Abstract
Immune checkpoint blockade produces clinical benefit in many patients. However better biomarkers of response are still needed, and mechanisms of resistance remain incompletely understood. To address this, we recently studied a cohort of melanoma patients treated with sequential checkpoint blockade against cytotoxic T lymphocyte antigen-4 (CTLA-4) followed by programmed death receptor-1 (PD-1), and identified immune markers of response and resistance. Building on these studies, we performed deep molecular profiling including T-cell receptor sequencing (TCR-seq) and whole exome sequencing (WES) within the same cohort, and demonstrated that a more clonal T cell repertoire was predictive of response to PD-1 but not CTLA-4 blockade. Analysis of copy number alterations identified a higher burden of copy number loss in non-responders to CTLA-4 and PD-1 blockade and found that it was associated with decreased expression of genes in immune-related pathways. The effect of mutational load and burden of copy number loss on response was non-redundant, suggesting the potential utility of a combinatorial biomarker to optimize patient care with checkpoint blockade therapy.
Introduction
Immune checkpoint blockade represents a major advancement in cancer therapy for advanced melanoma. However, durable clinical responses are seen in only a minority of patients treated with single-agent CTLA-4 (1) or PD-1 blockade (2, 3). Although higher response rates are achieved when CTLA-4 and PD-1 inhibitors are administered concurrently, this regimen also has greatly increased toxicity (3, 4). There is a clinical need to predict who will benefit from immunotherapy and to understand mechanisms of therapeutic resistance to improve patient management and outcomes. Recently, evidence has pointed to a role of tumor molecular features (such as mutational load) (5–8) and host immune infiltrates (9–12) in response to therapy, though complexities exist with the predictive power of these markers (13). Studies have also begun to uncover mechanisms of resistance, including expression of immune checkpoint molecules (10, 14–21), insufficient infiltration of CD8+ T cells (9, 10), oncogenic pathways (22–24), transcriptomic resistance signatures (25), lack of sensitivity to interferon signaling (26–30), defects in antigen processing and presentation (11, 30–32), diversity and abundance of bacteria within the gut microbiome (33, 34), and metabolism of cancer cells and T cells (35–37). However, additional insights are clearly needed for a more comprehensive understanding of resistance.
To further refine both host and tumor genomic contributions to resistance to checkpoint blockade, we assembled a cohort of longitudinal tissue samples from metastatic melanoma patients treated with sequential immune checkpoint blockade (CTLA-4 blockade followed by PD-1 blockade at time of progression). We previously performed deep immune profiling studies on these samples (via immunohistochemistry and gene expression profiling) and identified immune biomarkers of response and mechanisms of therapeutic resistance (38). To complement these studies, we report here the results of in-depth molecular analysis (via whole exome sequencing and T cell receptor sequencing) of these longitudinal samples. These studies have identified putative genomic and molecular biomarkers of response and resistance to immune checkpoint blockade, demonstrating the complex interplay of host and tumor in treatment response.
Results
T cell clonality predicts response to PD-1 blockade but not CTLA-4 blockade
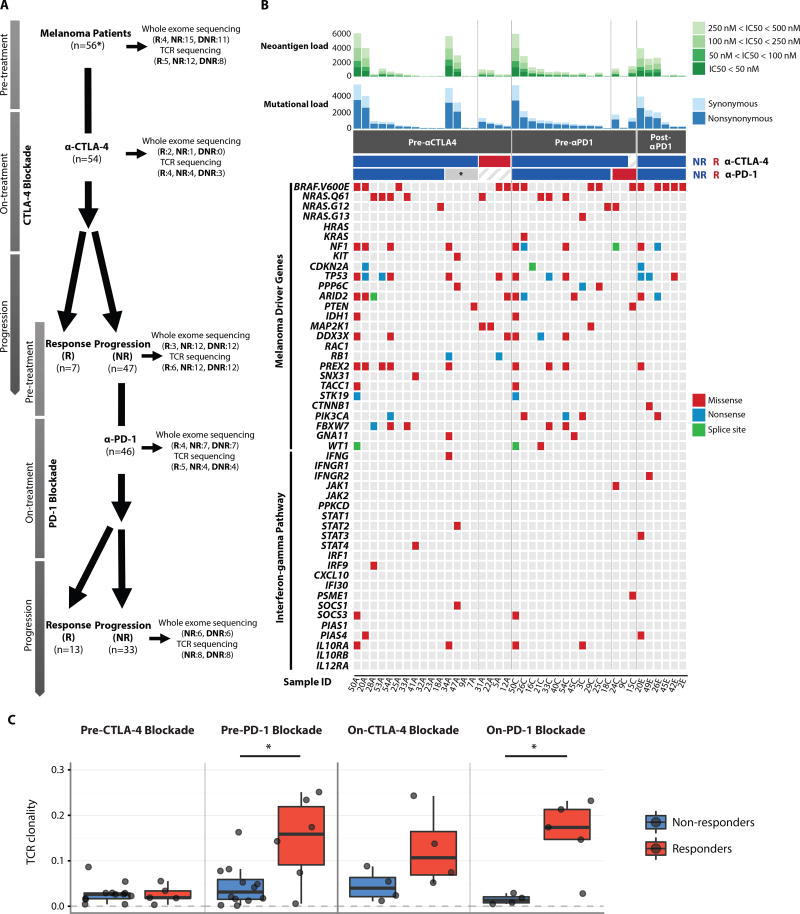
We studied a cohort of 56 patients who were first treated with CTLA-4 blockade, and then subsequently treated with PD-1 blockade at the time of progression, with longitudinal tumor samples collected as previously described (38) (Fig. 1A, table S1A–S1B, Table S2) by performing whole exome sequencing (WES) and TCR sequencing (TCR-seq) on DNA from available tumor samples (Fig. 1A, fig. S1–S2 table S3). Responders were defined as patients who had complete resolution or partial reduction in the size of tumors by CAT scan-based imaging (by at least 30%), or who had prolonged stable disease (for at least 6 months). Non-responders were defined as patients who had tumor growth of at least 20% on CAT scan, or had stable disease lasting less than 6 months. We first compared the mutation status of common melanoma driver genes (39, 40) in pre-treatment samples, and also assessed interferon-gamma pathway genes, given the importance of defects in interferon-gamma signaling in resistance to immune checkpoint blockade (30, 41–43), and found no significant differences between responders and non-responders to therapy with regard to somatic point mutations or indels (Fisher’s exact test with a false discovery rate threshold of 0.05) (Fig. 1B, table S4). Next, we compared the frequency of HLA somatic mutations (44) in pre-treatment samples and found that HLA somatic mutations were found in only one pre-treatment biopsy from a CTLA-4 blockade non-responder (table S5). No particular genes were enriched for mutations in post-PD-1 blockade samples except BRAF (V600E) (5 out of 6 patients), potentially due to prolonged survival with interim BRAF targeted therapy in 3 out of 5 patients (Fig. 1B and table S1B).
Fig. 1. Genomic landscape of serial tumor biopsies and genomic and immune correlates of treatment response.
