Abstract
Background
An increasing number of publications are drawing attention to the associations between six common polymorphisms in HOX transcript anti-sense RNA (HOTAIR) and the risk of cancers, while these results have been controversial and inconsistent. We conducted an up-to-date meta-analysis to pool eligible studies and to further explore the possible relationships between HOTAIR polymorphisms (rs920778, rs7958904, rs12826786, 4,759,314, rs874945, and rs1899663) and cancer risk.
Methods
A systematic retrieval was conducted up to 1 July 2017 in the PubMed, Web of Science, and CNKI databases. Eighteen eligible publications including 45 case-control studies with 58,601subjects were enrolled for assessing the associations between the 6 polymorphisms in HOTAIR and cancer risk. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were analyzed to reveal the polymorphisms and susceptibility to cancer. All the statistical analyses were performed using STATA 11.0 software.
Results
The pooled analyses detected significant associations between the rs920778 polymorphism and increased susceptibility to cancer in recessive, dominant, allelic, homozygous, and heterozygous models. For the rs7958904 polymorphism, we obtained the polymorphism significantly decreased susceptibility to overall cancer risk among five genetic models rather than recessive and homozygous models. For the rs12826786 polymorphism, we identified it significantly increased susceptibility to cancer risk in all genetic models rather than heterozygous models. However, no significant association was found between the rs1899663, rs874945, and rs4759314 polymorphisms and susceptibility of cancer.
Conclusion
These findings of the meta-analysis suggest that HOTAIR polymorphism may contribute to cancer susceptibility.
Electronic supplementary material
The online version of this article (10.1186/s12199-018-0697-0) contains supplementary material, which is available to authorized users.
Keywords: Cancer, HOTAIR, LncRNAs, Polymorphism, Susceptibility
Background
Long non-coding RNAs (lncRNAs) are a class of particular no-coding RNA molecules with lengths of more than 200 nucleotides (nt), which are mainly produced by RNA polymerase II (RNA pol II) transcription, lack of protein-encoding function, and an open reading frame [1, 2]. Several studies have demonstrated that lncRNAs not only act as intermediaries between DNA and proteins, but also as a kind of crucial substances with affecting cell function involved in the epigenetic modification, transcription, and post-transcription process to regulate gene expression ultimately [3, 4].Therefore, aberrant regulation of lncRNAs is often associated with a variety of diseases, especially cancers. In recent years, the whole genome cancer mutation analysis gradually identified lncRNAs properties, focusing on the complex regulation of lncRNA transcription how to contribute many important cancer phenotypes. Schmitt et al. revealed molecular mechanisms of the lncRNA interacting with DNA, RNA, and protein, and also generalized lncRNAs played vital role in intracellular signal transduction networks in the carcinogenesis and progression of various cancers as drivers of the cancer phenotypes [5]. Some cancer-related lncRNAs could affect the development and progression of cancer by means of p53, polycomb repressive complex 2 (PRC2), and other signaling pathways. In p53 complex signaling pathways, Grossi et al. reported that some lncRNAs can directly or indirectly regulate p53 activity leading to upregulation or downregulation of p53 expression [6]. In addition, to directly participate in the regulation of gene expression, lncRNAs also could compete with the same miRNA with other RNA transcripts as ceRNA (competing endogenous RNAs) to regulate expression of target genes at the posttranscriptional level [7].
HOX transcript anti-sense RNA (HOTAIR) is located on between the HOXC11 and HOXC12 coding region in the chromosome 12q13, which consists of five short exons and one long exon [8]. Significantly high expression of HOTAIR has been indicated in breast cancer [9], pancreatic cancer [10], liver cancer [11], colorectal cancer [12], lung cancer [13], and other malignant tumors, and with which tumor cells invasion and metastasis, tumor recurrence, and poor prognosis are closely related. It has been found that HOTAIR regulates the expression of genes by specifically binding different histone-modified complexes. As one of the regulators of oncogene transcription, HOTAIR remodels PRC2 and desmethylase LSD1 complexes by recruiting chromatin to cause demethylation of Histone H3K27em3, silence of HOXD gene expression, ultimately facilitate tumor proliferation or metastasis at the epigenetic level [14, 15]. HOTAIR also could form regulation network of ceRNAs with miRNA to participate in carcinogenesis of different cancers. Moreover, HOTAIR also acts as miRNA sponge in combination with miR-331-3p or miR-124 [16], leading to upregulation of HER2 and other relevant genes and activation of Akt signaling pathway, which could simultaneously interact with p53 pathway to promote proliferation, invasion, and metastasis of tumor cell [17]. In the final analysis, HOTAIR was intricately related to various cancers.
Several single nucleotide polymorphisms (SNPs) in HOTAIR are widely researched in cancer susceptibility, prognosis [18], and clinical outcome [19] and treatment response [20]. Recently, some published results have definitely shown the inconsistent and controversial associations of common SNPs (rs1899663, rs4759314, rs920778, rs874945, rs7958904, and rs12826786) in HOTAIR with the risk of various cancers including digestive cancers [21–29], estrogen-dependent cancers [18, 19, 30–33], papillary thyroid carcinoma [34], glioma [35], pancreatic cancer [36], prostate cancer [37], and osteosarcoma [38]. Herein, we indispensably conducted a meta-analysis to summarize all currently eligible case-control studies to more accurately elucidate the authentic associations between HOTAIR polymorphisms and cancer susceptibility, especially, for rs12826786 C>T, this is first meta-analysis to evaluate the relationship between the polymorphism and cancer risk in overall population.
