Significance
Pancreatic ductal adenocarcinoma is one of the most malignant human tumors for which there are no efficacious therapeutic strategies. This tumor type is characterized by an abundant desmoplastic stroma that promotes tumor progression. Yet recent studies have shown that physical or genetic elimination of the stroma leads to more aggressive tumor development. Here, we decided to reprogram the stromal tissue by identifying and subsequently targeting genes responsible for their protumorigenic properties. Comparative transcriptome analysis revealed several genes overexpressed in cancer-associated fibroblasts compared with those present in normal pancreata. We provide genetic evidence that one of these genes, Saa3, plays a key role on the protumorigenic properties of the stroma, opening the door to the design of future therapeutic strategies.
Keywords: CAFs, Saa3, PDAC, stroma, mouse models
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by the presence of abundant desmoplastic stroma primarily composed of cancer-associated fibroblasts (CAFs). It is generally accepted that CAFs stimulate tumor progression and might be implicated in drug resistance and immunosuppression. Here, we have compared the transcriptional profile of PDGFRα+ CAFs isolated from genetically engineered mouse PDAC tumors with that of normal pancreatic fibroblasts to identify genes potentially implicated in their protumorigenic properties. We report that the most differentially expressed gene, Saa3, a member of the serum amyloid A (SAA) apolipoprotein family, is a key mediator of the protumorigenic activity of PDGFRα+ CAFs. Whereas Saa3-competent CAFs stimulate the growth of tumor cells in an orthotopic model, Saa3-null CAFs inhibit tumor growth. Saa3 also plays a role in the cross talk between CAFs and tumor cells. Ablation of Saa3 in pancreatic tumor cells makes them insensitive to the inhibitory effect of Saa3-null CAFs. As a consequence, germline ablation of Saa3 does not prevent PDAC development in mice. The protumorigenic activity of Saa3 in CAFs is mediated by Mpp6, a member of the palmitoylated membrane protein subfamily of the peripheral membrane-associated guanylate kinases (MAGUK). Finally, we interrogated whether these observations could be translated to a human scenario. Indeed, SAA1, the ortholog of murine Saa3, is overexpressed in human CAFs. Moreover, high levels of SAA1 in the stromal component correlate with worse survival. These findings support the concept that selective inhibition of SAA1 in CAFs may provide potential therapeutic benefit to PDAC patients.
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies worldwide, and it is projected to be the second leading cause of cancer-related deaths by 2030 (1, 2). Despite extensive research efforts over the past decades, there have been few improvements in the diagnosis or treatment of the disease. Most of the patients are diagnosed at an advanced stage, when the tumor is unresectable and metastasis is already present. Furthermore, current therapies are based on chemotherapy agents and provide only a modest increase in survival, highlighting the need for new therapeutic strategies (3).
PDAC is characterized by an abundant desmoplasia that constitutes up to 90% of the total tumor volume. This stroma is composed mainly (around 90%) of cancer-associated fibroblasts (CAFs) (4, 5). CAFs secrete extracellular matrix (ECM) proteins as well as soluble factors (such as chemokines and cytokines) that stimulate cancer progression (6–8). Furthermore, it has also been reported that CAFs mediate drug resistance and immunosuppression (9–12). Hence, CAFs may represent an important target for anticancer therapy.
This concept has been recently challenged by two independent studies in which elimination of the stroma in genetically engineered mouse (GEM) PDAC models resulted in more aggressive tumors and reduced survival (13–15). However, CAFs are a heterogeneous population (6, 8); thus, it is conceivable that different CAFs may have differential pro- and antitumorigenic roles. Therefore, a better understanding of the role that these CAF subpopulations play in PDAC progression may allow their selective reprogramming to thwart their protumorigenic effects without having to resort to their physical elimination.
To this end, we have characterized the transcriptome of a protumorigenic subpopulation of CAFs defined by the expression of the platelet-derived growth factor receptor alpha (PDGFRα). These studies have revealed a series of highly overexpressed genes in these cells compared with those expressed in fibroblasts present in normal pancreata. The top overexpressed gene in our studies was Saa3, a member of the acute-phase serum amyloid A (SAA) apolipoprotein family found associated with high-density lipoproteins in plasma (16). SAA proteins are expressed mainly in the liver, although their expression has also been described in synovial tissue, placenta, adipocytes, and smooth muscle cells (17). Expression of SAA members is induced in injured tissues and cells, including atherosclerotic plaques and rheumatoid synovitis, as well as in the brain of Alzheimer’s patients and in certain tumor cells (18). Accumulation of these acute-phase proteins in the blood is also observed during chronic inflammation and cancer. Moreover, they are considered to be biomarkers whose expression is associated with tumor progression and reduced survival in many human cancers (18, 19).
Here, we report that Saa3 plays a key role in inducing the protumorigenic properties of PDGFRα+ CAFs. Unfortunately, Saa3 is also essential for the protumorigenic cross talk between these CAFs and their neighboring tumor cells, a property that may limit its potential therapeutic effect to those strategies that could specifically target Saa3 activity in CAFs. Importantly, we also report here that the protumorigenic activity of Saa3 is regulated by membrane palmitoylated protein 6 (Mpp6), a member of the peripheral membrane-associated guanylate kinases (MAGUK). Since this regulatory interaction between Saa3 and Mpp6 appears to take place only in CAFs, these observations open the door to the design of future therapeutic strategies by controlling the levels of expression of Mpp6.
Results
PDGFRα+ CAFs Promote Tumor Growth.
To characterize the populations of CAFs present in PDAC, we used a GEM tumor model previously generated by M. Barbacid's laboratory, the K-Ras+/LSLG12Vgeo;Trp53lox/lox;Elas-tTA/tetO-Cre compound strain, in which we can selectively induce the expression of a resident K-RasG12V oncogene and disable the p53 tumor suppressor in acinar cells during late embryonic development (20). In this study, we added a Rosa26LSLEYFP allele to have a color marker (EYFP) to identify those cells carrying the K-RasG12V/p53-null tumor-initiating mutations. These mice, designated as “KPeCY,” develop PDAC tumors with complete penetrance and an average latency of 3–4 mo. More importantly, they recapitulate the human disease, including the formation of a massive stromal desmoplasia made up of heterogeneous CAF populations. Furthermore, this stromal remodeling is characterized by a chronic inflammatory response. Therefore, we studied the functional implications of tumor-promoting inflammation mediated by CAFs by performing a comparative expression profiling of PDGFRα+ CAFs and PDGFRα+ normal pancreatic fibroblasts (NPFs) as previously described (21).
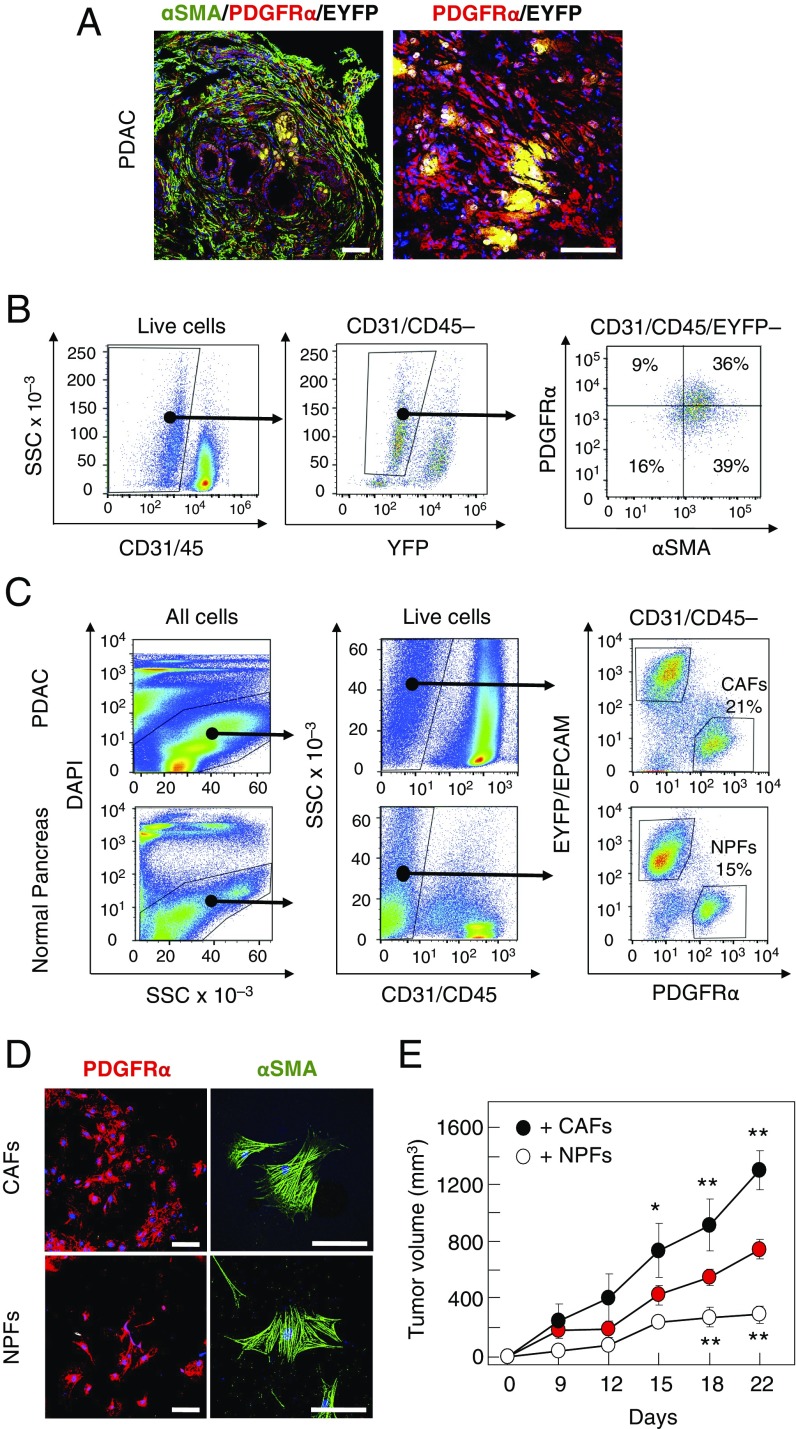
Immunofluorescence analysis of stromal tissue with antibodies elicited against α-smooth muscle actin (αSMA) and PDGFRα revealed that, whereas most stromal cells of KPeCY tumors expressed αSMA, only a subpopulation contained PDGFRα (Fig. 1A). FACS analysis of fresh tumors showed at least four distinct populations of stromal cells (Fig. 1B). Whereas one population only expressed αSMA (39% of the total), another population representing 36% of the cells contained both markers, αSMA and PDGFRα. We also identified two additional populations represented by those cells that expressed only PDGFRα (9%) and those that did not express either marker or expressed them at very low levels (16%) (Fig. 1B).