(A) Patients with metastatic melanoma were initially treated with CTLA-4 blockade (n=56*: * indicates that two of the 56 patients were CTLA-4 blockade naïve. Both responded to PD-1 blockade, and only pre-treatment samples were available for WES and TCR-seq). Non-responders to CTLA-4 blockade (n=47) were then treated with PD-1 blockade. Double non-responders progressed on CTLA-4 blockade first and then progressed on PD-1 blockade. Serial tumor biopsies were collected at multiple time points (pre-treatment, early on-treatment, and progression on CTLA-4 blockade and PD-1 blockade, respectively) when feasible. Whole exome sequencing and TCR sequencing were performed on these serial tumor biopsies. The numbers in parentheses indicate the number of samples available for responders and non-responders after quality control of WES and TCR-seq data. R: responders, NR: non-responders, DNR: double non-responders. (B) For each sample (columns), genomic profiles (rows) were characterized. Column annotations represent biopsy time (Pre-αCTLA4: pre-CTLA-4 blockade samples, Pre-αPD1: pre-PD-1 blockade samples, Post-αPD1: post-PD-1 blockade samples) and response status (red: responders indicated as R, blue: non-responders indicated as NR, *: failed CTLA-4 blockade but responded to PD-1 blockade) for each sample (Sample ID denotes patient ID followed by biopsy time: A=pre-αCTLA-4, C=post-CTLA-4/pre-PD-1, and E=post-PD-1). Shown at the top of the panel is mutational burden and neoantigen burden for each sample. Neoantigens were defined as having an IC50<500nM. Color scale shows the range of IC50 from 500nM to 50nM. Synonymous (light) and non-synonymous (dark) mutations are shown in different shades of blue. Additional genomic profiles included selected somatic point mutations, and indels. No indels were found among melanoma driver genes. When multiple mutations were found in one gene, the following precedence rule was applied: Nonsense mutation > Frame-shift indel > Splice site mutation > Missense mutation > In-frame indel. (C) Boxplots summarize TCR clonality by response status (blue: non-responders, red: responders) in pre-CTLA-4 blockade samples, pre-PD-1 blockade samples, on-CTLA-4 blockade samples, and on-PD-1 blockade samples, respectively; median values (lines) and interquartile range (whiskers) are indicated. P values were calculated using a two-sided Mann-Whitney U test (P > 0.05 for TCR clonality in pre-CTLA-4 blockade and on-CTLA-4 blockade samples, P = 0.041 for TCR clonality in pre-PD-1 blockade samples and P = 0.032 for TCR clonality in on-PD-1 blockade samples.).
In our cohort, we did not observe any statistically significant differences in mutational load or predicted neoantigen load in pre-treatment samples from responders versus non-responders to therapy by either CTLA-4 or PD-1 blockade (fig. S3A–S3B, table S6) (mutational load: P = 0.597 in pre-CTLA-4 blockade samples, P = 0.840 in pre-PD-1 blockade samples; neoantigen load: P = 0.411 in pre-CTLA-4 blockade samples, P = 0.942 in pre-PD-1 blockade samples), which is in contrast to published literature (5–8) and may be due to limited sample size. Further, no significant differences were observed in intratumor heterogeneity (ITH), estimated as the number of clones per tumor, between responders and non-responders to immune checkpoint blockade (fig. S4).
We next performed sequencing of the CDR3 variable region of the β-chain of the T cell receptor (TCR-seq) to understand the role of the T cell repertoire in response and resistance to checkpoint blockade (table S7). Although no significant differences were observed in TCR clonality when comparing responders to non-responders in the context of CTLA-4 blockade at the pre-treatment (P = 0.96) and on-treatment time points (P = 0.2) (Fig. 1C), an increase in clonality was noted in a subset of patients treated with CTLA-4 blockade (fig. S5A). Among 8 patients with matched longitudinal tumor samples (pre-CTLA-4 and pre-PD-1, n=8) available, all three PD-1 blockade responders showed an increase in TCR clonality on CTLA-4 blockade, whereas this was the case in only 1 out of 5 non-responders to PD-1 blockade (fig. S5A). The one patient (Patient 50) classified as a non-responder in the context of the trial criteria who demonstrated an increase in clonality appeared to have some clinical benefit from treatment with PD-1 blockade, as he continued on PD-1 blockade therapy for a total of 24 doses and had no evidence of disease at last follow up. Higher TCR clonality was observed in responders to PD-1 blockade at both pre- (P = 0.041) and on-treatment (P = 0.032) time points (Fig. 1C).
Next, we sought to investigate the association between TCR clonality and immune activation in the tumor microenvironment. To do so, we first calculated the immune score from gene expression profiling data (38) in our cohort (table S8). The immune score was defined as the geometric mean of gene expression in selected genes including cytolytic markers, HLA molecules, IFN-γ pathway, selected chemokines, and adhesion molecules related to tumor rejection in the context of the immunologic constant of rejection (45, 46) (table S9). Although no association was observed between TCR clonality and immune scores in pre-CTLA-4 blockade samples, a significant positive correlation was observed between TCR clonality and immune scores in pre-PD-1 blockade samples (P = 0.0018, fig. S5B).
High copy number loss burden is associated with poor response to immune checkpoint blockade
Given the lack of clear differences in point mutation and indel status in driver genes between responders and non-responders to CTLA-4 and PD-1 blockade, we then investigated whether copy number alterations (CNAs) may play a role in response and resistance to CTLA-4 and PD-1 blockade (table S10). With regards to specific genes, we did not find any significant association between copy number gain or loss status and response to therapy in pre-treatment biopsies for either therapy (Fisher’s exact test with a false discovery rate threshold of 0.05). Given a recent report demonstrating the impact of loss of HLA Class I and β2-microglobulin in resistance to cytolytic activity (11), we next examined the relevance of copy number loss in these genes within our cohort. In this study, although we observed no significant loss of HLA class I genes, loss of β2-microglobulin was detected in 4 non-responders to CTLA-4 blockade (with focal copy number loss in 2 patients and arm-level copy number loss in 2 patients). Focal copy number loss of β2-microglobulin was also observed in 1 pre-treatment sample from a CTLA-4 blockade naïve PD-1 blockade responder. To assess CNAs at the whole genome sample level, we defined burden of CNAs as the total number of genes with copy number gain or loss per sample (table S2). On testing the association between burden of CNAs and response to therapy in pre-treatment biopsies of patients on CTLA-4 or PD-1 blockade, we observed no significant differences in burden of copy number gain or loss (P > 0.05 for all comparisons) in the context of individual agent response (fig. S6). However, a trend toward higher burden of copy number loss was observed in pre-CTLA-4 blockade biopsies from CTLA-4 blockade responders compared to CTLA-4 blockade non-responders, though statistical significance was not attained (P = 0.077).
