Significance
We report the high-resolution structure of actin-bound myosin in the presence and absence of MgADP, revealing an allosteric pathway that links the release of MgADP to a rotation of the myosin lever arm. These results provide a mechanism by which myosin dynamically adjusts its power output in response to mechanical load. These results also provide the framework for understanding the diversity of force sensing among the members of the myosin superfamily.
Keywords: mechanochemistry, molecular motor, structural biology, cytoskeleton, cryoelectron microscopy
Abstract
Myosins adjust their power outputs in response to mechanical loads in an isoform-dependent manner, resulting in their ability to dynamically adapt to a range of motile challenges. Here, we reveal the structural basis for force-sensing based on near-atomic resolution structures of one rigor and two ADP-bound states of myosin-IB (myo1b) bound to actin, determined by cryo-electron microscopy. The two ADP-bound states are separated by a 25° rotation of the lever. The lever of the first ADP state is rotated toward the pointed end of the actin filament and forms a previously unidentified interface with the N-terminal subdomain, which constitutes the upper half of the nucleotide-binding cleft. This pointed-end orientation of the lever blocks ADP release by preventing the N-terminal subdomain from the pivoting required to open the nucleotide binding site, thus revealing how myo1b is inhibited by mechanical loads that restrain lever rotation. The lever of the second ADP state adopts a rigor-like orientation, stabilized by class-specific elements of myo1b. We identify a role for this conformation as an intermediate in the ADP release pathway. Moreover, comparison of our structures with other myosins reveals structural diversity in the actomyosin binding site, and we reveal the high-resolution structure of actin-bound phalloidin, a potent stabilizer of filamentous actin. These results provide a framework to understand the spectrum of force-sensing capacities among the myosin superfamily.
The myosin family of cytoskeletal motors performs diverse biological and mechanical functions, ranging from muscle contraction to intracellular motility (1). This versatility relies on the ability of myosin motors to adjust their power outputs in response to mechanical load in an isoform-dependent manner. However, the structural mechanism of this behavior is unknown (2).
Myosins generate force via the rotation of their lever arms in response to actin binding and phosphate release (3). Following these steps, a second rotation of the lever is a force-sensitive conformational transition that affects the rate of ADP release, which is often the step that limits that rate of motility (4–8). ADP release is thus an important mechanical control point affecting power output and is a source of the substantial kinetic diversity among myosin isoforms (2). Recent progress revealed subdomain motions within actin-bound myosin that accompany ADP release (9), but the mechanisms by which these changes are connected to the active site have not been resolved.
Here, we report high-resolution structures of actin-bound myosin-IB (myo1b), a motor characterized by its high force-sensitivity (7). The structures reveal an allosteric linkage between MgADP release from the myosin active site and the lever arm rotation during the force-sensitive transition, which would result in the inhibition of MgADP release upon a restraining force on the lever. Additionally, we provide the high-resolution structure of the actin binding interface of myo1b, finding that, despite the conserved spatial topology of actin-binding loops, the precise nature of actin binding among myosins is quite diverse. Finally, we provide a high-resolution structure of phalloidin bound to actin.
Results and Discussion
Cryo-EM of Actomyosin.
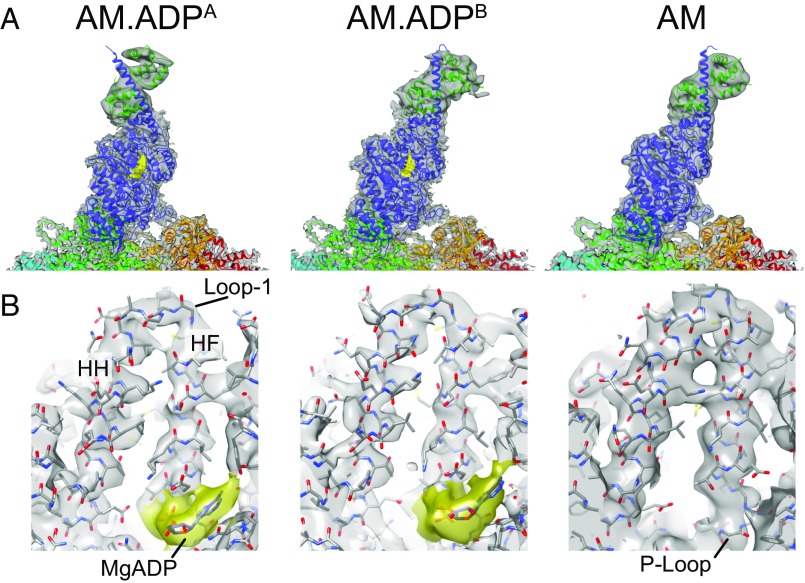
We generated complexes of actin filaments decorated with a myo1b construct containing a single IQ motif with calmodulin bound and solved near-atomic resolution structures both in the absence (rigor) and presence of 1 mM MgADP (SI Materials and Methods, Figs. S1–S5, and Movie S1). A high-resolution crystal structure of myo1b with sulfate in the active site was used to solve the actin-bound structures (SI Materials and Methods and Table S1). Consistent with previous cryo-EM studies of actomyosin, the resolution was highest in the core region proximal to the actin filament axis and progressively grew lower at larger radial distances, particularly in the distal region of the myosin lever, where secondary structure was barely visible (Figs. S2 and S5). The resolution for the motor domain (Fourier shell correlation, 0.143) was estimated as 3.3 Å for the ADP-bound state (AM.ADP) and 3.9 Å for the rigor state (AM), allowing us to trace the protein chains throughout most of the map, discriminate side chains in the best regions, visualize the MgADP, and determine the structure of actin-bound phalloidin (see below).
