Significance
Cytotoxic chemotherapy is frequently used in patients with triple-negative breast cancer (TNBC). Although patients initially respond to the treatment, the cancer often comes back and kills the patient. Recent studies have demonstrated that cancer cells express genes that protect them from killing by immune cells, but the stimulus that prompts this response is unknown. We show that when TNBC cells are treated with chemotherapy, the surviving cells turn on genes that enable them to escape killing by the immune system. We identify hypoxia-inducible factors (HIFs), which are known to promote metastasis of TNBC, as responsible for this countertherapeutic effect. We show that coadministration of an HIF inhibitor with chemotherapy blocks the ability of surviving TNBC cells to evade the immune system.
Keywords: HIF-1, effector T cells, regulatory T cells, adenosine, PD-1
Abstract
Triple-negative breast cancer (TNBC) is treated with cytotoxic chemotherapy and is often characterized by early relapse and metastasis. To form a secondary (recurrent and/or metastatic) tumor, a breast cancer cell must evade the innate and adaptive immune systems. CD47 enables cancer cells to evade killing by macrophages, whereas CD73 and PDL1 mediate independent mechanisms of evasion of cytotoxic T lymphocytes. Here, we report that treatment of human or murine TNBC cells with carboplatin, doxorubicin, gemcitabine, or paclitaxel induces the coordinate transcriptional induction of CD47, CD73, and PDL1 mRNA and protein expression, leading to a marked increase in the percentage of CD47+CD73+PDL1+ breast cancer cells. Genetic or pharmacological inhibition of hypoxia-inducible factors (HIFs) blocked chemotherapy-induced enrichment of CD47+CD73+PDL1+ TNBC cells, which were also enriched in the absence of chemotherapy by incubation under hypoxic conditions, leading to T cell anergy or death. Treatment of mice with cytotoxic chemotherapy markedly increased the intratumoral ratio of regulatory/effector T cells, an effect that was abrogated by HIF inhibition. Our results delineate an HIF-dependent transcriptional mechanism contributing to TNBC progression and suggest that combining chemotherapy with an HIF inhibitor may prevent countertherapeutic induction of proteins that mediate evasion of innate and adaptive antitumor immunity.
Triple-negative breast cancers (TNBCs), which lack expression of the estrogen receptor, progesterone receptor, and HER2, comprise ∼15% of all breast cancers (1). Targeted therapy is not available for TNBC, and cytotoxic chemotherapy is the primary systemic treatment (2). After an initial response to chemotherapy, many patients with TNBC have recurrent disease, which is drug-resistant and metastatic, leading to increased mortality compared with other breast cancer subtypes (1, 2). Thus, to reduce patient mortality, it is critical to understand the properties of TNBC cells that survive chemotherapy.
The ability of cancer cells to evade both the innate and adaptive immune systems plays a critical role in cancer relapse and metastasis (3–5). In breast cancer, the frequency of intratumoral immune infiltrates is significantly decreased in invasive breast cancer as compared with ductal carcinoma in situ (6). Among patients with TNBC, the absence of intratumoral immune infiltrates is associated with patient mortality (7). The aggressive nature of the recurrent, metastatic disease that follows chemotherapy in patients with TNBC suggests a further loss of antitumor immunity, which is supported by limited in vitro studies (8), but the underlying molecular mechanisms have not been established.
Several studies have implicated hypoxia-inducible factor 1α (HIF-1α) in the regulation of innate and adaptive immunity. Conditional knockout (cKO) of HIF-1α in the granulocyte-monocyte lineage impaired inflammatory responses by these cells (9). Defective B cell development and autoimmunity were found in chimeric KO mice lacking expression of HIF-1α in B and T lymphocytes (10). HIF-1α cKO in T cells was associated with increased levels of proinflammatory cytokines and augmented antibacterial immunity (11). HIF-1α cKO selectively in CD8+ T cells led to impaired tumor infiltration and tumor cell killing (12). HIF-1α cKO selectively in CD4+ T cells led to decreased regulatory T (Treg) cell antiinflammatory effects in models of autoimmune encephalitis (13) and colitis (14). However, the systemic effect of pharmacological inhibition of HIF-1 activity on antitumor immunity has not been determined.
A major mechanism mediating evasion of innate immunity by cancer cells is the expression of CD47, a cell-surface protein that interacts with signal regulatory protein α on macrophages to block phagocytosis (15–17). HIF-1 stimulates CD47 expression in hypoxic breast cancer cells (18). However, whether chemotherapy also affects the expression of CD47 has not been investigated.
Tumor cells escape from adaptive immunity by altering the expression of the B7 family of costimulatory ligands, which modulate effector T (Teff) cell responses (19, 20). There has been great interest in the programmed cell death-1 (PD1) receptor, which is expressed on T cells and interacts with its cognate ligand PDL1 (also known as CD274) on cancer cells (21). Remarkable therapeutic responses to anti-PD1 or anti-PDL1 antibodies have occurred in a minority of patients with melanoma, non-small cell lung cancer, and renal carcinoma (22, 23). However, these immunotherapies are neither effective against all cancer types nor effective in every patient with an immunogenic cancer type. Cell surface expression of PDL1 or defects in mismatch repair correlate with therapeutic response in some cancers (23, 24). PDL1 is expressed by solid tumors but is not expressed in normal epithelial tissues (5), and the CD274 gene encoding PDL1 is amplified in some cases of TNBC (6). Several other mechanisms leading to increased PDL1 expression have been reported (25–27). Hypoxia induces PDL1 expression in tumor and immune cells in an HIF-1–dependent manner (28, 29).
The failure of many tumors to respond to immune checkpoint inhibitors may reflect the multiple immunosuppressive mechanisms employed by cancer cells. Extracellular adenosine is a potent immunosuppressor that accumulates during tumor growth (30, 31). Extracellular ATP is converted to AMP by the enzyme CD39, and the subsequent dephosphorylation of AMP to adenosine is catalyzed by the 5′-ectonucleotidase CD73. Adenosine binds to cognate A2A receptors on Teff cells, leading to anergy or cell death. A2A receptor signaling reduces the cytotoxic activity of CD8+ T cells and natural killer (NK) cells (32–34). It also increases the number of immunosuppressive Treg cells and myeloid-derived suppressor cells (MDSCs). A2A receptor deletion or blockade impaired tumor growth and activated tumor-infiltrating lymphocytes (35). CD73 expression is induced by hypoxia in an HIF-dependent manner (30, 36). CD73 expression is increased in TNBC relative to other breast cancers and is associated with chemotherapy resistance, metastasis, and decreased patient survival (37, 38). Anti-CD73 antibody treatment enhanced the antitumor activity of anti-PD1 antibody treatment (39).
In addition to immune evasion, cancer cells must have the capacity for self-renewal to form secondary (recurrent or metastatic) tumors. We have previously demonstrated that exposure of breast cancer cells to chemotherapy enriches for cancer stem-like cells due to induction of HIF-dependent gene expression (40–42). In the present study, we investigated whether exposure to chemotherapy also induces HIF-dependent changes in CD47, CD73, and PDL1 gene expression that increase the ability of surviving cancer cells to evade innate and adaptive immunity.
Results
Chemotherapy Induces Expression of PDL1, CD47, and CD73 by TNBC Cells.