Methods
Literature search
Eligible literatures were systematically retrieved in several authoritative databases including PubMed, Web of Science, and CNKI databases to search comprehensive and systematic publications up to 1 July 2017, with the following keywords including “long non-coding RNA HOTAIR OR lncRNA HOTAIR OR HOTAIR,” “polymorphism OR variation OR mutation,” and “cancer OR carcinoma OR tumor OR malignancy OR neoplasm OR lymphoma OR leukemia.” Moreover, this study further retrieves the references lists of eligible studies to guarantee that all qualified studies are included in the meta-analysis.
Inclusion and exclusion criteria
Included criteria of eligible studies were as follows: (a) studies estimating the associations between lncRNA HOTAIR polymorphisms or genetic variations and the risk or susceptibility of various cancers, (b) case-control designed studies with genotypes distribution of controls followed Hardy–Weinberg equilibrium, (c) studies with available or adequate genotype data for calculation of the odds ratio (OR) with 95% confidence intervals (95% CIs). The main criteria for excluding unqualified researches were as follows: (1) did not focus on cancer risk or susceptibility, (2) did not involve the several HOTAIR SNPs (rs920778, rs4759314, rs1899663, rs7958904, rs874945, or rs12826786), (3) did not present the sufficient genotype frequency data of cases and controls. Ultimately, a total of 18 articles consist of 14,119 cases and 16,295 controls were included in this meta-analysis (presented in Table 1).
Table 1.
Characteristics of studies on HOTAIR polymorphism and cancer risk
| First author | Year | Country | Ethnicity | Source of control | Genotyping methods | Type of cancers | Case | Control | Included in meta-analysis |
|---|---|---|---|---|---|---|---|---|---|
| Zhang [21] | 2014 | china | Asian | PB | RFLP | ESCC | 1000 | 1000 | Yes |
| Bayram [33] | 2015 | Turkey | Caucasian | HB | TaqMan | BC | 123 | 122 | Yes |
| Bayram [28] | 2015 | Turkey | Caucasian | HB | TaqMan | GC | 104 | 209 | Yes |
| Du [29] | 2015 | china | Asian | HB | Taqman | GC | 753 | 1057 | Yes |
| Guo [25] | 2015 | china | Asian | PB | RFLP | GCA | 515 | 654 | Yes |
| Xue [22] | 2015 | china | Asian | HB | Taqman | CRC | 1147 | 1203 | Yes |
| Yan [31] | 2015 | china | Asian | PB | RFLP | BC | 502 | 504 | Yes |
| Bayram [19] | 2016 | Turkey | Caucasian | HB | TaqMan | BC | 123 | 122 | Yes |
| Guo [32] | 2016 | china | Asian | HB | MALDI-TOF-MS | CC | 510 | 713 | Yes |
| Pan [26] | 2016 | china | Asian | PB | RFLP | GC | 500 | 1000 | Yes |
| Qiu [18] | 2016 | china | Asian | HB | Taqman | Ovarian cancer | 190 | 380 | No |
| Qiu [24] | 2016 | china | Asian | NA | Taqman | CC | 215 | 430 | No |
| Wu [30] | 2016 | china | Asian | NA | MALDI-TOF-MS | EOC | 1000 | 1000 | Yes |
| Zhou [38] | 2016 | china | Asian | HB | MALDI-TOF-MS | Osteosarcoma | 500 | 500 | Yes |
| Zhu [34] | 2016 | china | Asian | NA | RFLP | PTC | 1000 | 1000 | Yes |
| Hu [36] | 2017 | china | Asian | PB | TaqMan | Pancreatic cancer | 416 | 416 | Yes |
| Jin [27] | 2017 | china | Asian | HB | TaqMan | CC | 1174 | 1304 | Yes |
| Taheri [37] | 2017 | Iran | Caucasian | HB | ARMS-PCR | Prostate cancer | 128 | 250 | Yes |
| Ulger [23] | 2017 | Turkey | Caucasian | HB | TaqMan | GC | 105 | 207 | Yes |
| Xavier‑Magalhaes [35] | 2017 | Portugal | Caucasian | PB | RFLP | Glioma | 177 | 199 | Yes |
BC Breast cancer, CRC Colorectal cancer, CC Cervical cancer, ESCC Esophageal squamous cell carcinoma, GC Gastric cancer, GCA Gastric cardia denocarcinoma, PTC Papillary thyroid carcinoma, HB Hospital-based, PB Population-based, RFLP restriction fragment length polymorphism, MOLDI-TOF-MS Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry
Data extraction
Two investigators (L.J. and L.X.) independently gathered the following information from each qualified publication: the first author’s name, year, country, and ethnicity of eligible publication, type of neoplasm, source of control, genotyping method, numbers of cases and controls, distribution of genotype frequencies as well as allele for cases and controls, P value of Hardy–Weinberg equilibrium (HWE) of control group, adjusted factors in the statistical analysis for each publication, and assessment of article quality. Two investigators reached a consensus on the basis of discussion when they have different opinions.
Quality score assessment
Two investigators individually assessed the quality of all included studies according to the Newcastle-Ottawa scale (NOS), and the scale totally comprises subject selection, comparability of cases, and controls as well as ascertainment of exposure.
Statistical analysis
Chi-square test was conducted to evaluate whether genotype distribution of the control group follow HWE. Q test and I2 test were used to examine the heterogeneity between each study, and the random effect model is used to combine the relevant studies when I2 is greater than 50%, otherwise, using a fixed-effect model and merged odds ratio (OR), and 95% confidence interval (95% CI) to evaluate the relationship between cancer risk and HOTAIR polymorphisms. In the current study, sensitivity analysis is to evaluate whether a single study would impact the overall effect value of the integrated researches. Begg’s funnel plots and Egger’s test was performed to uncover underlying publication bias by means of Stata 11. All calculations of the present meta-analysis were operated with Stata 11. P < 0.05 was uniformly regarded as a significant difference.