Fig. 1.
Protumorigenic properties of PDGFRα+ CAFs. (A, Left) Immunofluorescence staining with anti-αSMA (green) and anti-PDGFRα (red) antibodies and with EYFP (yellow) of a KPeCY PDAC tumor. (Right) Higher magnification of PDGFRα+ (red) and EYFP+ (yellow) cells. (Scale bars, 100 μm.) (B, Left) FACS analysis of fresh tumor samples with CD31/CD45 and EYFP markers. (Right) FACS analysis of CD31−/CD45−/EYFP− cells with anti-αSMA and anti-PDGFRα antibodies. The percentages of αSMA and PDGFRα single-positive cells as well as αSMA/PDGFRα double-negative and double-positive cells are indicated. (C) Cell sorting of KPeCY PDAC tumors and normal pancreata from control Elas-tTA/tetO-Cre;Rosa26+/LSLEYFP mice selected with DAPI, anti-CD31 and anti-CD45, anti-EpCAM, and anti-PDGFRα and EYFP. The percentages of NPFs and CAFs are indicated. (D) Immunofluorescence staining of sorted CAFs and NPFs after expansion in culture with anti-αSMA (green) and anti-PDGFRα (red) antibodies. (Scale bars, 50 μm.) (E) Growth of PDAC tumor cells (0.5 × 106) injected s.c. into immunocompromised mice either alone (red circles) or coinjected with the same amount of CAFs (black circles) or NPFs (open circles). *P < 0.05; **P < 0.001.
Next, we sorted PDGFRα+/EYFP−/CD45−/CD31− stromal cells from PDAC tumors of KPeCY mice as well as from normal pancreata of control Elas-tTA/tetO-Cre;Rosa26+/LSLEYFP animals (Fig. 1C and Fig. S1A). These cells represented 21% and 15% of the total cells, respectively. Cultures derived from cell population retained αSMA and PDGFRα expression and displayed the spindle shape characteristic of fibroblasts (Fig. 1D). The fibroblastic nature of these cells was further verified by the expression of fibroblast-specific genes including FAP, Vimentin, and PDGFRβ as well as by the lack of immune (CD45, CD68) and tumor (CK19, EpCAM) cell markers (Fig. 1C and Fig. S1B). These results indicate that the PDGFRα+ stromal cells isolated from KPeCY tumors represent CAFs, whereas those obtained from normal pancreata represent NPFs. The latter had slightly higher levels of αSMA than CAFs, a property that has been proposed to represent a marker of myofibroblast activation (Fig. S1C) (8). Herein, these PDGFRα+ cell populations are designated simply as “CAFs” and “NPFs.”
To determine whether these CAFs can promote tumorigenesis, we compared their tumor-supporting capabilities with those of NPFs using in vivo assays. EYFP+-sorted PDAC tumor cells (0.5 × 106) isolated from tumor-bearing KPeCY mice were s.c. inoculated alone (n = 4) or in combination with CAFs (0.5 × 106) (n = 4) or NPFs (0.5× 106) (n = 4) into the flanks of immunocompromised mice. Whereas CAFs stimulated tumor growth by as much as 76%, NPFs inhibited the proliferation of the pancreatic tumor cells by as much as 65% (Fig. 1E). These results illustrate the protumorigenic activity of the subpopulation of CAFs used in this study.
Comparative Transcriptional Profiles of CAFs vs. NPFs.
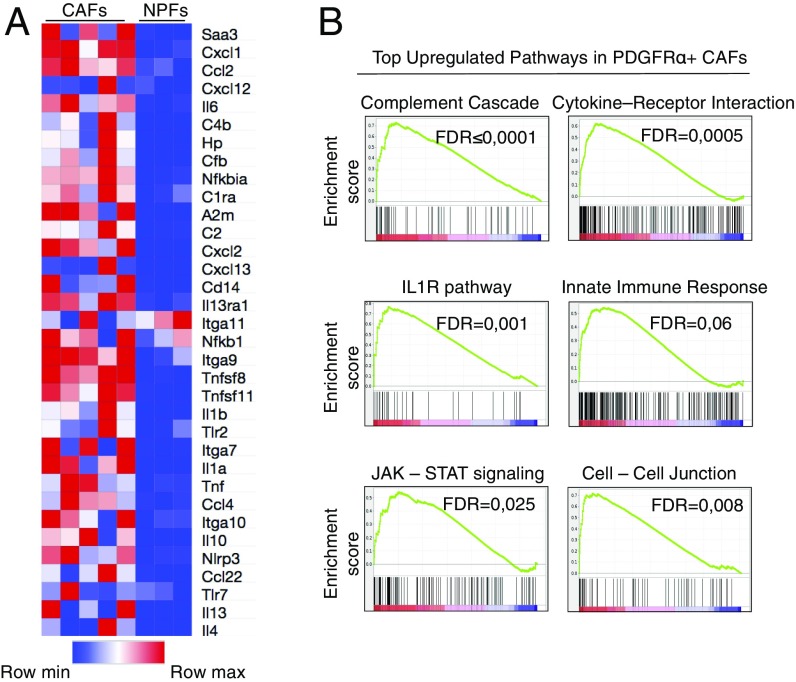
To understand the genetic bases responsible for the tumor-promoting activity of CAFs, we performed a comparative study of the transcriptome profiles of freshly isolated CAFs (n = 5) with those of NPFs (n = 3) by RNA sequencing (RNAseq) analysis. As illustrated in Fig. 2A, CAFs displayed a strong proinflammatory profile that included genes encoding competent cascade-related and acute-phase proteins (C4b, Hp, and A2m), cytokines (Il1b and Il6), chemokines (Cxcl1 and Ccl22), other interleukins (Il10), and members of the TNF family associated with the innate immune response. Furthermore, Gene Set Enrichment Analysis (GSEA) confirmed the up-regulation of the immune cell-recruiting complement cascade [false-discovery rate (FDR) ≤ 0001], IL1R (FDR = 0.001), and the Jak-Stat (FDR = 0.025) pathways in CAFs. This analysis also identified cytokine/receptor signaling as one of the most significantly up-regulated pathways in CAFs compared with NPFs (Fig. 2B and Fig. S2A). In addition, we observed a high enrichment in cell-to-cell junction pathways suggesting the presence of active contact-mediated signaling in CAFs (Fig. 2B and Fig. S2A), a property that may have an important role in tumor promotion (22). CAFs also displayed up-regulation of TGFβ- and Hedgehog-signaling pathways as well as ECM/receptor interactions (Fig. S2A), a set of transcriptional changes thought to result from their cross talk with the tumor cells (13, 23).
Fig. 2.
Transcriptional profiling of CAFs. (A) Heat map representing color-coded expression levels of differentially expressed inflammatory genes in CAFs (n = 5) vs. NPFs (n = 3). The heat map was generated from differential expression analysis, in which data were sorted by FPKM (fragments per kilobase of transcript per million mapped reads) expression value and log2 fold change with a q-value ≤0.05. The specific inflammatory gene set was selected from publicly available databases. (B) GSEA of CAFs significantly up-regulated in inflammatory and cell-adhesion pathways.
Down-regulated hallmarks include oxidative phosphorylation, suggesting that CAFs have undergone metabolic reprogramming and oxidative stress (Fig. S2A). The latter was further supported by the significant down-regulation of genes involved in peptide elongation, translation, and protein metabolism, hallmarks of stress responses indicating mitochondrial dysfunction. We also observed significant reduction of P53-signaling–related genes, suggesting cell death evasion (24, 25). These results, taken together, support the concept that the CAFs used in this study possess a unique secretory-inflammatory gene signature with emphasis on the activation of the innate immune response (Fig. 2) as well as impaired oxidative phosphorylation (Fig. S2A).
SAA3 Is Overexpressed in CAFs.
The top up-regulated gene in CAFs compared with NPFs was a gene encoding the SAA3 protein, Saa3 (Fig. 2A). These results were confirmed by qRT-PCR (Fig. S2B). Saa3 transcripts were also up-regulated compared with tumor cells as well as with other normal tissues such as pancreas or liver (Fig. S2B). Little is known regarding the putative role of Saa3 in cancer, although Hansen et al. (26) have suggested protumorigenic activity. During inflammation, Saa3 expression is induced by Il-1β, Tnfα, and Il-6 through NFκB signaling. In turn, Saa3 can also activate these cytokines (26, 27). Consistently, Il-1β, Tnfα, and Il-6, as well as the NFκB pathway, were found to be significantly up-regulated in our dataset (Fig. 2A and Fig. S2A).
PDAC Development in Saa3-KO Mice.
To interrogate the effect of ablating Saa3 expression in PDAC development, we incorporated Saa3-null alleles to the KPeCY strain. As previously reported, germline ablation of Saa3 does not induce detectable phenotypic alterations (28). Moreover, Saa3-null K-Ras+/LSLG12Vgeo;Elas-tTA/tetO-Cre mice displayed the same number of pancreatic intraepithelial neoplasia (PanIN) lesions and PDAC tumors as Saa3-competent animals (Fig. S3A). Likewise, KPeCY mice carrying either wild-type Saa3 alleles (n = 18) or Saa3-null alleles (n = 20) developed PDAC with 100% penetrance, succumbing to pancreatic tumors before 23 wk of age with median survivals of 15 and 16 wk, respectively. These observations, taken together, indicate that the absence of Saa3 expression from their germline has no significant effect on tumor development (Fig. S3B).
