show that variation among 30 angiosperm species, which have diverged for up to 140 million years, affects root bacterial diversity and composition. Greater similarity in root microbiomes between hosts leads to negative effects on plant performance through soil feedback, with specific microbial taxa in the endosphere and rhizosphere potentially affecting competitive interactions among plant species. Drought also shifts the composition of root microbiomes, most notably by increasing the relative abundance of the Actinobacteria. However, this drought response varies across host plant species, and host-specific changes in the relative abundance of endosphere Streptomyces are associated with host drought tolerance. Our results emphasize the causes of variation in root microbiomes and their ecological importance for plant performance in response to biotic and abiotic stressors.
The discovery that macroscopic organisms host unique assemblages of microorganisms has the potential to transform our understanding of ecology and evolution (1). In plants and animals, associated microbiomes contribute to host nutrition, development, and immunity (2–4), yet how they scale up to influence host ecological function and performance is largely unknown. For example, associated microbiota may alter the interactions between hosts and their environment. Here, we address how host plant evolution over macroevolutionary timescales shapes the assembly of root microbiomes, and in turn, how root microbiota mitigate biotic and abiotic environmental stressors experienced by host plants.
Land plants have formed symbioses with microorganisms since their colonization of terrestrial environments (5). Interactions between plants and microbes continue to benefit plants by increasing the acquisition of nutrients, producing growth hormones, and defending against enemies (6). Root microbiota can also reduce plant performance by competing for limited nutrients and attacking plants as pathogens (7). Recent work (8, 9) shows that plant roots assemble two distinct microbial compartments (i.e., microbiomes) from the pool of soil microbial diversity: the rhizosphere (microbes surrounding roots) and the endosphere (microbes within roots). Root microbiome assembly is a multistep process shaped by both soil type and host differences (6, 10). However, our understanding of how variation among host species shapes endosphere and rhizosphere assembly remains limited (6, 11–13), yet is essential for understanding how root microbiota contribute to the ecology and performance of their hosts.
Plants must contend with numerous environmental stressors throughout their lifetime. Competition between plants for shared resources is an important biotic stressor shaping both ecological and evolutionary outcomes (14, 15). Soil microbes have long been recognized as key components to plant competition (16, 17). For example, plants can indirectly compete with one another through recruitment of soil microbes (18), where microbial recruitment by one plant can feed back to affect the performance of a second plant. Competitive interactions among plant species mediated by these so-called “plant–soil feedbacks” (PSF) are known to affect fundamental terrestrial ecosystem processes, such as community assembly and succession, plant invasions, and primary productivity (19–22). The biotic drivers of PSF are not well understood but likely include the recruitment of assemblages of root microbiota across host plant species.
Drought represents one of the most important abiotic stressors that plants face in both natural and managed systems, negatively affecting plant growth and productivity worldwide (23–25). Due to their sessile nature, plants must employ a broad repertoire of phenotypic mechanisms to mitigate drought stress, including life history, morphological, physiological, and molecular changes (26, 27). Emerging evidence suggests that soil microbes may play an important yet poorly understood role in plant drought tolerance. For example, soil microbes can intercept hormones in plants leading to a dampened stress response to drought (28, 29), and drought-induced shifts in soil microbial communities can reduce the negative fitness effects of drought (30). Recent work shows that drought also shifts the composition of root microbial communities in numerous grass species (31, 32). However, whether variation in the diversity or composition of host plant root microbiota contributes to plant drought tolerance is unknown.
Here, we perform a comparative root microbiome study, characterizing the assembly of the endosphere and rhizosphere compartments of the root microbiome across phylogenetically diverse angiosperm species. We coupled our comparative study with manipulative experiments to understand the ecological function of the root microbiome. Specifically, we investigated how the root microbiome across a diverse set of host plant species mitigates biotic and abiotic stressors (SI Appendix, Fig. S1). Our study sought to answer four questions: (i) How do endosphere and rhizosphere microbiomes differ in diversity and composition across 30 phylogenetically diverse host plant species? (ii) Does evolutionary divergence among host plant species affect the assembly of the endosphere and rhizosphere microbiome? (iii) Does variation in the root microbiome between host plant species affect indirect competitive interactions via PSFs? (iv) Does the root microbiome influence drought tolerance across host plant species? Our results provide evidence of how evolution over long timescales shapes the root microbiome, and how root microbiota influence plant performance in response to variation in biotic and abiotic components of the environment.
Results
Endosphere and Rhizosphere Microbiomes Differ in Diversity and Composition Across Host Plant Species.
We grew 30 plant species that have diverged for up to 140 My (Fig. 1A and SI Appendix, Table S1). Plants were grown from surface-sterile seeds in a live soil inoculum collected from a naturalized field site where all species cooccur (Koffler Scientific Reserve). We measured a suite of morphological, physiological and performance traits from every plant (n = 10 per species) (SI Appendix, Table S2). After 16 wk, we partitioned root samples from each plant into endosphere and rhizosphere compartments (8, 9), extracted total DNA, and characterized the bacterial community by sequencing the V4 region of the 16S rRNA gene using Illumina MiSeq (SI Appendix, Fig. S1). We assembled quality-filtered reads into error-corrected amplicon sequence variants (ASVs) using DADA2 v1.4.0 (33), which represent unique bacterial taxa. We analyzed the effects of host plant species and root compartment on the diversity and composition of bacterial communities, as well as the abundance of individual bacterial taxa.
Fig. 1.