Results
Characteristics of the published studies
The main characteristics of 20 articles on relationship between 6 HOTAIR SNPs and cancer risk were presented in Table 1, and genotype distribution information of the 6 polymorphisms about case-control studies is shown Additional file 1: Table S1 in detail. A total of 18 eligible publications including 45 case-control studies with 58,601 subjects (comprising 27,016 cases and 31,585 controls) met the inclusion criteria and 6 SNPs were involved in the meta-analysis. Of the 45 case-control studies included in meta-analysis, the HOTAIR rs4759314 in 13 studies [21, 22, 25–27, 29, 31, 32, 34, 36, 37], rs920778 in 12 studies [21, 26, 28, 31–35], rs7958904 in 6 studies [22, 27, 29, 30, 38], rs874945 in 3 studies [22, 27, 29] rs1899663 in 6 studies [21, 26, 31, 32, 34, 37], and rs12826786 in 5 studies [19, 23, 25, 35, 37] were analyzed, respectively. Of the all case-control studies listed in Additional file 1: Table S1, 11 studies presented a significant deviation from HWE (2 studies on rs4759314 [30, 38], 4 on rs920778 [18, 24, 26], 1 study on rs7958904 [38], 1 study on rs12826786 [25], and 3 on rs874945 [30, 38]). The detailed flow diagram of the articles selection process is presented in Fig. 1. Genotyping method involved in PCR-RFLP, MALDI-TOF-MS, ARMS-PCR, and TaqMan methods. All of these studies mainly focused on the study of female-related cancers and digestive system cancer.
Fig. 1.
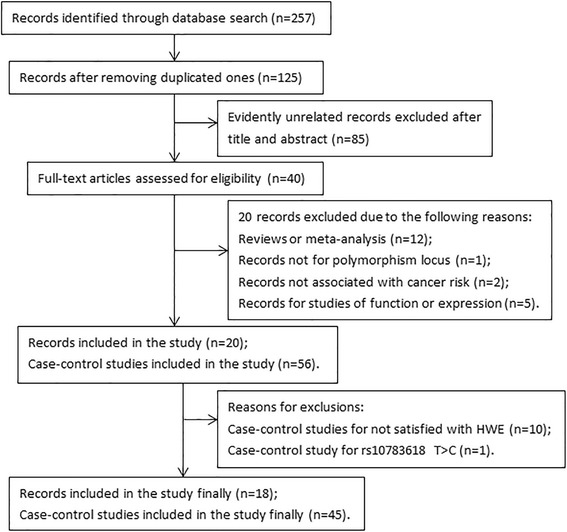
Flow diagram of articles identified with included and excluded criteria
Analysis of quantitative synthesis
Rs920778, rs12826786, and rs7958904 polymorphism and cancer susceptibility
There were a total of 12 eligible case-control studies with 6187 cases and 6897 controls that focused on the associations of rs920778 C>T and cancer risks after excluding the 4 studies (Qiu-a, b, and c and Pan-a) that their controls did not meet HWE as shown in Table 1. The pooled results for the association between rs920778(C>T) polymorphism and cancer risk are presented in Table 2. The pooled analyses indicated that rs920778 polymorphism significantly increased with susceptibility to overall cancer in all five genetic (CTvs.CC: OR = 1.298, 95% CI = 1.203–1.400, I2 = 13.0%; TT vs.CC: OR = 2.289, 95% CI = 1.815–2.887, I2 = 54.3%; CT + TT vs. CC: OR = 1.422, 95% CI = 1.322–1.529, I2 = 31.10%; TT vs. CT + CC: OR = 2.028, 95% CI = 1.641–2.506, I2 = 57.90%; T vs. C: OR = 1.414, 95% CI = 1.336–1.497, I2 = 48.2%) models. Additionally, the similar relations were obtained both in cancer type (including estrogen-dependent cancers and digestive cancers) and source of controls subgroup. The heterogeneity was conspicuously reduced in both Caucasian and Chinese population according to the results of Table 2 presented, and, in particular, this significant risk was observed in the Chinese population except Caucasian, and the decreased heterogeneity was also observed in the other subgroup.
Table 2.
