Significance
Immune cell activation is accompanied by dynamic changes in expression of genes related to inflammation. Concomitantly, immune reactions are tightly controlled to prevent harmful pathologies due to sustained inflammation. Gene expression is controlled at multiple checkpoints. Among these, the post-transcriptional regulation of the balance between Arid5a and Regnase-1 is important for the induction of inflammation. Our findings provide important insight into the role of the regulation of nucleocytoplasmic localization of Arid5a in the development of inflammation through the induction of a change in the ratio of Arid5a to Regnase-1.
Keywords: Arid5a, LPS, cytoplasm, inflammation, IL-6
Abstract
Adenine-thymine (AT)-rich interactive domain 5a (Arid5a) is an RNA-binding protein found in the cytoplasm and nucleus of normally growing cells. Although Arid5a is known to play an important role in immune regulation, whether and how Arid5a subcellular localization impacts immune regulation has remained unclear. In this study, we generated Arid5a transgenic (TG) mice to address this question. While ectopic Arid5a overexpression did not affect expression of inflammatory cytokines under unstimulated conditions, significantly higher levels of inflammatory cytokines, such as IL-6, were produced in response to lipopolysaccharide (LPS) stimulation. Consistent with this, TG mice were more sensitive to LPS treatment than wild-type mice. We also found that Arid5a is imported into the nucleus via a classical importin-α/β1–mediated pathway. On stimulation, nuclear Arid5a levels were decreased, while there was a concomitant increase in cytoplasmic Arid5a. Arid5a is associated with up-frameshift protein 1, and its nuclear export is regulated by a nuclear export receptor, chromosomal region maintenance 1. Taken together, these data indicate that Arid5a is a dynamic protein that translocates to the cytoplasm from the nucleus so as to properly exert its dual function in mRNA stabilization and transcriptional regulation during inflammatory conditions.
Adenine-thymine (AT)-rich interactive domain 5a (Arid5a) is a member of the Arid protein family and functions as a unique RNA-binding protein (1). At present, the Arid family is known to contain 15 members with various cellular functions, including roles in cell proliferation, cell growth, and cell cycle progression (2). Arid5a is highly expressed in macrophages in response to lipopolysaccharide (LPS) and controls posttranscriptional regulation of IL-6 by stabilizing Il6 mRNA by binding to its 3′ UTR (1). In addition, Arid5a stabilizes Stat3 and T-bet mRNAs in T cells (3, 4). Arid5a deficiency has been shown to inhibit the elevation of proinflammatory cytokine serum levels in LPS-treated mice (1, 4). Our group also has demonstrated that Arid5a-deficient mice are highly resistant to endotoxic shock, experimental autoimmune encephalomyelitis, and bleomycin-induced lung injury (1, 4, 5). Overall, these findings suggest that Arid5a plays important roles in RNA regulation and inflammatory disease control.
Regnase-1 (also known as Zc3h12a and Mcpip1) has been shown to destabilize Il6 mRNA by interacting with a conserved stem-loop motif in the 3′ UTR (6). Regnase-1 interacts with ribosome proteins and up-frameshift protein 1 (UPF1) (7).
UPF1 is essential for nonsense-mediated mRNA decay (NMD) and Regnase-1–mediated mRNA decay (7). NMD is a surveillance mechanism that degrades mRNAs containing premature termination codons. During NMD, UPF1 is recruited by UPF2, which in turn is recruited by UPF3 to create NMD ribonucleoprotein complexes on RNA (8). In addition, UPF1 shuttles between the nucleus and cytoplasm via chromosomal region maintenance 1 (CRM1), indicating its potential roles in both the nucleus and the cytoplasm (9, 10).
During nuclear pore complex (NPC) translocation, soluble transport factors are required to either bring cargo to the NPC or modulate cargo translocation across the NPC. Most of these soluble transport factors belong to the family of proteins known as karyopherins. Karyopherin-βs (also called importins, exportins, and tranportins) were the first family of shuttling transport factors discovered. Fourteen karyopherin-β family members are found in Saccharomyces cerevisiae, and at least 20 have been found in metazoans. A transport signal and shuttling receptor for the transport signal are the minimal requirements for facilitated translocation. The targeting of proteins into or out of the nucleus requires specific nuclear localization sequences (NLSs) or nuclear export sequences. The classical NLS (cNLS) is necessary for importin-α/β1 nuclear import (11). CRM1, a member of the importin-β superfamily, exports both proteins and mRNAs from the nucleus and is the main nuclear exporter (12–15). Leptomycin B (LMB) inhibits the function of CRM1 (16).
In this study, we investigated whether Arid5a-transgenic (TG) mice showed altered production of inflammatory cytokines under unstimulated and LPS-treated conditions. Interestingly, overproduction of Arid5a in TG mice did not elicit increases in IL-6 or inflammation under unstimulated conditions. In addition, we investigated the dynamics and subcellular localization of Arid5a. Our results provide important insight into the role of the regulation of nucleocytoplasmic localization of Arid5a in the development of inflammation through the induction of a change in the ratio of Arid5a to Regnase-1.
Results
Generation of Arid5a TG Mice.
Deregulated overexpression of IL-6 is associated with autoimmune inflammatory diseases (17). Significant therapeutic effects of the humanized anti–IL-6 receptor antibody tocilizumab have been demonstrated in various inflammatory diseases, including rheumatoid arthritis, demonstrating that abnormal production of IL-6 is responsible for the pathogenesis of these diseases. Indeed, Arid5a levels are significantly higher in untreated patients with rheumatoid arthritis than in healthy controls (18).