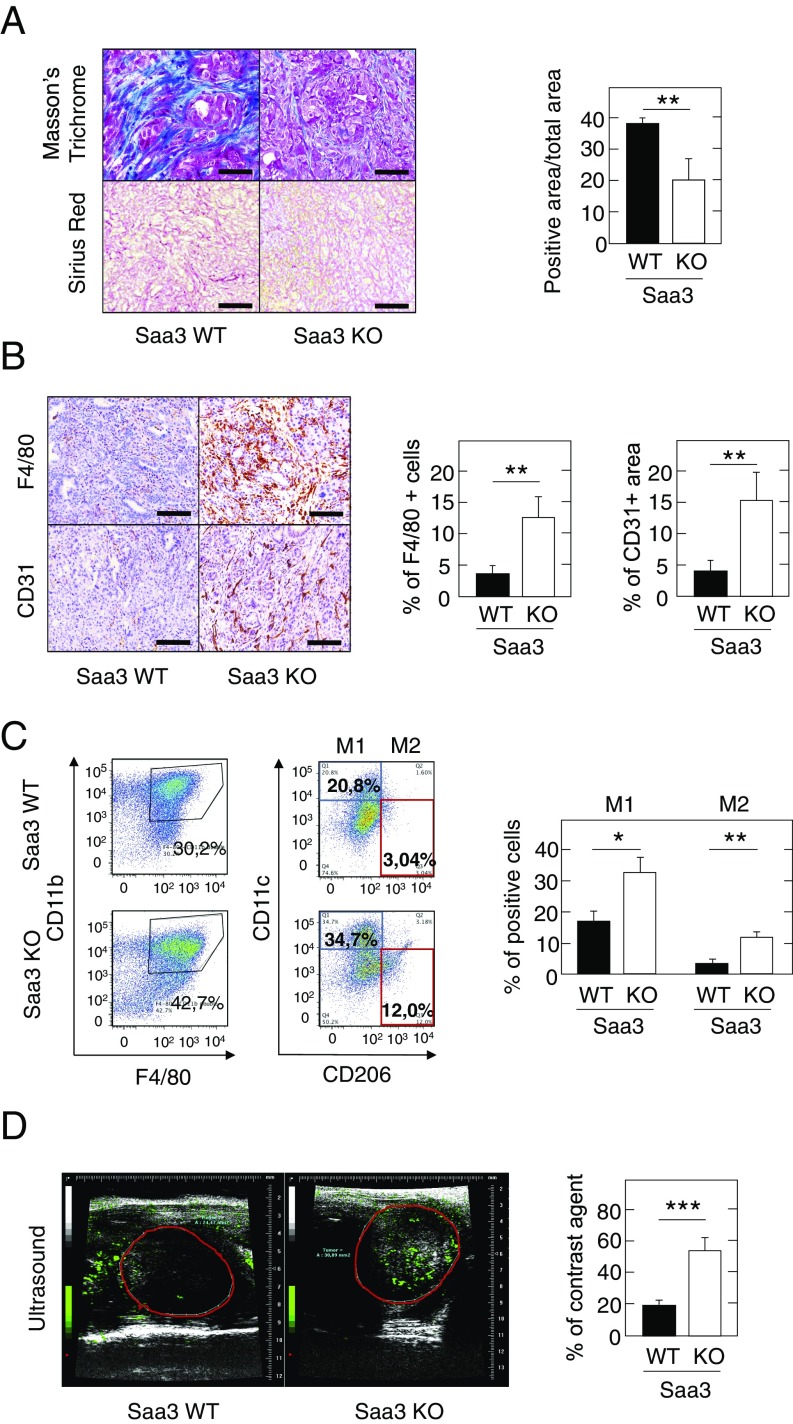
However, Saa3-null tumors had a higher proportion of EYFP+ tumor cells and displayed less dense fibrotic stroma, albeit these differences were not statistically relevant by FACS analysis (Fig. S3C). Histological analysis revealed that Saa3-null tumor cells were more packed than in control tumors and exhibit a significant reorganization of their ECM as revealed by their reduced levels of collagen content (Fig. 3A and Table 1). Interestingly, we did not observe a significant reduction in the PDGFRα+ population, indicating that the reduced stromal content is not due to the loss of this CAF subpopulation (Fig. S3D). Moreover, we observed a significant increase in the levels of macrophage infiltration in the Saa3-null tumors (12.5% vs. 3.8% of total area) (Fig. 3B and Table 1). The increase in tumor-infiltrating macrophages was observed in both the antitumorigenic M1 and the protumorigenic M2 populations (Fig. 3C). However, this increase appeared to be more pronounced in the M2 population, which has been associated with worse clinical outcome in PDAC patients (29). In contrast, there were no obvious differences in the amount or localization of neutrophils or T and B lymphocytes (Table 1 and Fig. S3E). Saa3-null tumors also displayed a significantly elevated number of endothelial CD31+ cells, indicating increased vessel density (Fig. 3B and Table 1). These vessels were functional as assessed by the greater perfusion observed upon injection of a contrast agent (Fig. 3D). Whether increased angiogenesis was promoted by the infiltrating macrophages, as previously suggested (30), remains to be determined.
Fig. 3.
Characterization of the stromal component of Saa3-null tumors. (A, Left) Masson’s trichrome and Sirius Red staining of collagen in Saa3-competent (WT) and Saa3-null (KO) tumors. (Scale bars, 50 μm.) (Right) Quantitative analysis of Masson’s trichrome-stained sections of Saa3-competent (WT) and Saa3-null (KO) tumors (n = 4). (B, Left) Representative images of F4/80 and CD31 immunostaining in Saa3-competent (WT) and Saa3-null (KO) tumors. (Scale bars, 50 μm.) (Right) Quantitative analysis of F4/80- and CD31-stained sections of Saa3-competent (WT) and Saa3-null (KO) tumors (n = 5). (C, Left) FACS analysis of fresh Saa3-competent (WT) and Saa3-null (KO) tumor samples with anti-F4/80 and anti-CD11b antibodies. (Center) FACS analysis of F4/80+/CD11b+ double-positive macrophages with anti-CD11c and anti-CD206 antibodies. The percentage of M1 (CD11chigh/CD206low) (blue square) and M2 (CD11clow/CD206high) (red square) macrophage populations is indicated for each tumor type. (Right) Quantitative analysis of M1 and M2 macrophages in Saa3-competent (WT) and Saa3-null (KO) tumors (n = 2). (D, Left) Micro-ultrasound images of Saa3-competent (WT) and Saa3-null (KO) tumors after injection of contrast agent. (Right) Quantitative analysis of vessel density in Saa3-competent (WT) and Saa3-null KO) tumors (n = 5). *P < 0.05; **P < 0.001; ***P < 0.001.
Table 1.
Most relevant features of Saa3-competent (WT) and Saa3-null (KO) KPeCY mice
| Features of KPeCY mice | Saa3 WT | Saa3 KO |
| Stroma reorganization | ||
| ECM | +++ | + |
| Vascularization | + | ++++ |
| Macrophage number and infiltration | + | ++++ |
| Other immune cell infiltration | + | + |
| CAFs | ||
| Tumor growth support | +++ | − |
| Wound healing property (in vitro) | ++ | +++ |
| Tumor cells | ||
| Differentiation | ++ | + |
| CSCs | + | +++ |
| Migratory properties (in vitro) | + | ++++ |
| Liver metastasis | ||
| Macrometastasis | ++ | − |
| Micrometastasis | ++ | + |
| Migratory tumor cells | + | ++++ |
Grading of the phenotypes is illustrated from not observed (–) to low (+), normal (++), high (+++), or very high (++++).
Next, we interrogated whether the effect of Saa3 ablation on stroma remodeling improved the therapeutic benefit of current drug treatments. As illustrated in Fig. S3F, Saa3-null mice exhibit a slight survival advantage when treated with either gemcitabine alone (50 mg/kg, i.p., twice weekly) or with gemcitabine in combination with clodronate, a macrophage-depleting agent. Similar results were obtained when we combined gemcitabine with B20.4.1.1, an anti-VEGF monoclonal antibody (Fig. S3F). However, the differences were not statistically significant.
Saa3-Null Tumor Cells Have Increased Migratory but Not Homing Properties.
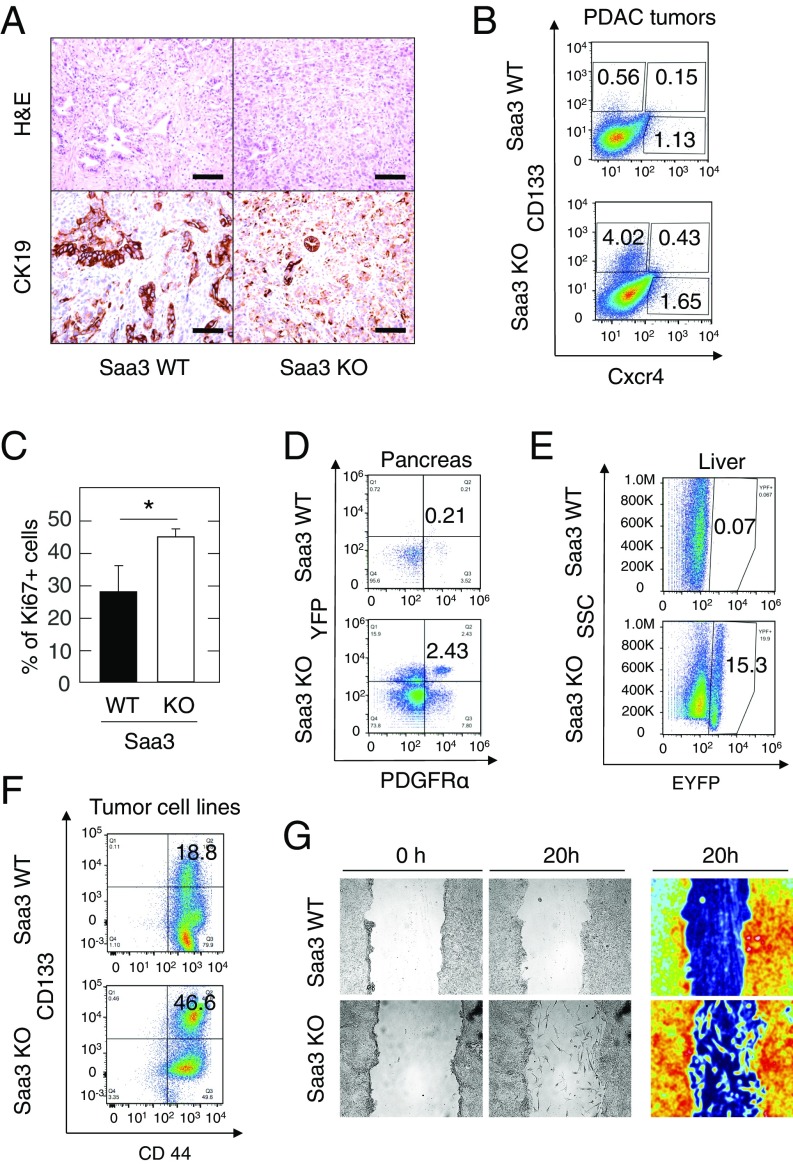
Tumors lacking Saa3 appeared less differentiated upon hematoxylin and eosin (H&E) and CK19 staining (Fig. 4A). The undifferentiated tumor phenotype has been associated with a cancer stem-like state in pancreatic cancer (31). Therefore, we assessed the cancer stem cell (CSC) compartment in Saa3-competent and Saa3-null tumors. As illustrated in Fig. 4B, we found a marked increase in CSC (CD133+) and metastatic CSC (CD133+/CXCR4+) populations in Saa3-competent PDAC tumors vs. those lacking Saa3 expression (0.56% vs. 4.02% CD133+ cells and 0.15% vs. 0.43% in CD133+/CXCR4+ cells, respectively) (Table 1). These results suggest that the absence of Saa3 confers a more invasive phenotype (32). Indeed, Saa3-null tumors showed a higher Ki67 proliferation index (Fig. 4C). In addition, EYFP+ tumor cells present in the pancreas of 8-wk-old Saa3-null mice displayed a considerably higher percentage of PDGFRα+ cells than those expressing Saa3 (2.43% vs. 0.21%, respectively) (Fig. 4D and Table 1). Since PDGFRα expression is a marker for epithelial-to-mesenchymal transition (EMT), these results suggest that the absence of Saa3 expression might promote the appearance of a migratory phenotype (33–35).