Results of meta-analysis the relationship between HOTAIR SNPs and cancer risk
| Subgroup | n | Heterozygote vs. Wild-type | Mutation homozygote vs. wild-type | Dominant model | Recessive model | Allelic model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P | I 2(%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | ||
| rs920778 | ||||||||||||||||
| Overall | 12 | 1.298(1.203-1.400) | 0.000 | 13.0 | 2.289(1.815-2.887) | 0.000 | 54.3 | 1.422(1.322-1.529) | 0.000 | 31.1 | 2.028(1.641-2.506) | 0.000 | 57.9 | 1.414(1.336-1.497) | 0.000 | 48.2 |
| Caucasian | 3 | 0.920(0.688-1.230) | 0.574 | 0.0 | 1.296(0.875-1.920) | 0.196 | 12.7 | 1.004(0.764-1.318) | 0.978 | 0.0 | 1.368(0.960-1.949) | 0.083 | 48.0 | 1.096(0.906-1.326) | 0.343 | 0.0 |
| Asian | 9 | 1.331(1.231-1.440) | 0.000 | 0.0 | 2.558(2.170-3.016) | 0.000 | 34.1 | 1.460(1.354-1.574) | 0.000 | 12.2 | 2.214(1.774-2.762) | 0.000 | 56.0 | 1.450(1.366-1.539) | 0.000 | 33.7 |
| Estrogen-Dependent | 3 | 1.233(0.995-1.528) | 0.056 | 22.7 | 2.397(1.684-3.412) | 0.000 | 0.0 | 1.435(1.171-1.759) | 0.001 | 0.0 | 1.969(1.291-3.005) | 0.002 | 63.1 | 1.449(1.269-1.654) | 0.000 | 0.0 |
| Digestive | 5 | 1.317(1.175-1.477) | 0.000 | 1.4 | 2.566(1.766-3.727) | 0.000 | 57.1 | 1.457(1.305-1.625) | 0.000 | 22.9 | 2.274(1.594-3.244) | 0.000 | 56.2 | 1.456(1.333-1.590) | 0.000 | 46.4 |
| HB | 4 | 1.310(1.116-1.539) | 0.001 | 35.9 | 2.237(1.422-3.518) | 0.000 | 55.7 | 1.470(1.262-1.713) | 0.000 | 30.4 | 2.090(1.380-3.167) | 0.001 | 54.1 | 1.470(1.305-1.655) | 0.000 | 46.5 |
| PB | 5 | 1.255(1.108-1.422) | 0.000 | 5.8 | 2.260(1.486-3.436) | 0.000 | 62.8 | 1.390(1.234-1.565) | 0.000 | 32.8 | 1.969(1.367-2.837) | 0.000 | 69.8 | 1.399(1.280-1.529) | 0.000 | 49.9 |
| TaqMan | 2 | 0.914(0.619-1.349) | 0.651 | 0.0 | 1.475(0.893-2.436) | 0.129 | 39.4 | 1.044(0.723-1.508) | 0.818 | 0.0 | 1.580(0.716-3.487) | 0.257 | 67.4 | 1.166(0.913-1.489) | 0.218 | 0.0 |
| RFLP | 9 | 1.314(1.211-1.425) | 0.000 | 11.6 | 2.385(1.829-3.109) | 0.000 | 56.7 | 1.431(1.324-1.548) | 0.000 | 38.1 | 2.061(1.619-2.624) | 0.000 | 61.0 | 1.425(1.300-1.562) | 0.000 | 52.9 |
| rs4759314 | ||||||||||||||||
| Overall | 13 | 1.089(0.996-1.191) | 0.063 | 47.1 | 1.084(0.735-1.599) | 0.683 | 0.0 | 1.088(0.997-1.188) | 0.059 | 47.7 | 1.087(0.738-1.602) | 0.674 | 0.0 | 1.082(0.996-1.175) | 0.062 | 47.4 |
| Asian | 12 | 1.104(1.008-1.209) | 0.034 | 45.9 | 0.985(0.650-1.493) | 0.943 | 0.0 | 1.099(1.005-1.202) | 0.040 | 49.5 | 0.976(0.644-1.479) | 0.908 | 0.0 | 1.092(0.964-1.237) | 0.168 | 51.4 |
| Digestive | 8 | 1.082(0.904-1.296) | 0.390 | 58.2 | 0.801(0.468-1.371) | 0.419 | 0.0 | 1.076(0.897-1.290) | 0.430 | 60.1 | 0.799(0.467-1.367) | 0.413 | 0.0 | 1.065(0.895-1.266) | 0.480 | 60.7 |
| Estrogen-dependent | 3 | 1.153(0.978-1.360) | 0.091 | 8.5 | 1.416(0.696-2.883) | 0.338 | 0.0 | 1.163(0.989-1.368) | 0.068 | 20.3 | 1.387(0.682-2.820) | 0.367 | 0.0 | 1.127(0.858-1.480) | 0.057 | 27.6 |
| PB | 5 | 0.975(0.829-1.146) | 0.759 | 0.0 | 0.892(0.424-1.878) | 0.763 | 0.0 | 0.972(0.829-1.140) | 0.726 | 0.0 | 0.903(0.430-1.899) | 0.788 | 0.0 | 0.971(0.835-1.129) | 0.699 | 0.0 |
| HB | 7 | 1.131(0.925-1.383) | 0.231 | 66.6 | 1.177(0.736-1.880) | 0.497 | 26.0 | 1.141(0.936-1.392) | 0.191 | 66.9 | 1.176(0.737-1.877) | 0.496 | 28.4 | 1.142(0.949-1.374) | 0.159 | 67.0 |
| TaqMan | 6 | 1.149(0.916-1.441) | 0.231 | 70.4 | 0.852(0.493-1.470) | 0.565 | 27.1 | 1.142(0.905-1.442) | 0.264 | 73.1 | 0.847(0.491-1.462) | 0.551 | 25.4 | 1.126(0.898-1.412) | 0.303 | 74.6 |
| RFLP | 5 | 1.047(0.887-1.236) | 0.589 | 0.0 | 0.953(0.349-2.601) | 0.925 | 0.0 | 1.044(0.886-1.231) | 0.604 | 0.0 | 0.946(0.347-2.582) | 0.914 | 0.0 | 1.040(0.887-1.218) | 0.629 | 0.0 |