Therefore, we investigated whether the overexpression of Arid5a induces inflammatory disease in Arid5a TG mice. A schematic showing the generation of Arid5a TG mice is presented in Fig. S1A. The presence of the transgene in the indicated tissues was detected by quantitative real-time reverse-transcription PCR (qRT-PCR), using transgene-specific primers. In tail samples, exogenous Flag-tagged Arid5a mRNA levels in TG mice were 32- or 28-fold higher than those in wild-type (WT) mice (Fig. S1 B and C). These observations confirmed higher Arid5a expression levels in TG mice compared with WT mice (Fig. S1D). To further confirm the expression of Flag-tagged Arid5a protein, we isolated peritoneal macrophages from WT and TG mice. The cells were stimulated with or without LPS for 2 h, and immunoprecipitation and Western blot analysis were then performed using anti-Flag beads and anti-Arid5a antibodies, respectively. As a result, Flag-tagged Arid5a protein was detected only in the stimulated cells of TG mice (Fig. S1E). Thus, Arid5a expression was higher in TG mice than in WT mice.
Arid5a Did Not Affect the Production of Inflammatory Cytokines Under Unstimulated Conditions.
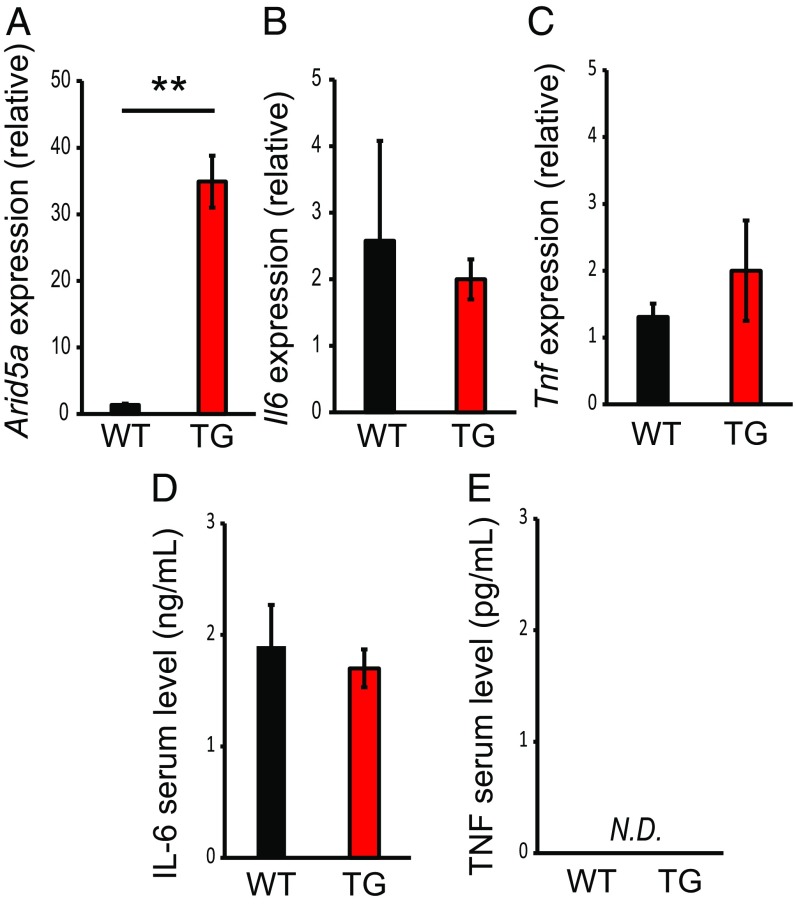
To analyze whether overexpression of Arid5a increased the mRNA and protein levels of IL-6 and tumor necrosis factor (TNF) in peritoneal macrophages or the serum, cells were isolated from WT and TG mice. qRT-PCR analysis showed that the mRNA levels of Arid5a were significantly up-regulated in TG peritoneal macrophages compared with WT peritoneal macrophages under unstimulated conditions (Fig. 1A). However, the levels of IL-6 and TNF in TG peritoneal macrophages and mice were not affected compared with those in WT peritoneal macrophages and mice (Fig. 1 B–E). Taken together, our results suggest that Arid5a did not affect the production of inflammatory cytokines under unstimulated conditions.
Fig. 1.
Arid5a did not affect inflammatory cytokine production under unstimulated conditions. (A–C) qRT-PCR analysis of Arid5a (A), Il6 (B), and Tnf (C) mRNAs in peritoneal macrophages, normalized to the expression of GAPDH. (D and E) Serum levels of IL-6 (D) and TNF (E) in WT and Arid5a TG mice were measured by ELISA. Data show the mean ± SEM of three independent experiments. Asterisks indicate statistically significant differences between WT and TG values. **P < 0.01, Student’s t test. N.D., not detected.
Arid5a TG Mice Were More Sensitive to Endotoxic Shock than WT Mice, and Overexpression of Arid5a Increased the Production of Proinflammatory Cytokines After LPS Stimulation.
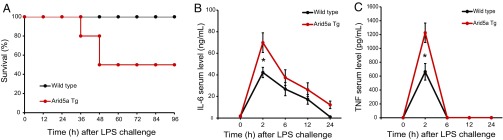
To determine the effects of Arid5a overexpression, WT and Arid5a TG mice were injected i.p. with LPS, and their survival was monitored. Results showed that 50% of TG mice died within 48 h of LPS administration, whereas all of the WT mice survived under the same conditions (Fig. 2A). To confirm the serum levels of proinflammatory cytokines, WT and Arid5a TG mice (6–8 wk old) were injected i.p. with LPS. IL-6 and TNF serum levels at 2 h after LPS challenge were significantly increased in Arid5a TG mice compared with WT mice (Fig. 2 B and C); thus, Arid5a contributed to augmentation of proinflammatory cytokine serum levels in vivo.