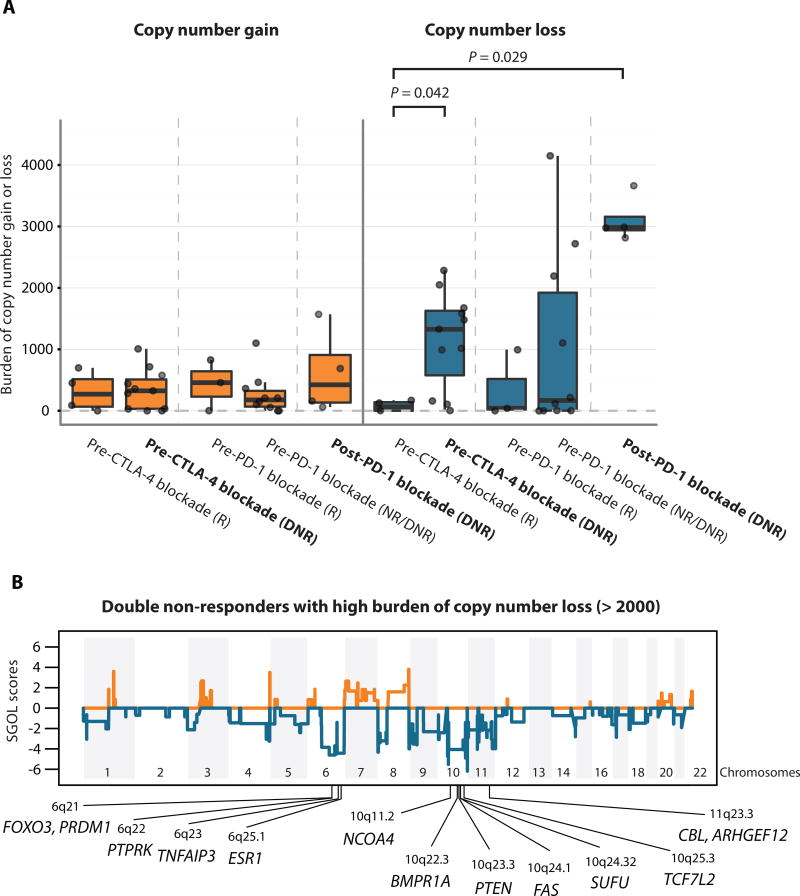
We next investigated the burden of copy number alterations in pre-CTLA-4 blockade biopsies from patients who progressed on CTLA-4 blockade first and then progressed on PD-1 blockade, termed double non-responders (DNRs) because we hypothesized that the association between burden of copy number alterations and resistance might be stronger in patients with potentially more resistant phenotype (failure on both treatments) than in patients who failed a single agent. We observed no significant differences in burden of copy number gain but significantly higher burden of copy number loss in pre-CTLA-4 blockade biopsies from DNRs compared to pre-CTLA-4 blockade biopsies from CTLA-4 blockade responders (P = 0.042) (Fig 2A, fig. S7). We noted a strikingly higher burden of copy number loss in post-PD-1 blockade biopsies compared to pre-CTLA-4 blockade biopsies from CTLA-4 blockade responders (P = 0.029) (Fig 2A, fig. S7). The burden of copy number loss was not correlated with mutational load at any of the time points studied (fig. S8), suggesting that the association is not readily attributable to decreased mutational burden.
Fig. 2. Copy number loss as a potential resistance mechanism.
(A) Boxplots summarize burden of copy number gain or loss in five groups of interest: responders to CTLA-4 blockade at pre-treatment, pre-CTLA-4 blockade double non-responders, responders to PD-1 blockade at pre-treatment, pre-PD-1 blockade double non-responders, and post-PD-1 blockade double non-responders; median values (lines) and interquartile range (whiskers) are indicated. P values were calculated using a two-sided Mann-Whitney U test. (P = 0.042 for pre-CTLA-4 blockade responders vs. double non-responders, P = 0.029 for pre-CTLA-4 blockade responders vs. post-PD-1 blockade double non-responders, and P > 0.05 for all others) DNR: double non-responders, NR: non-responders, R: responders. In bold are highlighted the Pre-CTLA-4 blockade and Post-PD-1 blockade double non-responder (DNR) groups. (B) Segment Gain or Loss (SGOL) scores were calculated for each copy number segments as sum of log2 copy ratios (tumor/normal) of each copy number segment across all double non-responder samples with burden of copy number loss higher than 2,000 (n=9). Higher positive SGOL scores indicate higher copy number gain of copy number segments and lower negative SGOL scores indicate higher copy number loss of copy number segments. Tumor suppressor genes with recurrent copy number loss are indicated in chromosome 6q, 10q, and 11q23.3.
To gain insight into mechanisms through which CNAs could influence response to therapy, we next investigated if there were any recurrent regions of copy number loss in double non-responders with high burden of copy number loss (> 2,000 genes with copy number loss). Recurrent copy number loss was observed at the arm level in chromosome 6q and 10q, and recurrent focal copy number loss was observed in 8p23.3, 11p15.5, 11q23, 11q24, and 11q25 (Fig. 2B, table S11). In these regions with recurrent copy number loss, tumor suppressor genes were located in chromosomes 6q (FOXO3, PRDM1, PTPRK, TNFAIP3, and ESR1), 10q (NCOA4, BMPR1A, PTEN, FAS, SUFU, and TCF7L2), and 11q23.3 (CBL, ARHGEF12). These data suggest that high burden of copy number loss in double non-responders is associated with recurrent copy number loss in tumor suppressor genes located in chromosomes 6q, 10q, and 11q23.3.
An independent patient cohort shows copy number loss as a putative resistance mechanism
To investigate the impact of higher burden of copy number loss on resistance in another cohort of patients on immune checkpoint blockade, we obtained WES SAM files from 110 melanoma patients and RNA-seq data from a subset of 42 melanoma patients (7) and reanalyzed the data utilizing the same informatics pipeline and calling criteria. We then tested the association between the burden of CNAs (table S12–S13) and response to therapy in pre-treatment biopsies on CTLA-4 blockade using the same response criteria (clinical benefit, long-term survival with no clinical benefit, and minimal or no clinical benefit) as previously described (7). Although the burden of copy number gain was not significantly associated with clinical benefit from CTLA-4 blockade, a lower burden of copy number loss was significantly associated with clinical benefit (P = 0.016) (Fig. 3A). As observed in our cohort, the burden of copy number loss once again did not correlate with mutational load (fig. S9). When examining the regions associated with recurrent copy number loss in the no clinical benefit subgroup, recurrent copy number loss was observed at arm level in chromosome 9p and 10q, and recurrent focal copy number loss was observed in 4q35.2, 6q25, 6q27, and 11p15.5 (fig. S10 and table S14). Among these regions, tumor suppressor genes were observed in 6q25.1 (ESR1) and 10q (NCOA4, BMPR1A, PTEN, FAS, and SUFU), as seen within our cohort. Of note, no recurrent copy number loss was observed in any tumor suppressor gene in the clinical benefit subgroup and long-term survival subgroup.
Fig. 3. Copy number loss as a potential resistance mechanism in an independent cohort.
(A) Boxplots summarize burden of copy number gain or loss in three patient subgroups from the Van Allen cohort: clinical benefit, long-term survival with no clinical benefit, and minimal or no clinical benefit; median values (lines) and interquartile range (whiskers) are indicated. P values were calculated using a two-sided Mann-Whitney U test (P = 0.016 for burden of copy number loss in clinical benefit vs. minimal or no clinical benefit, and P > 0.05 for all others). (B) Gene set enrichment analysis (GSEA) results show top enriched KEGG pathways from down-regulated genes (blue bars) and up-regulated genes (red bars) in high burden of copy number loss group versus low burden of copy number loss group (FDR-adjusted P < 0.001). (C) Proportions of patients with clinical benefit, long-term survival with no clinical benefit, and minimal or no clinical benefit were calculated within each of the four patient subgroups: high mutational load and low burden of copy number loss, high mutational load and high burden of copy number loss, low mutational load and low burden of copy number loss, and low mutational load and high burden of copy number loss. The numbers in parentheses indicate the number of patients with different levels of response (clinical benefit, long-term survival, and no clinical benefit) out of the total number of patients in each of the four patient subgroups.
Next, we investigated whether the recurrent region of copy number loss identified in our cohort (table S11) is also associated with CTLA-4 blockade resistance in this independent cohort (Van Allen cohort). To do so, we calculated the burden of copy number loss in this independent cohort as the total number of genes with copy number loss in the recurrent regions of copy number loss identified in our cohort. We observed a significantly higher burden of copy number loss in the minimal or no clinical benefit groups compared to the clinical benefit group (P = 0.0034) (fig. S11). This result suggests that the recurrent regions of copy number loss in our cohort are also associated with CTLA-4 blockade resistance in this independent cohort.