Fig. 4.
Phenotypic properties of Saa3-null tumor cells. (A) H&E staining (Upper) and CK19 immunostaining (Lower) of Saa3-competent (WT) and Saa3-null (KO) tumors. (Scale bars, 50 μm.) (B) FACS analysis of fresh tumor samples of Saa3-competent (WT) and Saa3-null (KO) tumors from KPeCY mice with anti-CD133 and anti-CXCR4. (C) Quantitative analysis of Ki67+ cells in Saa3-competent (WT) and Saa3-null (KO) tumor sections. (D) FACS analysis of EYFP-expressing PDGFRα+ pancreatic tumor cells in pancreas isolated from 8-wk-old Saa3-competent (WT) and Saa3-null (KO) KPeCY mice. (E) FACS analysis of EYPF-expressing cells in livers isolated from the same mice. (F) FACS analysis of pancreatic tumor cell lines with anti-CD133 and anti-CD44. (G, Left and Center) Migratory properties of Saa3-competent (WT) and Saa3-null (KO) tumor cells in an in vitro scratch assay. (Right) A color-enhanced picture for better visualization. *P < 0.05.
Since PDAC tumor cells most frequently metastasize to the liver, we examined the presence of Saa3-competent and Saa3-null EYFP+ pancreatic tumor cells in this tissue. As illustrated in Fig. 4E and Fig. S4A, we detected an unusually high number of disseminated tumor cells in the liver of KPeCY Saa3-null animals, representing as many as 15.3% of all liver cells. In contrast, the number of tumor cells in Saa3-competent animals represented only 0.07% of the liver cell population (Fig. 4E, Table 1, and Fig. S4A). However, these disseminated Saa3-null tumor cells did not proliferate and failed to propagate after colonization (Fig. S4B). Indeed, whereas 12 of 63 Saa3-competent mice (19%) displayed metastatic outgrowths, only 2 of 40 Saa3-null animals (5%) presented with metastatic lesions in their livers (P = 0.043) (Table 1). These reduced levels of metastatic outgrowth were not due to a reduction in the inflammatory cell population, CD11b+ and F4/80+ monocyte-derived immune cells, which are known to establish the metastatic niche (Fig. S4C) (36, 37), and thus suggest that the reduced metastatic potential of Saa3-null tumors is an intrinsic property of the tumor cells. Indeed, these infiltrated Saa3-null tumor cells were negative for Ki67 immunostaining, indicating that they have limited proliferative properties (Fig. S4B). However, it is also possible that absence of Saa3 expression in the liver may contribute to the limited proliferative and metastatic properties of these tumor cells.
It has been reported that Saa1 is a potent inducer of liver metastasis (26). In addition, SAA1 is among the top 50 up-regulated genes in the metastatic liver expression profile in a human PDAC dataset analyzed by Moffitt et al. (7), suggesting that SAA1 may be relevant in liver metastasis formation. Thus, we examined the levels of expression of other Saa family members in livers of Saa3-competent and Saa3-null tumor-bearing mice killed at the humane end point. As illustrated in Fig. S4D, the levels of expression of Saa1 and Saa2 are high in the livers of Saa3-competent, but not in Saa3-null, tumor-bearing mice. These results may also contribute to explaining why the abundant pancreatic tumor cells present in the livers of Saa3-null mice have limited metastatic potential.
To better characterize the effect of Saa3 ablation on the migratory properties of PDAC tumor cells, we generated cell lines from pancreatic tumors lacking this protein. FACS analysis confirmed that the number of CD133+/CD44+ CSCs was higher in Saa3-null EYFP+ tumor cell lines (46.6 vs. 18.8%) (Fig. 4F and Table 1), demonstrating a clear enrichment in this population (32, 38). Migration assays revealed that tumor cells lacking Saa3 displayed increased migratory properties (Fig. 4G and Table 1). While Saa3-expressing tumor cells advanced toward the scratch as a solid layer, cells lacking Saa3 moved freely throughout the scratch as individual cells (Fig. 4G). CAFs lacking Saa3 expression also had increased motility and closed the gap more efficiently than those expressing the protein (61.5% in Saa3-null vs. 40.8% in wild-type CAFs in a 16 h period) (Table 1 and Fig. S4E).
Saa3 Is Required for the Protumorigenic Properties of CAFs.
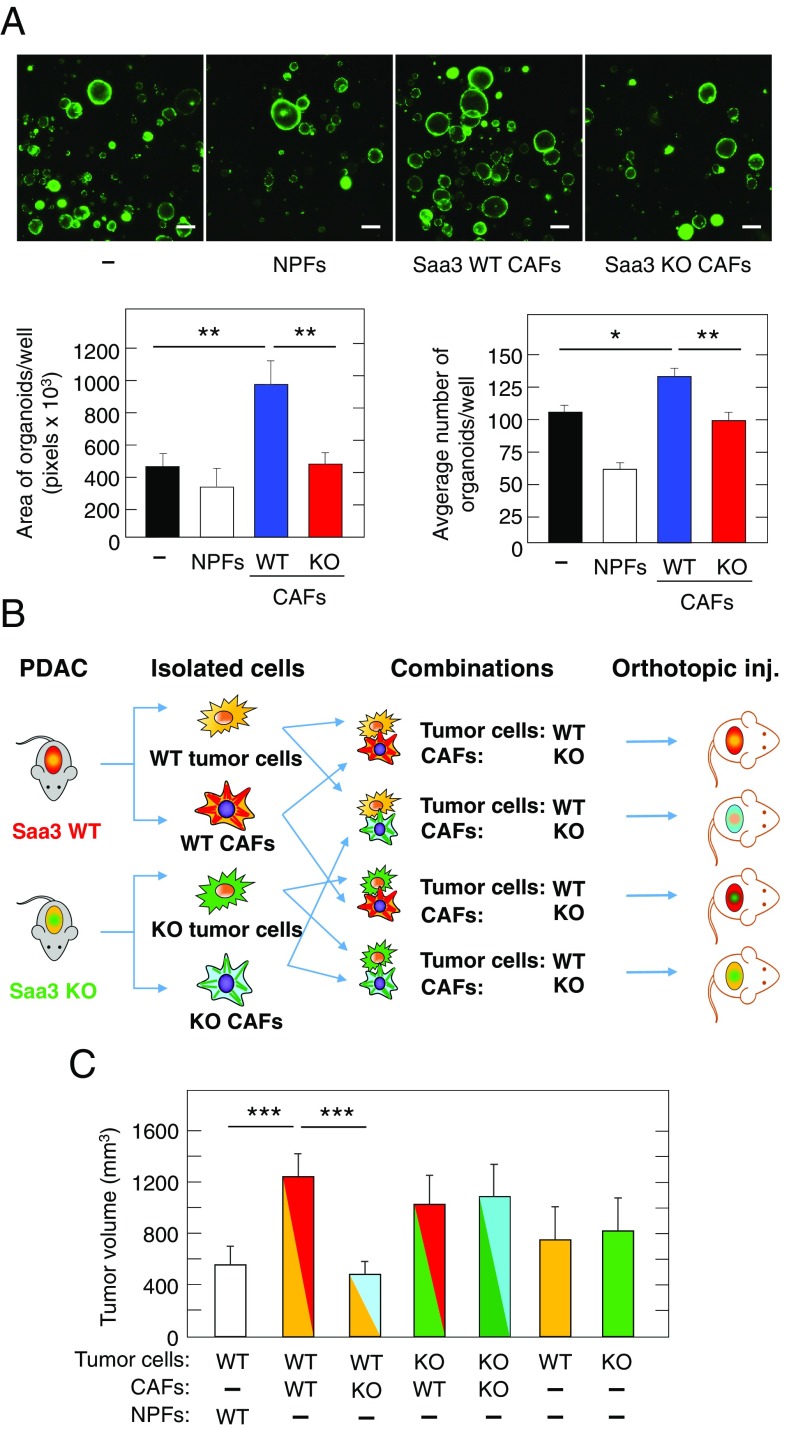
To characterize the effect of Saa3 on the interaction between pancreatic tumor cells and CAFs, we examined the growth properties of organoids generated from PDAC tumors of KPeCY mice cocultured with CAFs expressing or lacking Saa3. As illustrated in Fig. 5A, Saa3-expressing CAFs significantly increased the number and size of individual organoids (Fig. 5A). In contrast, Saa3-null CAFs failed to promote tumor growth resulting in organoid cultures similar to those grown in the absence of CAFs (Fig. 5A and Table 1). As expected, NPFs effectively reduced the growth of organoids. These results clearly indicate that Saa3 plays a key role on the ability of CAFs to support tumor cell growth.
Fig. 5.
Cross talk between pancreatic tumor cells and CAFs in the presence and absence of Saa3. (A, Upper) Cultures of EYFP tumor organoids grown in the presence of NPFs or Saa3-competent (WT) or Saa3-null (KO) CAFs. (Scale bars, 100 μm.) (Lower) Quantification of area (Left) and number of organoids (Right) under the indicated culture conditions. (B) Diagram depicting the in vivo orthotopic tumor assays in immunodeficient mice carried out to determine the protumorigenic properties of Saa3-competent (WT) (red) and Saa3-null KO) (light blue) CAFs on pancreatic tumor cells isolated from Saa3-competent (WT) (yellow) and Saa3-null (KO) (green) tumors. (C) Quantitative analysis of orthotopic tumor growth in immunodeficient mice inoculated with the indicated combinations of Saa3-competent (WT) and Saa3-null (KO) CAFs and pancreatic tumor cells. Color coding is as in B. NPFs (open bar) were used as a negative control. *P < 0.05; **P < 0.001; ***P < 0.001.