Consistent with previous EM and single-molecule studies (4, 5, 7, 10), the structures revealed a rotation in the lever, resulting in two well-defined end states, AM.ADP and AM (Fig. 1). Following the initial refinement, we used 3D classification to search for alternate conformations and found a second ADP-bound population, termed AM.ADPB and comprising ∼15% of the total population, with the predominant population termed AM.ADPA (Fig. 1 and SI Materials and Methods). While densities for ADP and associated Mg2+ cation are seen in the AM.ADPB map (Fig. 1), the lever rotated ∼25° relative to motor domain toward the barbed end of the actin filament compared with the AM.ADPA state and assumed a position similar to, but not identical to, that of the AM state (Figs. 1A and 2A). The three conformational states thus appear to give a mechanistic succession of states that occur after the initial power stroke that accompanies actin binding and phosphate release, proceeding from AM.ADPA to AM.ADPB to AM. The presence of two AM.ADP structural states is consistent with previous kinetic investigations that identified multiple AM.ADP biochemical intermediates (11–13).
Fig. 1.
Structural states of actin-bound myo1b in the presence and absence of MgADP. (A) Cryo-EM density maps (gray) and final fitted AM.ADPA, AM.ADPB, and AM models. The myo1b protein construct (blue) with a single IQ motif and bound calmodulin (dark green) are bound to actin subunits (cyan, green, light green, orange, and red). Cryo-EM Density for MgADP is highlighted in gold. (B) Cryo-EM density map showing the nucleotide binding site. The MgADP (gold) is bound to the P loop and framed by the HF and HH helices, which are connected by loop 1.
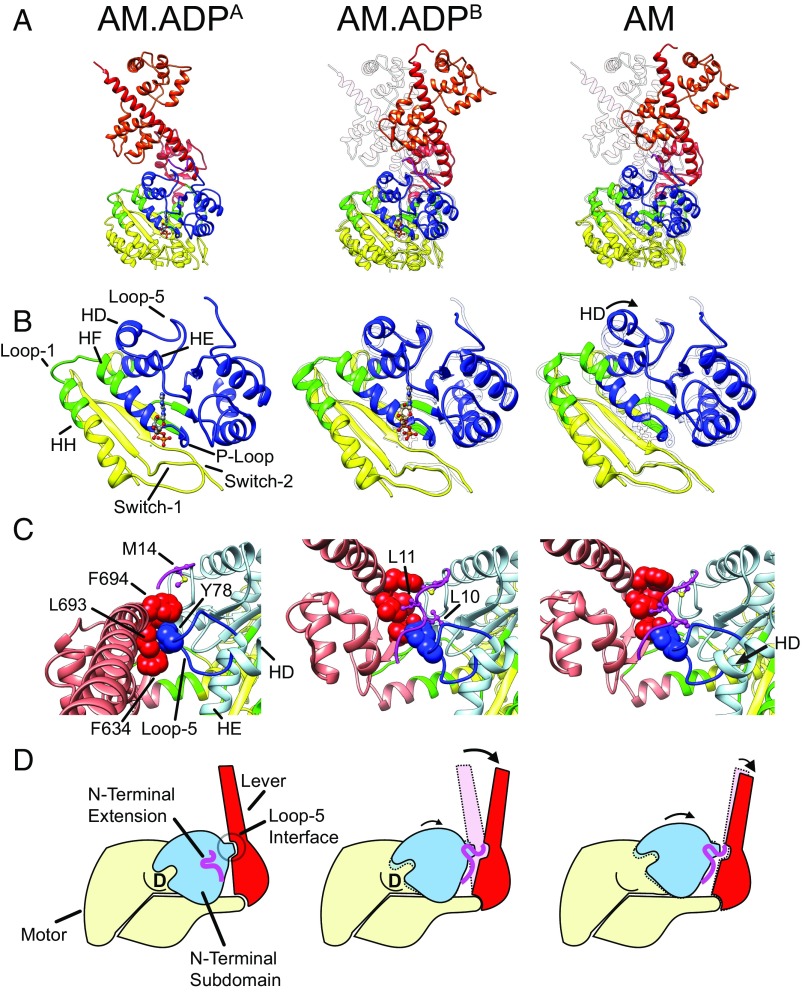
Fig. 2.