We next sought to determine the relative contribution of copy number loss burden from chromosome 10 in CTLA-4 blockade resistance. We were specifically interested in copy number loss from chromosome 10 because a recent study (47) showed functional evidence that recurrent loss of the entire chromosome 10 can result in collective repression of multiple tumor suppressor genes. This is also consistent with the observation that chromosome 10 harbored the largest number of tumor suppressor genes with recurrent copy number loss in both our cohort (Fig 2B) and the independent cohort (fig. S10). The logistic regression model showed that the odds of resistance to CTLA-4 blockade were exp(1.504) = 4.5 (95% CI: 1.56 – 13) times greater in patients with high burden of copy number loss in chromosome 10 than in patients with low burden of copy number loss in chromosome 10 (table S15A) and the odds of resistance were exp(1.069) = 2.91 (95% CI: 1.07 – 7.89) times greater in patients with high burden of copy number loss outside chromosome 10 than in patients with low burden of copy number loss outside chromosome 10 (table S15B). Therefore, the relative contribution of copy number loss burden from chromosome 10 in CTLA-4 blockade resistance was higher than copy number loss burden outside chromosome 10. We further investigated the extent to which PTEN loss in chromosome 10 is associated with CTLA-4 blockade resistance (24). In our data, the odds of resistance were 5.58 times greater in patients with PTEN loss than in patients with no PTEN loss (95% CI: 1.19 – 26.20) (table S15C), suggesting that PTEN loss is likely to be one of the driver resistance mechanisms exploited by tumors with high burden of copy number loss on chromosome 10.
Integrated analysis reveals putative mechanisms through which CNAs may influence response to therapy
In addition to studying the influence of copy number loss on molecular features such as tumor suppressor genes, we sought to study the relationship of these alterations with the immune tumor microenvironment. To do so, we examined the correlation between burden of copy number loss and immune scores. Although we observed no correlation between the burden of copy number loss and immune scores in pre-CTLA-4 blockade biopsies in our cohort (correlation coefficient = −0.13; Spearman rank correlation, P = 0.79), a moderate negative correlation between the burden of copy number loss and immune scores calculated by ESTIMATE (48) was identified in the Van Allen cohort (correlation coefficient = −0.41; Spearman rank correlation, P = 0.011) (7) (fig. S12A). In pre-PD-1 blockade biopsies in our cohort, we also observed a negative correlation between the burden of copy number loss and immune scores (correlation coefficient = −0.63; Spearman rank correlation, P = 0.091) (fig. S12B), although this could not be investigated in post-PD-1 blockade biopsies due to sample size. Our immune scores and those calculated by ESTIMATE (table S16) showed a strong positive correlation (correlation coefficient = 0.91; Spearman rank correlation, P < 2.2e-16) in the independent cohort (fig. S12C), suggesting concordance between immune scores.
We further sought to determine which pathways or gene ontologies (GO) were enriched in up/down-regulated genes at the mRNA expression level in the high burden of copy number loss (n=10; mean: 4149, range: 2815 to 6764) versus low burden of copy number loss (n=10; mean: 0) groups within the Van Allen cohort. Gene set enrichment analysis (GSEA) (49) showed that immune-related pathways were enriched among down-regulated genes, whereas cell cycle-related pathways were enriched among up-regulated genes (Fig. 3B, fig. S13A, table S17A, S17B, S17C). Similar results were found with GO terms (fig. S13B, table S17A, S17D–S17E). Collectively, these data suggest that high burden of copy number loss may be associated with down-regulation of immune-related pathways.
Mutational load and burden of copy number loss may allow better patient stratification for response than either correlate alone
Finally, we were interested in determining if the effect of mutational load and burden of copy number loss on clinical response was non-redundant. Using the reanalyzed data from the Van Allen cohort, we first stratified patients into four subgroups based on mutational load (high or low) and burden of copy number loss (low or high) (fig. S14). Within each subgroup, we then calculated the proportion of patients with clinical benefit, long-term survival, and no clinical benefit, respectively (Fig. 3C). The proportion of patients with clinical benefit was higher in the subgroup of patients with high mutational load and low burden of copy number loss (11 out of 26) compared to the subgroup of patients with low mutational load and high burden of copy number loss (4 out of 26) (P = 0.064, Fisher’s exact test). Similarly, the proportion of patients with no clinical benefit was significantly higher in the subgroup of patients with low mutational load and high burden of copy number loss (21 out of 26) compared to the subgroup of patients with high mutational load and low burden of copy number loss (13 out of 26) (P = 0.04, Fisher’s exact test). We then performed a logistic regression on response status (clinical response or no clinical response) with the log2-transformed mutational load and log2-transformed burden of copy number loss as covariates, and found an additive effect of mutational load (coefficient = 0.266, z = 1.939, P = 0.053) and burden of copy number loss (coefficient = −0.149, z = −2.55, P = 0.011) on clinical response (table S18). This suggests that the effects of mutational load and burden of copy number loss on clinical response are likely non-redundant. Collectively, the above data demonstrate the potential utility of a combinatorial biomarker using mutational load and copy number loss burden.
Discussion
There is now abundant evidence that both tumor- (5–8, 23, 24) and host-related factors (9–12) can influence heterogeneous response and resistance to immune checkpoint blockade. Here, we report genomic characterization of tumors from a cohort of metastatic melanoma patients in the context of sequential immune checkpoint blockade. This study builds on our prior immune profiling of tumors within the same cohort of metastatic melanoma patients (38), allowing for a more fully integrated analysis in this particular cohort.
In tumor compartment-specific analyses, we observed a higher burden of copy number loss in non-responders compared to responders on CTLA-4 blockade. This finding is in line with those in prior studies that have reported that the burden of copy number alterations increases in advanced melanoma and is implicated in melanoma progression (50–53). The association between burden of copy number loss and clinical response observed here suggests that melanoma progression may be associated with resistance to immune mediated tumor control. Furthermore, investigation of the findings reported here in a first line treatment setting will help delineate the value of these potential associations.
We also identified genomic regions of recurrent copy number loss in patients with high burden of copy number loss and determined that several tumor suppressor genes were located within these genomic regions. This suggests that loss of function in these tumor suppressor genes could potentially influence therapeutic resistance. In keeping with this suggestion, previous studies in preclinical models of melanoma with PTEN loss showed inhibition of T cell-mediated tumor killing and decrease in T cell trafficking into tumors (24). PTEN was one of the tumor suppressor genes with recurrent copy number loss from patients with high burden of copy number loss in this study as well. A correlation between copy number loss burden and down-regulation of immune-related gene expression was found, suggesting that there may be gene expression sequelae of extensive copy number loss, including PTEN loss. More extensive analyses on larger cohorts with matched WES and RNA-seq data are needed to expand on these findings and develop an integrated expression/copy number evaluation approach to validate and potentially exploit the correlation seen here.
We also observed that the effects of low copy number loss burden and high mutational load on clinical response are non-redundant, suggesting the possibility of a combinatorial biomarker using copy number loss burden and mutational load. From a clinical perspective, the optimal cutoffs for high or low copy number loss burden and mutational load will need to be further validated if they are to impact improved patient stratification in the clinical setting.
Our work also confirms previous reports that TCR clonality is correlated with response to PD-1 blockade (10). A combinatorial biomarker approach of TCR clonality and genomic correlates such as mutational load and copy number loss burden needs to be further tested in a large cohort with pre-PD-1 blockade biopsies available.