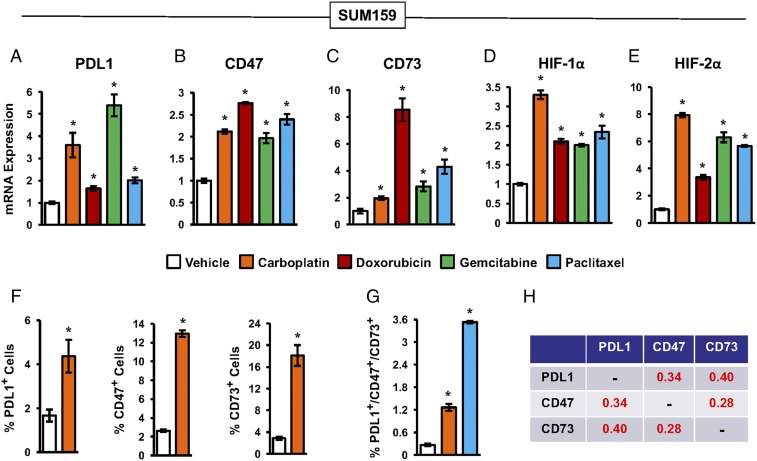
SUM159 human TNBC cells were exposed to each of four different chemotherapy drugs (carboplatin, doxorubicin, gemcitabine, and paclitaxel) for 4 d, at the drug concentration that inhibited growth by 50%, in a standard 95% air/5% CO2 incubator with an ambient O2 concentration of 20%. Reverse transcription-quantitative real-time PCR (RT-qPCR) analysis of total RNA isolated from chemotherapy-exposed TNBC cells revealed that each of the drugs increased the expression of PDL1, CD73, CD47, HIF-1α, and HIF-2α mRNA (Fig. 1 A–E). For example, gemcitabine increased PDL1 mRNA by 5.4-fold (Fig. 1A), doxorubicin increased CD73 mRNA by 8.5-fold (Fig. 1C), and carboplatin increased HIF-2α mRNA by 7.9-fold (Fig. 1E). Flow cytometry revealed that, compared with vehicle treatment, carboplatin increased the percentage of cells with surface expression of PDL1, CD47, or CD73 protein by 2.6-, 4.9-, and 6.3-fold, respectively (Fig. 1F). Chemotherapy also increased HIF-1α and HIF-2α protein levels (Fig. S1). We previously reported that chemotherapy induces MDR1 expression in an HIF-dependent manner (40), suggesting that there might be differential survival of cells with high HIF activity due to increased drug efflux mediated by MDR1. However, the observed increases in mRNA and protein expression cannot be due solely to enhanced chemoresistance, since only 50% of the cells were killed by chemotherapy, resulting in a maximum enrichment of twofold due to differential survival.
Fig. 1.
Chemotherapy induces expression of PDL1, CD73, and CD47. (A–E) SUM159 cells were treated with 50 μM carboplatin, 50 nM doxorubicin, 10 nM gemcitabine, 10 nM paclitaxel, or vehicle for 4 d. RT-qPCR was performed to quantify PDL1 (A), CD47 (B), CD73 (C), HIF-1α (D), and HIF-2α (E) mRNA levels relative to 18S rRNA and normalized to vehicle-treated cells (mean ± SEM; n = 3). *P < 0.001 compared with vehicle (by one-way ANOVA with a Bonferroni posttest). (F) Cells were treated with vehicle or carboplatin. After 4 d, the percentage of cells with surface expression of PDL1 (Left), CD47 (Center), or CD73 (Right) was determined by flow cytometry (mean ± SEM; n = 3). *P < 0.001 compared with vehicle (by Student’s t test). (G) Cells were treated with vehicle, carboplatin, or paclitaxel as described above. After 4 d, the percentage of cells that were triple-positive for PDL1, CD73, and CD47 was determined (mean ± SEM; n = 3). *P < 0.001 compared with vehicle (by one-way ANOVA with a Bonferroni posttest). All experiments in this figure were performed using cells exposed to 20% O2 in a standard 95% air/5% CO2 incubator. (H) Analysis of gene expression data from primary human breast cancers. The Pearson correlation test was performed to compare coexpression of PDL1, CD73, and CD47 mRNA using data from 1,215 breast cancer samples from The Cancer Genome Atlas (TCGA) database. Pearson’s correlation (r) is shown; P < 0.0001 for all comparisons.
Treatment with carboplatin or paclitaxel increased the percentage of triple-positive (PDL1+/CD73+/CD47+) SUM159 cells by 4.7- and 13-fold, respectively (Fig. 1G). Furthermore, the observed frequency of carboplatin-treated cells expressing all three proteins (1.27%) is 12.7-fold higher than would be expected if the expression of these proteins was independent of each other (0.10%). Thus, chemotherapy coordinately induces the expression of PDL1, CD73, and CD47 by surviving TNBC cells.
Next, we investigated whether PDL1, CD47, and CD73 mRNAs were expressed in a coordinated manner in 1,215 human breast cancer specimens from The Cancer Genome Atlas (TCGA) database (43, 44) using the Pearson correlation test. PDL1, CD73, and CD47 mRNA levels were significantly correlated with each other (P < 0.0001 for all pairwise comparisons) (Fig. 1H). These results are consistent with the coordinate expression of PDL1, CD73, and CD47 in human breast cancer, which implies that these genes are subject to similar regulatory mechanisms.
Chemotherapy Induces HIF-Dependent Expression of PDL1, CD73, and CD47.
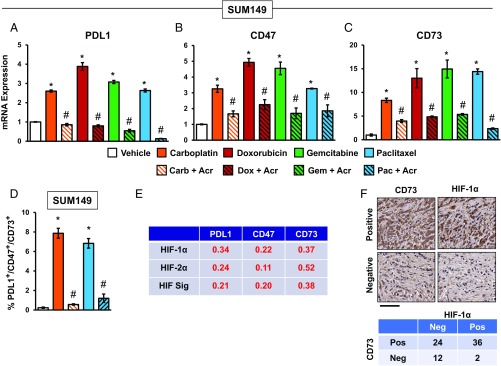
To investigate the role of HIFs, we exposed SUM149 TNBC cells to chemotherapy in the absence or presence of the HIF inhibitor acriflavine, which binds to HIF-1α or HIF-2α and blocks its heterodimerization with HIF-1β (45). Induction of PDL1, CD47, and CD73 mRNA expression in response to chemotherapy was blocked by acriflavine (Fig. 2 A–C). Similar results were obtained using SUM159 cells (Fig. S2). Acriflavine also blocked the coordinate cell surface expression of PDL1, CD73, and CD47 protein in response to chemotherapy, as determined by detection of triple-positive cells by flow cytometry (Fig. 2D). Remarkably, carboplatin and paclitaxel induced 34-fold and 29-fold increases in the percentage of PDL1+/CD47+/CD73+ cells (from 0.23% among vehicle-treated cells to 7.9% and 6.8%, respectively, among cells surviving chemotherapy); these effects of chemotherapy were abrogated by coadministration of acriflavine.
Fig. 2.
HIFs mediate chemotherapy-induced PDL1, CD73, and CD47 expression. (A–C) SUM149 cells were treated with vehicle or chemotherapy, either alone or in combination with 2 μM acriflavine, for 4 d. RT-qPCR was performed to quantify PDL1, CD73, and CD47 mRNA levels relative to 18S rRNA and normalized to vehicle-treated cells (mean ± SEM; n = 3). *P < 0.01 compared with vehicle; #P < 0.01 compared with chemotherapy alone (by one-way ANOVA with a Bonferroni posttest). Acr, acriflavine; Carb, carboplatin, Dox, doxorubicin; Gem, gemcitabine; Pac, paclitaxel. (D) Cells were exposed to carboplatin or paclitaxel for 4 d, and flow cytometry was performed to determine the percentage of triple-positive (PDL1+/CD73+/CD47+) cells (mean ± SEM; n = 3). *P < 0.01 compared with vehicle; #P < 0.01 compared with chemotherapy alone (by one-way ANOVA with a Bonferroni posttest). (E) Analysis of gene expression data from primary human breast cancers. The Pearson correlation test was performed to compare expression of HIF-1α, HIF-2α, and HIF signature (HIF Sig) with expression of PDL1, CD47, and CD73 mRNA, using data from 1,215 breast cancer samples in the TCGA database. Pearson’s correlation (r) is shown; P < 0.0001 for all comparisons. (F) Immunohistochemistry was performed on breast cancer biopsies using anti-CD73 and anti–HIF-1α antibodies. (Upper) Representative positive and negative staining (biopsies from patients 2 and 3, respectively) is shown. (Scale bar, 100 μm.) (Lower) Summary of CD73 and HIF-1α expression in 74 human breast cancer biopsies is shown. Neg, negative; Pos, positive.