Coupling of nucleotide-dependent changes at the active site to lever rotation. (A) Structural models showing changes in the myo1b active site and lever position. Coloring of subdomains is as defined by DynDom (SI Materials and Methods): yellow, motor; blue, N-terminal subdomain, which includes sequences in the N and C termini and the transducer; red, lever, which includes the converter domain and bound calmodulin. The N-terminal extension (magenta) is docked and resolved in the density maps for AM.ADPB and AM structures, but not AM.ADPA. Structural models are aligned by aligning bound actin subunits. (B) Enlarged region showing the nucleotide binding site and conformational rearrangements in the N-terminal subdomain. Coloring and orientation are identical to A. The AM.ADPA structure is shown as an outline in Center (AM.ADPB) and Right (AM) of A and B. (C) Interaction of loop 5 (dark blue) in the N-terminal subdomain (light blue) with the lever (red) and N-terminal extension (magenta). The side chains in loop 5 are not resolved in the cryo-EM density and were placed based on MD. (D) Cartoon model showing the conformational changes in the structural subdomains that accompany lever rotation.
Connection Between MgADP Release and Lever Rotation.
A comparison of the AM.ADPA and AM structures revealed details of the pathway that couples MgADP release with a lever-arm swing (Fig. 1 and Movie S2), which is distinct from the pathway that drives the power stroke that is linked to closing of the actin-binding cleft, phosphate release, and movement of the SH1, SH2, and relay helices (3, 14). Using DynDom (15), we identified myosin subdomains (motor, lever, and N-terminal subdomains) that rotate and translate semiindependently about hinge axes and bending residues (Fig. 2, Fig. S6, and Table S2). During the AM.ADPA to rigor transition, the N-terminal subdomain (blue) pivots to open the nucleotide cleft, moving ∼4 Å away from the upper 50-KDa region of the motor (Fig. 2B and Movie S3). A similar rotation of the N-terminal subdomain was also observed in the 8-Å cryo-EM structure of myosin V (9). Opening of the nucleotide cleft involves substantial axial and lateral shifts of the P loop and adjoining helix (HF; K114–V128) in the N-terminal subdomain, relative to the HH helix (A133–F152; connected to HF via loop 1; C129–G132). The lever position in AM.ADPA is accompanied by unwinding of the C terminus of the relay helix relative to AM by 24°, resulting in a break in the alpha-helical hydrogen bonds. This disruption in the helix is not present in AM.ADPB and AM, and it is in the position of the relay helix that kinks in the pre-power-stroke state of myosin (14, 16, 17).
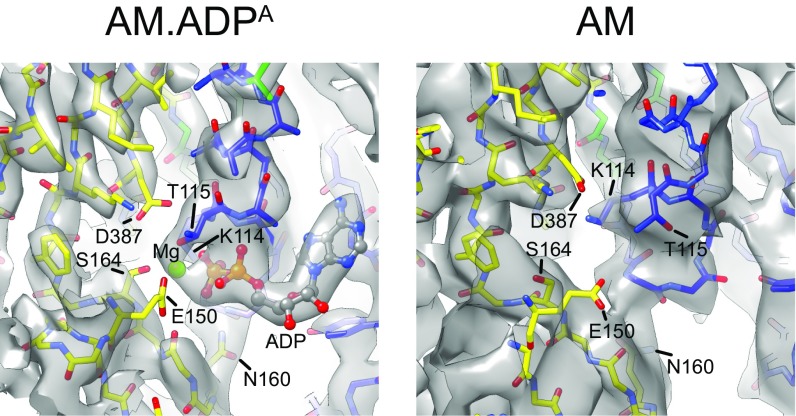
The quality of the AM.ADPA and AM density maps is sufficiently high to visualize the density of some key side chains that directly and indirectly interact with MgADP (Fig. 3 and Movie S4). Nucleotide cleft opening and MgADP release disrupts a conserved hydrogen-bonding pair, T115–D387, that coordinates Mg in most nucleotide-bound structures of myosin. These changes are accompanied by the formation of a salt bridge between K114 (P loop) and D387 (which precedes switch 2) and the loss of coordination of the MgADP phosphates by K114. Consequently, the network of interactions holding the HH helix and switch 1 to the HF helix and P loop is lost, facilitating repositioning of the N-terminal subdomain.
Fig. 3.
MgADP-induced conformational changes at the active site. Shown is the nucleotide binding site of myo1b in the AM.ADPA (Left) and AM (Right) states, showing side chains that interact directly or indirectly with MgADP. Obscured residues are seen clearly in Movie S4. Cryo-EM density for the E150 side chain is not resolved, but is included as a nucleotide-interacting residue based on MD (Movie S5). Coloring is as in Fig. 2.
MgADP Changes the Interface Between the Lever, N-Terminal Subdomain, and N-Terminal Extension.
We identified an interface between the AM.ADPA lever and a region within the N-terminal subdomain which we named loop 5 (Y72–H83), located between the HD (P66–D71) and HE helices (I84–Q98) (18) (Fig. 2 B and C). Residue Y78 in loop 5, conserved in the myosin I family, is nestled within a cluster of hydrophobic side chains from the converter and the base of the lever to stabilize the lever position (Fig. 2C and Movie S6). As the N-terminal subdomain rotates toward the lever in AM, Y78 is pulled out of its position in the lever cavity and occludes the AM.ADPA position of the lever via steric clashes between Y78 and L693 of the lever. Consistent with this observation, multiple rounds of structural classification of the AM cryo-EM dataset failed to yield evidence that the lever can assume the AM.ADPA-like orientation. These observations indicate that the ADP-like position of the lever and opening of the nucleotide cleft (to release ADP) are mutually exclusive.