Additionally, we observed increased TCR clonality after CTLA-4 blockade treatment in all PD-1 blockade responders with paired pre-CTLA-4 and pre-PD-1 blockade biopsies available. Prior work has shown that TCR clonality at the pre-PD-1 time point was not significantly different (P = 0.1604) between anti-CTLA-4-treated PD-1 blockade responders and anti-CTLA-4-naïve PD-1 blockade responders (10). Therefore, CTLA-4 blockade treatment may increase TCR clonality to a level high enough to mediate response to subsequent PD-1 blockade in certain patients. This result suggests that responders to PD-1 blockade may derive clinical benefit from prior CTLA-4 blockade, substantiating the utility of sequential CTLA-4 and PD-1 blockade. From a clinical perspective, sequential CTLA-4 and PD-1 blockade treatment might be able to increase the number of patients with high baseline TCR clonality prior to PD-1 blockade compared with PD-1 blockade monotherapy.
What emerges from this and other work regarding immune checkpoint responder/non-responder identification is a complex picture likely involving the interplay of tumor genomic characteristics, tumor modulation of the local microenvironment, and the extent of immune surveillance in the tumor milieu at the time of initiation of therapy. Furthermore, several intriguing questions emerge from this and other work. What is the effect of CTLA-4 blockade on the molecular profile of anti-PD-1 responders? Do the data reported hold true when applied to CTLA-4 blockade treatment-naïve patients? To what extent do the data emerging from melanoma studies apply to other tumor treatment contexts? There will likely be a need to develop integrated molecular phenotyping approaches to more accurately delineate responders/non-responders and develop tractable predictive models for these promising therapies.
Materials and Methods
Study design
Serial tumor biopsies were collected from patients with metastatic melanoma treated with CTLA-4 blockade and/or PD-1 blockade through the Expanded Access Program for MK-3475 at the UT MD Anderson Cancer Center. From serial tumor biopsies, we generated multidimensional profiling data (whole exome sequencing data, TCR sequencing data, and NanoString gene expression profiling). Multidimensional profiling data were analyzed to identify genomic and immune correlates of treatment response and resistance mechanisms of immune checkpoint blockade.
Patient cohort and tumor samples
A cohort of 56 patients with metastatic melanoma were included in this study (38). These patients were treated at the UT MD Anderson Cancer Center between October 2011 and March 2015 and had tumor samples collected with appropriate written informed consent and analyzed (IRB LAB00-063; LAB03-0320; 2012-0846; PA13-0291; PA12-0305). All tumor measurements were performed by a physician formally trained in tumor metrics, specifically RECIST 1.1 as it applies to the cohort. All metrics used CT scan imaging of measurable lesions (5 lesions total and 2 per organ max.) that met measurability based on strict RECIST 1.1 criteria (i.e > 10mm long axis per target lesion or > 15mm short axis for target lymph nodes). The sum of these respective diameters were compared to the sum at baseline. Per RECIST 1.1 criteria a lymph node < 10mm short axis was considered non-pathologic. As such patients were first defined at those having either a (1) complete response (disappearance of ALL target lesions, reduction in any pathological lymph nodes (whether target or not) in short axis to <10 mm, and the appearance of NO new lesions), (2) partial response (at least a 30% decrease in the sum of diameters of target lesions, no PD in non-target lesions and the appearance of NO new lesions), (3) progressive disease (at least a 20% increase in the sum of diameters of target lesions, taking as reference the smallest sum or baseline, with a minimum absolute increase of 5mm, and/or the development of any new lesions, or (4) stable disease [neither sufficient decrease to designate CR/PR nor increase to qualify as progressive disease (again using as a reference the smallest sum of appropriate diameters)]. All image responses were vetted with ≥2 serial images over a ≥6 month interval between baseline and assignment of response. RECIST 1.1 quantification of response was then used to assign patient designation as responder (i.e. CR, PR, or SD ≥6 months) or non-responder (PD or SD < 6 months duration). All specimens were excisional biopsies or resection specimens. Tumor samples were assessed by two pathologists for adequate tumor tissue for whole exome sequencing (table S2).
Sample processing
After fixation and mounting, 5 to 10 slices of 5 µm thickness were obtained from formalin-fixed, paraffin-embedded (FFPE) tumor blocks. Tumor-enriched tissue was macrodissected, and Xylene (EMD Millipore) was used for deparaffinization, followed by two ethanol washes. Reagents from the Qiagen QIAamp DNA FFPE Tissue Kit (#56404) were used in conjunction with an overnight incubation at 55°C to complete tissue lysis. Next, samples were incubated at 90°C for one hour to reverse formaldehyde modification of nucleic acids. After isolation by QIAamp MinElute column, variable amounts of buffer ATE were added to each column to elute the DNA. Germline DNA was obtained from peripheral blood mononuclear cells (PBMCs).
Whole exome sequencing
The initial genomic DNA input into the shearing step was 250 ng in 55 µL of low Tris-EDTA buffer. Forked Illumina paired-end adapters with random 8 base pair indexes were used for adapter ligation. All reagents used for end repair, A-base addition, adapter ligation, and library enrichment PCR were from the KAPA Hyper Prep Kit (#KK8504). Unligated adapter and/or adapter-dimer molecules were removed from the libraries before cluster generation using SPRI bead cleanup. The elution volume after post-ligation cleanup was 25 µL. Library construction was performed following manufacturer's instructions. Sample concentrations were measured after library construction using the Agilent Bioanalyzer. Each hybridization reaction contained 650–750 ng of the prepared library in a volume of 3.4 µL. Samples were lyophilized and reconstituted to bring the final concentration to 221 ng/µL. After reconstitution, the Agilent SureSelect-XT Target Enrichment (#5190–8646) protocol was followed, according to manufacturer guidelines. The libraries were then normalized to equal concentrations using an Eppendorf Mastercycler EP Gradient instrument and pooled to equimolar amounts on the Agilent Bravo B platform. Library pools were quantified using the KAPA Library Quantification Kit (#KK4824). Based on qPCR quantification, libraries were then brought to 2 nM and denatured using 0.2N NaOH. After denaturation, libraries were diluted to 14–20 pM using Illumina hybridization buffer. Next, cluster amplification was performed on denatured templates according to manufacturer’s guidelines (Illumina), HiSeq v3 cluster chemistry and flow cells, as well as Illumina’s Multiplexing Sequencing Primer Kit. The pools were then added to flow cells using the cBot System and sequenced using the HiSeq 2000/2500 v3 Sequencing-by-Synthesis method, then analyzed using RTA v.1.13 or later. Each pool of whole exome libraries was subjected to paired 76 bp runs. An 8-base index-sequencing read was used to meet coverage and to demultiplex the pooled samples. Mean coverage for exome data was 177X in tumors and 91X in germline. Mean sequencing coverage and tumor purities were similar across groups, with the exception of on-treatment biopsies given the presence of lower tumor content and enriched immune infiltrates (fig. S2). Therefore, whole exome sequencing data from on-treatment samples were excluded from downstream analysis.
Mutation calling and intratumor heterogeneity analysis
Exome sequencing data was processed using SaturnV, the NGS data processing and analysis pipeline developed and maintained by the Bioinformatics group of the Institute for Applied Cancer Science and Department of Genomic Medicine at UT MD Anderson Cancer Center. BCL files (raw output of Illumina HiSeq) were processed using Illumina CASAVA (Consensus Assessment of Sequence and Variation) software (v1.8.2) for demultiplexing/conversion to FASTQ format. The FASTQ files were then aligned to the hg19 human genome build using BWA (v0.7.5) (54). The aligned BAM files were subjected to mark duplication, realignment, and recalibration using the Picard tool and GATK software tools (55–57). The BAM files were then used for downstream analysis. MuTect (v1.1.4) (58) was applied to identify somatic point mutations, and Pindel (v0.2.4) (59) was applied to identify small insertions and deletions. Somatic mutations in HLA genes were called by POLYSOLVER (v1.0) (44). EXPANDS (v1.6.1) (60) and SciClone (v1.0.7) (61) were applied with only LOH-free regions to estimate the number of clones per tumor.