We next analyzed TCGA data to determine whether expression of PDL1, CD47, and CD73 was correlated with expression of HIF-1α or HIF-2α mRNA, or with an HIF signature, which comprised HIF-1α mRNA and 13 HIF target-gene mRNAs (PLOD1, VEGFA, LOX, P4HA2, NDRG1, SLC2A1, ERO1L, ADM, LDHA, PGK1, ANGPTL4, SLC2A3, and CA9) in 1,215 breast cancer specimens (43, 44), using the Pearson correlation test. The expression levels of PDL1, CD73, and CD47 mRNAs were significantly correlated with expression of HIF-1α, HIF-2α, and the HIF signature (P < 0.0001 in each case; Fig. 2E).
Next, we performed immunohistochemistry on adjacent sections from breast cancer biopsies to analyze CD73 and HIF-1α expression. Sections with >5% stained cells were classified as positive for CD73 or HIF-1α (Fig. 2F, Upper). CD73 was expressed in 36 of 38 tumors in which HIF-1α overexpression was detected, compared with 24 of 36 tumors in which HIF-1α overexpression was not detected (Fig. 2F, Lower).
HIFs Directly Activate PDL1, CD73, and CD47 Gene Transcription.
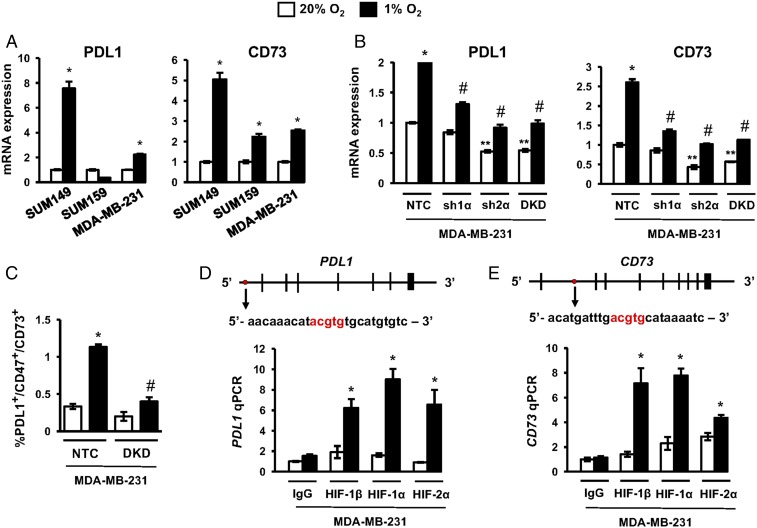
We previously demonstrated that HIF-1 directly activated CD47 gene transcription when breast cancer cells were exposed to hypoxia (18). Hypoxia-induced expression of CD73 and PDL1 has also been reported in various cell types (28, 29). To test whether HIFs regulate PDL1 and CD73 expression in human TNBC, we exposed SUM149, SUM159, and MDA-MB-231 cells to 20% or 1% O2 for 24 h. Hypoxia induced the expression of PDL1 in two of the three cell lines and CD73 in all three TNBC lines (Fig. 3A).
Fig. 3.
HIFs transactivate the PDL1 and CD73 genes. (A) TNBC cell lines were exposed to 20% or 1% O2 for 24 h, and the expression of mRNAs encoding PDL1 (Left) and CD73 (Right) was analyzed by RT-qPCR. The expression of each mRNA was quantified relative to 18S rRNA and then normalized to the result obtained from cells at 20% O2 (mean ± SEM; n = 3). *P < 0.01 versus 20% O2 (by two-way ANOVA with a Bonferroni posttest). (B) Analysis of PDL1 and CD73 mRNA expression in MDA-MB-231 subclones, which expressed an NTC shRNA or shRNA targeting HIF-1α (sh1α), HIF-2α (sh2α), or both HIF-1α and HIF-2α (DKD). Cells were exposed to 20% or 1% O2 for 24 h. Data were normalized to NTC at 20% O2 (mean ± SEM; n = 3). *P < 0.01 versus NTC at 20% O2; **P < 0.01 versus NTC at 20% O2; #P < 0.001 versus NTC at 1% O2 (by two-way ANOVA with a Bonferroni posttest). (C) MDA-MB-231 subclones were exposed to 20% or 1% O2 for 72 h, and the percentage of triple-positive cells was determined by flow cytometry (mean ± SEM; n = 3). *P < 0.01 versus NTC at 20% O2; #P < 0.001 versus NTC at 1% O2 (by two-way ANOVA with a Bonferroni posttest). (D and E) MDA-MB-231 cells were exposed to 20% or 1% O2 for 24 h and chromatin immunoprecipitation assays were performed using IgG or antibodies against HIF-1α, HIF-1β, or HIF-2α. Primers flanking the candidate HIF binding sites were used for qPCR, and results were normalized to lane 1 (mean ± SEM; n = 3). *P < 0.05 versus 20% O2 (by one-way ANOVA with a Bonferroni posttest). The nucleotide sequence (noncoding strand) of HIF binding sites (in red), which are located 5.7 kb 5′ to the transcription start site of the PDL1 gene (D) and within intron 1 of the CD73 gene (E), respectively, are shown. Exons and introns are not drawn to scale.
To determine whether HIF-1α or HIF-2α was required for PDL1 and CD73 expression in TNBC cells, we analyzed MDA-MB-231 subclones, which were stably transfected with an expression vector encoding short hairpin RNA (shRNA) targeting HIF-1α (sh1α), HIF-2α (sh2α), or both HIF-1α and HIF-2α [double knockdown (DKD)], or a nontargeting control (NTC) shRNA, which have been extensively validated and used to investigate the role of HIFs in breast cancer progression (46). Hypoxia-induced expression of PDL1 and CD73 was decreased in the sh1α, sh2α, and DKD subclones (Fig. 3B), indicating a requirement for both HIF-1α and HIF-2α. These results suggest that intratumoral hypoxia may be responsible for the correlation of CD47, CD73, and PDL1 mRNA expression with the HIF transcriptome in TCGA data (Fig. 2E), which was derived from treatment-naive human breast cancers (44). Hypoxia increased the percentage of NTC cells, but not DKD cells, that were PDL1+/CD47+/CD73+ (Fig. 3C). Thus, hypoxia induces expression of genes that enable TNBC cells to evade innate and adaptive immunity.
We next investigated whether HIF-1 or HIF-2 binds directly to the PDL1 and CD73 genes. Matches to the HIF binding-site sequence 5′-(A/G)CGTG-3′ were identified, and HIF binding was evaluated by chromatin immunoprecipitation assays performed in MDA-MB-231 cells. Hypoxia induced the binding of HIF-1α, HIF-2α, and HIF-1β to sites located in the 5′-flanking region of PDL1 (Fig. 3D) and in the first intron of CD73 (Fig. 3E). The HIF binding sites in the CD73 and PDL1 genes identified in MDA-MB-231 cells differ from sites identified in other cell lines (28–30). Taken together with our previous study demonstrating HIF-1 binding to the CD47 gene (18), these data indicate that HIFs directly activate PDL1, CD47, and CD73 transcription in human breast cancer cells, providing a molecular mechanism for coordinate regulation of these genes and their protein products in TNBCs.
Chemotherapy Promotes HIF-Dependent Immune Evasion by TNBC Cells.