Fig. 2.
Arid5a TG mice were more sensitive to endotoxic shock than WT mice. (A) The 6- to 8-wk-old WT (n = 10) and Arid5a TG mice (n = 10) were injected with E. coli LPS (12.5 mg/kg), and survival was monitored for 96 h. (B and C) WT and Arid5a TG mice (6–8 wk old) were injected with LPS (5 mg/kg). Serum levels of IL-6 (B) or TNF (C) measured by ELISA at the indicated time points after LPS challenge. Data are the mean ± SEM of three independent experiments. Asterisks indicate statistically significant differences between WT and TG values (*P < 0.05, Student’s t test).
To further analyze whether overexpression of Arid5a increased the mRNA and protein levels of inflammatory cytokines in peritoneal macrophages, cells were isolated from WT and TG mice and stimulated with LPS. qRT-PCR analysis showed that the mRNA levels of cytokines were significantly up-regulated in TG peritoneal macrophages compared with WT peritoneal macrophages (Fig. S2 A–C). Moreover, ELISA showed significantly higher secretion of IL-6 and TNF in Arid5a TG cells compared with WT cells (Fig. S2 D and E). Consistent with the in vivo data, we observed that Arid5a TG cells secreted higher levels of inflammatory cytokines than WT cells. Taken together, our results suggest that Arid5a is associated with the proinflammatory response following LPS stimulation.
Arid5a Was Actively Imported into the Nucleus.
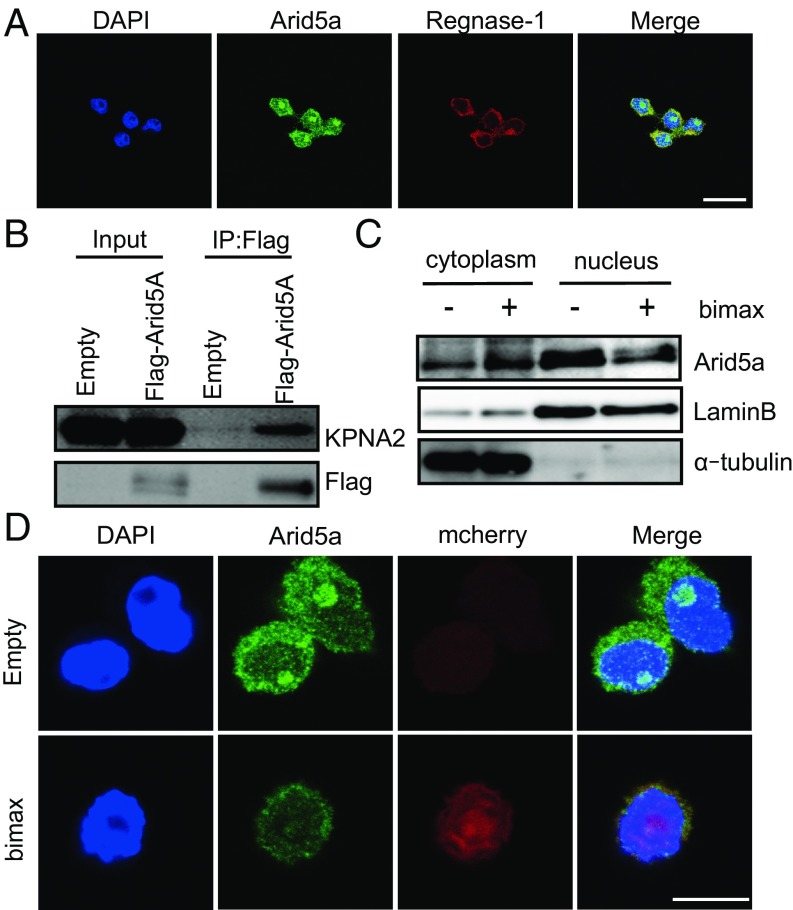
Arid5a interacts with nuclear proteins (18–20), whereas Regnase-1 localizes to the cytoplasm (6). We confirmed the localization of Arid5a and Regnase-1 by immunofluorescence staining in RAW264.7 cells. Our data indicated that Arid5a was localized mainly in the nucleus, whereas Regnase-1 was localized to the cytoplasm under unstimulated conditions (Fig. 3A).
Fig. 3.
Arid5a was actively imported into the nucleus. (A) Immunofluorescence staining of RAW264.7 cells was performed. The cells were incubated with indicated antibodies, and nuclei were visualized with DAPI. (Scale bar: 10 µm.) (B) HEK293T cells were transfected with Flag-tagged Arid5A or empty vector. Whole-cell lysates were prepared. Immunoprecipitation was performed with anti-Flag beads, followed by Western blotting with anti-Flag and anti-KPNA2 antibodies. (C) RAW264.7 cells were transfected with an empty or bimax vector. After 24 h, the cells were fractionated into cytoplasmic and nuclear extracts and analyzed by Western blot analysis using the indicated antibodies. (D) RAW264.7 cells were transfected with empty or bimax vector. After 24 h, cells were subjected to immunofluorescence staining. The cells were incubated with anti-Arid5a antibodies. The nuclei were visualized with DAPI. (Scale bar: 10 µm.)