Accompanying the lever swing, a structural element, which we call the N-terminal extension (residues V6–I15), becomes sandwiched between the motor and lever in AM, contributing side chains L10 and L11 to the hydrophobic cluster of residues (Fig. 2C and Fig. S7), as found in the apo crystal structure (18). The binding site of the N-terminal extension is sterically occluded in AM.ADPA, and residues preceding L11 are unresolved. The N-terminal extension of myo1b is important for activation of ADP release, as its deletion slows the rate of ADP release 10-fold, while the force dependence of the step is unchanged (18). However, changing the sequence of this region impacts myosin-I force sensing (19). Therefore, docking of the N-terminal extension likely facilitates the movement of the N-terminal subdomain to the AM position and opening of the nucleotide-binding site, allowing for ADP release.
The AM.ADPB structure represents an intermediate state in which the lever swing is nearly finalized, but is incompletely coupled to opening of the nucleotide cleft. The lower resolution of the AM.ADPB state does not allow the same precision of modeling as the other two structures, but the interface created by the lever, the N-terminal subdomain, and the N-terminal extension is clearly similar to AM (Fig. 2C). However, the N-terminal subdomain in AM.ADPB rotates only halfway (4°) toward the AM position (9° total rotation). Consequently, the AM.ADPB nucleotide binding site remains mostly closed, except for a ∼1.5-Å axial shift of helix HF and the adjoining P loop compared with AM.ADPA (Fig. 2). The similarity of the AM.ADPB nucleotide cleft to AM.ADPA suggests a similar coordination of the nucleotide in both states. Upon the transition from AM.ADPB to AM, the additional rotation of the N-terminal subdomain results in an oblique rotation of the lever due to these elements being linked to each other by the N-terminal extension sandwich configuration (Fig. S8). These observations indicate that the structure of the ADP-occupied active site is not tightly coupled to the position of the lever, in contrast to previous suggestions (9).
The AM structure of the myo1b motor domain is largely similar to that found for the ∼8-Å rigor structure of Dictyostelium Myo1e (20); however, differences are apparent in the orientation of the HF and relay helices and in the N-terminal subdomain. These differences may be due to experimental limitations of the map resolution and modeling, differences in the myosin isoform structures, or to the absence of the lever and light chains in the Myo1e construct.
Mechanism of Myosin Force Sensing.
As a mechanism for force sensing, we propose that mechanical loads that prevent the rotation of the myosin lever toward the AM position block the translation and rotation of the N-terminal subdomain required for breaking the network of interactions that hold MgADP in place. This inhibition is not only due to the steric blocking of the N-terminal subdomain rotation, but is also due to interactions between the lever and N-terminal subdomain that stabilize the AM.ADPA state (Fig. 2 C and D), holding the P loop, HF and HH helices, and switch 1 in a position to coordinate the MgADP. It is likely that all myosin isoforms require the rotation of the N-terminal subdomain to release MgADP, but different isoforms have structural modifications that differentially link N-terminal subdomain rotation to the magnitude of lever rotation. These modifications may include the variable angular positions of the lever, as in myo1b and myosin V (9), as well as the sequence of an N-terminal extension that interacts with the N-terminal subdomain, as found in myosin I (18, 19) and II (21). We propose that myosins in which the N-terminal subdomain is able to rotate without requiring a large amplitude rotation of the lever, or lack stabilizing interactions between the lever and the N-terminal subdomain, are less force-dependent than those that have N-terminal subdomain rotations linked to large lever rotations.
The Actin-Binding Site of Myosin Is Structurally Diverse.
The myo1b actin-binding site is well resolved and is nearly identical in the AM.ADPA and AM states. Thus, myo1b binding to actin during ADP release, as found for myosin V (9), holds fixed the geometric relationship between the upper and lower 50-kDa domains helping orchestrate the above-described coupling pathway between the nucleotide pocket and the lever, in particular by constraining the relative position of the lever and the N-terminal subdomain.
Like myosins II and V (9, 22), a helix–loop–helix motif (L457–T485) in the lower 50-KDa region of myosin fits in a hydrophobic pocket between two actin subunits, which includes a well-ordered DNase I binding loop (D loop) of actin (Fig. S9). Modeling within this region shows interactions between myosin E477 and the side chain of actin K50, myosin Q481 side-chain oxygen with the backbone hydrogen of actin G48, and myosin E462 with S350 and T351 in actin. Finally, the backbone of myosin G468 hydrogen-bonds with actin G146. Also within the lower 50-KDa region, residues within the myosin activation loop (N452 and T453) interact with S350 of actin, but density for an interaction with the actin N terminus as predicted for myosin II (22) and observed for myosin V (9) was not resolved.