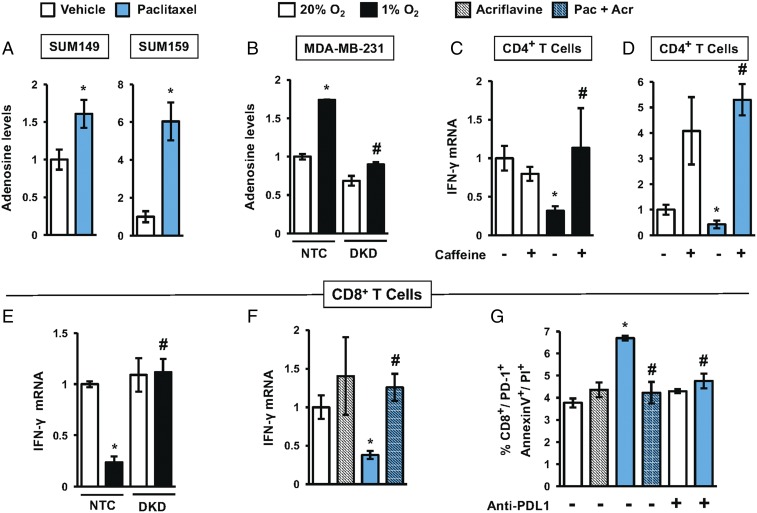
We next investigated whether adenosine production in TNBC cells leads to evasion of adaptive immunity. Cells were exposed to hypoxia or chemotherapy, culture media were collected, and adenosine levels were assayed and normalized to the number of cells counted at the end of the experiment. Paclitaxel treatment of SUM149 and SUM159 cells increased adenosine production, particularly in SUM159 cells (Fig. 4A). Hypoxia also increased adenosine production in the NTC subclone of MDA-MB-231, whereas in the DKD subclone, hypoxia-induced adenosine production was abrogated (Fig. 4B).
Fig. 4.
Exposure of TNBC cells to chemotherapy or hypoxia enables evasion of antitumor T cells. (A) Adenosine levels were measured in CM of SUM149 and SUM159 cells treated with paclitaxel (5 nM for SUM149 and 10 nM for SUM159) for 4 d. The values were corrected for cell number at the end of the experiment and then normalized to vehicle-treated cells (mean ± SEM; n = 3). *P < 0.05 versus vehicle (by Student’s t test). (B) Adenosine levels were measured in CM of MDA-MB-231 NTC and DKD subclones exposed to 20% or 1% O2 for 72 h. The values were normalized to NTC at 20% O2 (mean ± SEM; n = 3). *P < 0.05 versus NTC at 20% O2; #P < 0.05 versus NTC at 1% O2 (by two-way ANOVA with a Bonferroni posttest). (C and D) IFN-γ mRNA levels were determined (mean ± SEM; n = 3) in activated CD4+ T cells incubated for 24 h with CM (from hypoxic or paclitaxel-treated TNBCs) in the presence or absence of 4 mM caffeine. *P < 0.05 versus NTC at 20% O2; #P < 0.05 versus NTC at 1% O2 (by two-way ANOVA with a Bonferroni posttest). (E) IFN-γ mRNA levels (mean ± SEM; n = 3) were determined in activated CD8+ T cells cultured with CM from MDA-MB-231 subclones exposed to 20% or 1% O2 for 24 h. *P < 0.05 versus NTC at 20% O2; #P < 0.05 versus NTC at 1% O2 (by two-way ANOVA with a Bonferroni posttest). (F) IFN-γ mRNA levels (mean ± SEM; n = 3) were determined in activated CD8+ T cells incubated for 24 h with CM (from TNBC cells exposed to paclitaxel with or without 2 μM acriflavine for 4 d). *P < 0.05 versus vehicle-treated cells; #P < 0.05 versus paclitaxel-treated cells (by one-way ANOVA with a Bonferroni posttest). (G) CD8+ T cells were incubated with 4T1 cells (which were pretreated for 4 d with vehicle, acriflavine, paclitaxel, or paclitaxel + acriflavine) in presence or absence of anti-PDL1 blocking antibody. Cell death was assessed by staining with Annexin V and PI. The percentage of CD8+/PD1+ cells that were Annexin V+ and PI+ was quantified by flow cytometry (mean ± SEM; n = 3). *P < 0.05 versus vehicle-treated cells; #P < 0.05 versus paclitaxel-treated cells (by two-way ANOVA with a Bonferroni posttest).
Adenosine inhibits antitumor immunity through binding to A2A receptors on T cells (47, 48). To test whether exposure of TNBC cells to hypoxia or chemotherapy inhibits antitumor T cells through adenosine signaling, we cultured activated T cells with conditioned media (CM) from TNBC cells that were exposed to hypoxia vs. normoxia or vehicle vs. paclitaxel. We quantified IFN-γ mRNA levels as a standard measure of T cell activation (49). T cells cultured with CM from TNBC cells that were exposed to hypoxia (Fig. 4C) or paclitaxel (Fig. 4D) exhibited 3.1-fold and 2.3-fold decreased IFN-γ expression, respectively, indicating immunosuppression. Addition of the adenosine receptor antagonist caffeine (50) blocked the decrease in IFN-γ expression induced by CM from TNBC cells exposed to hypoxia (Fig. 4C) or paclitaxel (Fig. 4D), demonstrating that the immunosuppression was mediated by adenosine produced in response to hypoxia or chemotherapy.
To test whether the decrease in T cell activation induced by CM from TNBC cells was HIF-dependent, we prepared CM from MDA-MB-231 NTC and DKD subclones that were exposed to 20% or 1% O2 and from SUM159 cells treated with vehicle, acriflavine, paclitaxel, or paclitaxel + acriflavine. IFN-γ expression was inhibited 4.2-fold when activated CD8+ T cells were incubated with CM from hypoxic NTC cells, but not from hypoxic DKD cells (Fig. 4E). Incubation of T cells with CM from paclitaxel-treated SUM159 cells decreased IFN-γ expression by 2.6-fold, whereas CM from cells exposed to paclitaxel + acriflavine did not (Fig. 4F). Thus, HIF-dependent adenosine production by TNBC cells exposed to hypoxia or chemotherapy inhibits antitumor immunity mediated by both CD4+ (Fig. 4 C and D) and CD8+ (Fig. 4 E and F) T cells.
Chemotherapy Induces T Cell Apoptosis by Increasing PDL1 Expression in TNBC Cells.
The interaction of PDL1 on cancer cells with PD1 on T cells leads to anergy or cell death. To investigate whether chemotherapy-induced PDL1 expression by TNBC cells leads to T cell killing, we exposed mouse 4T1 TNBC cells to vehicle, acriflavine, paclitaxel, or paclitaxel + acriflavine. The 4T1 cells were then cocultured with activated CD8+ T cells, which were subsequently analyzed for apoptosis by propidium iodide (PI) and Annexin V staining. Coculture with paclitaxel-treated 4T1 cells led to a 1.8-fold increase in apoptosis of CD8+/PD1+ T cells, and the effect of paclitaxel was completely blocked by coadministration of acriflavine or by coculture in the presence of anti-PDL1 antibody (Fig. 4G). Taken together, the results presented in Fig. 4 demonstrate that exposure of TNBC cells to hypoxia or chemotherapy promotes evasion of adaptive immunity by inducing HIF-dependent adenosine and PDL1 signaling that mediate increased T cell anergy or apoptosis.
Acriflavine Blocks Chemotherapy-Induced Immune Evasion in a Syngeneic Mouse Model.