| rs7958904 | ||||||||||||||||
| Overall | 6 | 0.887(0.819-0.961) | 0.003 | 0.0 | 0.747(0.529-1.056) | 0.098 | 79.2 | 0.869(0.806-0.938) | 0.000 | 47.4 | 0.787(0.571-1.086) | 0.145 | 77.0 | 0.869(0.769-0.981) | 0.024 | 73.8 |
| Estrogen-dependent | 2 | 0.920(0.813-1.042) | 0.191 | 43.0 | 0.910(0.312-2.653) | 0.863 | 94.5 | 0.911(0.676-1.227) | 0.538 | 84.0 | 0.944(0.345-2.577) | 0.910 | 94.1 | 0.928(0.650-1.325) | 0.682 | 92.6 |
| Digestive | 3 | 0.870(0.776-0.975) | 0.017 | 0.0 | 0.717(0.579-0.888) | 0.002 | 0.0 | 0.842(0.756-0.939) | 0.002 | 0.0 | 0.758(0.615-0.933) | 0.009 | 0.0 | 0.852(0.781-0.930) | 0.000 | 0.0 |
| HB | 5 | 0.899(0.823-0.982) | 0.018 | 0.0 | 0.804(0.552-1.172) | 0.258 | 78.4 | 0.891(0.819-0.969) | 0.007 | 48.3 | 0.845(0.596-1.198) | 0.344 | 75.9 | 0.890(0.778-1.019) | 0.091 | 73.4 |
| TaqMan | 4 | 0.907(0.826-0.997) | 0.043 | 0.0 | 0.866(0.568-1.321) | 0.503 | 81.1 | 0.897(0.782-1.028) | 0.118 | 55.5 | 0.904(0.611-1.336) | 0.611 | 79.0 | 0.912(0.783-1.062) | 0.234 | 76.9 |
| MALDI-TO F-MS | 2 | 0.838(0.720-0.975) | 0.022 | 0.0 | 0.538(0.399-0.726) | 0.000 | 0.0 | 0.783(0.678-0.905) | 0.001 | 0.0 | 0.577(0.430-0.774) | 0.000 | 0.0 | 0.779(0.693-0.876) | 0.000 | 0.0 |
| rs1899663 | ||||||||||||||||
| Overall | 6 | 0.958(0.862-1.066) | 0.431 | 0.0 | 0.810(0.596-1.101) | 0.179 | 0.0 | 0.942(0.850-1.045) | 0.260 | 0.0 | 0.818(0.608-1.100) | 0.184 | 0.0 | 0.939(0.858-1.027) | 0.166 | 0.0 |
| Asian | 5 | 0.950(0.852-1.058) | 0.350 | 0.0 | 0.726(0.512-1.028) | 0.071 | 0.0 | 0.932(0.839-1.036) | 0.193 | 0.0 | 0.735(0.520-1.040) | 0.082 | 0.0 | 0.925(0.842-1.016) | 0.102 | 0.0 |
| Estrogen-dependent | 2 | 1.001(0.832-1.205) | 0.991 | 0.0 | 0.746(0.430-1.293) | 0.296 | 0.0 | 0.979(0.818-1.173) | 0.822 | 18.0 | 0.752(0.435-1.301) | 0.308 | 0.0 | 0.961(0.821-1.125) | 0.620 | 23.1 |
| Digestive | 2 | 0.973(0.831-1.138) | 0.729 | 0.0 | 0.773(0.463-1.290) | 0.325 | 0.0 | 0.957(0.821-1.116) | 0.576 | 0.0 | 0.775(0.465-1.292) | 0.328 | 0.0 | 0.948(0.826-1.087) | 0.443 | 0.0 |
| HB | 2 | 1.106(0.883-1.387) | 0.381 | 0.0 | 1.080(0.638-1.828) | 0.774 | 0.0 | 1.099(0.881-1.369) | 0.403 | 0.0 | 1.022(0.632-1.651) | 0.930 | 0.0 | 1.067(0.892-1.277) | 0.475 | 0.0 |
| PB | 3 | 0.956(0.834-1.095) | 0.512 | 0.0 | 0.737(0.487-1.114) | 0.147 | 0.0 | 0.937(0.821-1.069) | 0.334 | 0.0 | 0.746(0.494-1.125) | 0.162 | 0.0 | 0.928(0.825-1.043) | 0.208 | 0.0 |
| RFLP | 4 | 0.920(0.816-1.038) | 0.176 | 0.0 | 0.701(0.480-1.024) | 0.066 | 0.0 | 0.903(0.803-1.015) | 0.086 | 0.0 | 0.715(0.491-1.043) | 0.081 | 0.0 | 0.899(0.810-0.997) | 0.044 | 0.0 |
| rs87494 | ||||||||||||||||
| Overall | 3 | 1.100((0.989-1.223) | 0.079 | 0.0 | 1.161(0.891-1.512) | 0.269 | 0.0 | 1.106(0.999-1.225) | 0.053 | 0.0 | 1.127(0.867-1.465) | 0.372 | 0.0 | 1.091((1.000-1.191) | 0.050 | 0.0 |
| Digestive | 2 | 1.092(0.955-1.250) | 0.199 | 0.0 | 1.171(0.839-1.635) | 0.353 | 0.0 | 1.101(0.967-1.253) | 0.148 | 0.0 | 1.140(0.819-1.588) | 0.436 | 0.0 | 1.090(0.975-1.218) | 0.132 | 0.0 |
| rs12826786 | ||||||||||||||||
| Overall | 5 | 1.155(0.971-1.374) | 0.104 | 22.8 | 1.670(1.244-2.242) | 0.001 | 48.2 | 1.233((1.044-1.456) | 0.013 | 29.5 | 1.551(1.186-2.027) | 0.001 | 40.5 | 1.237(1.092-1.401) | 0.001 | 34.9 |
| Caucasian | 4 | 1.092(0.851-1.400) | 0.489 | 36.9 | 1.507(0.903-2.515) | 0.116 | 53.9 | 1.174(0.928-1.485) | 0.181 | 43.2 | 1.433(1.059-1.938) | 0.020 | 45.6 | 1.196(1.017-1.406) | 0.030 | 47.8 |
| Digestive | 2 | 1.158(0.929-1.443) | 0.191 | 0.0 | 1.595(0.796-3.196) | 0.188 | 58.2 | 1.225(0.991-1.514) | 0.060 | 14.4 | 1.569(1.034-2.378) | 0.034 | 45.0 | 1.224(1.034-1.448) | 0.019 | 29.1 |