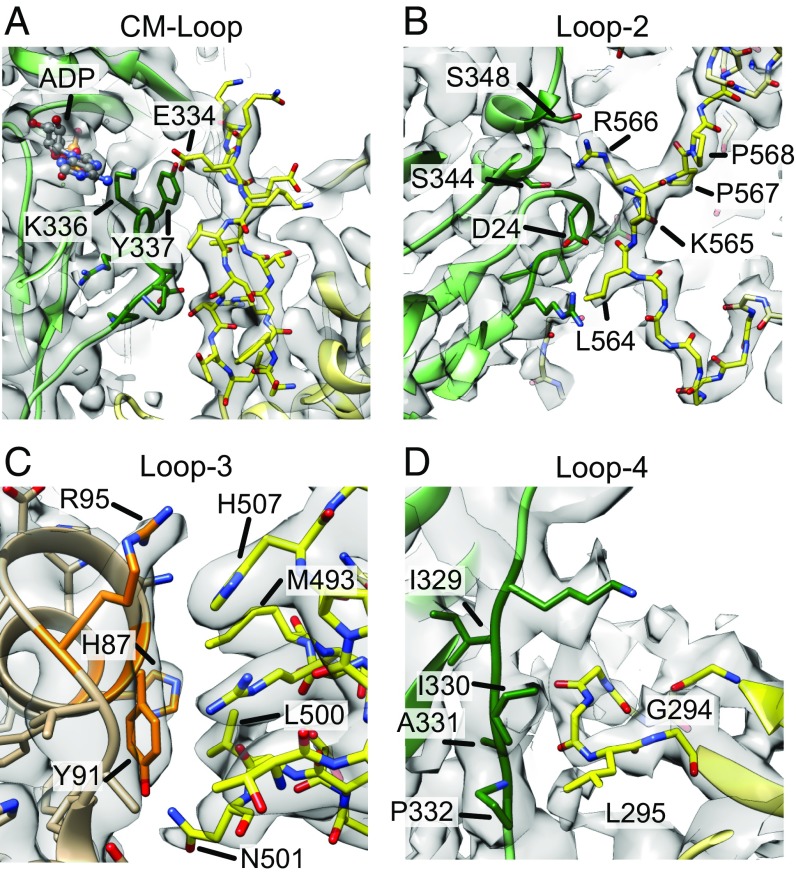
Although the spatial topology of the cardiomyopathy (CM) loop and loops 2, 3, and 4 is also conserved in all myosins, our myo1b structures show that the precise nature of actin binding among myosins is quite diverse (Fig. 4).
Fig. 4.
Myo1b has a unique actin-binding interface. Cryo-EM densities (gray) and structural models of myo1b loops (yellow) binding to actin are shown. The relationship of actin subunits in the filament to the myosin binding site is shown by the actin coloring (green and orange) in Fig. 1A. Actin residues that interact with myosin are shown in darker colors. (A) CM loop binding to actin. E334 is the TEDS site that interacts with K336 and Y337. The actin-bound ADP molecule is shown and interacts with E334 through K336. (B) Loop 2 binding to actin (green). (C) Loop 3 becomes ordered upon actin binding and interacts through a group of hydrophobic interactions clustered around Y91. (D) Loop 4 interacts with actin (green) via a network of hydrophobic interactions. The cryo-EM map is less resolved than in panels A, B, or C, suggesting a weaker interaction for Loop 4.
The CM loop (T328–S337) forms a key interface with actin, important for establishing tight binding, that involves a cluster of hydrophobic interactions with Y337 of actin (Fig. 4A). However, in myo1b, these interactions are attenuated compared with myosin II (22), and the electrostatic interactions differ (Fig. 4A). Notably, E334 in the CM loop is a highly conserved position that contains a T, E, D, or S residue in nearly all myosins (i.e., TEDS site; ref. 23) and is important for ATPase regulation in some myosin I isoforms (24, 25). In contrast to the corresponding residue in myosin II, which does not contact the actin surface (22), the E334 side chain interacts with the side chains of K336 and Y337 in subdomain 1 of actin. The former of these actin side chains also interacts with the base of the actin-bound ADP. It will be of interest to determine if this interaction between the CM loop and the actin-bound nucleotide is of regulatory significance (26).
Loop 2 (P555–P568), which is variable in sequence and length among myosin isoforms, mediates myosin’s initial attachment to actin. In the majority of high-resolution structures, loop 2 is disordered. The high-resolution structure of myosin II bound to actin indicates that only the N-terminal segment of loop 2 becomes ordered upon actin binding due to hydrophobic contact between a short, nonconserved segment at its base and the actin surface (22). In striking contrast, loop 2 is well resolved in available X-ray structures of the myosin I family (including myo1b), and its conformation changes very little upon binding to actin (Fig. 2B and ref. 18) These differences appear to be due to shortening in the length of loop 2 in the myosin I family, as well as the presence of either one or two adjacent conserved, class-specific prolines at the loop 2 (P568–P567). By restricting the geometry of loop 2, these modifications substantially restrict the ability of loop 2 to contact the actin surface. Hence, in contrast to myosin II where loop 2 is anchored on the actin surface by a small hydrophobic cluster near its C terminus, in myo1b, the actin interface of loop 2 is mainly formed by a pair of basic amino acids conserved among myosins (K565 and R566) important for activating phosphate release (ref. 27 and Fig. 4), as well as the adjacent, nonconserved L564. These differences compared with other myosins likely result in the low affinity of the myosin I pre-power-stroke states for actin (28).