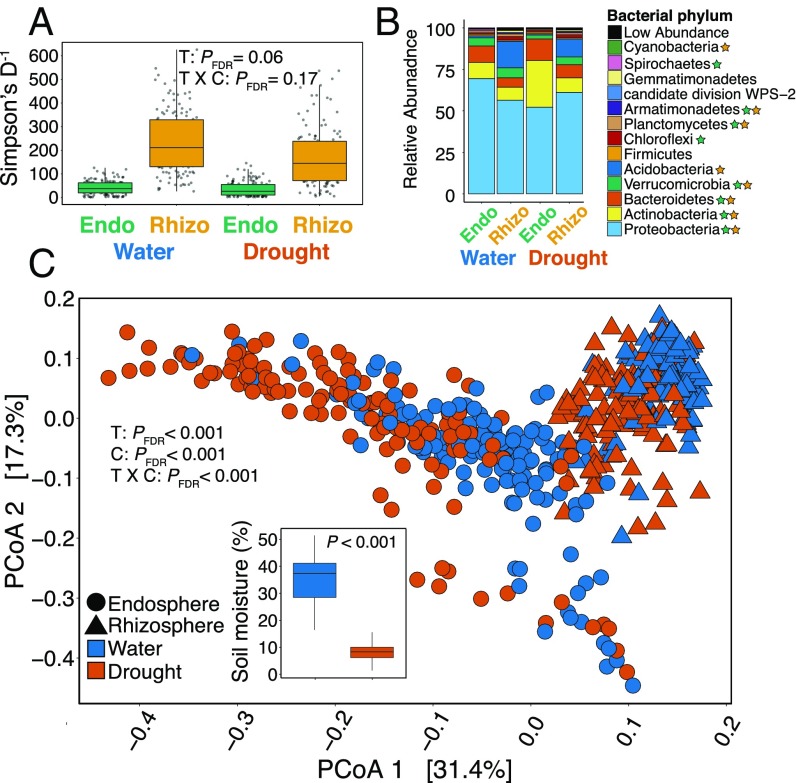
The diversity and composition of endosphere and rhizosphere compartments across plant species. (A) The endosphere exhibited less than one-quarter of the diversity found in the rhizosphere [F(1, 56) = 64.62, PFDR < 0.001, P-value adjusted using the FDR]. (B) The abundance of bacterial phyla were significantly affected (GLM: PFDR < 0.05) by compartment (black star) and host plant species (green star, endosphere; yellow star, rhizosphere). (C) Endosphere diversity exhibited greater variation across host plants than rhizosphere diversity (χ2 = 17.72, PFDR < 0.001). Endosphere diversity was also correlated with the underlying plant phylogeny, while rhizosphere diversity was not. (D) Plant species varied more in the composition of their endosphere versus rhizosphere compartments (χ2 = 20.06, PFDR < 0.001). Mantel tests revealed a significant correlation between endosphere (but not rhizosphere) compositional similarity and phylogenetic relatedness.
Across plant species, the rhizosphere exhibited higher diversity and greater evenness in abundance than the endosphere [Simpson’s D−1 mean SE: rhizosphere, 202 1.8, endosphere, 38 8.2, F(1, 56) = 64.62, P < 0.001; evenness: rhizosphere, 0.32 0.01, endosphere, 0.13 0.01, F(1, 56) = 73.89, P < 0.001] (Fig. 1A and SI Appendix, Fig. S2 and Table S3). We quantified microbiome community composition using weighted UniFrac distances with principal coordinates analysis and found clear differences in the composition of endosphere and rhizosphere compartments (SI Appendix, Fig. S3 and Table S4). Nearly 90% of bacterial phyla and 55% of bacterial ASVs exhibited significant differential abundance between endosphere and rhizosphere compartments [generalized linear model (GLM): PFDR < 0.05 after false-discovery rate (FDR) correction] (SI Appendix, Fig. S4A). In the endosphere, Actinobacteria and Bacteroidetes exhibited higher relative abundance, while Acidobacteria were significantly reduced (Fig. 1B). Additionally, we found a higher number of ASVs that were unique to the endosphere (65 ASVs) versus those that were only found in the rhizosphere (46 ASVs) or live bulk soil (8 ASVs) (SI Appendix, Fig. S5).
Our comparative framework uncovered larger effects of host plant species on endosphere than rhizosphere compartments (Fig. 1 C and D and SI Appendix, Figs. S6 and S7 and Tables S3 and S4). Host species varied much more in their endosphere (Simpson’s D−1 range: 6–87; SE: 8.2) than rhizosphere diversity [range: 111–315; SE: 1.8; Levene’s test: F(1, 58) = 18.55, P < 0.001] (Fig. 1C and SI Appendix, Table S3). Similarly, host plant species explained 40% of the total variation in endosphere composition [PERMANOVA: pseudoF(1, 29) = 7.57, P < 0.001], but only 17% in rhizosphere composition [PERMANOVA: pseudoF(1, 29) = 1.90, P < 0.001]. Consequently, large proportions of bacterial taxa at all taxonomic ranks in the endosphere varied significantly in abundance among host plant species (bacterial phyla: 65%; ASVs: 12%), whereas far fewer taxa in the rhizosphere were affected (bacterial phyla: 19%; ASVs: 1%; GLM: PFDR < 0.05) (SI Appendix, Fig. S4D). Additionally, only a fraction of the responsive bacterial taxa in the endosphere were also influenced by host plant species in the rhizosphere (bacterial phyla: 36%; ASVs: 17%). Several phyla in particular were strongly affected by variation among host plant species, including Proteobacteria, Actinobacteria, and Bacteroidetes (GLM: PFDR < 0.05) (Fig. 1D). Across host plant species, we found little correlation between endosphere and rhizosphere diversity (Simpson’s D−1: r = 0.06, P = 0.09), despite a significant correlation between endosphere and rhizosphere community composition (weighted UniFrac distances: rMantel = 0.26, P = 0.04). Finally, we identified 133 endosphere and 334 rhizosphere ASVs found in all host plant species (Dataset S1), suggesting the existence of a prevalent core microbial assemblage despite tremendous variability occurring among host plant species. Of these ASVs, 59% in the endosphere and 40% in the rhizosphere that make up the core microbiome were found at intermediate (two to three individuals per host species) or high prevalence (five individuals per host species).
Evolutionary Divergence Among Host Plant Species Affects the Assembly of the Endosphere and Rhizosphere Microbiome.
To understand how plant evolution has shaped root microbial communities, we tested whether close relatives share similar endosphere and rhizosphere microbiomes. Microbial diversity in the endosphere (Blomberg’s K = 1.08, P = 0.001), but not the rhizosphere (Blomberg’s K = 0.67, P = 0.94), exhibited significant phylogenetic signal (Fig. 1C and SI Appendix, Table S6). We used Mantel tests of phylogenetic relatedness versus root microbial community similarity among plant species to understand whether plant evolution shapes the community composition of the root microbiome. Again, endosphere similarity (rMantel = 0.15, P = 0.004), but not rhizosphere (rMantel = 0.05, P = 0.15), was positively correlated with phylogenetic relatedness between plant species (Fig. 1D and SI Appendix, Table S7). We used phylogenetic generalized least-squares regression (PGLS) to investigate the relationship between experimentally measured plant traits and root microbial diversity and composition (SI Appendix, Table S8). Root microbial diversity was associated with numerous host plant traits; however, the importance of individual traits varied between endosphere and rhizosphere compartments (SI Appendix, Table S8). Endosphere diversity was positively associated with increasing root hair density, while rhizosphere diversity was positively associated with host plant productivity and negatively associated with root length (PGLS: PFDR < 0.05). Endosphere and rhizosphere composition were also associated with numerous plant traits, including host plant productivity, physiology, and root architectural traits (SI Appendix, Table S8) (PGLS: PFDR < 0.05).
Variation in the Root Microbiome Between Host Plant Species Affects Indirect Competitive Interactions via PSFs.