Neoantigen prediction
HLA class I neo-epitopes were predicted for each patient as previously described (62). In short, patient HLA-A, -B, and -C variants were identified using ATHLATES (v2014_04_26) (63). Next, all possible 9- to 11-mer peptides flanking a nonsynonymous exonic mutation were generated computationally, and binding affinity was predicted based on patient HLA and compared to that of the wild-type peptide counterpart using NetMHCpan (v2.8) algorithm (64). Mutated peptides with predicted IC50 < 500 nM were considered as predicted neoantigens. TCGA melanoma (40) gene expression data were used to further filter out predicted neoantigens with mean gene expression values below 5 (mean RSEM < 5).
Copy number alteration analysis
Sequenza (v2.1.2) (65) was applied to obtain copy number segments of log2 copy ratios (tumor/normal) for each tumor sample. R package ‘CNTools’ (v1.24.0) (66) was used to identify copy number gain (log2 copy ratios > log21.5) or loss (log2 copy ratios < −log21.5) at the gene level. The burden of copy number gain or loss was then calculated as the total number of genes with copy number gain or loss per sample. For recurrent copy number alteration analysis, R package ‘cghMCR’ (v1.26.0) (67) was applied to log2 copy ratios (tumor/normal) obtained from ‘exomecn’ (in-house copy number caller). Segment Gain or Loss (SGOL) scores of copy number segments or genes were calculated as sum of log2 copy ratios of each copy number segment or gene across all samples within a group of interest. Copy number segments with both copy number gain and copy number loss present within a group were excluded. We identified genomic regions of recurrent copy number alterations (MCRs: minimum common regions) using cghMCR function with the following parameters: gapAllowed=500, alteredLow=−log2(1.5), alteredHigh=log2(1.5), recurrence=60, spanLimit=2+e+07, thresholdType=”value” (recurrent copy number loss was defined as copy number loss observed in more than 60% of samples in a group of interest). Tumor suppressor genes annotated in recurrent copy number loss plots were obtained as cancer genes present in both the COSMIC (v77) (68) and TSGene databases (69). Five samples were excluded from analysis due to unusable copy number profiles (45C, 40C, 19D, 45E, and 20E).
TCR sequencing and clonality analysis
T cell receptor sequencing of the CDR3 variable region of the beta chain was performed by ImmunoSeq hsTCRB Kit as described previously (Adaptive Biotechnologies) (70, 71). In brief, DNA was extracted from FFPE tumor tissues, and CDR3 regions were amplified prior to sequencing by MiSeq 150X (Illumina). Data were then transferred to Adaptive Technologies for deconvolution of CDR3 beta sequences. For each sample, Shannon entropy and TCR clonality were calculated using the ImmunoSeq Analyzer (10).
NanoString gene expression profiling
NanoString was performed using a custom codeset of 795 genes as previously described (38). In brief, RNA was extracted using the RNeasy Mini Kit (QIAGEN) from FFPE blocks, after initial confirmation of tumor presence and content by two pathologists by H&E. For gene expression studies, 1 µg of RNA was used per sample. Hybridization was performed for 16–18 hours at 65°C, and samples were loaded onto the nCounter Prep Station for binding and washing prior to scanning and capture of 600 fields using the nCounter. Preprocessing of NanoString data was performed as previously described (38). Immune scores were calculated as geometric mean of gene expression of cytolytic markers (GZMA, GZMB, PRF1, GNLY), HLA molecules (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G, HLA-H, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DRA, HLA-DRB1), IFN-γ pathway genes (IFNG, IFNGR1, IFNGR2, IRF1, STAT1, PSMB9), chemokines (CCR5, CCL3, CCL4, CCL5, CXCL9, CXCL10, CXCL11), and adhesion molecules (ICAM1, ICAM2, ICAM3, ICAM4, ICAM5, VCAM1).
Independent (Van Allen) cohort analysis
Mutational load was obtained from nonsynonymous mutational load in an earlier study (7). WES data (SAM files) from 110 melanoma patients and RNA-seq data from 42 patients (FASTQ files) were downloaded through the dbGaP (accession number phs000452.v2.p1). Copy number alterations were identified from the same computational pipeline as described above. For the Van Allen cohort, recurrent copy number loss was defined as copy number loss observed in more than 40% of samples in a group of interest. Twelve samples were excluded from analysis due to unusable copy number profiles (Pat06, Pat73, Pat78, Pat81, Pat92, Pat106, Pat121, Pat132, Pat165, Pat166, Pat171, and Pat175). In the minimal or no clinical benefit group, samples with low burden of copy number loss (<100) were excluded from recurrent copy number alteration analysis. The relatively low cutoff of 100 was chosen to capture the majority of recurrent events. For RNA-seq analysis, all the RNA-seq samples were first aligned to the human reference genome (hg19, GRCh37.75) with Bowtie2 (v2.2.5). RSEM (v1.2.12) was used to quantify transcript expression at the gene level in FPKM. Immune scores for the independent cohort were calculated by ESTIMATE (48). eBayes-moderated t-test was performed to compare the high burden of copy number loss (n=10) and low burden of copy number loss groups (n=10). Rank metric was then calculated as the sign of log2 fold changes multiplied by inverse of P values. Gene set enrichment analysis (GSEA) (49) was performed on the rank metric-sorted list of genes (table S17A).
Statistical analysis
Statistical analyses were performed using R 3.2.2. Statistical tests included two-sided Fisher’s exact tests and two-sided Mann-Whitney tests.
Supplementary Material
Table S1. Clinical and demographic characteristics of patient cohort.
Table S2. Sample information, tumor purity, mutational load, neoantigen load, number of clones per tumor, and burden of copy number alterations
Table S3. Summary measure of the data across clinical subgroups
Table S4. List of somatic point mutations and indels
Table S5. List of HLA somatic mutations
Table S6. List of neoantigens and predicted MHC binding affinity
Table S7. TCR clonality
Table S8. Log2-transformed NanoString normalized count data in our cohort
Table S9. Immune scores (NanoString) in our cohort
Table S10. Log2 copy number ratio segments file by Sequenza in our cohort
Table S11. Recurrent copy number alterations in double non-responders with burden of copy number loss higher than 2,000.
Table S12. Log2 copy number ratio segments file by Sequenza in the Van Allen cohort
Table S13. Burden of copy number alterations in the Van Allen cohort
Table S14. Recurrent copy number alterations in the Van Allen cohort.
Table S15. Relative contribution of copy number loss burden on chromosome 10 in CTLA-4 blockade resistance.
Table S16. Immune scores (RNA-seq) in the Van Allen cohort
Table S17. Pre-ranked gene list for GSEA, KEGG pathway and Gene Ontology analysis in the Van Allen cohort.
Table S18. Additive effect of high mutational load and low burden of copy number loss on response to CTLA-4 blockade.
Fig. S1. Workflow diagram of multidimensional profiling analysis.
Fig. S2. Distribution of sequencing coverage and tumor purities across samples.
Fig. S3. Mutational load, neoantigen load and clinical response.
Fig. S4. Intratumor heterogeneity and clinical response.
Fig. S5. The effects of previous CTLA-4 blockade exposure on the baseline TCR clonality of pre-PD-1 blockade samples.