Loop 3 (S491–S508) also exhibits substantially different interactions with actin than myosin II. This loop is significantly longer in the myosin I family than other family members and is not visualized in myo1b crystal structures; however, loop 3 becomes well-ordered upon binding actin to subdomain 1 of actin (Fig. 4C). Compared with myosin II, loop 3 in myo1b interacts much more extensively with actin through a group of hydrophobic interactions clustered around Y91 of actin. Specifically, L500 contacts aliphatic groups on both Y91 and H87 in actin, and M493 also contacts actin Y91. There is a probable cation–pi interaction between H507 and actin R95, and a single hydrogen bond between N501 and actin Y91. Densities are consistent with H-bond interactions between N501 and actin Y91; L500 and actin H87; and H507 and actin R95. Potentially relevant to regulation, phosphorylation sites have been identified in the actin-binding interface of loop 3 (S504 and S508; ref. 29), although they have not been physiologically and mechanistically examined. Loop 4 (I284–K301) is less well resolved in the EM map than the other features at the actin-binding site, suggesting a weaker interaction (Fig. 4D). However, structural modeling suggests a hydrophobic interaction between G294–L295 of myosin and I329, I330, A331, and P332 in actin.
The CM loop and loops 2 and 4 bind to actin across the same interface as tropomyosin occupies in the “blocked” state, which inhibits myosin activation (30). However, myo1b is not predicted to clash sterically with the position of tropomyosin found in the AM myosin-II–actin–tropomyosin structure (Protein Data Bank ID code 5JLH). Importantly, myo1b does not interact with tropomyosin-bound actin in cells, and tropomyosin inhibits myo1b-based motility in vitro (31). Therefore, tropomyosin inhibition must occur before the formation of the strong-binding state of the myosin during ATPase cycling.
High-Resolution Structure of Actin-Bound Phalloidin.
The cryo-EM datasets contain regions of excess density at the interface of three actin monomers. We assume these densities represent phalloidin, the bicyclic heptapeptide [cyclic(l-Ala1-d-Thr2-l-Cys3-cis-4-hydroxy-Pro4-l-Ala5-2-mercapto-l-Trp6-4,5-dihydroxy-l-Leu7) cyclic (3->6) sulfide], which was used to stabilized the actin filaments.
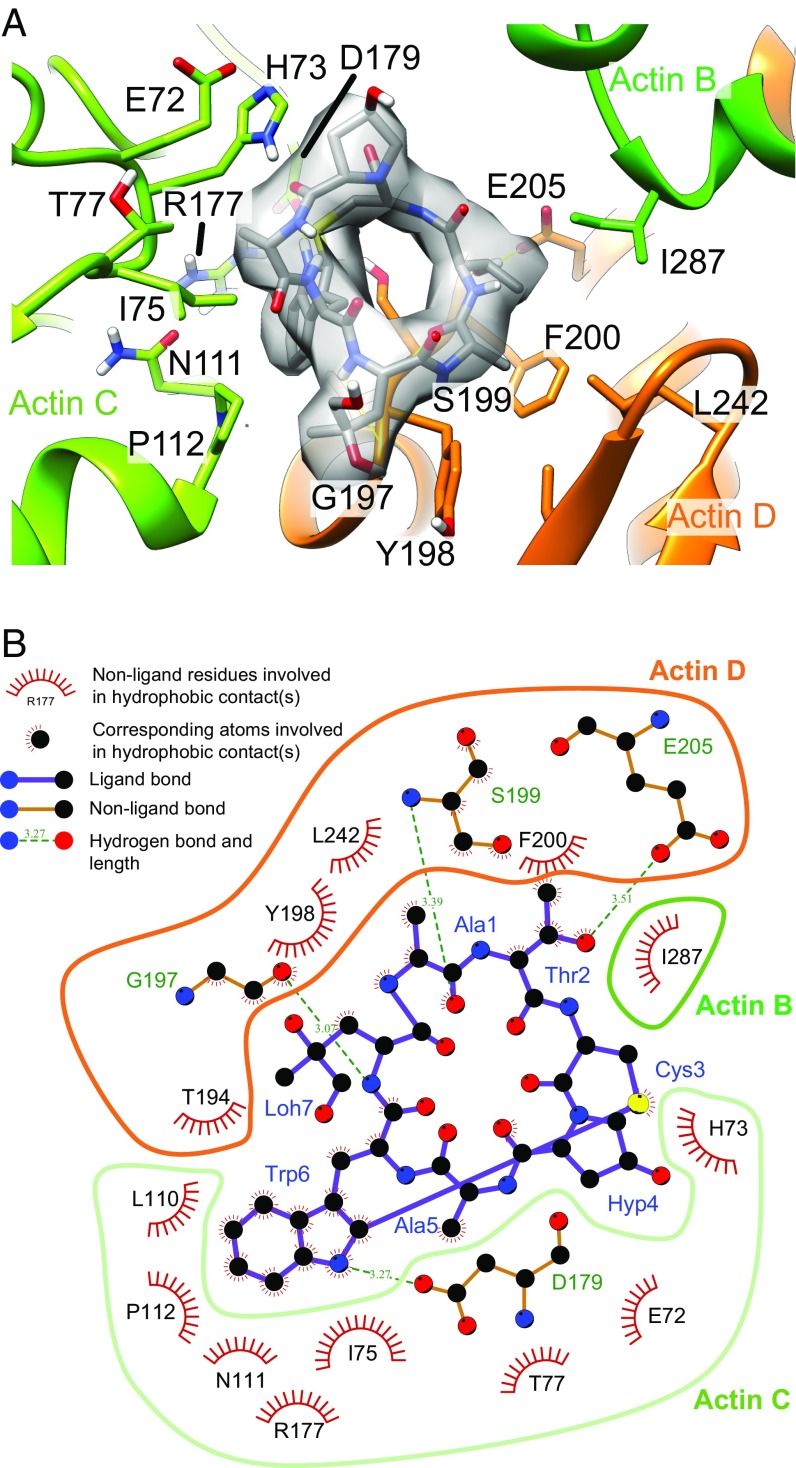
The position and orientation of phalloidin was near that predicted from X-ray fiber diffraction (32, 33) and negative-stained EM of gold-labeled phalloidin (34) with the dihydroxyleucine 7 facing the outside of the filament. The phalloidin–actin interactions predicted from the fit are shown in the atomic model (Fig. 5A and Movie S7) and the schematic determined by LigPlot (Fig. 5B) (35). The phalloidin interactions primarily couple adjacent actin monomers of the one-start short-pitch helix (actin C and D) or equivalently laterally (transversely) between strands of the two-start long-pitch helices. The additional