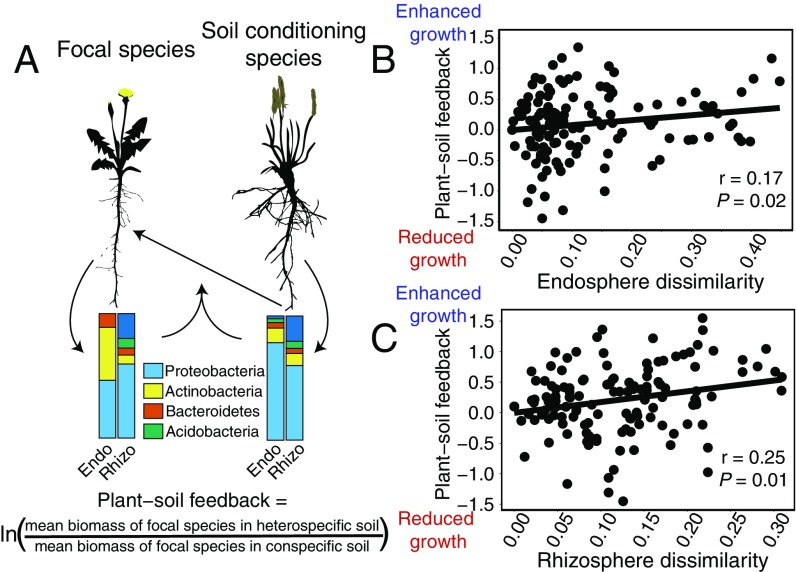
Using a multigeneration PSF experiment, we investigated how patterns of root microbial recruitment among host plant species can feed back to affect competitive interactions (SI Appendix, Fig. S1). In the first generation, we grew each of the 30 plant species in a homogenous soil mixture collected from the same field site as our comparative microbiome study. In the second generation, we grew replicate individuals of five focal species, representative of our host plant phylogenetic diversity, in bulk and rhizosphere soil collected and preserved from each of the 30 plant species from the previous generation. The net effect of soil conditioning in the first generation on plant performance in the second generation is the PSF. PSF can be caused by modification to both biotic and abiotic soil properties, including the alteration of soil bacterial communities, as well as the depletion of soil nutrients. We calculated the PSF as: loge [(focal species biomass in heterospecific soil)/(focal species biomass in conspecific soil)]; positive values indicate that a focal species performed better in soil conditioned by a different species from the focal plant relative to soil conditioned by the same species as the focal plant, whereas negative values indicate the opposite (34). We observed strong positive and negative soil feedback occurring among pairs of plant species.
We sought to understand how root microbiota assembled by different plant species contributes to their experimentally measured PSF (Fig. 2A). We correlated the root microbiome similarity (weighted and unweighted UniFrac distances) between host plant species with their PSF measured in our multigeneration experiment. Remarkably, the effect of inoculation with soil conditioned by heterospecific plants depended on the degree of similarity between the root microbiomes assembled by the focal and soil-conditioning plant species. On average, highly dissimilar microbiomes had more positive effects on focal plant growth than highly similar ones (Fig. 2 B and C and SI Appendix, Fig. S8). This pattern was consistent for both the endosphere and rhizosphere, although it depended on the particular measure of community similarity used.
Fig. 2.
Root microbial composition is related to PSF. (A) PSF occurs when the soil microbes recruited by one plant influence the growth of other plants. Positive values indicate that a focal species performed better in soil conditioned by a heterospecific plant relative to a conspecific plant, whereas negative values indicate the opposite. (B) Plants exhibit enhanced growth when inoculated with soil conditioned by a heterospecific species with dissimilar endosphere (measured as weighted UniFrac distance) and (C) rhizosphere compartments (measured as unweighted UniFrac distance).
Next, we investigated how specific bacterial taxa contributed to the effect of the root microbiome on PSF. First, we used GLMs to calculate the log2 fold-change (i.e., doublings) of each taxon abundance between all pairs of focal and soil-conditioning host plant species (35). We identified bacterial taxa across all taxonomic ranks that exhibited significant differential abundance across host plant species in either the endosphere or rhizosphere (e.g., Fig. 3 A and B). We correlated the differential abundance between host plant species of each bacterial taxon with the experimentally measured host plant pairs’ PSF (Dataset S2).
Fig. 3.
Differential abundance of root bacterial taxa and PSF. Host plant species exhibit differential abundance for numerous root bacterial taxa in either the endosphere or rhizosphere, including: (A) an endosphere Streptomyces ASV and (B) the genus Pseudoxanthomonas, found in the rhizosphere (GLM: PFDR < 0.05). We estimated the log2 fold-change of differentially abundant root bacterial taxa among all unique pairs of focal (starred taxa on the host plant phylogeny) and soil-conditioning host plant species and correlated this with their measured PSF. Negative log2 fold-changes indicate a higher taxon abundance in soil-conditioning host plant species, while positive values indicate a higher taxon abundance in focal host plant species. (C) The differential abundance of the endosphere Streptomyces ASV between focal and soil-conditioning host plant species was positively related to their PSF. (D) However, the differential abundance of rhizosphere Pseudoxanthomonas between focal and soil-conditioning host plant species was negatively related to their PSF. (E and F) PSF between host plant species was significantly associated with the differential abundance of 66 endosphere taxa and 33 rhizosphere taxa. (E) In the endosphere, we observed a high proportion (35%) of PSF-related taxa exhibiting the association depicted in C (green lines illustrate significant trend lines between differential abundance of endosphere taxa and PSF at PFDR < 0.05). (F) While in the rhizosphere, a greater proportion (88%) of taxa exhibited the association depicted in D (yellow lines illustrate significant trend lines between rhizosphere taxa and PSF at PFDR < 0.05). See Dataset S2 for a full list of PSF-related taxa.
Numerous bacterial taxa were strongly associated with positive and negative PSF occurring between plant species (hereafter, PSF-related taxa), including a number of endosphere and rhizosphere ASVs found across all host species (representative taxa shown in Fig. 3 C and D; for full list, see Dataset S2). Differential abundance of particular ASVs explained up to 15% of the total variation in the measured PSF between host plant species (e.g., Fig. 3C and Dataset S2). Although bacterial phyla—such as Proteobacteria, Bacteroidetes, and Actinobacteria—are well represented in the list of PSF-related taxa in both the endosphere and rhizosphere, we found little overlap at lower taxonomic ranks (Dataset S2). In general, when the abundance of a bacterial taxon in the focal host species was less than the soil-conditioner host species, we observed enhanced growth of the focal plant. In contrast, when the abundance of the bacterial taxon was greater in the focal host species than the soil-conditioner plant species, we observed reduced growth of the focal plant (e.g., Fig. 3D and Dataset S2, r values < 0, unshaded rows). However, we noticed that for some microbial taxa the association was reversed (e.g., Fig. 3C and Dataset S2, r values > 0, blue-shaded rows). Furthermore, of the microbial taxa significantly related to PSF, a greater proportion in the endosphere (35% of taxa) versus the rhizosphere (12% of taxa) exhibited this opposite association (Fig. 3 E and F) (Fisher’s exact test for the difference in proportion: P = 0.01) (Dataset S2, r values > 0, blue-shaded rows).