| HB | 4 | 1.183(0.978-1.430) | 0.084 | 37.9 | 1.929(1.392-2.674) | 0.000 | 16.0 | 1.289(1.074-1.546) | 0.006 | 31.4 | 1.741(1.299-2.333) | 0.000 | 4.3 | 1.299(1.134-1.489) | 0.000 | 2.1 |
| TaqMan | 2 | 0.875(0.597-1.283) | 0.494 | 0.0 | 1.512(0.919-2.487) | 0.104 | 47.3 | 1.018(0.711-1.456) | 0.923 | 0.0 | 1.665(0.786-3.527) | 0.183 | 62.4 | 1.169(0.914-1.495) | 0.212 | 19.7 |
| RFLP | 2 | 1.169(0.947-1.443) | 0.147 | 0.0 | 1.409(0.549-3.616) | 0.476 | 75.8 | 1.211(0.989-1.484) | 0.064 | 20.4 | 1.344(0.558-3.236) | 0.510 | 73.8 | 1.145(0.856-1.532) | 0.363 | 61.8 |
OR Odds ratio, CI Confidence interval. If P<0.05, the results are in bold. If I2>50%, the results were calculated by random model. HB Hospital-based, PB Population-based, RFLP restriction fragment length polymorphism, MOLDI-TOF-MS Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry
For the rs12826786 C>T, this is first meta-analysis to evaluate the relationship in whole population by pooling 5 published studies comprising 1048 cases and 1432 controls. The results of the pooled analyses distinctly indicated that rs12826786 genetic variation was increased with susceptibility of cancer in recessive, dominant, allelic, and homozygous(TTvs.CC: OR = 1.670, 95% CI = 1.244–2.242, I2 = 48.20%; CT + TT vs. CC: OR = 1.233, 95% CI = 1.044–1.456, I2 = 29.50%; TT vs. CT + CC: OR = 1.551, 95% CI = 1.186–2.027, I2 = 40.50%; T vs. C: OR = 1.237, 95% CI = 1.092–1.401, I2 = 34.90%) models, and the associations were similar with the results of hospital-based control subgroup. Moreover, we reanalyze after excluded one study of non-satisfying HWE, the heterogeneity of the merged studies is not improved.
For the rs7958904 G>C, 6 eligible studies in Chinese population, totally consisting of 5123 cases and 5701 controls after excluded 1 study (Zhou-b) that genotype distribution of their controls did not conformed to HWE. Overall, we identified a significantly decreased susceptibility to overall cancer risk in all genetic (GC vs. GG: OR = 0.887, 95% CI = 0.819–0.961, I2 = 0%; CC + GC vs. GG: OR = 0.869, 95% CI = 0.806–0.938, I2 = 47.40%; C vs. G: OR = 0.869, 95% CI = 0.769–0.981, I2 = 73.8%) models rather than recessive and homozygous models, and a similar results was obtained in digestive cancers, hospital-based control and MALDI-TOF-MS subgroup. In addition, the heterogeneity of both digestive cancers and MALDI-TOF-MS subgroup was significantly lower than the other subgroups.
Rs4759314, rs1899663, and rs874945 polymorphism and cancer susceptibility
For the rs4759314 genetic variation, a total of 13 eligible case-control studies, comprising 8350 cases and 9940 controls were enrolled. For rs1899663 polymorphism, 6 eligible studies with 3239 cases and 4067 controls were included. For rs874945 genetic variation, 3 eligible studies consisted of 3069 cases and 3548 controls were included. Overall, the general OR with its 95% CI did not reveal a significant risk in all genetic models for rs4759314, rs1899663 and rs874945, and subsequent subgroup analyses also did not show the statistical associations rather than the significant risk of cancers correlated with rs4759314 polymorphism in heterozygous and dominant models for Chinese subgroup and rs874945 had a significantly increased risk of overall cancer was shown in allelic model. We detected that the heterogeneity of these subgroups (including estrogen-dependent cancers, RFLP, and population-based control group) was observed significantly decreased compared with other subgroups. In the study, Fig. 2 presented forest plots in heterozygous model for six common polymorphisms.
Fig. 2.