predicted axial (longitudinal) coupling is the hydrophobic interaction of I287 of actin B with actin D through phalloidin. The lateral contacts of native filaments are composed of a small number of electrostatic interactions which are expected to be comparatively weak (36, 37). On the other hand, the phalloidin-coupled lateral interactions are composed of extensive hydrophobic contacts and five additional hydrogen bonds which are expected to be strong. The van der Waals surfaces of phalloidin and its binding site suggests that Trp6 provides a large measure of its binding strength, while the remainder of the molecule provide its specificity. Interestingly, the pocket in the actin filament in which Trp6 of phalloidin binds is a major site of jasplakinolide binding (38). Our results are consistent with mutational analysis of Drubin et al. (39) that predicted the importance of R177 and D179 as phalloidin binding residues.
Fig. 5.
Phalloidin structure and actin-binding site. (A) The phalloidin model (gray) in cryo-EM density and its interactions with residues in three actin subunits, actin B (green), C (light green), and D (orange). The pointed end of the filament is at the top and barbed end is at the bottom of the image. (B) The 2D interaction map of phalloidin with actin subunits as calculated by using Ligplot (35). Actin and phalloidin residues are denoted by one- or three-letter amino acid abbreviations, respectively. Phalloidin contains dihydroxy-l-leucine (Loh7) and 4-hydroxy-proline (Hyp4) amino acids.
Summary.
We identified an allosteric mechanism that couples the conformation of the nucleotide-binding site with the position of the myosin lever that reveals how mechanical load inhibits MgADP release by preventing the lever movements necessary to transition from AM.ADPA to AM, and ultimately power output. The current high-resolution structures of the myosin–actin interface also highlight the diversity of actin-docking interactions within the myosin family.
Materials and Methods
Details regarding protein expression and purification and experimental conditions and methods for structural determination by cryo-EM are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Purdue Cryo-EM facility for the use of their Titan Krios microscope and assistance with data collection; staff in the Yale Cryo-EM facility and High-Performance Computing facility for their maintenance of these facilities; and National Science Foundation Extreme Science and Engineering Discovery Environment computing resources (TG-MCB160065). This work was supported by NIH Grant R37 GM057247.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R.S. is a guest editor invited by the Editorial Board.
Data deposition: The cryo-EM maps have been deposited with the EMDataBank (accession codes EMD-7329, EMD-7330, and EMD-7331). The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5V7X, 6C1D, 6C1G, and 6C1H).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718316115/-/DCSupplemental.