To investigate the role of chemotherapy in immune evasion in an immunocompetent model of TNBC, we utilized 4T1 mouse mammary carcinoma cells, which form primary tumors and metastases similar to human TNBC after implantation into the mammary fat pad of syngeneic BALB/c mice (51, 52). Exposure of 4T1 cells to chemotherapy in vitro induced expression of PDL1, CD73, and CD47 (Fig. 5 A–C), as well as HIF-1α and HIF-2α mRNA (Fig. S3A). Exposure of 4T1 cells to paclitaxel in vitro increased the percentage of PDL1+/CD73+/CD47+ cells 7.1-fold (Fig. 5D). We verified that acriflavine blocked expression of an HIF target gene (PDK1) in hypoxic 4T1 cells in a dose-dependent manner but had no effect on the expression of RPL13A, which is not an HIF target gene (Fig. S3B).
Fig. 5.
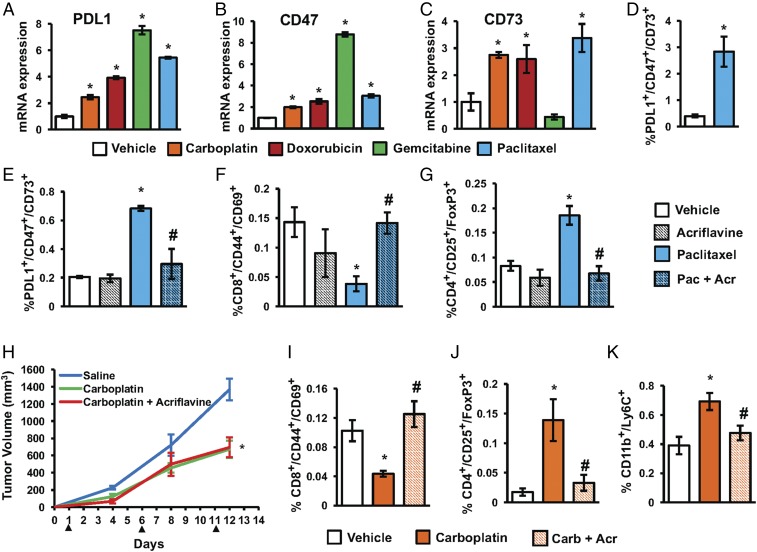
Acriflavine blocks paclitaxel-induced enrichment of PDL1+/CD73+/CD47+ TNBC cells and makes the tumor environment less immunosuppressive. (A–C) Mouse 4T1 mammary carcinoma cells were cultured for 4 d in the presence of vehicle, 50 μM carboplatin, 250 nM doxorubicin, 15 nM gemcitabine, or 10 nM paclitaxel. PDL1, CD47, and CD73 mRNA levels were normalized to vehicle-treated cells (mean ± SEM; n = 3). *P < 0.001 compared with vehicle (by one-way ANOVA with a Bonferroni posttest). (D) The 4T1 cells were treated with vehicle or paclitaxel in vitro, and flow cytometry was performed to quantify PDL1+/CD73+/CD47+ cells (mean ± SEM; n = 3). *P < 0.001 compared with vehicle (by Student’s t test). (E–G) The 4T1 cells were implanted into the mammary fat pad of female BALB/c mice. When tumor volume reached 200 mm3, mice were treated with: vehicle (saline), acriflavine (4 mg/kg; days 1–13), paclitaxel (10 mg/kg; days 5 and 10), or paclitaxel and acriflavine. Tumors were harvested on day 13, and the percentage of CD45−/PDL1+/CD73+/CD47+ (E), CD8+/CD44+/CD69+ (F), and CD4+/CD25+/FoxP3+ (G) cells was determined (mean ± SEM; n = 4). *P < 0.05 versus vehicle-treated mice; #P < 0.05 versus paclitaxel-treated mice (by one-way ANOVA with a Bonferroni posttest). Acr, acriflavine; Pac, paclitaxel. (H–K) The 4T1 cells were implanted into the mammary fat pad of female BALB/c mice. When tumors were palpable, mice were treated with: vehicle (saline), acriflavine (4 mg/kg; days 1–13), carboplatin (Carb; 20 mg/kg; days 1, 6, and 11), or carboplatin and acriflavine. (H) Tumor volumes were measured. Tumors were harvested on day 13, and the percentage of CD8+/CD44+/CD69+ (I), CD4+/CD25+/FoxP3+ (J), and CD11b+/Ly6C+ (K) cells was determined (mean ± SEM; n = 6). *P < 0.05 versus vehicle-treated mice; #P < 0.05 versus paclitaxel-treated mice (by one-way ANOVA with a Bonferroni posttest).
To investigate whether paclitaxel elicited similar effects in vivo, we implanted 4T1 cells in the mammary fat pad of female BALB/c mice. When the tumors reached a volume of 200 mm3 (designated day 1), mice were randomized to receive i.p. injections of saline, acriflavine (4 mg/kg on days 1–13), paclitaxel (10 mg/kg on days 5 and 10), or both acriflavine and paclitaxel. Tumors were harvested on day 13. There was no effect of paclitaxel, acriflavine, or paclitaxel + acriflavine on tumor growth or body weights of the mice (Fig. S4). Freshly harvested tumors were dissociated into single-cell suspensions and subjected to flow cytometry. Compared with vehicle, paclitaxel increased CD45−/PDL1+/CD73+/CD47+ tumor cells by 3.3-fold, and this effect was blocked by acriflavine (Fig. 5E). Thus, TNBC cells that survive cytotoxic chemotherapy manifest increased expression of genes encoding immunosuppressive proteins, whereas coadministration of an HIF inhibitor blocks chemotherapy-induced expression of gene products that mediate immune evasion.
Next, we analyzed immune cells within the tumor. Paclitaxel treatment decreased intratumoral CD8+/CD44+/CD69+ Teff cells by 3.7-fold, whereas acriflavine coadministration restored Teff cells to levels that were not significantly different from tumors in saline-treated mice (Fig. 5F). Paclitaxel treatment significantly increased CD4+/CD25+/FoxP3+ Treg cells by 2.2-fold, and this effect was completely blocked by acriflavine (Fig. 5G).
We next investigated whether similar effects would be observed if tumor-bearing mice were treated as soon as tumors became palpable, rather than after extensive tumor growth, and were treated with carboplatin, instead of paclitaxel (Fig. 5H and Fig. S5 A and B). As in the case of paclitaxel, we observed that compared with vehicle, carboplatin treatment significantly decreased the percentage of intratumoral CD8+/CD44+/CD69+ Teff cells (Fig. 5I) and increased the percentage of intratumoral CD4+/CD25+/Foxp3+ Treg cells (Fig. 5J). The intratumoral Teff/Treg cell ratio fell from 5.92 in vehicle-treated mice to 0.31 in carboplatin-treated mice, a 19-fold decrease. In contrast, coadministration of acriflavine abrogated the carboplatin-induced Teff cell decrease and Treg cell increase in the tumor, restoring a 3.8-fold excess of Teff cells over Treg cells. In addition, carboplatin induced a 1.8-fold increase in intratumoral CD11b+/Ly6C+ MDSCs, which was also abrogated by coadministration of acriflavine (Fig. 5K).
In contrast to the effects on T cells and MDSCs, carboplatin treatment had no effect on the percentage of intratumoral CD3−/NK1.1+ NK cells, CD3−/CD19+ B cells, or CD11b+F4/80+ tumor-associated macrophages (Fig. S5 C–E). Taken together, the results presented in Fig. 5 indicate that cytotoxic chemotherapy induces HIF-dependent enrichment of PDL1+/CD73+/CD47+ TNBC cells in vivo and makes the tumor microenvironment markedly less immunogenic by specifically increasing both Treg cells and MDSCs, as well as decreasing Teff cells. Coadministration of acriflavine is sufficient to block all of the observed countertherapeutic effects of cytotoxic chemotherapy on antitumor innate and adaptive immunity.