We next examined how Arid5a was imported into the nucleus. First, we predicted the cNLS of Arid5a using cNLS mapper. Interestingly, cNLS prediction algorithms detected cNLS in the Arid5a protein (Table S1), suggesting that Arid5a was imported by a classical nuclear import pathway using importin-α/β1. To confirm the association between Arid5A and KPNA2, a member of the importin-α family, we performed a coimmunoprecipitation assay using expressed tagged-proteins. HEK293T cells were transfected with the empty vector or Flag-Arid5A plasmid, and anti-FLAG immunoprecipitates were analyzed by Western blot analysis with the indicated antibodies. The coimmunoprecipitation assay demonstrated that Arid5A was associated with KPNA2 (Fig. 3B).
Next, we monitored the effects of bimax peptides, which inhibit cNLS-dependent nuclear import via high-affinity interactions with NLS-binding sites of importin-α (21). We transfected RAW264.7 cells with empty vector or bimax-expressing plasmid and then performed Western blotting and immunofluorescence staining. The results showed that the nuclear import of Arid5a was inhibited by bimax (Fig. 3 C and D), suggesting that nuclear import of Arid5a was mediated by an importin-α/β1–mediated pathway.
The Nucleocytoplasmic Ratio of Arid5a Protein Changed After LPS Stimulation.
Despite the potential presence of a cNLS in Arid5a, our immunofluorescence results demonstrated that a substantial fraction of Arid5a was also localized in the cytoplasm. Thus, we hypothesized that Arid5a shuttles between the nucleus and cytoplasm and plays different roles in the regulation of Il6 mRNA depending on its subcellular (nuclear or cytoplasmic) localization. Therefore, we next examined the nucleocytoplasmic dynamics and RNA-binding abilities of the protein.
We first performed immunofluorescence staining with RAW264.7 cells stimulated with or without LPS for 4 h (Fig. 4A), and then measured the nuclear and cytoplasmic ratios of Arid5a (Fig. 4B). The data indicated that Arid5a was localized mainly in the nucleus before LPS stimulation; however, after LPS stimulation, nuclear Arid5a protein was decreased, whereas cytoplasmic Arid5a was increased. Moreover, Western blot analysis of the fractionated extracts showed that in both RAW264.7 cells and isolated peritoneal macrophages, Arid5a protein was gradually increased in the cytoplasm after LPS stimulation, whereas nuclear Arid5a was decreased (Fig. 4 C and D). These results suggest that a fraction of Arid5a translocated from the nucleus to the cytoplasm after LPS stimulation.
Fig. 4.
Nucleocytoplasmic ratio of Arid5a protein changes after LPS stimulation (A) RAW264.7 cells were stimulated with or without LPS (500 ng/mL) for 4 h and then analyzed by immunofluorescence staining. The cells were incubated with anti-Arid5a antibodies, and nuclei were visualized with DAPI. (Scale bar: 10 µm.) (B) Mean nuclear/cytoplasm ratios (n = 50; **P < 0.01; Student’s t test). (C and D) Cells were stimulated with LPS (RAW264.7 cells: 500 ng/mL, peritoneal macrophages: 100 ng/mL) at the indicated times and were then fractionated into cytoplasmic and nuclear extracts and analyzed by Western blot analysis using the indicated antibodies. (E) RAW264.7 cells were stimulated with LPS (100 ng/mL) at the indicated times. Cytoplasmic and nuclear extracts were isolated, and Il6 mRNA was evaluated by qRT-PCR, with normalization to GAPDH expression. The data show the mean ± SD of three independent experiments. Asterisks indicate statistically significant differences between time point values (*P < 0.05; **P < 0.01, Student’s t test). (F) After LPS stimulation for 4 h, total RNA isolated from Arid5a immunoprecipitation of each compartment was subjected to qRT-PCR analysis. RNA levels in the Arid5a-bound fraction of each compartment were normalized to input levels and then compared with those in cells without stimulation. Data show the mean ± SD of three independent experiments. Asterisks indicate statistically significant differences between unstimulated and stimulated Arid5a-Il6 mRNA values (**P < 0.01, Student’s t test).
We then examined whether nuclear and cytoplasmic Arid5a have the potential to bind Il6 mRNA. To do so, we first monitored the amounts of nuclear and cytoplasmic Il6 mRNA in RAW264.7 cells. The cells were fractionated into nuclear and cytoplasmic fractions before and after LPS stimulation. qRT-PCR demonstrated that nuclear Il6 mRNA was increased after 1–4 h of LPS stimulation. In contrast, cytoplasmic Il6 mRNA was increased only after 4 h (Fig. 4E).
Next, to examine whether the nuclear and/or cytoplasmic Il6 mRNA bound to Arid5a, we performed RNA immunoprecipitation (RIP) assays using anti-Arid5a antibodies. The RIP assays showed a 3.9-fold increase in the amount of Il6 mRNA bound to cytoplasmic Arid5a and a 2.6-fold increase in nuclear Arid5a after LPS stimulation, compared with in cells without stimulation (Fig. 4F). Thus, these data indicate that both nuclear and cytoplasmic Arid5a exhibits the ability to bind to Il6 mRNA after LPS stimulation.
Arid5a Was Associated with UPF1.
A previous study demonstrated that Arid5a competes with Regnase-1 (1, 3). UPF1 is essential for NMD and Regnase-1–mediated mRNA decay (7); therefore, we next investigated whether Arid5a is associated with the UPF family.