To explore the effect of Saa3 in the cross talk between tumor cells and CAFs in vivo, we orthotopically inoculated immunocompromised mice with CAFs as well as with tumor cells (0.5 × 106 each) either expressing or lacking Saa3 in (Fig. 5B). As illustrated in Fig. 5C, ablation of Saa3 in tumor cells (n = 6) had no effect on their ability to induce tumors. As expected, based on the results described above using in vitro assays, coinjection of Saa3-expressing tumor cells with NPFs (n = 6) reduced tumor growth whereas coinjection with Saa3-expressing CAFs (n = 8) led to a significant increase in tumor volume. Interestingly, when these tumor cells were coinjected with CAFs lacking Saa3 (n = 8), tumor growth was significantly reduced to levels even lower than those observed with NPFs, indicating that Saa3 is essential for the ability of CAFs to stimulate tumor growth in vivo. However, this effect was not observed when we coinjected Saa3-null tumor cells along with Saa3-null CAFs (n = 6) (Fig. 5C and Table 1). No significant differences were observed in the proliferation (Ki67 and Phospho-HistoneH3) or apoptosis (cleaved caspase 3) levels that could explain the differences in tumor volume induced by Saa3-competent vs. Saa3-null CAFs (Fig. S5). These observations indicate that, whereas Saa3 provides protumorigenic properties to CAFs, the antitumorigenic effect of Saa3-null CAFs requires that the corresponding tumor cells express the Saa3 protein. Likewise, we also observed a protumorigenic effect when we coinjected Saa3-null tumor cells with Saa3-null CAFs, suggesting that when both cell types are deficient in Saa3 expression there is an alternative cross talk that promotes tumor progression. These results provide an explanation as to why tumor development in Saa3-null mice is not affected, since the potential tumor-inhibitory effect of Saa3 ablation in CAFs does not take place when their neighboring tumor cells also lack Saa3.
Saa3 Ablation Alters the Transcriptional Profile of CAFs and Tumor Cells.
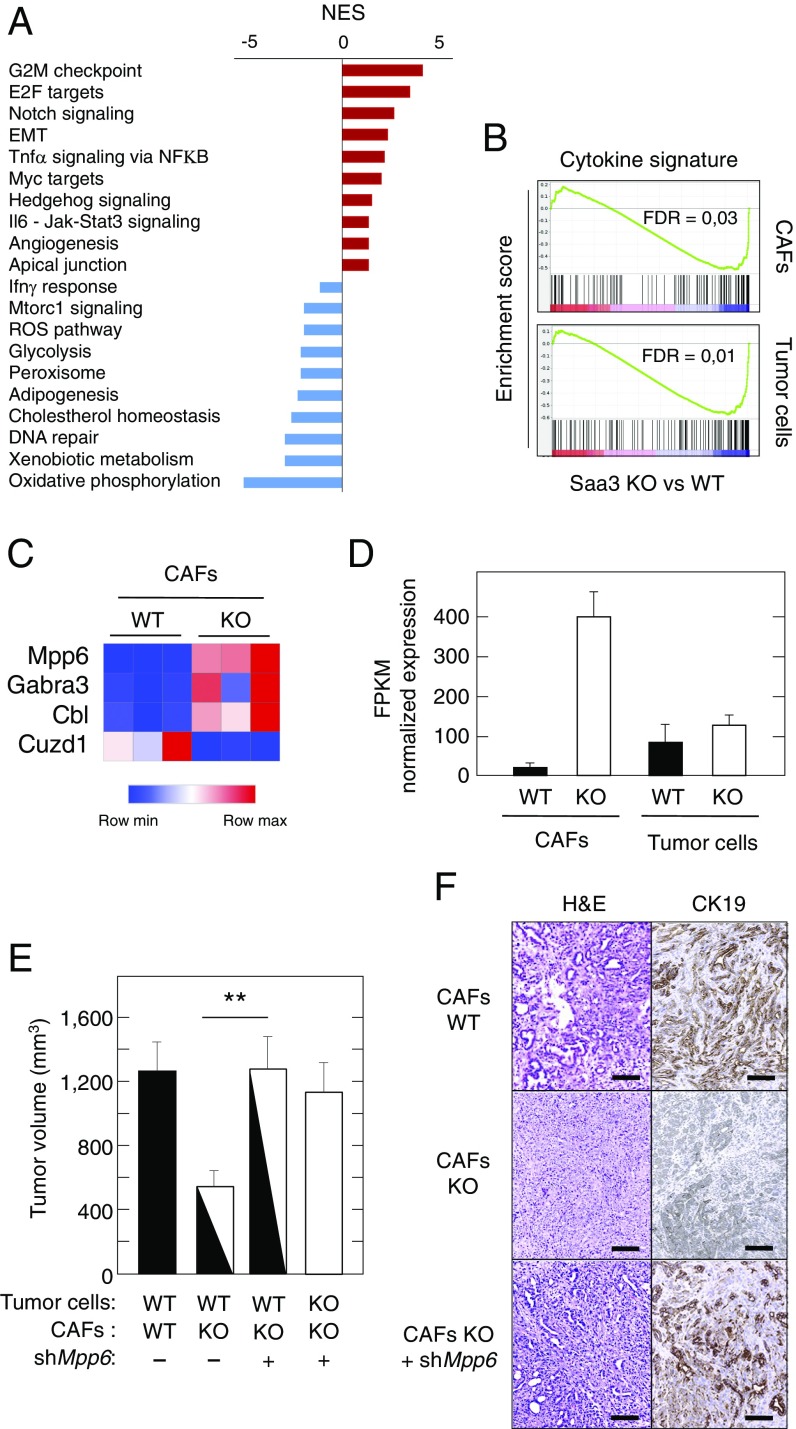
To dissect the mechanism by which Saa3 confers tumor stimulatory properties to CAFs, we used RNAseq to compare the transcriptome of Saa3-null and Saa3-proficient CAFs. GSEA pathway analysis of Saa3-null CAFs revealed a significant enrichment in Proliferation and Angiogenesis hallmarks, as well as up-regulation of Sonic Hedgehog, TNFα/NFκB, and IL-6 pathways. Moreover, we observed enrichment in genes implicated in EMT, suggesting increased plasticity of Saa3-null CAFs as well as their potential effect in inducing an undifferentiated phenotype in their neighboring tumor cells (39). In addition, the Saa3-null CAFs displayed up-regulation of the apical junction pathway, a property that predicts increased physical contact between stromal and tumor cells (22).
The most down-regulated gene sets included the oxidative phosphorylation and drug metabolism and cholesterol homeostasis (Fig. 6A). Moreover, loss of Saa3 expression also down-regulated other metabolic pathways such as Glycolysis. This pathway is activated in Saa3-competent CAFs, possibly playing a role in providing metabolites to their neighboring tumor cells (25). These observations suggest that loss of Saa3 might induce metabolic reprogramming of CAFs along with a reduction in the production of nutritional metabolites available to the adjacent tumor cells.
Fig. 6.
Transcriptional profiling of Saa3-null CAFs and pancreatic tumor cells. (A) GSEA pathway analysis of Saa3-null vs. Saa3-proficient CAFs. The normalized enrichment score (NES) ranking was generated by the GSEA. (B) GSEA analysis of a specific cytokine signature (144 cytokines) in Saa3-null (KO) CAFs and pancreatic tumor cells. (C) Heat map of the differentially expressed genes in Saa3-null (KO) CAFs compared with Saa3-competent (WT) CAFs. (D) RNAseq analysis of Mpp6 expression in Saa3-competent (WT, solid bars) and Saa3-null (KO, open bars) CAFs and tumor cells. (E) Tumor growth of orthotopic allografts of immunocompromised mice of Saa3-competent (WT) and Saa3-null (KO) pancreatic tumor cells in the presence of Saa3-competent (WT) and Saa3-null (KO) CAFs treated (+) or nontreated (−) with a shRNA against Mpp6. Tumor volume is indicated by solid (WT cells), open (KO cells), and mixed solid/open (WT and KO cells) bars. (F) Images of H&E and CK19 staining of orthotopic tumors illustrated in E. (Scale bars, 100 μm.) **P < 0.001.
We also studied the transcriptomes of Saa3-competent and Saa3-null tumor cells (Fig. S6A). This analysis revealed that loss of Saa3 expression in pancreatic tumor cells results in a significant enrichment of cell cycle- and metabolism-related gene sets, thus confirming the increased proliferative capacity of the Saa3-null tumor cells. In addition, these mutant tumor cells displayed up-regulation of the tight junction pathway, suggesting deregulated cell-to-cell contact properties. On the other hand, we observed significant down-regulation in ECM reorganization-related pathways, suggesting a decrease in the levels of extracellular collagen, a feature that might explain the higher migratory properties of Saa3-null tumor cells (Fig. S6A).
Saa3 is involved in the regulation of several inflammatory cytokines (26, 27). Thus, we examined the profile of cytokine enrichment changes by GSEA analysis utilizing a specific signature of 144 cytokines. As illustrated in Fig. 6B, elimination of Saa3 down-regulated the global cytokine profile not only in CAFs but also in the tumor cell compartment. Taken together, these results indicate that elimination of Saa3 induces proliferation, metabolic reprogramming, and cell-to-cell contact in both CAFs and tumor cell population. On the other hand, lack of Saa3 reduces overall cytokine secretion with the exception of the TNFα and IL-6 pathways in CAFs.
Mpp6 Down-Regulation Reverts the Antitumorigenic Properties of Saa3-Null CAFs.
Differential expression analysis of the above dataset revealed the presence of three significantly up-regulated genes in Saa3-null CAFs. In particular, the gene encoding MPP6 (fold change = 15.8), a member of the palmitoylated membrane protein subfamily of peripheral membrane-associated MAGUK guanylate kinases (Fig. 6 C and D). The other up-regulated genes included those encoding the γ-aminobutyric acid receptor 3 (GABRA3; fold change = 3.2) and Cbl, an E3 ubiquitin–protein ligase involved in cell signaling and protein ubiquitination (fold change = 2.3) (Fig. 6C). These observations were validated for Mpp6 using qRT-PCR analysis of Saa3-null and -competent CAFs (Fig. S6B).
To determine whether Mpp6 up-regulation was functionally responsible for the antitumorigenic effect of Saa3-null CAFs, we knocked down Mpp6 expression using specific shRNAs that resulted in a significant decrease of its expression levels. Mpp6–down-regulated Saa3-null CAFs were coinjected orthotopically with Saa3-competent PDAC tumor cells (n = 4). These tumor cells grew significantly faster than those coinjected with Saa3-null CAFs (n = 8), reaching proliferation levels similar to those observed with Saa3-competent CAFs (Fig. 6E). Down-regulation of Mpp6 also reverted the undifferentiated phenotype of tumor cells in the presence of Saa3-null CAFs (Fig. 6F). These observations, taken together, indicate that the growth-inhibitory activity of Saa3-null CAFs on their adjacent tumor cells is mediated by the up-regulation of the tight junction protein Mpp6. Interestingly, Saa3 ablation did not alter the levels of expression of Mpp6 in pancreatic tumor cells (Fig. 6D), indicating that Saa3 selectively controls the expression of Mpp6 in CAFs. These observations open the door to future therapeutic strategies aimed at up-regulating Mpp6 expression.