Discussion
Recent studies have demonstrated the role of the immune system in promoting cancer cell death in response to chemotherapy (3). In the present study, however, we have focused on the cancer cells that survive chemotherapy. Prior studies reported that hypoxia induces CD47, CD73, and PDL1 gene expression in an HIF-dependent manner (18, 28–30, 32). We previously demonstrated that treatment of tumor-bearing mice with HIF inhibitor blocked recruitment of MDSCs to orthotopic TNBCs (50) and that knockdown of HIFs or CD47 led to increased phagocytosis of breast cancer cells by macrophages (18). In the present study, we have demonstrated effects of chemotherapy on immune cells that are due to HIF-mediated gene expression in TNBC cells. Our results indicate that for multiple chemotherapy agents and multiple TNBC cell lines, chemotherapy induces HIF-dependent, coordinate transcriptional activation of PDL1, CD47, and CD73 expression. Cell surface expression of PDL1 and/or CD73 enables TNBC cells to induce anergy or apoptosis of Teff cells with a concomitant increase in Treg cells in the tumor microenvironment, thereby impairing adaptive antitumor immunity.
One limitation of in vitro exposure of cancer cells to chemotherapy is that it excludes the role of different immune cell types in either enhancing or inhibiting chemotherapy-induced cancer cell death (53). Another caveat is that serum in tissue culture media contains high levels of adenosine deaminase, which may result in an underestimation of cellular adenosine production in vitro. However, despite these caveats, there was a striking concordance of results from in vitro and in vivo studies demonstrating that chemotherapy induces CD47, CD73, and PDL1 expression and an immunosuppressive tumor microenvironment in an HIF-dependent manner.
Although we demonstrated surprisingly strong inhibitory effects of chemotherapy on antitumor immunity in vivo, a caveat of these studies is that intratumoral immune cell populations were analyzed using a single mouse strain, cancer cell line, chemotherapy dose, and time point. Thus, BALB/c mice were treated with 10 mg/kg of paclitaxel every 5 d, and 4T1 orthotopic tumors were analyzed 3 d after the last dose of paclitaxel, revealing a markedly increased percentage of Treg cells compared with tumors from vehicle-treated mice. In a previously published study, C57BL/6 mice bearing 3LL Lewis lung tumor xenografts were treated with paclitaxel at a dose of 10 mg/kg each day for 3 d and analyzed 1 d later, revealing a significant decrease in Treg cells (54). Further studies are required to investigate which of the many differences between these two studies were responsible for the divergent outcomes. We obtained remarkably similar results when we repeated the experiment using a different chemotherapy agent (carboplatin) and different tumor size (first palpable rather than 200 mm3). In both experiments, chemotherapy-induced increases in Treg cells were accompanied by reciprocal decreases in Teff cells, and all of the effects were blocked by coadministration of the HIF inhibitor acriflavine. In addition to inhibiting CD73 and PDL1 expression in TNBC cells, acriflavine administration may affect the Treg/Teff cell ratio by inhibiting HIF-dependent expression of FOXP3 (14), which is a critical determinant of Treg cell differentiation.
The coordinate induction of PDL1, CD47, and CD73 in response to chemotherapy endows TNBC cells with the ability to evade both innate and adaptive immune systems (Fig. 6). Our results provide a rationale for combining chemotherapy and immunotherapy (anti-PD1 or anti-PDL1) to improve the outcome of patients with TNBC. Our results also offer a potential explanation for the limited response of cancer patients to immune checkpoint inhibitors (55). If multiple mechanisms of immune evasion are coordinately regulated by HIFs, then targeting any single pathway (e.g., by anti-PD1, anti-PDL1, or adenosine receptor antagonist therapy) may be insufficient to restore antitumor immunity in a tumor with high HIF activity. An alternative approach is to combine cytotoxic chemotherapy with immune checkpoint inhibitors. However, an improvement in the therapeutic response of mice bearing Brca1−/− TNBC tumors (which have a high mutation load due to defective DNA repair) to cisplatin required combined treatment with both anti-CTLA4 and anti-PD1 antibodies (56), which is a treatment regimen that has been associated with considerable toxicity (57).
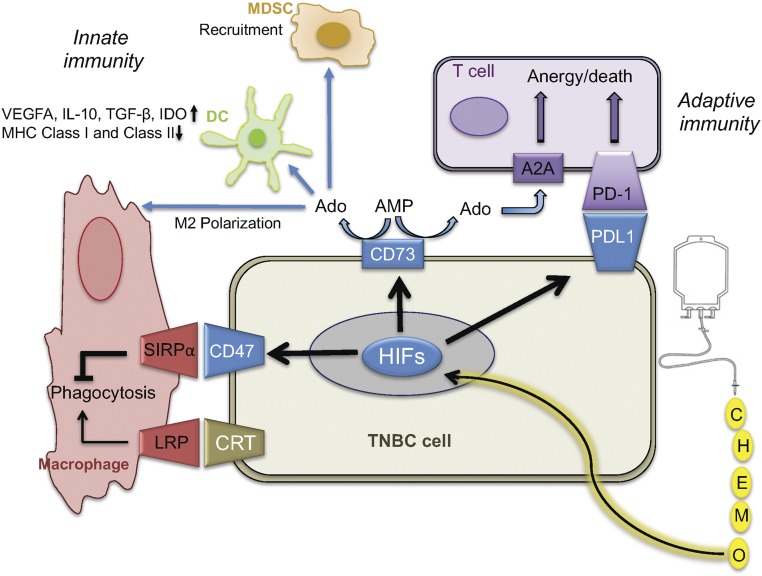
Fig. 6.
Chemotherapy promotes immune evasion phenotype in surviving TNBC cells. Exposure of TNBC cells to cytotoxic chemotherapy (or hypoxia) induces expression of HIF-1α and HIF-2α, leading to the HIF-mediated expression of PDL1, CD73, and CD47, which promote suppression of innate antitumor immunity mediated by macrophages, dendritic cells (DCs), and MDSCs, and suppression of adaptive antitumor immunity mediated by T cells. Ado, adenosine; IDO, indoleamine-2,3-dioxygenase.
Prior studies delineated multiple HIF-dependent pathways leading to the induction of tumor-initiating cells [also known as the breast cancer stem cell-like (BCSC) phenotype] in response to chemotherapy (40–42). CD47 and CD73 have been reported to promote the BCSC phenotype through mechanisms that are not understood (18, 58). Given that patients with TNBC in remission after chemotherapy may harbor millions of cancer cells or more, the finding that 0.7% of TNBC cells that survived paclitaxel therapy in vivo expressed proteins that mediate both immune evasion and tumor initiation provides a molecular mechanism for TNBC recurrence, metastasis, and patient mortality. Coadministration of HIF inhibitor blocks chemotherapy-induced expression of genes encoding the BCSC phenotype (40–42) and immune evasion as demonstrated in the present study. Thus, coadministration of HIF inhibitors with chemotherapy might improve the survival of patients who have TNBC.
Our results demonstrate that HIF inhibitors block the countertherapeutic effect of paclitaxel and other cytotoxic chemotherapy agents in promoting immune evasion by TNBC cells. HIF inhibitors might serve as broad-spectrum inducers of antitumor immunity, even in tumors such as 4T1 that express a very limited number of mutant epitopes (59). Further studies are required to determine whether, by blocking the expression of multiple proteins that mediate evasion of the adaptive and innate immune systems, HIF inhibitors might also improve responses to immune checkpoint inhibitors, adoptive cell transfer, or drugs targeting the adenosine signaling pathway.
Materials and Methods
Cell Lines.