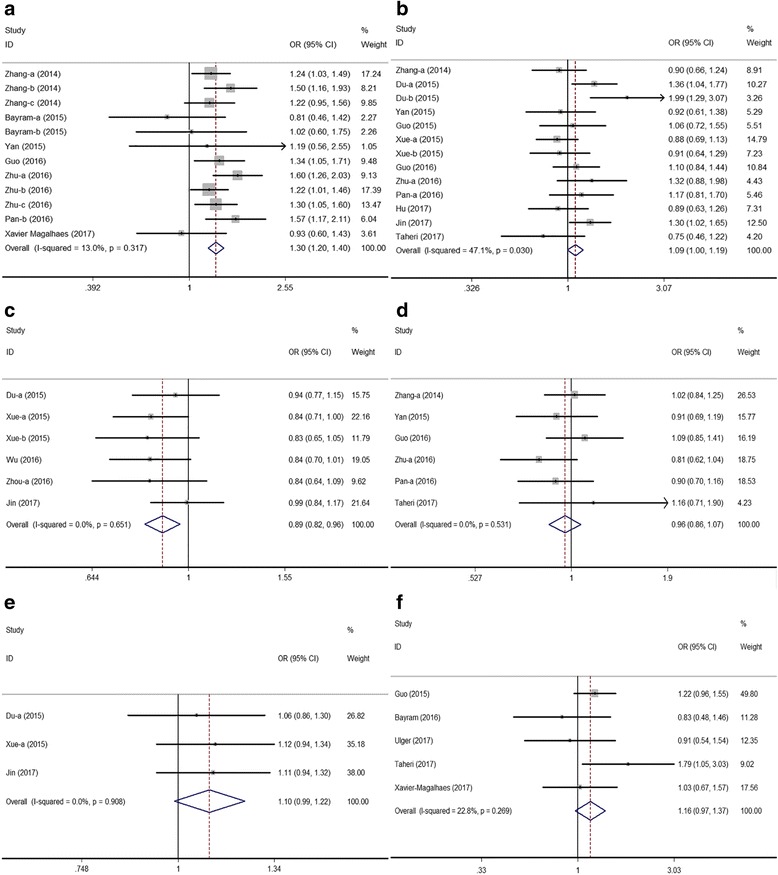
Forests plots for HOTAIR polymorphisms and cancer risk in heterozygous model. a rs920778, b rs4759314, c rs7958904, d rs1899663, e rs874945, f rs12826786
Sensitivity analysis and publication bias
Results of sensitivity analysis of six polymorphisms identified that any single study not qualitatively changed the all pooled ORs in five genetic models, which indicated that all the results of our meta-analysis remained robust and stable in both Asian and Caucasian population, the results of the heterozygous model are shown in Additional file 2: Figure S1. Then, we evaluated published bias by performing Begg’s funnel plots and Egger’s test. As illustrated in all funnel plots, the shapes of plots were no obvious asymmetry under dominant, recessive, allelic, heterozygous, and homozygous models for six genetic variations, and Fig. 3 only presented in heterozygous model. Additionally, all statistical results of Egger’s test illustrated the all corresponding P values of t test > 0.05, and the all relevant 95% CIs, including 0, were not listed. These statistical results further supported for the absence of publication bias in five genetic models, and so our results remained credibly and reliably.
Fig. 3.
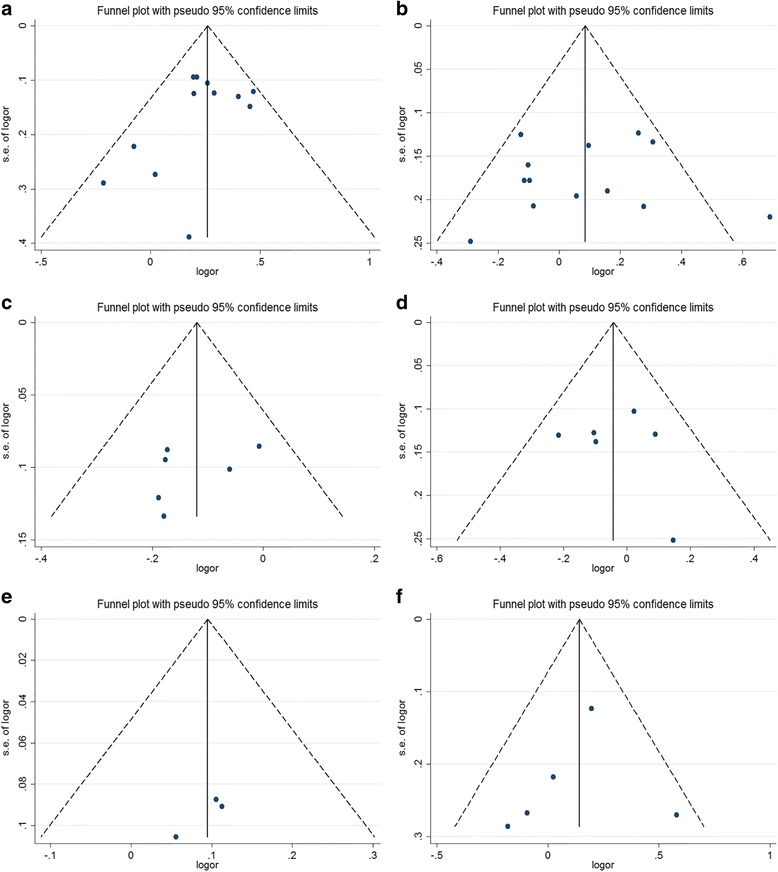
Funnel plots for HOTAIR polymorphisms and cancer risk in heterozygous model. a rs920778, b rs4759314, c rs7958904, d rs1899663, e rs874945, f rs12826786
Discussion
Previously, there have been several meta-analysis [39–41] to pool eligible studies to examine the relationship between cancer risk and HOTAIR polymorphisms (including rs920778, rs4759314, rs7958904, rs874945, and rs1899663), and most of which included all studies and did not take into account the rejection of studies that the genotype distribution of controls did not conform to the HWE except Lv et al.’s study [42]. Among the 18 studies included in our updated meta-analysis, 12 include Asian populations and 6 include Caucasian populations. In addition, this is the first time to assess the effect of rs12826786 polymorphism on cancer susceptibility and also evaluate the relationships of abovementioned polymorphisms in HOTAIR with cancer risk. Overall, the results provided that rs920778, rs7958904, and rs12826786 but not rs4759314, rs1899663, and rs874945 loci are related to cancer risk among Caucasian and Asian populations, among of which rs920778 and rs12826786 increase and rs7958904 decreases cancer risk, respectively. To some extent, these findings denoted that the polymorphisms in HOTAIR may be related to the development of varieties of cancers and offered a novel and compelling evidence for functional analysis of the effects on susceptibility loci to diseases.