The Root Microbiome Is Associated with Drought Tolerance Across Host Plant Species.
During our comparative root microbiome study, we imposed a chronic drought treatment on replicate individuals from each host plant species, which resulted in a fourfold difference in soil moisture compared with well-watered control plants. We investigated how this abiotic stressor affects the diversity and composition of the root microbiome across 30 host plant species. We also included pots without plants that were filled with the same soil mixture in each watering treatment and identically treated, nonliving structures (bamboo toothpicks) that were analogous to plant roots (9). Comparing the bacterial communities in living roots to nonliving root analogs allowed us to understand the host-mediated effects of drought on the root microbiome.
Drought reduced microbial diversity in the endosphere and rhizosphere by 15% and 27%, respectively [F(1, 53) = 5.56, PFDR = 0.06] (Fig. 4A). Drought also caused large changes in bacterial community composition (Fig. 4 B and C and SI Appendix, Table S3). Surprisingly, the effect of drought was stronger on the endosphere than the rhizosphere microbiome, suggesting large indirect effects of drought through changes in host plant physiology or immune status (26, 27, 36). Consistent with this result, drought caused changes in the relative abundance of 65% of bacterial phyla in the endosphere versus only 43% in the rhizosphere (GLM: PFDR < 0.05) (SI Appendix, Fig. S4C). In particular, the abundance of Actinobacteria in the drought endosphere increased over twofold, whereas the abundance of Proteobacteria decreased nearly 2.5-fold (GLM: PFDR < 0.05) (Fig. 4B, SI Appendix, Fig. S9 and Dataset S3). However, host plants varied in the magnitude of the shift in their endosphere microbiome during drought (drought × host species: χ2 = 7.15, PFDR = 0.03) (SI Appendix, Table S4), which included the enrichment/depletion of bacteria found in well-watered plants and the recruitment of new taxa into their roots (SI Appendix, Figs. S5 and S7). The drought-induced changes in the microbiota of living plant roots were distinct from those occurring in the microbiota of root analogs and soil (SI Appendix, Fig. S15 and Table S9 and Dataset S3).
Fig. 4.
The effects of drought on root microbial communities. (A) The drought treatment (denoted by T) caused small reductions in the diversity of the endosphere and rhizosphere compartments (denoted by C), and (B) had large effects on the relative abundance of major bacterial phyla; starred phyla were significantly affected (GLM: PFDR < 0.05) by drought (green, endosphere; orange, rhizosphere). (C) Drought also had strong effects on the overall composition of the endosphere and rhizosphere microbiomes, although endosphere compartments exhibited a greater response. (Inset) Plants under drought experienced fourfold lower soil moisture than well-watered plants.
We sought to understand whether drought-induced shifts in the root microbiome were related to drought tolerance across plant species. This question was motivated by the prediction that plastic responses in the root microbiome may maintain host functions and ultimately plant fitness in response to stress (37, 38). We measured drought tolerance as the proportional difference in total biomass between drought and well-watered conditions. On average, plants exhibited a 35% reduction in total biomass in response to drought; however, species varied between an 80% reduction to a 127% increase in biomass in response to drought (Fig. 5A and SI Appendix, Fig. S10). We found no evidence of phylogenetic signal in drought tolerance across the plant phylogeny. Changes in overall root microbiome composition or diversity under drought were not associated with drought tolerance. However, coarse estimates of overall composition have a poor ability to detect the ecological effects of particular bacterial clades or ASVs.
Fig. 5.
The relationship between drought tolerance and Streptomycetaceae. (A) On average, the drought treatment (denoted by T) caused a 35% reduction in biomass compared with well-watered conditions [F(1, 44) = 17.37, P < 0.001], and plant species (denoted by S) varied significantly in their response to drought (represented by dots connected by individual lines). (B) Drought caused a sixfold increase in the mean relative abundance of endosphere Streptomycetaceae (Actinobacteria), but this effect varied among plant species. (C) Plant species with greater relative increases in an endosphere Streptomyces ASV under drought conditions had greater drought tolerance. See Dataset S4 for a full list of drought-related taxa.
To further examine whether the root microbiome affects drought tolerance, we investigated how individual bacterial taxa were related to drought tolerance across plant species. First, for each host plant species, we used GLMs to calculate the log2 fold-change of each drought-responsive taxon between watering treatments (35). We identified bacterial taxa at all taxonomic ranks in the endosphere and rhizosphere that were differentially abundant between well-watered and drought conditions. Next, we correlated patterns of differential abundance for each bacterial taxon with drought tolerance across host plant species. We detected two striking results. First, the Streptomycetaceae (all ASVs combined, three of which were found in the endosphere of every host plant species), exhibited a threefold increase abundance within the endosphere, but not rhizosphere, under drought (SI Appendix, Fig. S11). However, the magnitude of Streptomycetaceae enrichment varied between 0 to fourfold across plant species (Fig. 5B and SI Appendix, Fig. S13). Second, the relative enrichment of one Streptomyces ASV, found in the endosphere of every host plant species in the experiment, explained nearly 40% of the variation in drought tolerance among host plant species (Fig. 5C and Dataset S4). A number of other endosphere taxa exhibited strong correlations (r > 0.4) with host drought tolerance, including another Streptomyces ASV at high host prevalence (Dataset S4). However, with the exception of the Streptomyces ASV depicted in Fig. 5C, these correlations were nonsignificant after multiple test correction (Dataset S4). Importantly, after examination of the nonliving wood samples, all of the endosphere ASVs related to drought tolerance in living roots were not enriched in the endosphere of these root analogs under drought (Dataset S3).
Discussion
We demonstrate that plant evolution over long timescales shapes root microbiome assembly, which in turn influences how host plants respond to biotic and abiotic environmental stressors. Our results successfully address our four research questions. First, the diversity and composition of the root microbiome was markedly different between the endosphere and rhizosphere compartments across host species. Second, host plant species explained much of the variation in the diversity and composition of the root microbiome. Variation in the endosphere microbiome exhibited strong correspondence with the underlying host plant phylogeny, although this was not the case for the rhizosphere. Third, patterns of root microbial recruitment among host plants in both the endosphere and rhizosphere influence indirect competitive interactions among plant species through PSFs. Fourth, under drought stress the root endosphere dynamically responds, and these changes correspond to variation in host plant tolerance to drought. Below, we discuss how these results inform our understanding of the factors that shape root microbiomes and their ecological importance.
Root Microbiome Assembly Across Angiosperm Species.