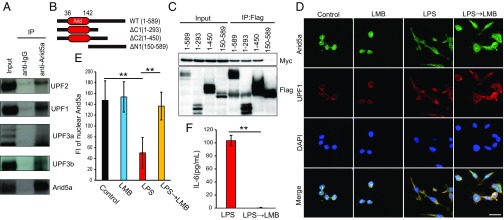
To this end, cell lysates were prepared from RAW264.7 cells, and anti-Arid5a immunoprecipitates were analyzed by Western blot analysis with the indicated antibodies. The data indicate that Arid5a indeed bound to UPF family members (Fig. 5A). To further confirm the association between Arid5A and UPF1, we performed a coimmunoprecipitation assay using ectopically expressed tagged proteins. HEK293T cells were transfected with the Flag-Arid5A– and/or Myc-UPF1–expressing plasmids, and anti-FLAG immunoprecipitates were analyzed by Western blot analysis with the indicated antibodies. The coimmunoprecipitation assay demonstrated the Arid5A was associated with UPF1 (Fig. S3).
Fig. 5.
Arid5a was associated with UPF1. (A) RAW264.7 cells were lysed with NETN buffer, and whole-cell lysates were then prepared. Immunoprecipitation was performed with anti-Arid5a antibodies and protein G Sepharose, followed by Western blot analysis with the indicated antibodies. (B) Schematic representation of truncated Arid5A. The Arid domain is shown in red. The amino acids present in each mutant are indicated. (C) Flag-tagged full-length (1–589) or truncated (1–293, 1–450, or 150–589) Arid5a was expressed with Myc-tagged UPF1 in HEK293T cells, and cell lysates were prepared. Immunoprecipitation was performed with anti-Flag beads, followed by Western blot analysis with anti-Flag and anti-Myc antibodies. (D) RAW264.7 cells were treated with or without LMB (10 nM) for 2 h, LPS (500 ng/mL) for 4 h, or LPS for 2 h + LMB for 2 h, and then analyzed by immunofluorescence staining. The nuclei were visualized with DAPI. (Scale bar: 10 µm.) (E) Fluorescence intensity of nuclear Arid5a. Asterisks indicate statistically significant differences between samples (n = 50; **P < 0.01; Student’s t test). (F) Levels of IL-6 in the medium from cultures of RAW264.7 cells were measured by ELISA. Data show the mean ± SEM of three independent experiments. Asterisks indicate statistically significant differences between LPS and LPS → LMB values (**P < 0.01, Student’s t test).
Next, we attempted to determine which domain of Arid5A was required for association with UPF1. A coimmunoprecipitation assay with truncated mutants of Arid5A (Fig. 5B) revealed that the ΔC1 and ΔC2 mutants, but not the ΔN1 mutant, bound to UPF1 (Fig. 5C). Thus, our results indicate that the 1- to 150-aa region (i.e., N-terminal region) of Arid5A, containing the Arid domain, is required for its association with UPF1.
UPF1 shuttles between the nucleus and cytoplasm via CRM1 (9, 10). To examine whether inhibition of the CRM1 pathway by LMB affects the nuclear export of Arid5a, we performed immunofluorescence staining with RAW264.7 cells and measured the fluorescence intensity of nuclear Arid5a. We found that Arid5a decreased in the nucleus after LPS stimulation; however, LMB treatment inhibited Arid5a nuclear export after LPS stimulation (Fig. 5 D and E). Taken together, these data indicate that Arid5a was associated with UPF1, and that LMB inhibited CRM1-mediated Arid5a nuclear export after LPS stimulation. Finally, we investigated whether LMB inhibits IL-6 production after LPS stimulation. ELISA showed that the secretion of IL-6 was significantly down-regulated by LMB treatment in RAW264.7 cells; this result was consistent with findings observed in U937 human promyeloid cells (22) (Fig. 5F).
Collectively, these data suggest that while Arid5a protein normally localizes to both the nucleus and the cytoplasm to stabilize Il6 mRNA, cytoplasmic Arid5a was increased to a greater extent after LPS stimulation. This increase may lead to more efficient translation of Il6 mRNA and other inflammatory cytokines.
Discussion
Here we have examined dynamics and subcellular localization of Arid5a in response to inflammation. It was not clear how Arid5a in the nucleus competes with Regnase-1 localized in the cytoplasm; therefore, we investigated the subcellular localization of Arid5a and Regnase-1. We found that Arid5a translocates to the cytoplasm from the nucleus during the inflammatory response. Arid5a localizes mainly to the nucleus with nuclear protein in resting cells. In contrast, Regnase-1 localizes to the cytoplasm (Fig. 3A). We predicted the cNLSs of Arid5a and Regnase-1; the results showed that Arid5a has a cNLS (Table S1), whereas Regnase-1 does not. Our results suggest that Arid5a is imported into the nucleus via a cNLS-dependent pathway with importin α/β. Under unstimulated conditions, Regnase-1 negatively regulates c-Jun N-terminal kinase (JNK) and nuclear factor-kappa B (NF-κB) signaling (23) and prevents unwanted production of cytokines (24). Given that Regnase-1–deficient mice spontaneously developed inflammatory disease (6), a dual locking system of inhibition of transcription and degradation of mRNA seems to be essential for the maintenance of homeostasis (24). However, under stimulated conditions, IL-6 was necessary for the immune response, and Il6 mRNA had to be protected from destabilization. Therefore, Arid5a translocates to the cytoplasm from the nucleus to inhibit Regnase-1–mediated mRNA decay. Thus, these findings suggest that Il6 mRNA is highly regulated by Arid5a in a compartment-specific manner; Regnase-1–mediated mRNA decay is inhibited by cytoplasmic translocation of Arid5a.