References
- 1.Heissler SM, Sellers JR. Kinetic adaptations of myosins for their diverse cellular functions. Traffic. 2016;17:839–859. doi: 10.1111/tra.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenberg MJ, Arpağ G, Tüzel E, Ostap EM. A perspective on the role of myosins as mechanosensors. Biophys J. 2016;110:2568–2576. doi: 10.1016/j.bpj.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houdusse A, Sweeney HL. How myosin generates force on actin filaments. Trends Biochem Sci. 2016;41:989–997. doi: 10.1016/j.tibs.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 5.Veigel C, et al. The motor protein myosin-I produces its working stroke in two steps. Nature. 1999;398:530–533. doi: 10.1038/19104. [DOI] [PubMed] [Google Scholar]
- 6.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 7.Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capitanio M, et al. Two independent mechanical events in the interaction cycle of skeletal muscle myosin with actin. Proc Natl Acad Sci USA. 2006;103:87–92. doi: 10.1073/pnas.0506830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wulf SF, et al. Force-producing ADP state of myosin bound to actin. Proc Natl Acad Sci USA. 2016;113:E1844–E1852. doi: 10.1073/pnas.1516598113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittaker M, et al. A 35-A movement of smooth muscle myosin on ADP release. Nature. 1995;378:748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- 11.Jontes JD, Milligan RA, Pollard TD, Ostap EM. Kinetic characterization of brush border myosin-I ATPase. Proc Natl Acad Sci USA. 1997;94:14332–14337. doi: 10.1073/pnas.94.26.14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geeves MA, Perreault-Micale C, Coluccio LM. Kinetic analyses of a truncated mammalian myosin I suggest a novel isomerization event preceding nucleotide binding. J Biol Chem. 2000;275:21624–21630. doi: 10.1074/jbc.M000342200. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld SS, et al. Kinetic and spectroscopic evidence for three actomyosin:ADP states in smooth muscle. J Biol Chem. 2000;275:25418–25426. doi: 10.1074/jbc.M002685200. [DOI] [PubMed] [Google Scholar]
- 14.Münnich S, Taft MH, Manstein DJ. Crystal structure of human myosin 1c–The motor in GLUT4 exocytosis: Implications for Ca2+ regulation and 14-3-3 binding. J Mol Biol. 2014;426:2070–2081. doi: 10.1016/j.jmb.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Hayward S, Kitao A, Berendsen HJ. Model-free methods of analyzing domain motions in proteins from simulation: A comparison of normal mode analysis and molecular dynamics simulation of lysozyme. Proteins. 1997;27:425–437. doi: 10.1002/(sici)1097-0134(199703)27:3<425::aid-prot10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 16.Smith CA, Rayment I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 17.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 18.Shuman H, et al. A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning. Proc Natl Acad Sci USA. 2014;111:2116–2121. doi: 10.1073/pnas.1321022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg MJ, Lin T, Shuman H, Ostap EM. Mechanochemical tuning of myosin-I by the N-terminal region. Proc Natl Acad Sci USA. 2015;112:E3337–E3344. doi: 10.1073/pnas.1506633112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behrmann E, et al. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 22.von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- 23.Bement WM, Mooseker MS. TEDS rule: A molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil Cytoskeleton. 1995;31:87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- 24.Ostap EM, Lin T, Rosenfeld SS, Tang N. Mechanism of regulation of Acanthamoeba myosin-IC by heavy-chain phosphorylation. Biochemistry. 2002;41:12450–12456. doi: 10.1021/bi0262193. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg MJ, Ostap EM. Regulation and control of myosin-I by the motor and light chain-binding domains. Trends Cell Biol. 2013;23:81–89. doi: 10.1016/j.tcb.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann D, Santos A, Kovar DR, Rock RS. Actin age orchestrates myosin-5 and myosin-6 run lengths. Curr Biol. 2015;25:2057–2062. doi: 10.1016/j.cub.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joel PB, Trybus KM, Sweeney HL. Two conserved lysines at the 50/20-kDa junction of myosin are necessary for triggering actin activation. J Biol Chem. 2001;276:2998–3003. doi: 10.1074/jbc.M006930200. [DOI] [PubMed] [Google Scholar]
- 28.El Mezgueldi M, Tang N, Rosenfeld SS, Ostap EM. The kinetic mechanism of myo1e (human myosin-IC) J Biol Chem. 2002;277:21514–21521. doi: 10.1074/jbc.M200713200. [DOI] [PubMed] [Google Scholar]
- 29.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von der Ecken J, et al. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang N, Ostap EM. Motor domain-dependent localization of myo1b (myr-1) Curr Biol. 2001;11:1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz M, Popp D, Holmes KC. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- 33.Oda T, Namba K, Maéda Y. Position and orientation of phalloidin in F-actin determined by X-ray fiber diffraction analysis. Biophys J. 2005;88:2727–2736. doi: 10.1529/biophysj.104.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinmetz MO, et al. Evaluating atomic models of F-actin with an undecagold-tagged phalloidin derivative. J Mol Biol. 1998;276:1–6. doi: 10.1006/jmbi.1997.1529. [DOI] [PubMed] [Google Scholar]
- 35.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 36.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pospich S, et al. Near-atomic structure of jasplakinolide-stabilized malaria parasite F-actin reveals the structural basis of filament instability. Proc Natl Acad Sci USA. 2017;114:10636–10641. doi: 10.1073/pnas.1707506114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drubin DG, Jones HD, Wertman KF. Actin structure and function: Roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.