Our results provide clear insight into how host plants affect the assembly of root microbiomes. Large differences in endosphere and rhizosphere diversity and composition (Fig. 1 A and B) are likely a conserved feature in plants, reflecting general rules for the assembly of root microbiomes across angiosperm species. For example, we found a significant correlation between endosphere and rhizosphere community composition, indicating that the host-specific factors shaping composition, but not diversity, are at least partly shared between endosphere and rhizosphere compartments. Despite a broad conservation of root microbiome assembly, we also uncovered tremendous variation in microbiome communities occurring across host plant species. Plant species varied much more in their endosphere diversity (Fig. 1C) and composition (Fig. 1D) than in their rhizosphere microbiome compartment, which supports the idea of greater host plant importance in the assembly of the endosphere microbiome (6). Several plant lineages exhibited pronounced differences in their endosphere microbiota, including the Fabaceae, which have an elevated proportion of Proteobacteria, and the Poaceae, which are enriched in Actinobacteria.
We find support for the emerging view that plant evolution influences the root microbiome (12, 31, 39). Pronounced effects of host plant species in the face of recruitment of microbiota from the surrounding environment suggest that plants have evolved traits that govern root microbiome assembly. We found a particularly strong association between host plant evolutionary relatedness and endosphere diversity and composition, which indicates that host traits underlying endosphere assembly covary with phylogenetic relatedness among hosts. In contrast, rhizosphere assembly exhibited no clear relationship with host plant phylogeny (Fig. 1 C and D and SI Appendix, Tables S6 and S7), despite host plant species having a strong effect on the rhizosphere microbiome (SI Appendix, Tables S3–S5). This result suggests that the plant traits, which shape the rhizosphere compartment, are themselves uncorrelated with host plant phylogeny.
Our analysis of plant traits revealed that plant productivity and physiology are associated with variation in root microbiome diversity and composition, similar to a recent study of leaf bacterial communities in tropical tree species (40). These physiological traits are often correlated with broad resource acquisition strategies employed across plant species (41), suggesting that plant resource consumption and turnover are correlated with root microbiota. Several host traits were associated with microbial composition in both the endosphere and rhizosphere; however, this was not the case for microbial diversity (SI Appendix, Table S8). These results support our previous finding that host plant factors associated with root microbial composition, but not diversity, are partially shared between endosphere and rhizosphere compartments. We speculate that finer insight into how host plant variation and evolution affects root microbiome assembly, particularly the endosphere compartment, will require characterization of root metabolites and exudates as well as the microbial-triggered immune responses across plant species (4, 42–44).
The Ecological Importance of the Plant Root Microbiome.
Plants evolved the ability to colonize land at a time when the terrestrial environment already contained microorganisms. Interactions between plants and soil microbes were key to the colonization and persistence of land plants (5), and they continue to play essential roles in host plant evolution and ecology. How assemblages of root microbiota contribute to the interaction between host plants and their biotic and abiotic environment is poorly understood and was a central focus of our study.
Root microbiota and PSF.
Biotic interactions via PSFs are a form of plant competition that have far-reaching importance for terrestrial ecosystems (19, 20, 22). Soil microbes are generally recognized as the main factors driving PSF, but beyond this, general theories addressing how microbial taxa contribute to the strength of PSF among plants remain limited (45, 46). Our results lead to several important conclusions. First, the PSF between host plants depends on overall compositional differences of root microbiota (Fig. 2). On average, highly similar root microbiomes lead to negative PSF between host plant species. Increasing root microbial similarity between plant species could directly reduce plant performance due to shared pathogenic bacteria transferred through soil. If host immunity shapes associated microbiota (4, 42, 43), or if host-microbiota affect immunity (43, 47), then host plants with similar root microbiomes may exhibit increased susceptibility to, and coinfection with, the same pathogens. This hypothesis is indirectly supported by studies reporting higher coinfectivity rates of pathogens between close versus distant plant relatives, presumably driven by variation in pathogen-specific resistance across the plant phylogeny (48). Alternatively, root microbiota may influence plant–plant interactions through soil resource partitioning. In our PSF experiment, the major source of mineral nutrients for focal plants was the inoculum from soil conditioned in the previous generation. Differential association with particular soil microorganisms is thought to increase soil resource partitioning between plant species (16). Focal plants with similar root microbiota to soil-conditioning plants may exhibit reduced growth due to a shared microbial mutualist, involved in the acquisition of a limiting soil resource depleted during generation one. Future work is required to understand the relative importance of antagonistic versus beneficial microorganisms in driving the correlation between root microbial similarity and PSF.
PSF between host plant species also depended on the differential abundance of particular root bacterial taxa (Fig. 3). This suggests a dynamic interplay between the root microbiota of interacting host plant species. In general, a higher abundance of PSF-related taxa in the rhizosphere of soil-conditioning host plants led to increased focal plant performance (e.g., Fig. 3 D and F and Dataset S2, unshaded rows). Greater abundance of mutualistic bacterial taxa in the rhizosphere of a host plant could enhance soil quality for future generations of plants by increasing abundance of the bacterial mutualist or through a fertilization effect (46). In the endosphere, we observed a high proportion of bacterial taxa exhibiting the opposite association, whereby increased abundance in the endosphere of the soil-conditioning host plant lead to reduced focal plant performance (e.g., Fig. 3 C and E and Dataset S2, blue shaded rows). This opposing association suggests that greater abundance of specific bacterial taxa in the endosphere reduces soil quality for the next generation of plants. This pattern is consistent with root bacteria recruited by a tolerant host plant acting as a plant pathogen in subsequent generations, but could also be driven by the depletion of mutualistic bacteria from the soil environment reducing subsequent host plant performance (46, 49).
The opposing effects of differential abundance illustrate that microbial members of either the endosphere or rhizosphere may have very different roles in plant competitive interactions mediated through soil feedback, potentially related to their relative importance as either pathogens or mutualists. Additionally, the effects of root microbiota on PSF include compositional differences of entire root microbial compartments and the unique effects of individual bacterial taxa. Overall, our results raise the possibility that patterns of root microbial recruitment among plant species, through their effects on PSF, may contribute to fundamental terrestrial ecology, such as the mechanisms underlying species coexistence (50) and ecosystem processes (21).
Drought and the root microbiome.
In natural and managed ecosystems, water availability is a strong determinant of plant performance. We investigated how drought shapes the root microbiome and whether or not drought-induced changes in root microbiota are associated with drought tolerance across host plant species. Drought substantially altered the composition of the root microbiome and marginally reduced microbial diversity, with larger effects on the endosphere than the rhizosphere microbiome (Fig. 4). Our results suggest that the effects of drought on microbiota are indirectly mediated by host plant responses (SI Appendix, Fig. S13 and Table S9 and Dataset S3). A number of drought-induced plant responses, including physiological and molecular changes, could be responsible for these effects of plants on the endosphere microbiome. Interestingly, one of the chief regulators of drought stress response in plants is the hormone abscisic acid (ABA), which exhibits negative cross-talk with a number of defense hormones (36). A dampening of host plant immunity during drought could facilitate large shifts in endosphere colonization by microorganisms, otherwise restricted by the plant immune system (42). Indeed, a number of bacterial pathogens exploit this cross-talk by producing metabolites that mimic ABA (51). Two recent studies have shown that drought alters the root microbiome of cereal crop species (31, 32). Our findings extend these results to a wider phylogenetic diversity of plant species and demonstrate that large effects of host plants on the root endosphere under drought are a general pattern shared among angiosperms.