Arid5a physically interacts with the SRY-box 9 (Sox9) in the nucleus and up-regulates the chondrocyte-specific action of Sox9. Sox9 is an essential transcription factor in chondrocyte lineage determination and differentiation (19). Moreover, Arid5A also associates with retinoic acid receptor-related orphan nuclear gamma t (RORγT) (18), which functions as a lineage-specifying transcription factor in Th17 cells (25), as well as estrogen receptor (ER)α, which regulates genes involved in cell proliferation, differentiation, and migration. In addition, Arid5A suppresses RORγT and ERα functions (18, 20). In this study, we have shown that the N-terminal region of Arid5A is required for the association with UPF1. Thus, these data suggest that Arid5a function differs depending on localization; cytoplasmic Arid5a regulates mRNA stabilization by suppressing the function of UPF1, whereas nuclear Arid5a regulates the function of transcription factors.
UPF1 is essential for NMD and Regnase-1–mediated mRNA decay (7). Moreover, UPF1 shuttles between the nucleus and cytoplasm via CRM1, indicating that the protein has potential roles in both the nucleus and the cytoplasm (9, 10). CRM1 is the only exportin that mediates the transport of more than 230 proteins, including tumor suppressors, growth regulators/proinflammatory proteins, and antiapoptotic proteins (26). A previous study showed that NF-κB/IκBα complexes shuttle between the cytoplasm and nucleus through cNLS-dependent nuclear import and CRM1-dependent nuclear export. In addition, LMB sequesters NF-κB/IκBα complexes in the nucleus (27). Thus, LMB induces NF-κB inactivation. Accordingly, these findings suggested that the inhibition of Arid5a nuclear export may be induced by NF-κB inactivation. However, NF-κB was found to be activated by LPS stimulation because Il6 mRNA was produced in the nucleus after 1 h (Fig. 4E). Furthermore, in this study, LMB was added after LPS stimulation for 2 h (Fig. 5 D–F). Therefore, LMB inhibited the nuclear export of Arid5a, which interacted with Il6 mRNA and UPF1, after LPS stimulation. Thus, Arid5a may be translocated via UPF1/Il6 mRNA/CRM1-dependent export after LPS stimulation (Fig. 6).
Fig. 6.
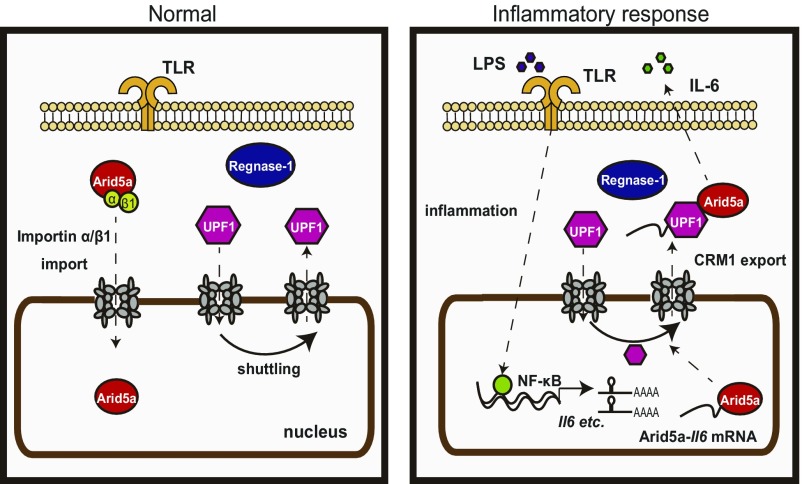
Model of inflammatory response by Arid5a/Regnase-1/UPF1. Regnase-1 localizes to the cytoplasm. Arid5a is imported via an importin-α/β1 pathway. UPF1 shuttles between the nucleus and the cytoplasm. In response to inflammation, the Toll-like receptor (TLR) is activated by LPS, and Il6 mRNA is induced by NF-κB. After that, Arid5a interacts with Il6 mRNA. Arid5a is exported to the cytoplasm via CRM1 pathway with UPF1.
RNA-binding proteins are associated with various diseases (28). Notably, tat activating regulatory DNA-binding protein 43 is essential for Il6 mRNA processing and stability in a stem loop/intron-dependent manner (29), This protein functions as another predominantly nuclear RNA-binding protein that translocates into the cytoplasm and aggregates as inclusion bodies in patients afflicted with amyotrophic lateral sclerosis or frontotemporal lobar degeneration with ubiquitin-positive inclusions (30). Interestingly, our previous study showed that inhibition of Arid5a phosphorylation and degradation increases the production of Il6 mRNA (31); thus, the inhibition of Arid5a protein degradation in the cytoplasm also may be related to neurodegeneration, cancer, or immune-related diseases.
In summary, our findings suggest that Arid5a is associated with cytokine regulation during the inflammatory response. Arid5a is exported into the cytoplasm after LPS stimulation. Moreover, Arid5a is imported into the nucleus via an importin-α/β1 pathway, but is exported into the cytoplasm via a CRM1-dependent mechanism. The precise molecular mechanism underlying nucleocytoplasmic Arid5a transport requires further study. Inhibition of Arid5a nuclear export is expected to facilitate the development of autoimmune therapeutic strategies.
Materials and Methods
Mice.