MDA-MB-231 cells were maintained in high-glucose (4.5 mg/mL) Dulbecco’s modified Eagle medium (DMEM) with 10% (vol/vol) FBS and 1% penicillin/streptomycin. SUM159 and SUM149 cells were maintained in DMEM/F12 (50:50) with 10% FBS, hydrocortisone, insulin, and 1% penicillin/streptomycin. MDA-MB-231 shRNA subclones were cultured in the presence of 0.5 μg/mL puromycin. The 4T1 cells were maintained in RPMI-1640 with 10% FBS and 1% penicillin/streptomycin. Cell line identity and absence of mycoplasma infection were validated by PCR-based assays. Cells were maintained at 37 °C in a 5% CO2/95% air incubator (20% O2). Hypoxic cells were maintained at 37 °C in a modular incubator chamber (Billups–Rothenberg) flushed with a gas mixture containing 1% O2, 5% CO2, and 94% N2. Paclitaxel, doxorubicin, acriflavine, and gemcitabine were obtained from Sigma–Aldrich and dissolved in DMSO at 1,000× relative to final concentration in tissue culture medium. Caffeine was obtained from Sigma–Aldrich and dissolved in deionized water.
RT-qPCR.
Total RNA was extracted from cells using TRIzol (Invitrogen) and treated with DNase I (Ambion). One microgram of total RNA was used for first-strand DNA synthesis with the iScript cDNA Synthesis system (BioRad). qPCR was performed using SYBR Green qPCR Master Mix (BioRad). For each primer pair (nucleotide sequences are shown in Table S1), the annealing temperature was optimized by gradient PCR. The expression of each target mRNA relative to 18S rRNA was calculated based on the threshold cycle (Ct) as 2−Δ(ΔCt), where ΔCt = Cttarget − Ct18S and Δ(ΔCt) = ΔCttest − ΔCtcontrol.
Immunoblot Assays.
Whole-cell lysates were prepared in radioimmunoprecipitation assay lysis buffer. Blots were probed with antibodies against HIF-1α and HIF-2α (Table S2). HRP-conjugated anti-rabbit (Roche) and anti-mouse (Santa Cruz Biotechnology) secondary antibodies were used. The chemiluminescent signal was detected using ECL Plus (GE Healthcare). Blots were stripped and reprobed with antibody against actin (Santa Cruz Biotechnology) to confirm equal protein loading.
Flow Cytometry.
Cultured cells were trypsinized, whereas tumor tissues were minced, digested with 1 mg/mL type 1 collagenase (Sigma) at 37 °C for 30 min, and filtered through 70-μm cell strainers. Cells were incubated with Fc Block (BD Pharmingen). PDL1+/CD47+/CD73+ cells were identified by staining with fluorescein isothiocyanate (FITC)-conjugated anti-PDL1, phycoerythrin-conjugated anti-CD47, and allophycocyanin (APC)-conjugated anti-CD73 antibodies, and quantified by flow cytometry. Activated Teff cells were identified by staining with APC-conjugated anti-CD8, FITC-conjugated anti-CD69, and AF405-conjugated anti-CD44 antibodies, and subjected to flow cytometry. Treg cells were identified by staining with APC-conjugated anti-CD4, FITC-conjugated anti-CD25, and AF405-conjugated anti-FoxP3 antibodies, and quantified by flow cytometry. All fluorescent antibodies were from Novus Biologicals (Table S2). Unstained control and single-stained cells were prepared in every experiment for gating. Dead cells were gated out by side-scatter and forward-scatter analysis.
Chromatin Immunoprecipitation.
MDA-MB-231 cells were incubated at 20% or 1% O2 for 24 h, cross-linked in 3.7% formaldehyde for 15 min, quenched in 0.125 M glycine for 5 min, and lysed with SDS lysis buffer. Chromatin was sheared by sonication, and lysates were precleared with salmon sperm DNA/protein A agarose slurry (Millipore) for 1 h and incubated with IgG or antibody against HIF-1α, HIF-1β, or HIF-2α (Table S2) in the presence of protein A-agarose beads overnight. After washes of the agarose beads with low-salt, high-salt, and LiCl buffer, DNA was eluted in 1% SDS with 0.1 M NaHCO3, and cross-links were reversed by addition of 0.2 M NaCl. DNA was purified by phenol-chloroform extraction and ethanol precipitation, and analyzed by qPCR (Table S3).
Adenosine Measurements.
Cells were counted, plated, and exposed to hypoxia for 3 d or to chemotherapy for 4 d. CM were collected and centrifuged to pellet cell debris. Adenosine levels were determined based on a standard curve according to the manufacturer’s instructions (BioVision).
T Cell Isolation.
Naive CD4+ T cells (CD4+/CD25−/CD62L+) or CD8+ T cells were isolated from wild-type C57BL/6 mice by magnetic immunoseparation (catalog nos. 130-104-453 and 130-104-075, respectively; Miltenyi) and activated with anti-CD3 and anti-CD28 (Bio Legend) antibodies in a 24-well plate (1 μg and 4 μg per well, respectively) overnight.
IFN-γ mRNA Expression.
CM from MDA-MB-231 cells exposed to hypoxia for 3 d and SUM159 cells treated with chemotherapy for 4 d were collected. Five hundred microliters of CM was lyophilized. CD4+ or CD8+ T cells were activated and then incubated for 24 h with the lyophilized CM dissolved in fresh media with or without 4 mM caffeine. At the end of the experiment, RNA was extracted and RT-qPCR was performed.
Coculture Assays.
SUM159 cells were treated with vehicle, paclitaxel, acriflavine, or paclitaxel + acriflavine for 4 d. The surviving cells were counted, and an equal number of live cells were plated for each condition. The next day, activated CD8+ T cells were plated in the wells containing the cancer cells and cocultured for 48 h, and cell death was quantified by staining with FITC-conjugated anti-Annexin V antibody and PI, along with AF405-conjugated anti-CD8 and APC-conjugated anti-PD1 antibodies. Unstained control and single-stained cells were prepared in every experiment for gating. The percentage of CD8+/PD1+/AnnexinV+/PI+ cells was determined by flow cytometry.
Mouse Studies.
Animal protocols were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (60) and were approved by The Johns Hopkins University Animal Care and Use Committee. Female 5- to 7-wk-old BALB/c mice (Charles River Laboratories) were studied. Carboplatin, paclitaxel, and saline for injection were obtained from the research pharmacy of The Johns Hopkins Hospital. Cultured 4T1 cells were harvested by trypsinization, rinsed with PBS, and resuspended at 5 × 105 cells per milliliter in a 1:1 solution of PBS/Matrigel. The 4T1 cells were injected into the mammary fat pad. Primary tumors were measured in three dimensions (a, b, and c), and volume (V) was calculated as V = abc × 0.52.
Immunohistochemistry.
We utilized a tissue microarray of invasive breast carcinomas that were represented by triplicate punched core samples (0.6 mm in diameter) selected from paraffin tissue blocks of surgically resected primary cancers. An exemption for the use of deidentified human tumor tissue was approved by the Johns Hopkins Institutional Review Board. The following antibodies and staining methods were used: for CD73, Novus antibody NBP1-85740 was incubated at a 1:1,000 dilution for 1 h at room temperature; for HIF-1α, Novus antibody NB100-105 was incubated at a 1:100 dilution at 4 °C overnight after target antigen retrieval in citrate buffer (pH 6) in a pressure cooker for 3 min on high.
Statistical Analysis.
Data are expressed as mean ± SEM. Differences between two groups or multiple groups were analyzed by a Student’s t test or one-way ANOVA followed by a Bonferroni posttest, respectively. The Pearson correlation test was performed to compare mRNA expression data from 1,215 TCGA breast cancer samples (44). The Graphpad Prism (Version 6.0) software package was used.
Supplementary Material
Acknowledgments
We thank Karen Padgett of Novus Biologicals for providing IgG and antibodies against PDL1, CD73, CD47, CD45, CD4, CD8, CD44, CD69, and FOXP3. This work was supported by the Emerson Collective Cancer Research Fund, American Cancer Society, and Cindy Rosencrans Fund for Triple-Negative Breast Cancer. G.L.S. is an American Cancer Society Research Professor and the C. Michael Armstrong Professor at The Johns Hopkins University School of Medicine.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718197115/-/DCSupplemental.