Fig. S6. Burden of copy number alterations in responders versus non-responders.
Fig. S7. Copy number profiles of responders and double non-responders.
Fig. S8. Correlation between burden of copy number loss and mutational load.
Fig. S9. Correlation between burden of copy number loss and mutational load in the Van Allen cohort.
Fig. S10. Segment Gain or Loss (SGOL) scores in three patient subgroups from the Van Allen cohort.
Fig. S11. Burden of recurrent CNAs in patients in Van Allen cohort.
Fig. S12. Correlation between burden of copy number loss and immune scores.
Fig. S13. Up and down-regulated pathways in patients with high versus low copy number loss in the Van Allen cohort.
Fig. S14. Patient stratification based on mutational load and burden of copy number loss in the Van Allen cohort.
Acknowledgments
Funding:
JAW acknowledges the Melanoma Research Alliance Team Science Award, the Kenedy Memorial Foundation grant #0727030, U54CA163125, STARS award, UT Regents, and the generous philanthropic support of several families whose lives have been affected by melanoma. This work was supported by National Institutes of Health grants 1K08CA160692-01A1 (JAW), U54CA163125 (ZAC, JAW, and LC), T32CA009599 (PAP), T32CA163185 (PLC, WSC), and R01 CA187076-02 (MAD, PH). JPA, PS, and JAW are members of the Parker Institute for Cancer Immunotherapy (PICI) at MD Anderson Cancer Center. WR is supported by the CPRIT Graduate Scholar Award. ICG holds a Conquer Cancer Foundation ASCO Young Investigator Award. LC is a CPRIT Scholar in Cancer Research and was supported by a grant from the Cancer Prevention Research Institute of Texas (R1204). PAF holds CPRIT funding (R1205 01), a Robert Welch Distinguished University Chair (G-0040), and STARS award. RNA has received research support from Merck, Novartis/Array and Bristol-Myers Squibb. WJH has received research support from Bristol-Myers Squibb, Merck, GSK and MedImmune. This work was supported by MD Anderson’s Institutional Tissue Bank Award (2P30CA016672) from the National Cancer Institute. This study was also supported by philanthropic contributions to The University of Texas MD Anderson Cancer Center Melanoma Moon Shot Program and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
JAW has honoraria from speakers’ bureau of Dava Oncology, Illumina and is an advisory board member for GlaxoSmithKline, Roche/Genentech, Novartis, and Bristol-Myers Squibb. MAD is an advisory board member for GlaxoSmithKline, Roche/Genentech, Novartis and Sanofi-Aventis and has received research support from GlaxoSmithKline, Roche/Genentech, Sanofi-Aventis, Oncothyreon, Myriad, and AstraZeneca. JEG is on the advisory board of Merck, and receives royalties from Mercator Therapeutics. SPP has honoraria from speakers’ bureau of Dava Oncology and Merck and is an advisory board member for Amgen and Roche/Genentech. PH serves on the advisory board of Lion Biotechnologies and Immatics US. R.N. WJH serves on the advisory board of Merck’s melanoma advisory board. RNA has received research support from Merck, Novartis and Bristol-Myers Squibb. PS is a consultant for Bristol-Myers Squibb, Jounce Therapeutics, Helsinn, and GlaxoSmithKline as well as a stockholder from Jounce Therapeutics. JPA is a consultant and stockholder for Jounce Therapeutics, receives royalties from Bristol-Myers Squibb, and has intellectual property with Bristol-Myers Squibb and Merck. ZAC is an employee in MedImmune and owns stock or options in AstraZeneca.
Footnotes
Author contributions: WR, PLC, ZAC, JAW, and PAF designed experiments. WR, PLC, AR, JAW, and PAF wrote the manuscript. All authors edited the manuscript. WR, PLC, AR, JR, JPM, ZAC, JAW, and PAF analyzed data. WR, JR, FW, and JZ ran computational pipelines. WR and JH performed statistical analyses. PLC, CNS, PAP, VG, MAD, JEG, PH, SPP, AD, ICG, HT, WJH, SEW, RNA, and JAW acquired and compiled clinical and response data. CNS, PLC, MTT, MAD, JEG, SPP, AD, ICG, HT, AJL, PH, WJH, SEW, RNA, VGP, and JAW enrolled subjects and contributed samples. PLC, WSC, and MTT performed pathological assessment. PLC, JPM, CG, LL, QC, KW, MPDM, EC, and WSC performed experiments. JAW and PAF supervised the study.
Competing interests: No other potential conflicts of interest were disclosed.
Data and materials availability: WES data (BAM files) from melanoma patients from our cohort are available at dbGaP (Bioproject ID PRJNA369259). WES data (SAM files) and RNA-seq data (FASTQ files) from melanoma patients from the Van Allen cohort are available at dbGaP (accession number phs000452.v2.p1).
References
- 1.Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, Patt D, Chen TT, Berman DM, Wolchok JD. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol. 2015;33:1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. New England Journal of Medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, Ferrucci PF, Hill A, Wagstaff J, Carlino MS, Haanen JB, Maio M, Marquez-Rodas I, McArthur GA, Ascierto PA, Long GV, Callahan MK, Postow MA, Grossmann K, Sznol M, Dreno B, Bastholt L, Yang A, Rollin LM, Horak C, Hodi FS, Wolchok JD. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA., Jr PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, Dronca R, Gangadhar TC, Patnaik A, Zarour H, Joshua AM, Gergich K, Elassaiss-Schaap J, Algazi A, Mateus C, Boasberg P, Tumeh PC, Chmielowski B, Ebbinghaus SW, Li XN, Kang SP, Ribas A. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik B, Galon J, Trajanoski Z. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher TN, Kesmir C, van Buuren MM. Biomarkers in cancer immunotherapy. Cancer Cell. 2015;27:12–14. doi: 10.1016/j.ccell.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, Eppolito C, Qian F, Lele S, Shrikant P, Old LJ, Odunsi K. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci U S A. 2010;107:7875–7880. doi: 10.1073/pnas.1003345107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Wu K, Tao K, Chen L, Zheng Q, Lu X, Liu J, Shi L, Liu C, Wang G, Zou W. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology. 2012;56:1342–1351. doi: 10.1002/hep.25777. [DOI] [PubMed] [Google Scholar]
- 17.Woo S-R, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, Bettini ML, Gravano DM, Vogel P, Liu CL. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer research. 2012;72:917–927. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, Shen X, Boyd Z, Hegde PS, Chen DS, Vogelzang NJ. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 19.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, Eaton DL, Grogan JL. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Chauvin JM, Pagliano O, Fourcade J, Sun Z, Wang H, Sander C, Kirkwood JM, Chen TH, Maurer M, Korman AJ, Zarour HM. TIGIT and PD-1 impair tumor antigen-specific CD8(+) T cells in melanoma patients. J Clin Invest. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF–MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. The Journal of experimental medicine. 2006;203:1651–1656. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 24.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, Williams LJ, Deng W, Chen G, Mbofung R, Lazar AJ, Torres-Cabala CA, Cooper ZA, Chen PL, Tieu TN, Spranger S, Yu X, Bernatchez C, Forget MA, Haymaker C, Amaria R, McQuade JL, Glitza IC, Cascone T, Li HS, Kwong LN, Heffernan TP, Hu J, Bassett RL, Jr, Bosenberg MW, Woodman SE, Overwijk WW, Lizee G, Roszik J, Gajewski TF, Wargo JA, Gershenwald JE, Radvanyi L, Davies MA, Hwu P. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Science translational medicine. 2012;4:127ra137–127ra137. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taube JM, Young GD, McMiller TL, Chen S, Salas JT, Pritchard TS, Xu H, Meeker AK, Fan J, Cheadle C, Berger AE, Pardoll DM, Topalian SL. Differential Expression of Immune-Regulatory Genes Associated with PD-L1 Display in Melanoma: Implications for PD-1 Pathway Blockade. Clin Cancer Res. 2015;21:3969–3976. doi: 10.1158/1078-0432.CCR-15-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayers MD, Nebozhyn M, Hirsch HA, Cristescu R, Murphy EE, Kang SP, Ebbinghaus SW, McClanahan TK, Loboda A, Lunceford JK. Assessment of gene expression in peripheral blood from patients with advanced melanoma using RNA-Seq before and after treatment with anti-PD-1 therapy with pembrolizumab (MK-3475) Cancer Research. 2015;75:1307–1307. [Google Scholar]