C57BL/6J WT mice (6–8 wk old) were obtained from Japan SLC, Inc. For generation of Arid5a TG mice, the DNA sequence encoding the Flag tag was attached to the 5′ end of Arid5a cDNA (1), and the resulting fragments were inserted between the NotI and XbaI sites of the pCAGGS vector, containing the chicken beta-actin promoter and a CMV immediate early enhancer to allow ubiquitous expression. A total of 223 pronuclear eggs were injected with a SacI-KpnI digested fragment (4.2 kb) and then transferred into the oviducts of pseudopregnant females the next day. Sixty-four pups were born, and PCR genotyping was performed using tail tips. Eleven pups were found to carry the CAG-Flag-Arid5a transgene. TG mice were identified by qRT-PCR with specific primers (forward: ACGATGATGACAAGGGCGAAT; reverse: TGGCTTGGGGGATACAGGAA). All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Osaka University’s Graduate School of Frontier Bioscience.
Cell Culture, Transfection, and Reagents.
RAW264.7, HEK293T, and HeLa cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% (vol/vol) FCS. Peritoneal macrophages were prepared as described previously (1). Thioglycolate-induced peritoneal macrophages were cultured in RPMI 1640 (Sigma-Aldrich) with 10% (vol/vol) FCS, 100 μg/mL streptomycin, and 100 U/mL penicillin G (Nacalai Tesque). Cells were transfected using the Nucleofector system (Lonza) and Fugene HD (Promega) according to the manufacturer’s recommendations. We treated the cells with LPS from Escherichia coli and LMB (Sigma-Aldrich).
qRT-PCR Analysis.
TRIzol (Invitrogen) was used for the isolation of total RNA, and ReverTra Ace (Toyobo) was used according to the manufacturer’s instructions for reverse transcription. For qRT-PCR, DNA fragments were amplified using KOD-SYBR qPCR Mix (Toyobo). qRT-PCR was carried out in an ABI PRISM 7900 HT (Applied Biosystems). Cycling conditions were 98 °C for 2 min, followed by 40 cycles of 98 °C for 10 s, 60 °C for 10 s, and 68 °C for 30 s. We applied the comparative ΔΔCt (ΔCt target − ΔCt control) method normalized to GAPDH for Il6, Tnf, and Arid5a mRNA quantitative analyses. The sequences of the PCR primers are shown in Table S2.
Immunoprecipitation and Western Blot Analysis.
Whole-cell lysates were prepared, and immunoblotting was performed as described previously (32).
Antibodies.
Anti-Flag (F1804; Sigma-Aldrich), anti-Arid5a (customized antibody; Sigma-Aldrich), anti–Regnase-1 (sc-515275; Santa Cruz Biotechnology), anti-KPNA2 (ab84440; Abcam), anti–Lamin-B (PM064; MBL), anti–α-tubulin (M175; MBL), anti-UPF1 (sc-166092; Santa Cruz Biotechnology), anti-UPF2 (D3B10; Cell Signaling Technology), anti-UPF3A (MBS712426; My Bio Source), anti-UPF3B (HPA001882; Atlas), and anti–c-Myc (Nacalai Tesque) antibodies were used in this study.
Cytokine Measurement by ELISA.
The levels of mouse IL-6 and TNF in serum were measured by ELISA using commercial kits according to the manufacturer’s protocol (R&D Systems). Serum was prepared from blood collected from the tail veins at various time points after LPS treatment.
Plasmids.
The oligonucleotides for the bimax peptide sequence (RRRRPRKRPLEWDEDEEPPRKRKRLW) (21, 33) were cloned into the pmCherry-C1 vector (Takara Bio). Human WT, truncated Arid5A, and UPF1 were generated by PCR amplification from total RNA and subsequently subcloned into pcDNA3.1 (Invitrogen) using BamHI and XhoI sites.
Immunofluorescence Staining.
Cells fixed with 3.7% paraformaldehyde in PBS were permeabilized by incubating with 0.5% Triton X-100 in PBS for 5 min. After blocking with 3% skim milk in PBS for 30 min, the cells were incubated with antibodies at 4 °C overnight, washed five times with PBS, and then incubated with Alexa Fluor 488- or 594-conjugated anti-mouse or anti-rabbit antibodies (1: 100 dilution) at 25 °C for 1 h. Nuclei were counterstained with 100 ng/mL DAPI (Dojindo Laboratories) for 10 min. The cells were observed under a Leica Microsystems TCS SP8 microscope.
Isolation of Nuclear and Cytoplasmic Proteins.
Nuclear and cytoplasmic proteins were isolated using an NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Protease inhibitor (Nacalai Tesque) was added to the CERI and NER extraction reagents before use.
Isolation of Nuclear and Cytoplasmic RNA and RNA Immunoprecipitation.
To isolate distinct pools of RNA from nuclear and cytoplasmic fractions, cells were collected on ice and resuspended in ice-cold fractionation buffer (10 mM Tris⋅HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.5% vol/vol Nonidet P-40) supplemented with 1 mM DTT, 100 U/mL RNase OUT (Invitrogen), and protease inhibitor cocktail (Nacalai Tesque) for 10 min on ice. Cell lysates were centrifuged at 1,000 × g for 5 min, and the supernatant (cytoplasmic fraction) was separated from the nuclear pellet. On isolation and separation of the cytoplasmic lysates, the nuclear fraction was rinsed once with ice-cold TSE buffer (10 mM Tris, 300 mM sucrose, 1 mM EDTA, and 0.1% Nonidet P-40 pH 7.5), and then passed through a syringe. Nuclear samples were incubated on ice for 20 min and then centrifuged for 10 min at 15,600 × g. The supernatant (nuclear fraction) was separated from the pellet (34). After preclearance with protein G-Sepharose (GE Healthcare), the lysates were incubated with anti-Arid5a or anti-IgG at 4 °C for 8 h. The resin was washed three times with RNA immunoprecipitation buffer (50 mM Tris⋅HCl pH 7.4, 100 mM NaCl, and 0.5% Nonidet P-40 in diethylpyrocarbonate-treated water). Arid5a/RNA complexes were eluted and isolated with TRIzol reagent and purified according to the manufacturer’s instructions.