References
- 1.Newman LA, Reis-Filho JS, Morrow M, Carey LA, King TA. The 2014 Society of Surgical Oncology Susan G. Komen for the Cure Symposium: Triple-negative breast cancer. Ann Surg Oncol. 2015;22:874–882. doi: 10.1245/s10434-014-4279-0. [DOI] [PubMed] [Google Scholar]
- 2.Gadi VK, Davidson NE. Practical approach to triple-negative breast cancer. J Oncol Pract. 2017;13:293–300. doi: 10.1200/JOP.2017.022632. [DOI] [PubMed] [Google Scholar]
- 3.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 4.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gil Del Alcazar CR, et al. Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 2017;7:1098–1115. doi: 10.1158/2159-8290.CD-17-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, Su DM, Liang M, Fu J. Chemopreventive agents induce programmed death-1-ligand 1 (PD-L1) surface expression in breast cancer cells and promote PD-L1-mediated T cell apoptosis. Mol Immunol. 2008;45:1470–1476. doi: 10.1016/j.molimm.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Cramer T, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kojima H, et al. Abnormal B lymphocyte development and autoimmunity in hypoxia-inducible factor 1α-deficient chimeric mice. Proc Natl Acad Sci USA. 2002;99:2170–2174. doi: 10.1073/pnas.052706699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiel M, et al. Targeted deletion of HIF-1α gene in T cells prevents their inhibition in hypoxic inflamed tissues and improves septic mice survival. PLoS One. 2007;2:e853. doi: 10.1371/journal.pone.0000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palazon A, et al. An HIF-1α/VEGF-A axis in cytotoxic T cells regulates tumor progression. Cancer Cell. 2017;32:669–683.e5. doi: 10.1016/j.ccell.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clambey ET, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci USA. 2012;109:E2784–E2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kershaw MH, Smyth MJ. Immunology. Making macrophages eat cancer. Science. 2013;341:41–42. doi: 10.1126/science.1241716. [DOI] [PubMed] [Google Scholar]
- 16.Gardai SJ, et al. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123:321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 17.Chao MP, et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med. 2010;2:63ra94. doi: 10.1126/scitranslmed.3001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, et al. HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc Natl Acad Sci USA. 2015;112:E6215–E6223. doi: 10.1073/pnas.1520032112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 20.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 21.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott PA, Hodi FS, Kaufman HL, Wigginton JM, Wolchok JD. Combination immunotherapy: A road map. J Immunother Cancer. 2017;5:16. doi: 10.1186/s40425-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortez MA, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst. 2015;108:djv303. doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ota K, et al. Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin Cancer Res. 2015;21:4014–4021. doi: 10.1158/1078-0432.CCR-15-0016. [DOI] [PubMed] [Google Scholar]
- 27.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noman MZ, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74:665–674. doi: 10.1158/0008-5472.CAN-13-0992. [DOI] [PubMed] [Google Scholar]
- 30.Hatfield SM, et al. Systemic oxygenation weakens the hypoxia and hypoxia inducible factor 1α-dependent and extracellular adenosine-mediated tumor protection. J Mol Med (Berl) 2014;92:1283–1292. doi: 10.1007/s00109-014-1189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young A, Mittal D, Stagg J, Smyth MJ. Targeting cancer-derived adenosine: New therapeutic approaches. Cancer Discov. 2014;4:879–888. doi: 10.1158/2159-8290.CD-14-0341. [DOI] [PubMed] [Google Scholar]
- 32.Beavis PA, et al. Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc Natl Acad Sci USA. 2013;110:14711–14716. doi: 10.1073/pnas.1308209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatfield SM, et al. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med. 2015;7:277ra30. doi: 10.1126/scitranslmed.aaa1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittal D, et al. Antimetastatic effects of blocking PD-1 and the adenosine A2A receptor. Cancer Res. 2014;74:3652–3658. doi: 10.1158/0008-5472.CAN-14-0957. [DOI] [PubMed] [Google Scholar]
- 35.Cekic C, Linden J. Purinergic regulation of the immune system. Nat Rev Immunol. 2016;16:177–192. doi: 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
- 36.Synnestvedt K, et al. Ecto-5′-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest. 2002;110:993–1002. doi: 10.1172/JCI15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stagg J, et al. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci USA. 2010;107:1547–1552. doi: 10.1073/pnas.0908801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loi S, et al. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc Natl Acad Sci USA. 2013;110:11091–11096. doi: 10.1073/pnas.1222251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allard B, Pommey S, Smyth MJ, Stagg J. Targeting CD73 enhances the antitumor activity of anti-PD-1 and anti-CTLA-4 mAbs. Clin Cancer Res. 2013;19:5626–5635. doi: 10.1158/1078-0432.CCR-13-0545. [DOI] [PubMed] [Google Scholar]
- 40.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci USA. 2014;111:E5429–E5438. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Lu H, et al. Chemotherapy triggers HIF-1-dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc Natl Acad Sci USA. 2015;112:E4600–E4609. doi: 10.1073/pnas.1513433112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu H, et al. Chemotherapy-induced Ca2+ release stimulates breast cancer stem cell enrichment. Cell Rep. 2017;18:1946–1957. doi: 10.1016/j.celrep.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Goldman M, et al. The UCSC Cancer Genomics Browser: Update 2013. Nucleic Acids Res. 2013;41:D949–D954. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K, et al. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci USA. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Zhang H, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31:1757–1770. doi: 10.1038/onc.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Haskó G, Linden J, Cronstein B, Pacher P. Adenosine receptors: Therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 2008;7:759–770. doi: 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linden J. Molecular approach to adenosine receptors: Receptor-mediated mechanisms of tissue protection. Annu Rev Pharmacol Toxicol. 2001;41:775–787. doi: 10.1146/annurev.pharmtox.41.1.775. [DOI] [PubMed] [Google Scholar]
- 49.Ohta A, et al. A2A adenosine receptor may allow expansion of T cells lacking effector functions in extracellular adenosine-rich microenvironments. J Immunol. 2009;183:5487–5493. doi: 10.4049/jimmunol.0901247. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, et al. Caffeine inhibits the activation of hepatic stellate cells induced by acetaldehyde via adenosine A2A receptor mediated by the cAMP/PKA/SRC/ERK1/2/P38 MAPK signal pathway. PLoS One. 2014;9:e92482. doi: 10.1371/journal.pone.0092482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 52.Chaturvedi P, Gilkes DM, Takano N, Semenza GL. Hypoxia-inducible factor-dependent signaling between triple-negative breast cancer cells and mesenchymal stem cells promotes macrophage recruitment. Proc Natl Acad Sci USA. 2014;111:E2120–E2129. doi: 10.1073/pnas.1406655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coffelt SB, de Visser KE. Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends Immunol. 2015;36:198–216. doi: 10.1016/j.it.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Liu N, Zheng Y, Zhu Y, Xiong S, Chu Y. Selective impairment of CD4+CD25+Foxp3+ regulatory T cells by paclitaxel is explained by Bcl-2/Bax mediated apoptosis. Int Immunopharmacol. 2011;11:212–219. doi: 10.1016/j.intimp.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 55.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nolan E, et al. Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med. 2017;9:eaal4922. doi: 10.1126/scitranslmed.aal4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hassel JC, et al. Combined immune checkpoint blockade (anti-PD-1/anti-CTLA-4): Evaluation and management of adverse drug reactions. Cancer Treat Rev. 2017;57:36–49. doi: 10.1016/j.ctrv.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Yu J, et al. A preliminary study of the role of extracellular -5′- nucleotidase in breast cancer stem cells and epithelial-mesenchymal transition. In Vitro Cell Dev Biol Anim. 2017;53:132–140. doi: 10.1007/s11626-016-0089-y. [DOI] [PubMed] [Google Scholar]
- 59.Kim K, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc Natl Acad Sci USA. 2014;111:11774–11779. doi: 10.1073/pnas.1410626111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.