SAA1, the Functional Human Ortholog of Saa3, Is Up-Regulated in a Subpopulation of Human CAFs Associated with Poor Clinical Outcome.
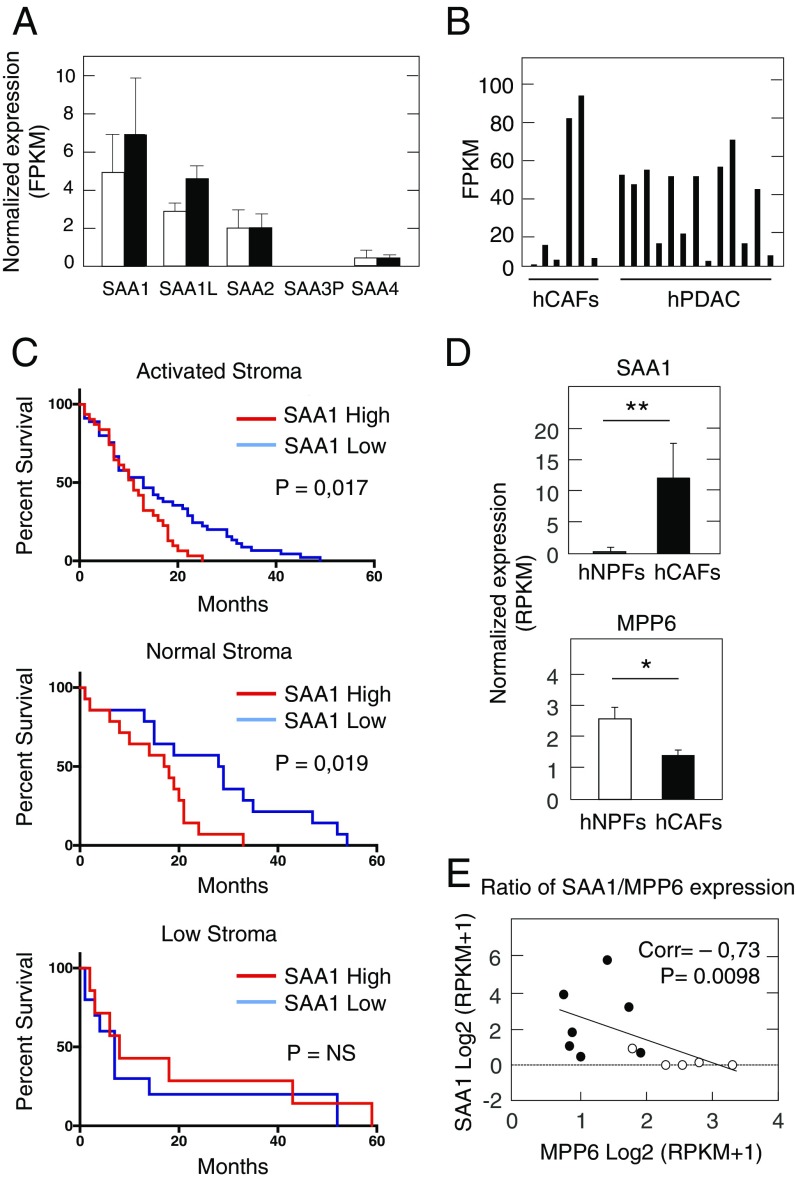
The human genome contains three genes encoding highly related SAA family members (40% amino acid identity): SAA1, SAA2, and SAA4 (40). It also contains a nonfunctional SAA3 pseudogene (SAA3P) (41). Among the three SAA functional genes, SAA1 is the most similar in structure and function to murine Saa3. The SAA1 protein is expressed in several stromal cell types including activated synovial fibroblasts (42). Moreover, SAA1 has been shown to control neutrophil plasticity and has anti- and protumorigenic inflammatory properties in melanoma (43). SAA1 is highly expressed in a variety of tumors including PDAC [The Cancer Gene Atlas (TCGA) database]. Moreover, SAA1 is the SAA family member most highly expressed in CAFs of PDAC tumors (Fig. 7A). To further confirm these observations, we examined the SAA1 expression levels in the PDAC RNAseq and microarray dataset recently published by Moffitt et al. (7). RNAseq data revealed high but variable expression of SAA1 both in tumors and in CAFs isolated from PDAC samples (Fig. 7B). Moffitt’s report described two types of PDAC-associated stroma, “normal” and “activated,” based on stromal signatures considering high αSMA expression or an inflammatory signature, respectively (7). Although, SAA1 was found to be expressed primarily in tumor samples with an “activated” stroma signature, high levels of SAA1 expression correlated with significantly worse survival in tumor samples containing either “normal” or “activated” stroma (Fig. 7C). In those tumor samples that contained low amounts of stroma, high SAA1 expression correlated with a slight increase in survival, suggesting that the protumoral effect of SAA1 overexpression is primarily mediated by the stromal cells. In addition, SAA1 was identified among the top 50 genes of Moffitt’s liver-specific metastatic signature in PDAC.
Fig. 7.
SAA1 and MPP6 expression in human PDAC tumors. (A) Normalized gene-expression values of SAA family members in human NPFs (open bars) and CAFs (solid bars) by RNAseq. (B) FPKM values of SAA1 expression by RNAseq in human CAFs and PDAC samples obtained from Moffitt’s dataset (7). (C) Kaplan–Meier survival analysis of PDAC patients with high (red) or low (blue) SAA1 expression levels classified by the presence of activated (Top) or normal (Middle) stroma signatures and in PDAC tumors with low stroma content (Bottom) based on microarray data from Moffitt’s dataset (7). (D) SAA1 (Upper) and MPP6 (Lower) expression in freshly sorted human NPFs (open bars) (n = 5) and CAFs (solid bars) (n = 7). (E) Correlation of SAA1 and MPP6 expression in freshly sorted human CAFs (solid circles) (n = 7) and human NPFs (open circles) (n = 5). Spearman’s correlation (Corr) and the P value are indicated. *P < 0.05;**P < 0.001.
Analysis of freshly isolated CAFs from tumor samples of PDAC patients (n = 7) and from adjacent normal pancreas (n = 5) revealed that SAA1 is up-regulated in the CAF samples compared with those obtained from normal pancreata (log2 fold change = 3.74; Padj < 0.005). In contrast, the levels of MPP6 expression were lower in CAFs than in normal pancreatic fibroblasts (Fig. 7D). That is, the levels of both genes, SAA1 and MPP6, inversely correlated in both types of fibroblasts (Fig. 7E). Similar results were obtained with Moffitt’s dataset (Fig. S6C). These results support the concept that SAA1 may play a role in human PDAC similar to that described for Saa3 in GEM tumors.
Discussion
PDAC is characterized by a rich desmoplastic stroma composed mainly of a heterogeneous population of CAFs. A CAF subpopulation characterized by expressing PDGFRα is thought to mediate an inflammatory response (21). However, their putative protumorigenic activity has not been properly documented. Here, we show that PDGFRα+ CAFs possess protumorigenic properties in vivo based on their ability to promote the growth of coinjected pancreatic tumor cells in immunocompromised mice. This property is specific for PDGFRα+ fibroblasts isolated from PDAC tumors, since the corresponding PDGFRα+ fibroblasts isolated from normal pancreata inhibited tumor growth.
Recent studies have described distinct populations of CAFs (6, 8, 44). A subpopulation designated as “myCAFs” is characterized by elevated expression of αSMA and appears to localize immediately adjacent to the neoplastic cells. A distinct subpopulation, iCAFs, is located more distantly from the neoplastic cells and expresses low levels of αSMA. Instead, these cells display higher levels of secreted IL-6 and other inflammatory mediators (44). The CAFs isolated in our study, based on the expression of PDGFRα, also have high levels of IL-6, suggesting that they may represent iCAFs. Other similarities between these iCAFs and the PDGFRα+ CAFs isolated here include significant up-regulation of cytokine/chemokine receptor-signaling pathways and JAK-STAT signaling. However, the PDGFRα+ CAFs characterized in this study display significant overexpression of innate immune response-related signaling and high enrichment in cell-to-cell junction pathways, two properties not reported in iCAFs, thus, suggesting that the PDGFRα+ CAFs described here might represent yet another subpopulation of inflammatory CAFs.
Transcriptome analysis of the PDGFRα+ CAFs studied here revealed a series of selectively up-regulated genes compared with those fibroblasts present in normal pancreata. The top-scoring gene was Saa3, a member of the gene family encoding Saa proteins. In humans, SAA1 and SAA2 are secreted during the acute phase of inflammation and have been implicated in several chronic inflammatory diseases, such as rheumatoid arthritis, atherosclerosis, and amyloidosis. Another member of this gene family, SAA3, is not expressed in human cells but has been shown to be a major acute-phase reactant in other species such as rabbits and rodents (16, 45). Murine Saa3 has been shown to be expressed in macrophages (46) and adipose tissue (47). During inflammatory processes, Saa3 expression is effectively induced by Il-1β, Tnfα, and Il-6 through NFκB signaling. Interestingly, these cytokines, as well as the NFκB pathway, were found to be significantly up-regulated in our CAF dataset. Finally, a fourth member of this gene family, SAA4, appears to be expressed constitutively in the liver (48).
Germline elimination of Saa3 had no effect of PDAC development, as reflected by the similar number of lesions observed in Saa3-null mice and by the lack of benefit in survival. However, Saa3-null tumors exhibit stroma remodeling, including reduced fibrosis and ECM, infiltrating macrophages, and increased vessel density. Indeed, Saa3-null CAFs had an elevated angiogenesis signature as revealed by GSEA pathway analysis. It has been suggested that increased vessel density along with a reduction in fibrosis may improve the efficacy of chemotherapy treatments (23, 49, 50). However, we did not observe a significant increase in the therapeutic benefit of tumor-bearing Saa3-null mice treated with gemcitabine alone, with gemcitabine in combination with clodronate, or with an anti-VEGF monoclonal antibody.