Statistical Analysis.
The paired Student’s t test was used to analyze data for statistically significant differences. Differences with a P value <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank nonprofit organization Biotechnology Research and Development for technical assistance in generating the Arid5a TG mice. This work was funded by the Kishimoto Foundation and Immunology Frontier Research Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719921115/-/DCSupplemental.
References
- 1.Masuda K, et al. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc Natl Acad Sci USA. 2013;110:9409–9414. doi: 10.1073/pnas.1307419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin C, et al. Recent advances in the ARID family: Focusing on roles in human cancer. Onco Targets Ther. 2014;7:315–324. doi: 10.2147/OTT.S57023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuda K, et al. Arid5a regulates naive CD4+ T cell fate through selective stabilization of Stat3 mRNA. J Exp Med. 2016;213:605–619. doi: 10.1084/jem.20151289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaman MM-U, et al. Arid5a exacerbates IFN-γ-mediated septic shock by stabilizing T-bet mRNA. Proc Natl Acad Sci USA. 2016;113:11543–11548. doi: 10.1073/pnas.1613307113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubey PK, et al. Arid5a-deficient mice are highly resistant to bleomycin-induced lung injury. Int Immunol. 2017;29:79–85. doi: 10.1093/intimm/dxx004. [DOI] [PubMed] [Google Scholar]
- 6.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 7.Mino T, et al. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 2015;161:1058–1073. doi: 10.1016/j.cell.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendell JT, ap Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 10.Ajamian L, et al. HIV-1 recruits UPF1 but excludes UPF2 to promote nucleocytoplasmic export of the genomic RNA. Biomolecules. 2015;5:2808–2839. doi: 10.3390/biom5042808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 14.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 15.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 16.Kudo N, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 17.Kishimoto T. Interleukin-6: From basic science to medicine 40 years in immunology. Annu Rev Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 18.Saito Y, et al. AT-rich interactive domain-containing protein 5a functions as a negative regulator of ROR gamma t-induced Th17 cell differentiation. Arthritis Rheum. 2014;66:1185–1194. doi: 10.1002/art.38324. [DOI] [PubMed] [Google Scholar]
- 19.Amano K, et al. Arid5a cooperates with Sox9 to stimulate chondrocyte-specific transcription. Mol Biol Cell. 2011;22:1300–1311. doi: 10.1091/mbc.E10-07-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgescu SP, et al. Modulator recognition factor 1, an AT-rich interaction domain family member, is a novel corepressor for estrogen receptor alpha. Mol Endocrinol. 2005;19:2491–2501. doi: 10.1210/me.2004-0311. [DOI] [PubMed] [Google Scholar]
- 21.Tsujii A, et al. Retinoblastoma-binding protein 4-regulated classical nuclear transport is involved in cellular senescence. J Biol Chem. 2015;290:29375–29388. doi: 10.1074/jbc.M115.681908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh CC, et al. Gene-specific repression of proinflammatory cytokines in stimulated human macrophages by nuclear IκBα. J Immunol. 2010;185:3685–3693. doi: 10.4049/jimmunol.0902230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang J, et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwasaki H, et al. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12:1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 26.Ishizawa J, Kojima K, Hail N, Jr, Tabe Y, Andreeff M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol Ther. 2015;153:25–35. doi: 10.1016/j.pharmthera.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang TT, Kudo N, Yoshida M, Miyamoto S. A nuclear export signal in the N-terminal regulatory domain of IkBa controls cytoplasmic localization of inactive NF-kB/IkBa complexes. Proc Natl Acad Sci USA. 2000;97:1014–1019. doi: 10.1073/pnas.97.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, et al. Identification of a subnuclear body involved in sequence-specific cytokine RNA processing. Nat Commun. 2015;6:5791. doi: 10.1038/ncomms6791. [DOI] [PubMed] [Google Scholar]
- 30.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 31.Nyati KK, et al. TLR4-induced NF-κB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res. 2017;45:2687–2703. doi: 10.1093/nar/gkx064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higa M, Zhang X, Tanaka K, Saijo M. Stabilization of ultraviolet (UV)-stimulated scaffold protein A by interaction with ubiquitin-specific peptidase 7 is essential for transcription-coupled nucleotide excision repair. J Biol Chem. 2016;291:13771–13779. doi: 10.1074/jbc.M116.724658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosugi S, et al. Design of peptide inhibitors for the importin α/β nuclear import pathway by activity-based profiling. Chem Biol. 2008;15:940–949. doi: 10.1016/j.chembiol.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Wigington CP, Morris KJ, Newman LE, Corbett AH. The polyadenosine RNA-binding protein, zinc finger Cys3His protein 14(ZC3H14), regulates the pre-mRNA processing of a key ATP synthase subunit mRNA. J Biol Chem. 2016;291:22442–22459. doi: 10.1074/jbc.M116.754069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.