- 29.Minn AJ, Wherry EJ. Combination Cancer Therapies with Immune Checkpoint Blockade: Convergence on Interferon Signaling. Cell. 2016;165:272–275. doi: 10.1016/j.cell.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 30.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. New England Journal of Medicine. 2016 doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, Hundal J, Wendl MC, Demeter R, Wylie T, Allison JP, Smyth MJ, Old LJ, Mardis ER, Schreiber RD. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre M-L. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, Tonc E, Schreiber RD, Pearce EJ, Pearce EL. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, Kaech SM. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W, Bai Y, Xiong Y, Zhang J, Chen S, Zheng X, Meng X, Li L, Wang J, Xu C, Yan C, Wang L, Chang CC, Chang TY, Zhang T, Zhou P, Song BL, Liu W, Sun SC, Liu X, Li BL, Xu C. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531:651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V, Wani K, Petaccia De Macedo M, Austin-Breneman JL, Jiang H, Chang Q, Reddy SM, Chen WS, Tetzlaff MT, Broaddus RJ, Davies MA, Gershenwald JE, Haydu L, Lazar AJ, Patel SP, Hwu P, Hwu WJ, Diab A, Glitza IC, Woodman SE, Vence LM, Wistuba, Amaria RN, Kwong LN, Prieto V, Davis RE, Ma W, Overwijk WW, Sharpe AH, Hu J, Futreal PA, Blando J, Sharma P, Allison JP, Chin L, Wargo JA. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.N. Cancer Genome Atlas. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon γ-dependent tumor surveillance system in immunocompetent mice. Proceedings of the national academy of sciences. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunn GP, Sheehan KC, Old LJ, Schreiber RD. IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer research. 2005;65:3447–3453. doi: 10.1158/0008-5472.CAN-04-4316. [DOI] [PubMed] [Google Scholar]
- 43.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE. Loss of IFN-g Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:1–8. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, Stevens J, Lane WJ, Dellagatta JL, Steelman S, Sougnez C, Cibulskis K, Kiezun A, Hacohen N, Brusic V, Wu CJ, Getz G. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang E, Worschech A, Marincola FM. The immunologic constant of rejection. Trends Immunol. 2008;29:256–262. doi: 10.1016/j.it.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39:11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Kwong LN, Chin L. Chromosome 10, frequently lost in human melanoma, encodes multiple tumor-suppressive functions. Cancer research. 2014;74:1814–1821. doi: 10.1158/0008-5472.CAN-13-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 50.Bastian BC, LeBoit PE, Hamm H, Bröcker E-B, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer research. 1998;58:2170–2175. [PubMed] [Google Scholar]
- 51.Bauer J, Bastian BC. Distinguishing melanocytic nevi from melanoma by DNA copy number changes: comparative genomic hybridization as a research and diagnostic tool. Dermatologic therapy. 2006;19:40–49. doi: 10.1111/j.1529-8019.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- 52.Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, Dummer R, North J, Pincus L, Ruben B, Rickaby W, D'Arrigo C, Robson A, Bastian BC. The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med. 2015;373:1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 53.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16:345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Current protocols in bioinformatics. 2013:11.10. 11–11.10. 33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andor N, Harness JV, Muller S, Mewes HW, Petritsch C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics. 2014;30:50–60. doi: 10.1093/bioinformatics/btt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, Vij R, Tomasson MH, Graubert TA, Walter MJ, Ellis MJ, Schierding W, DiPersio JF, Ley TJ, Mardis ER, Wilson RK, Ding L. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10:e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snyder A, Chan TA. Immunogenic peptide discovery in cancer genomes. Curr Opin Genet Dev. 2015;30:7–16. doi: 10.1016/j.gde.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu C, Yang X, Duffy B, Mohanakumar T, Mitra RD, Zody MC, Pfeifer JD. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 2013;41:e142. doi: 10.1093/nar/gkt481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, Szallasi Z, Eklund AC. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J. CNTools: Convert segment data into a region by sample matrix to allow for other high level computational analyses. R package (Version 1.6. 0.) 2014 [Google Scholar]
- 67.Zhang J, Feng B, Zhang MJ. C. biocViews Microarray, Package ‘cghMCR’. 2013 [Google Scholar]
- 68.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, Wu D, Wood BL, Rieder MJ, Robins H. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinical and demographic characteristics of patient cohort.
Table S2. Sample information, tumor purity, mutational load, neoantigen load, number of clones per tumor, and burden of copy number alterations
Table S3. Summary measure of the data across clinical subgroups
Table S4. List of somatic point mutations and indels
Table S5. List of HLA somatic mutations
Table S6. List of neoantigens and predicted MHC binding affinity
Table S7. TCR clonality
Table S8. Log2-transformed NanoString normalized count data in our cohort
Table S9. Immune scores (NanoString) in our cohort
Table S10. Log2 copy number ratio segments file by Sequenza in our cohort
Table S11. Recurrent copy number alterations in double non-responders with burden of copy number loss higher than 2,000.
Table S12. Log2 copy number ratio segments file by Sequenza in the Van Allen cohort
Table S13. Burden of copy number alterations in the Van Allen cohort
Table S14. Recurrent copy number alterations in the Van Allen cohort.
Table S15. Relative contribution of copy number loss burden on chromosome 10 in CTLA-4 blockade resistance.
Table S16. Immune scores (RNA-seq) in the Van Allen cohort
Table S17. Pre-ranked gene list for GSEA, KEGG pathway and Gene Ontology analysis in the Van Allen cohort.
Table S18. Additive effect of high mutational load and low burden of copy number loss on response to CTLA-4 blockade.
Fig. S1. Workflow diagram of multidimensional profiling analysis.
Fig. S2. Distribution of sequencing coverage and tumor purities across samples.
Fig. S3. Mutational load, neoantigen load and clinical response.
Fig. S4. Intratumor heterogeneity and clinical response.
Fig. S5. The effects of previous CTLA-4 blockade exposure on the baseline TCR clonality of pre-PD-1 blockade samples.
Fig. S6. Burden of copy number alterations in responders versus non-responders.
Fig. S7. Copy number profiles of responders and double non-responders.
Fig. S8. Correlation between burden of copy number loss and mutational load.
Fig. S9. Correlation between burden of copy number loss and mutational load in the Van Allen cohort.
Fig. S10. Segment Gain or Loss (SGOL) scores in three patient subgroups from the Van Allen cohort.
Fig. S11. Burden of recurrent CNAs in patients in Van Allen cohort.
Fig. S12. Correlation between burden of copy number loss and immune scores.
Fig. S13. Up and down-regulated pathways in patients with high versus low copy number loss in the Van Allen cohort.
Fig. S14. Patient stratification based on mutational load and burden of copy number loss in the Van Allen cohort.