Saa3-null tumors were also less differentiated and more invasive, as suggested by a higher proliferation index and increased numbers of pancreatic CD133+ CSCs (32, 38). In addition, Saa3-null tumor cells showed an enhanced migratory phenotype. We observed an unexpected abundance of Saa3-null tumor cells in the liver, constituting as much as 15% of all liver cells, during the early stages of pancreatic tumor development. However, these migrating tumor cells did not elicit metastatic outgrowths, possibly due to their observed lack of proliferative capacity within the Saa3-null liver microenvironment. Whether this migratory phenomenon is an intrinsic property of the Saa3-null tumor cells or is a consequence of the absence of this protein in liver tissue and/or in prometastatic macrophages remains to be determined. Interestingly, we observed that the absence of Saa3 in liver tissue of tumor-bearing mice inhibits the expression of the Saa1 and Saa2 isoforms. Since SAA1 has been described as a potent inducer of liver metastasis (7, 26), the reduce levels of expression of Saa1/Saa2 in Saa3-null livers could explain why the abundant disseminated pancreatic tumor cells cannot form metastatic foci. Conditional ablation of Saa3 expression in specific cell populations, including tumor cells, CAFs, macrophages, and possibly other immune cells, should help to better define the role of Saa3 during the various stages of tumor development.
Saa3 is required for the protumorigenic properties of CAFs. Indeed, orthotopic coinjection of Saa3-null CAFs with Saa3-competent tumor cells in the pancreas of nude mice significantly reduced tumor size. The inhibitory effect of Saa3-null CAFs was even more pronounced than that induced by NPFs. However, this inhibitory effect was not observed when we used Saa3-null tumor cells. Since loss of Saa3 expression had no significant effect on the tumorigenic properties of pancreatic tumor cells, the observed lack of an antitumorigenic effect of Saa3-null CAFs on Saa3-null tumor cells must be due to defective cross talk between Saa3-null tumor cells and Saa3-null CAFs. These results were also observed in vitro using tumor organoids, ruling out a putative role of a third cellular partner in this cross talk. These reconstruction experiments recapitulate the results obtained with Saa3-null mice in which both CAFs and tumor cells are devoid of Saa3, thus explaining why PDAC development is unaffected in Saa3-null mice. Taken together, these results underscore the critical role for Saa3 in mediating the interaction between CAFs and tumor cells and predict that selective elimination of Saa3 in CAFs might provide significant therapeutic benefit. Future studies using conditional ablation of Saa3 should help better define the protumorigenic role of Saa3 in CAFs.
Comparative transcriptional profiling of tumor cells and CAFs expressing or lacking Saa3 revealed that Saa3-null cells display an increased proliferative signature and metabolic reprogramming and could suggest altered heterotypic cell-to-cell contact. Moreover, Saa3-deficient CAFs show reduced overall cytokine secretion, with the exception of the TNFα and IL-6 pathways. This result was unexpected, since acute-phase SAA apolipoproteins have been reported to enhance the expression of inflammatory cytokines, including IL-1β, IL-6, TNFα, IL-8, and G-CSF (51–53). Likewise, proinflammatory cytokines such as IL-1β, IL-6, and TNFα induce the synthesis of these SAA proteins (54). Therefore, up-regulation of TNFα, NFκB, and IL-6 pathways may result from an effort by the Saa3-null cells to induce the expression of Saa3, thereby generating an inflammatory loop. Finally, the reduction of glycolysis and the down-regulation of cholesterol homeostasis in Saa3-null CAFs, shown by GSEA pathway analysis, could contribute to their inhibitory activity on tumor cells observed in orthotopic coinjection experiments, by reducing nutrient transport.
Comparative analysis of Saa3-competent and Saa3-null CAFs revealed minor changes in their transcriptome. However, we identified three overexpressed genes in Saa3-deficient CAFs. Of particular interest was Mpp6, a member of the peripheral MAGUK family of proteins primarily involved in controlling epithelial cell polarity (55, 56). Mpps also function in tumor suppression and receptor clustering by forming multiprotein complexes containing distinct sets of transmembrane, cytoskeletal, and cytoplasmic signaling proteins (57). Interestingly, Mpp6 overexpression appears to be responsible for the loss of the protumorigenic effect of Saa3-null CAFs. Indeed, knockdown of Mpp6 expression in these mutant CAFs restored their protumorigenic properties, as determined in coinjection studies with Saa3-proficient pancreatic tumor cells in nude mice. Interestingly, the expression levels of Mpp6 were unaffected by the presence or absence of Saa3 in tumor cells, suggesting that the functional relationship between Saa3 and Mpp6 might be limited to CAFs. Understanding the molecular pathways implicated in the inhibitory role of Mpp6 on the protumorigenic effect of CAFs should unveil novel therapeutic opportunities.
Finally, we have interrogated whether our observations in GEM PDAC tumor models could be translated to a human scenario. As indicated above, the SAA3 locus is a nonexpressed pseudogene (41, 58). On the other hand, the acute-phase SAA1 protein has structural and functional characteristics that closely resemble those of murine Saa3, suggesting that SAA1 and Saa3 could be ortholog proteins (59). Indeed, SAA1 is overexpressed in human CAFs compared with NPFs. Moreover, high levels of SAA1 expression in the stromal component of human PDAC tumors correlate with significantly worse survival regardless of whether the tumor samples contain “normal” or “activated” stroma. However, high SAA1 expression in tumor samples with low stroma content also correlate with slightly increased survival, suggesting that SAA1 may have a protumoral effect when highly expressed in stroma and a possible antitumor effect when overexpressed in tumor cells. A dual role for SAA proteins depending on cellular context has already been reported by other investigators (18, 60). Finally, MPP6 levels inversely correlated with SAA1 expression in human PDAC samples. Therefore, our results support the concept that murine Saa3 may serve as a model to study the role of the human acute-phase SAA1 protein in pancreatic cancer and to develop novel therapeutic strategies against this potential target. Since SAA1 is a secreted protein, it is conceivable that its protumorigenic properties could be dwarfed by specific monoclonal antibodies, providing that they are primarily delivered to the desmoplastic stroma, avoiding as much as possible their interaction with the tumor cells. In addition, a better understanding of the signaling pathways driven by SAA1 should provide clues about differential signaling mechanisms in stromal vs. tumor cells.
Materials and Methods
Mice.
The K-Ras+/LSLG12Vgeo;Trp53lox/lox;Elas-tTA/tetO-Cre strain has been previously described (20). Rosa26+/LSLEYFP mice were obtained from The Jackson Laboratory (MGI: 2449038). Saa3-null sperm [Saa3tm1(KOMP)Vlcg] was obtained from the KOMP Repository (https://www.komp.org/geneinfo.php?geneid=78011) and was used to generate Saa3-null mice by in vitro fertilization of KPeCY females at the Spanish National Cancer Research Centre (CNIO) Transgenic Unit. Mice were maintained in a mixed 129/Sv-C57BL/6 background. Immunodeficient NU-Foxn1nu mice (5-wk-old females) were purchased from Harlan Laboratories. All animal experiments were approved by the Ethical Committees of the CNIO, the Carlos III Health Institute, and the Autonomous Community of Madrid and were performed in accordance with the guidelines stated in the International Guiding Principles for Biomedical Research Involving Animals, developed by the Council for International Organizations of Medical Sciences. All strains were genotyped by Transnetyx.
Human Samples.
Primary tumors were obtained from the Virgen de la Arrixaca Hospital. Specific informed consent for tumor implantation in mice was obtained from all patients, and the study received the approval of the CNIO Ethics Committee. Additional tumors used to isolate the different cell populations by cell sorting were obtained from patients at the University Hospital. This part of the study was approved by the ethical committee of the University of Heidelberg and was conducted in accordance with the Helsinki Declaration. Written informed consent was obtained from all patients.
Isolation of CAFs, NPFs, and Tumor Cells.
Mouse NPFs, CAFs, and tumor cells were obtained by mechanical dissociation of freshly dissected tumor tissue as previously described (61). Single-cell suspensions were immunostained with APC anti-mouse CD31 (1:200; clone MEC 13.3; BD Biosciences), APC anti-mouse CD45 (1:200; clone 30-F11; BD Biosciences), FITC anti-mouse CD326 (EpCAM; 1:200; clone: G8.8; BioLegend), and PE anti-mouse CD140a (PDGFRα; 1:100; clone: APA5; eBioscience). Samples were applied to an Influx cell sorter (BD Pharmingen) and were separated directly into TRIzol (RNA isolation) or PBS (cell culture). Analysis was performed by FlowJo software (Tree Star). Human CAFs and NPFs were obtained from fresh primary PDAC specimens and adjacent normal pancreas by cell sorting and were immunostained with FITC anti-human CD326 (EpCAM; 1:11; clone: AC128; Miltenyi Biotec); VioBlue anti-human CD45 (1:11; clone: 5B1; Miltenyi Biotec); and APC anti-human CD31 (1:11; clone: HEA-125; Miltenyi Biotec). Fibroblasts were defined as the EpCAM−/CD45−/CD31− population. RNA extraction and library preparation were performed as described elsewhere (62).
RNAseq, Gene-Expression Profiling, and GSEA Analysis.
Total RNA extracted from mouse cells by the Qiagen RNeasy Mini Kit enriched by polyA pull-down was used to prepare a cDNA library using the Illumina TruSeq RNA Library Preparation Kit. Libraries were sequenced with Illumina HiSeq 2000 at Columbia Genome Center. Reads were analyzed with the Nextpresso pipeline (63). Sequencing quality was checked with FastQC v0.11.0 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the human genome (GRCh37/hg19) with TopHat 2.0.10 using Bowtie 1.0.0 (64) and SAMtools 0.1.1.9 (65), allowing two mismatches and 20 multihits. GSEA was performed with GSEAPreranked (66), setting 1,000 gene set permutations. RNAseq data are available online in the National Center for Biotechnology Information (NCBI) Geo Database (GSE106901). Only gene sets with significant enrichment levels (FDR q-value <0.25) were considered. Human RNA extraction and library preparation were performed as described elsewhere (62).
S.c. and Orthotopic Allograft Models.
Tumor (0.5 × 106) cells only or in combination with 0.5 × 106 CAFs or NPFs were injected in PBS:Matrigel (1:1) into the dorsal flanks of the mice. Growth was measured every 3 d until the humane end point. Orthotopic injection was performed by surgery under anesthesia (4% isofluorane) using the same number of cells for injection as in the s.c. model. Tumor growth was monitored by micro-ultrasound (Vevo 770; VisualSonics). Mice were killed 3 wk postinjection. Tumor volume was limited to 1,500 mm3. Tumors were measured by caliper and calculated as length × width2/2.