We found compelling evidence that increases in endosphere Actinobacteria—and especially members of the Streptomycetaceae—are associated with increased drought tolerance (Fig. 5). The Streptomycetaceae exhibit traits of potential benefit to host plants, including the production of antimicrobial compounds, thick-walled spores resilient to environmental perturbation, and inducible exploratory behavior (52), all of which may increase colonization rates of plant tissue under stressful environments. Another study investigating Streptomyces isolated from wheat roots found a potential benefit to host plants under drought stress, possibly through production of plant hormones and biochemical activity that help mitigate water stress (53). Members of the Actinobacteria were also enriched in root analogs (toothpicks) under drought, but these were not the same ASVs associated with drought tolerance in living host plants (Dataset S3). Moreover, the ASVs enriched in the endosphere of root analogs under drought represented only 3% of the total ASVs enriched in living plant roots. Surprisingly, we failed to find any rhizosphere taxa that were related to host drought tolerance, despite numerous reports of drought-related rhizobacteria (54). While features of the Actinobacteria make them particularly suited to persist in stressful abiotic conditions like drought, our findings and others’ point to the existence of lineages enriched only in roots of living plants under drought (31).
Our results present the intriguing hypothesis that changes in the host-microbiome under abiotic stress may be adaptive for the host (37, 38). If true, this would represent a form of adaptive phenotypic plasticity mediated by a plant’s extended microbial community. More work is required to unravel the genetic and physiological mechanisms underlying host plant effects on the root microbiome, as well as any fitness benefits of increased Streptomycetaceae abundance under drought. Recent findings may provide some insight into possible mechanisms regulating adaptive host–microbiome interactions. This work suggests that hosts modify their associated microbiota through regulating innate immunity (e.g., ref. 4) or by interfering with quorum-sensing in bacteria (55, 56). How animal and plant hosts modify their associated microbiota in response to environmental perturbations, and whether these modifications represent adaptations, are important questions for understanding the ecological and evolutionary importance of host microbiota.
The examination of both biotic and abiotic stressors in our study uncovered several important findings. Different compartments of the root microbiome (endosphere and rhizosphere) are uniquely associated with a plant’s response to environmental stress. For example, while the endosphere and rhizosphere microbiome were both associated with PSF, different taxa in each compartment were related to the strength of PSF (Dataset S2). In contrast, only the endosphere compartment was related to drought tolerance (Dataset S4). We also found three bacterial ASVs in the root endosphere that were strongly associated with plant responses to both biotic and abiotic host plant stress (Datasets S2 and S4, green-, orange-, and purple-shaded ASVs). We speculate that some members of the root microbiome may benefit host plants across a wide range of biotic and abiotic stressors. Finally, many of the PSF-related and drought-related endosphere ASVs were found in all host plant species (Datasets S2 and S4), which points to the importance of widespread root bacterial symbionts for plant ecology.
Caveats.
Linking ecological functions across host plant species with root microbial diversity and composition derived from deep-amplicon sequencing data has several important limitations. First, although strongly suggestive of an important role for root bacterial communities in mitigating interactions between host plants and their biotic and abiotic environment, our results are correlative. Future research requires controlled experiments using synthetic communities or single inoculations to understand the mechanisms underlying the patterns uncovered here. Second, measures of relative abundance are unable to detect absolute increases in bacterial abundance. Using qPCR, Naylor et al. (31) recently confirmed that relative increases in Actinobacteria abundance in plant roots reflect absolute increases. Thus, the results from our drought study likely reflect absolute changes in the abundance of Streptomycetaceae. Third, characterization based on the 16S rRNA gene yields little functional information about microbial communities. Genomic analyses of root microbiota in addition to ecological assays of individual taxa or synthetic communities will elucidate the functional importance of root microbiota (57–59). Despite these limitations, our results reveal important effects of plant evolution and stress on root microbiota, and how the root microbiome is tightly related to the ecological response of plants to environmental stressors.
Conclusions
Host-associated microbiota are essential for nutrition, development, and immunity across plant and animal hosts (2), yet our understanding of their broader ecological importance remains limited. Our study provides evidence of the causes of variation in the host microbiome across a wide range of host plant species, as well as the general ecological importance of this variation for biotic and abiotic stressors. This study may also inform future efforts to engineer the root microbiome in diverse agricultural systems to increase plant performance in the face of competition and drought stress (60, 61).
Materials and Methods
To understand the assembly and ecological function of the angiosperm root bacterial microbiome, we combined a comparative study of 30 phylogenetically diverse plant species with manipulative experiments (Fig. 1A and SI Appendix, Table S1). First, we characterized the endosphere and rhizosphere microbiome of replicate individuals grown from surface-sterile seeds in a common environment. Seeds were planted in a live soil inoculum collected from a naturalized field site where all species cooccur (Koffler Scientific Reserve). We measured a suite of morphological, physiological, and performance traits from every plant (SI Appendix, Table S2). After 16 wk, we partitioned standard root samples from each plant into endosphere and rhizosphere compartments (8, 9) and extracted total DNA. We characterized the bacterial community by sequencing amplicons of the V4 region of the 16S rRNA gene using Illumina MiSeq (SI Appendix, Fig. S1). To reduce host contamination, we used peptide nucleic acids designed to block amplification of host plant plastid and mitochondrial sequences (62). We assembled quality-filtered reads into error-corrected ASVs using DADA2 v1.4.0 (33), which represent unique bacterial taxa. ASVs exhibit fewer false-positive taxa and reveal cryptic diversity, otherwise undetected by traditional OTU approaches (33). In total, we profiled 271 endosphere communities, 255 rhizosphere communities, and 58 soil and control samples (SI Appendix, Table S1), and assembled 56,063 ASVs.
Assembled ASVs were assigned taxonomy (phylum to genus) using the Ribosomal Database Project (RDP) naïve Bayesian classifier (implemented in DADA2) and the “RDP training set 14” (63). We used PASTA to align ASV sequences and build a maximum-likelihood phylogenetic tree (64). Next, using the R package “phyloseq” (65), we removed any ASVs without a bacterial phylum assignment, assigned to Archaea, chloroplast, or mitochondrial origin. To simplify downstream analyses, we applied a prevalence and abundance threshold for bacterial ASVs, where taxa were kept only if they were found in 1% of samples (seven samples) and at a frequency of 25 reads per sample. This yielded 2,799 ASVs, which accounted for 94% of the total number of sequences in the dataset (SI Appendix, Fig. S14). For downstream composition analyses, we performed proportional abundance normalization (relative abundance) on this common set of ASVs, where the sequencing reads for an ASV in a given sample were divided by the total number of sequencing reads in that sample (66). As an additional set of analyses, we used the traditional approach of rarefaction (to 800 reads) to normalize our full dataset before any threshold, which yielded ∼13,000 ASVs and accounted for less than 2% of the total read count (SI Appendix, Fig. S14). Both methods (rarefaction on the full dataset and relative abundance normalization on the simplified dataset) yielded qualitatively identical results; we therefore present the nonrarefied data because it retained a larger portion of our data.