Supplementary Material
Acknowledgments
We thank Beatriz Jiménez, María del Carmen G. Lechuga, Marta San Roman, Raquel Villar, and Silvia Jiménez for excellent technical assistance; Jaime Muñoz and Sagrario Ortega (CNIO Transgenic Unit) for help in generating Saa3-null mice; Isabel Aragón, Mayte Lamparero, Alejandra López, Patricia Villanueva, and Isabel Blanco (CNIO Animal Facility) for mouse work; Gloria Visdomine, Cristina Peñalba, and Francisca Mulero (CNIO Molecular Imaging Unit) for ultrasound studies; Manuel Pérez, Jesús Gómez, and Diego Megías (CNIO Confocal Microscopy Unit) for confocal imaging; Ultan Cronin and Lola Martínez (Flow Cytometry Unit) for FACS analysis; Nuria Cabrera and Alba de Martino (Histopathology Unit) for histopathological analysis; Manuel Morente for his advice with immunohistochemical (CNIO Tumor Bank); Corinna Klein for advice concerning the orthotopic studies; and the German Cancer Research Center-Heidelberg Center for Personalized Oncology (DKFZ-HIPO) for technical support and funding through the Grant HIPO-015. This work was supported by European Research Council Grants ERC-AG/250297-RAS AHEAD and ERC-AG/695566-THERACAN, Spanish Ministry of Economy and Competitiveness Grant SAF2014-59864-R, and Asociación Española contra el Cáncer Grant GC16173694BARB (to M. Barbacid). M.D. was supported by a fellowship from La Caixa International Fellowship Program. M. Barbacid is the recipient of an Endowed Chair from the AXA Research Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717802115/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, et al. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 4.Xing F, Saidou J, Watabe K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci (Landmark Ed) 2010;15:166–179. doi: 10.2741/3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol. 2017;10:76. doi: 10.1186/s13045-017-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffitt RA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 9.Crawford Y, et al. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell. 2009;15:21–34. doi: 10.1016/j.ccr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Garrido-Laguna I, et al. Tumor engraftment in nude mice and enrichment in stroma- related gene pathways predict poor survival and resistance to gemcitabine in patients with pancreatic cancer. Clin Cancer Res. 2011;17:5793–5800. doi: 10.1158/1078-0432.CCR-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feig C, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Özdemir BC, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JJ, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci USA. 2014;111:E3091–E3100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhim AD, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 17.Urieli-Shoval S, Cohen P, Eisenberg S, Matzner Y. Widespread expression of serum amyloid A in histologically normal human tissues. Predominant localization to the epithelium. J Histochem Cytochem. 1998;46:1377–1384. doi: 10.1177/002215549804601206. [DOI] [PubMed] [Google Scholar]
- 18.Malle E, Sodin-Semrl S, Kovacevic A. Serum amyloid A: An acute-phase protein involved in tumour pathogenesis. Cell Mol Life Sci. 2009;66:9–26. doi: 10.1007/s00018-008-8321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urieli-Shoval S, et al. Expression of serum amyloid a in human ovarian epithelial tumors: Implication for a role in ovarian tumorigenesis. J Histochem Cytochem. 2010;58:1015–1023. doi: 10.1369/jhc.2010.956821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell. 2010;17:135–147, erratum (2010) 17:523. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Labernadie A, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol. 2017;19:224–237. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lujambio A, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D, et al. Metabolic reprogramming of cancer-associated fibroblasts by IDH3α downregulation. Cell Rep. 2015;10:1335–1348. doi: 10.1016/j.celrep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Hansen MT, et al. A link between inflammation and metastasis: Serum amyloid A1 and A3 induce metastasis, and are targets of metastasis-inducing S100A4. Oncogene. 2014;34:424–435. doi: 10.1038/onc.2013.568. [DOI] [PubMed] [Google Scholar]
- 27.Reigstad CS, Lundén GO, Felin J, Bäckhed F. Regulation of serum amyloid A3 (SAA3) in mouse colonic epithelium and adipose tissue by the intestinal microbiota. PLoS One. 2009;4:e5842. doi: 10.1371/journal.pone.0005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.den Hartigh LJ, et al. Deletion of serum amyloid A3 improves high fat high sucrose diet-induced adipose tissue inflammation and hyperlipidemia in female mice. PLoS One. 2014;9:e108564. doi: 10.1371/journal.pone.0108564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, et al. Tumor-associated macrophages correlate with the clinicopathological features and poor outcomes via inducing epithelial to mesenchymal transition in oral squamous cell carcinoma. J Exp Clin Cancer Res. 2016;35:12. doi: 10.1186/s13046-015-0281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riabov V, et al. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front Physiol. 2014;5:75. doi: 10.3389/fphys.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zagorac S, et al. DNMT1 inhibition reprograms pancreatic cancer stem cells via upregulation of the miR-17-92 cluster. Cancer Res. 2016;76:4546–4558. doi: 10.1158/0008-5472.CAN-15-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Jechlinger M, et al. Autocrine PDGFR signaling promotes mammary cancer metastasis. J Clin Invest. 2006;116:1561–1570. doi: 10.1172/JCI24652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steller EJA, et al. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia. 2013;15:204–217. doi: 10.1593/neo.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekpe-Adewuyi E, Lopez-Campistrous A, Tang X, Brindley DN, McMullen TP. Platelet derived growth factor receptor alpha mediates nodal metastases in papillary thyroid cancer by driving the epithelial-mesenchymal transition. Oncotarget. 2016;7:83684–83700. doi: 10.18632/oncotarget.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan HH, et al. Gr-1+CD11b+ myeloid cells tip the balance of immune protection to tumor promotion in the premetastatic lung. Cancer Res. 2010;70:6139–6149. doi: 10.1158/0008-5472.CAN-10-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simeone DM. Pancreatic cancer stem cells: Implications for the treatment of pancreatic cancer. Clin Cancer Res. 2008;14:5646–5648. doi: 10.1158/1078-0432.CCR-08-0584. [DOI] [PubMed] [Google Scholar]
- 39.Baulida J. Epithelial-to-mesenchymal transition transcription factors in cancer-associated fibroblasts. Mol Oncol. 2017;11:847–859. doi: 10.1002/1878-0261.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Brien KD, Chait A. Serum amyloid A: The “other” inflammatory protein. Curr Atheroscler Rep. 2006;8:62–68. doi: 10.1007/s11883-006-0066-0. [DOI] [PubMed] [Google Scholar]
- 41.Kluve-Beckerman B, Drumm ML, Benson MD. Nonexpression of the human serum amyloid A three (SAA3) gene. DNA Cell Biol. 1991;10:651–661. doi: 10.1089/dna.1991.10.651. [DOI] [PubMed] [Google Scholar]
- 42.De Buck M, et al. Structure and expression of different serum amyloid A (SAA) variants and their concentration-dependent functions during host insults. Curr Med Chem. 2016;23:1725–1755. doi: 10.2174/0929867323666160418114600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Santo C, et al. Invariant NKT cells modulate the suppressive activity of IL-10-secreting neutrophils differentiated with serum amyloid A. Nat Immunol. 2010;11:1039–1046. doi: 10.1038/ni.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Öhlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell TI, Coon CI, Brinckerhoff CE. Serum amyloid A (SAA3) produced by rabbit synovial fibroblasts treated with phorbol esters or interleukin 1 induces synthesis of collagenase and is neutralized with specific antiserum. J Clin Invest. 1991;87:1177–1185. doi: 10.1172/JCI115116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ather JL, et al. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J Immunol. 2011;187:64–73. doi: 10.4049/jimmunol.1100500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommer G, et al. The adipokine SAA3 is induced by interleukin-1β in mouse adipocytes. J Cell Biochem. 2008;104:2241–2247. doi: 10.1002/jcb.21782. [DOI] [PubMed] [Google Scholar]
- 48.de Beer MC, et al. Characterization of constitutive human serum amyloid A protein (SAA4) as an apolipoprotein. J Lipid Res. 1995;36:526–534. [PubMed] [Google Scholar]
- 49.Jacobetz MA, et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut. 2013;62:112–120. doi: 10.1136/gutjnl-2012-302529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Provenzano PP, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furlaneto CJ, Campa A. A novel function of serum amyloid A: A potent stimulus for the release of tumor necrosis factor-alpha, interleukin-1beta, and interleukin-8 by human blood neutrophil. Biochem Biophys Res Commun. 2000;268:405–408. doi: 10.1006/bbrc.2000.2143. [DOI] [PubMed] [Google Scholar]
- 52.Koga T, et al. Serum amyloid A-induced IL-6 production by rheumatoid synoviocytes. FEBS Lett. 2008;582:579–585. doi: 10.1016/j.febslet.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 53.Jijon HB, Madsen KL, Walker JW, Allard B, Jobin C. Serum amyloid A activates NF-kappaB and proinflammatory gene expression in human and murine intestinal epithelial cells. Eur J Immunol. 2005;35:718–726. doi: 10.1002/eji.200425688. [DOI] [PubMed] [Google Scholar]
- 54.Jensen LE, Whitehead AS. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem J. 1998;334:489–503. doi: 10.1042/bj3340489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gosens I, et al. MPP1 links the Usher protein network and the Crumbs protein complex in the retina. Hum Mol Genet. 2007;16:1993–2003. doi: 10.1093/hmg/ddm147. [DOI] [PubMed] [Google Scholar]
- 56.Quinn BJ, et al. Erythrocyte scaffolding protein p55/MPP1 functions as an essential regulator of neutrophil polarity. Proc Natl Acad Sci USA. 2009;106:19842–19847. doi: 10.1073/pnas.0906761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng T-C, et al. VAM-1: A new member of the MAGUK family binds to human Veli-1 through a conserved domain. Biochim Biophys Acta. 2001;1518:249–259. doi: 10.1016/s0167-4781(01)00191-9. [DOI] [PubMed] [Google Scholar]
- 58.Larson MA, Wei SH, Weber A, Weber AT, McDonald TL. Induction of human mammary-associated serum amyloid A3 expression by prolactin or lipopolysaccharide. Biochem Biophys Res Commun. 2003;301:1030–1037. doi: 10.1016/s0006-291x(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 59.Lu J, Yu Y, Zhu I, Cheng Y, Sun PD. Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis. Proc Natl Acad Sci USA. 2014;111:5189–5194. doi: 10.1073/pnas.1322357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siegmund SV, et al. Serum amyloid a induces inflammation, proliferation and cell death in activated hepatic stellate cells. PLoS One. 2016;11:e0150893. doi: 10.1371/journal.pone.0150893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharon Y, Alon L, Glanz S, Servais C, Erez N. Isolation of normal and cancer-associated fibroblasts from fresh tissues by fluorescence activated cell sorting (FACS) J Vis Exp. 2013:e4425. doi: 10.3791/4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cabezas-Wallscheid N, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 63.Graña O, Rubio-Camarillo M, Fernandez-Riverola F, Pisano DG, Glez-Peña D. Nextpresso: Next generation sequencing expression analysis pipeline. Curr Bioinform. 2017;12 [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.