We investigated the ecological importance of root microbiota for both biotic and abiotic stressors. As a biotic stressor, we measured how patterns of root microbial recruitment among host plant species can feed back to affect indirect competitive interactions via PSFs (SI Appendix, Fig. S1). In the first generation of our PSF experiment, we grew each of our 30 plant species in a homogenous soil mixture collected from the same field site as our comparative microbiome study. Pots were filled with 800 mL of sterilized soil [mixture of potting soil and sand (2:3 v/v) and 200 mL of live inoculum collected from KSR]. We preserved bulk and rhizosphere soil collected and pooled from five individuals from each of the 30 plant species and used it to inoculate replicate individuals of five focal species, representative of our host plant phylogenetic diversity. In this second generation, we mixed live soil inoculum preserved from the previous generation with the same sterile soil mix in the same ratio as the first generation. The effect of soil conditioning in the first generation on plant performance in the second generation is the PSF. Operationally, we measured the PSF as: loge [(focal species biomass in heterospecific soil)/(focal species biomass in conspecific soil)]; positive values indicate that a focal species performed better in soil conditioned by a different species from the focal plant relative to soil conditioned by the same species as the focal plant, whereas negative values indicate the opposite (34).
As an abiotic stressor, we manipulated drought and measured how water limitation affected patterns of root microbial recruitment among host plant species and host plant drought tolerance. We used a drip irrigation system to impose a chronic drought stress on replicate individuals from each host plant species during the comparative root microbiome study, as well as an equal number of well-watered control plants. Our manipulation resulted in a fourfold difference in soil moisture and a mean biomass reduction of 35% across host plant species in the drought treatment compared with the control, although host plant species varied widely in their tolerance to drought. Alongside living plants we also included bare soil pots and pots planted with structurally similar root analogs (toothpicks). Differences in drought responses between living root microbial communities and root analogs or soil indicate the effects of living host plants on microbial dynamics.
We analyzed the effects of host plant species, root compartment, and watering treatment on the diversity [observed ASV richness, Simpson’s D−1, and evenness (Simpson’s D−1/observed ASV richness)], and composition [weighted UniFrac dissimilarity (67)] of bacterial communities using linear mixed models (SI Appendix, Tables S3 and S4). We also analyzed the effects of host plant species, root compartment, and watering treatment on the differential abundance of bacterial taxa using DESeq2 (35). DESeq2 uses negative binomial models and ASV read counts to test whether individual bacterial taxa are differentially abundant across experimental factors. To understand how plant evolution has shaped root microbial communities, we calculated phylogenetic signal (Blomberg’s K and Pagel’s λ) present in diversity estimates and used Mantel tests to determine the correlation between host plant evolutionary relatedness and root microbial compositional similarity (SI Appendix, Tables S6 and S7). Finally, we used PGLS to determine the relationship between plant traits and root microbial diversity and composition (SI Appendix, Table S8).
We investigated the ecological importance of root microbiota by correlating patterns of root microbial composition and differential abundance among host plant species with experimentally measured PSF and drought tolerance. Correlations between root microbial composition and ecological processes indicate an importance of broad patterns of root microbiome assembly, whereas correlations with individual taxa indicate particular individual bacterial taxa are associated with host plant performance. First, we analyzed how endosphere and rhizosphere compositional differences (weighted and unweighted UniFrac dissimilarity) among pairs of host plant species was correlated with PSF. Next, we identified those bacterial taxa that were differentially abundant among host plant species and correlated their log2 fold-change between focal and soil-conditioning plant species with the experimentally measured PSF (Dataset S2). In the drought experiment, we correlated endosphere and rhizosphere compositional differences (weighted and unweighted UniFrac dissimilarity) between watering treatments within a host plant species with their measured drought tolerance. To understand the potential role of individual taxa, we first identified drought-responsive bacterial taxa and correlated their log2 fold-change between watering treatments within a host plant species with host species’ drought tolerance (Dataset S4). All analyses were carried out in R v3.3.3 (68). For detailed materials and methods, see SI Appendix, SI Materials and Methods. Sequence files associated with individual samples are available on the NCBI Sequence Read Archive (accession no. SRP128025). All data and R code used in the analyses are available on the Dryad digital repository (https://doi.org/10.5061/dryad.5p414).
Supplementary Material
Acknowledgments
We thank M. Kalich and B. Pitton for greenhouse support; A. Chen, K. Cory, D. Filice, L. Gehant, J. Lee, A. Longley, M. Middleton, L. Rawofi, H. Sekhon, A. Severino, K. Vigmond, and members of the Centre for the Analysis of Genome Evolution and Function for providing assistance with experiments and data collection; members of the M.T.J.J. and D.S.G. laboratories, J. L. Dangl, D. S. Lundberg, J. R. Stinchcombe, and M. R. Wagner for helpful discussion; and two anonymous reviewers for providing thoughtful evaluation, which greatly improved the manuscript. This work was supported by Ontario Graduate Scholarship and Queen Elizabeth II awards (to C.R.F.), and funding from the University of Toronto, University of Toronto Mississauga Office of the Vice Principal Research, and Natural Sciences and Engineering Research Council (to D.S.G., P.M.K., and M.T.J.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper has been deposited in the Dryad database (https://doi.org/10.5061/dryad.5p414). Sequence files associated with individual samples are available on the NCBI Sequence Read Archive (accession no. SRP128025).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717617115/-/DCSupplemental.
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hacquard S, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castrillo G, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. Symbiotic options for the conquest of land. Trends Ecol Evol. 2015;30:477–486. doi: 10.1016/j.tree.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 7.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 10.Edwards J, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA. 2014;111:585–592. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouffaud M-L, Poirier M-A, Muller D, Moënne-Loccoz Y. Root microbiome relates to plant host evolution in maize and other Poaceae. Environ Microbiol. 2014;16:2804–2814. doi: 10.1111/1462-2920.12442. [DOI] [PubMed] [Google Scholar]
- 13.Zgadzaj R, et al. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc Natl Acad Sci USA. 2016;113:E7996–E8005. doi: 10.1073/pnas.1616564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorpe AS, Aschehoug ET, Atwater DZ, Callaway RM. Interactions among plants and evolution. J Ecol. 2011;99:729–740. [Google Scholar]
- 15.Tilman D. Resource Competition and Community Structure. Princeton Univ Press; Princeton, NJ: 1982. [PubMed] [Google Scholar]
- 16.Bever JD, et al. Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol. 2010;25:468–478. doi: 10.1016/j.tree.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodge A, Fitter AH. Microbial mediation of plant competition and community structure. Funct Ecol. 2012;27:865–875. [Google Scholar]
- 18.Bever JD. Feedback between plants and their soil communities in an old field community. Ecology. 1994;75:1965–1977. [Google Scholar]
- 19.Kardol P, Bezemer TM, van der Putten WH. Temporal variation in plant-soil feedback controls succession. Ecol Lett. 2006;9:1080–1088. doi: 10.1111/j.1461-0248.2006.00953.x. [DOI] [PubMed] [Google Scholar]
- 20.Klironomos JN. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature. 2002;417:67–70. doi: 10.1038/417067a. [DOI] [PubMed] [Google Scholar]
- 21.Schnitzer SA, et al. Soil microbes drive the classic plant diversity-productivity pattern. Ecology. 2011;92:296–303. doi: 10.1890/10-0773.1. [DOI] [PubMed] [Google Scholar]
- 22.Callaway RM, Thelen GC, Rodriguez A, Holben WE. Soil biota and exotic plant invasion. Nature. 2004;427:731–733. doi: 10.1038/nature02322. [DOI] [PubMed] [Google Scholar]
- 23.Choat B, et al. Global convergence in the vulnerability of forests to drought. Nature. 2012;491:752–755. doi: 10.1038/nature11688. [DOI] [PubMed] [Google Scholar]
- 24.Siepielski AM, et al. Precipitation drives global variation in natural selection. Science. 2017;355:959–962. doi: 10.1126/science.aag2773. [DOI] [PubMed] [Google Scholar]
- 25.Farooq M, Wahid A, Kobayashi N, Fujita D. Plant drought stress: Effects, mechanisms and management. Agron Sustain Dev. 2009;29:185–212. [Google Scholar]
- 26.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 27.Brunner I, Herzog C, Dawes MA, Arend M, Sperisen C. How tree roots respond to drought. Front Plant Sci. 2015;6:547. doi: 10.3389/fpls.2015.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timmusk S, Wagner EG. The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: A possible connection between biotic and abiotic stress responses. Mol Plant Microbe Interact. 1999;12:951–959. doi: 10.1094/MPMI.1999.12.11.951. [DOI] [PubMed] [Google Scholar]
- 29.Mayak S, Tirosh T, Glick BR. Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci. 2004;166:525–530. [Google Scholar]
- 30.Lau JA, Lennon JT. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc Natl Acad Sci USA. 2012;109:14058–14062. doi: 10.1073/pnas.1202319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017;11:2691–2704. doi: 10.1038/ismej.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos-Medellín C, Edwards J, Liechty Z, Nguyen B, Sundaresan V. Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. mBio. 2017;8:e00764-17. doi: 10.1128/mBio.00764-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pernilla Brinkman E, van der Putten WH, Bakker E-J, Verhoeven KJF. Plant-soil feedback: Experimental approaches, statistical analyses and ecological interpretations. J Ecol. 2010;98:1063–1073. [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 37.Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol Evol. 2016;31:689–699. doi: 10.1016/j.tree.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Goh C-H, Veliz Vallejos DF, Nicotra AB, Mathesius U. The impact of beneficial plant-associated microbes on plant phenotypic plasticity. J Chem Ecol. 2013;39:826–839. doi: 10.1007/s10886-013-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeoh YK, et al. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat Commun. 2017;8:215. doi: 10.1038/s41467-017-00262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kembel SW, et al. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci USA. 2014;111:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comas LH, Eissenstat DM. Linking fine root traits to maximum potential growth rate among 11 mature temperate tree species. Funct Ecol. 2004;18:388–397. [Google Scholar]
- 42.Lebeis SL, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 43.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay between innate immunity and the plant microbiota. Annu Rev Phytopathol. 2017;55:565–589. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
- 44.van Dam NM, Bouwmeester HJ. Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends Plant Sci. 2016;21:256–265. doi: 10.1016/j.tplants.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 45.van der Putten WH, et al. Plant-soil feedbacks: The past, the present and future challenges. J Ecol. 2013;101:265–276. [Google Scholar]
- 46.Bever JD, Platt TG, Morton ER. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu Rev Microbiol. 2012;66:265–283. doi: 10.1146/annurev-micro-092611-150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel C, Bodenhausen N, Gruissem W, Vorholt JA. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016;212:192–207. doi: 10.1111/nph.14036. [DOI] [PubMed] [Google Scholar]
- 48.Gilbert GS, Parker IM. The evolutionary ecology of plant disease: A phylogenetic perspective. Annu Rev Phytopathol. 2016;54:549–578. doi: 10.1146/annurev-phyto-102313-045959. [DOI] [PubMed] [Google Scholar]
- 49.Bever JD, Mangan SA, Alexander HM. Maintenance of plant species diversity by pathogens. Annu Rev Ecol Evol Syst. 2015;46:305–325. [Google Scholar]
- 50.Chung YA, Rudgers JA. Plant-soil feedbacks promote negative frequency dependence in the coexistence of two aridland grasses. Proc Biol Sci. 2016;283:20160608. doi: 10.1098/rspb.2016.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mine A, et al. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc Natl Acad Sci USA. 2017;114:7456–7461. doi: 10.1073/pnas.1702613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones SE, et al. Streptomyces exploration is triggered by fungal interactions and volatile signals. eLife. 2017;6:e21738. doi: 10.7554/eLife.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yandigeri MS, et al. Drought-tolerant endophytic Actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Growth Regul. 2012;68:411–420. [Google Scholar]
- 54.Yang J, Kloepper JW, Ryu C-M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009;14:1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Pietschke C, et al. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc Natl Acad Sci USA. 2017;114:E8488–E8497. doi: 10.1073/pnas.1706879114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao M, Teplitski M, Robinson JB, Bauer WD. Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol Plant Microbe Interact. 2003;16:827–834. doi: 10.1094/MPMI.2003.16.9.827. [DOI] [PubMed] [Google Scholar]
- 57.Bai Y, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 58.Cole BJ, et al. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 2017;15:e2002860. doi: 10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haney CH, Samuel BS, Bush J, Ausubel FM. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants. 2015;1:15051. doi: 10.1038/nplants.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finkel OM, Castrillo G, Herrera Paredes S, Salas González I, Dangl JL. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155–163. doi: 10.1016/j.pbi.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meena KK, et al. Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front Plant Sci. 2017;8:172. doi: 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat Methods. 2013;10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 63.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mirarab S, et al. PASTA: Ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol. 2015;22:377–386. doi: 10.1089/cmb.2014.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMurdie PJ, Holmes S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.