HOTAIR, a functional trans-acting lncRNA specifically transcribed from the HOXC gene, which is situated the antisense strand within the intergenic region between HOXC11 and HOXC12 on chromosome 12 [8, 43], and more and more studies focused on the mechanism of HOTAIR, and all these evidences have confirmed that HOTAIR expression lead to malignant transformation of normal cells in varieties of cancers such as ovarian cancers [44, 45], pancreatic tumors [10], hepatocellular carcinoma [11, 46–48], and ESCC [49, 50] to a certain extent. Various researches have previously demonstrated SNPs in several lncRNAs may be involved in carcinogenesis by influencing the secondary structure of the corresponding mRNA or altering its interacting partners [51, 52], which implied that functional susceptibility loci play a crucial role in occurrence and development of cancer. Therefore, we speculated polymorphisms in HOTAIR may also be related to cancer risk by modifying the secondary structure of HOTAIR then indirectly involved the relevant signaling pathways. Recently, several lines of published studies have researched the associations of HOTAIR polymorphisms with different cancers susceptibility, whereas their conclusions are discordant. Regarding the HOTAIR rs920778, Zhang et al. identified the SNP impacts HOTAIR expression via the gene intronic enhancer, which is located in between + 1719 bp and + 2353 bp from the transcriptional start site, with higher HOTAIR expression among T allele carriers, through reporter assays, which might be a potential genetic basis or mechanism for altering susceptibility of ESCC [21].These conclusions of Zhang et al. and other studies were in line with our results that the genetic variation of rs920778 increased cancer risk. But, our results showed the rs920778 SNP was significantly associated with an increased risk of cancer in Chinese rather than in Caucasian, according to the stratified analyses by ethnicity. The possible reasons for no statistically significant results in Caucasians are as follows. First, differences of two subgroups may be due to differences of genetic or inherited background in different population. For example, based on the dbSNP data from NCBI, the allele frequencies of the rs920778 polymorphisms are diverse between Chinese and Caucasian populations. Second, it is likely that there is not enough statistical power to obtain a convincing result in Caucasian population on the grounds that the sample size in Asian population (12,150n) is about 13 times the of Caucasian (934n). Finally, the different types of cancers as well as other unknown and uncontrollable factors may also be potential reasons for the differences of the findings between Asians and Caucasians, and differences of other subgroups maybe ascribe to the same reason mentioned above. Rs12826786 SNP, located on the promoter region of HOTAIR gene, Guo et al. reported that subjects carrying the rs12826786 TT genotype existed a remarkably elevated level of HOTAIR expression than those with the CC genotype in GCA tumor tissue [25], the results was consistent with our findings that rs12826786 polymorphism significantly increased susceptibility to cancer risk in all genetic models except heterozygous models, indicated that C-to-T transition of rs12826786 may first influence the transcription of HOTAIR then affecting the expression of the gene and ultimately impacts the susceptibility of various cancers. Rs7958904 genetic polymorphism, which is located on the exon of HOTAIR gene, we identified the SNP significantly decreased susceptibility to overall cancer risk in all genetic models rather than recessive and homozygous models. Jin et al. also obtained that the rs7958904 CC genotype increased risk of cervical cancer compared with the GG genotypes on the grounds of functional assays, which showed patients with rs7958904 CC genotype existed higher HOTAIR expression than with GG genotype [27]. Additionally, Xue et al. revealed that rs7958904 G/C variation strikingly altered the secondary structure by silico analyses, which indicated that the genetic variation may participate in influencing the susceptibility of colorectal cancer through changing for HOTAIR structure [22]. Besides, Taheri et al. and his colleagues found that rs1899663 and rs12826786 may alter the affinity for binding of some relevant transcription factors, which be related to the occurrence and progression of various cancers including prostate cancer [37]. Moreover, Du et al. attained that HOTAIR and HOXC11 expression levels were higher in subjects with the rs4759314 AG genotype than with AA genotype in GC tissues. Simultaneously, they further explored the SNP, in an intronic promoter region, influenced the activity of this promoter and contributed to the expression of HOTAIR and its downstream gene HOXC11 in a genotype-specific way by The Cancer Genome Atlas (TCGA) database, which is an underlying mechanism for GC susceptibility [29]. However, our meta-analysis found that rs1899663 (intron), rs8749459 (3′ near gene), and rs4759314 polymorphisms were not significantly associated with susceptibility to cancer. Therefore, it is possible that genetic polymorphisms as inherited basis that may modify the function or expression of the involved genes, ultimately contribute to cancer risk based on the above evidence.
In the present meta-analysis, there existed a few limitations potentially. First, the study presented latent language bias due to all published studies only restricted in English. Second, analysis of stratification resulted in smaller sample size in the subgroup that affected statistical efficacy. For example, compared to Caucasian population, most of the studies included in the meta-analysis focused on Chinese population to estimate HOTAIR polymorphisms how to contribute to cancer susceptibility, and therefore need to broaden the sample size in the Caucasian to further verify how HOTAIR polymorphism affects the susceptibility of the associated cancer. Third, we cannot further calculate gene-environment interactions or cumulative effects of genetic polymorphisms on account of not getting original data. Fourth, after excluding those that did not follow the HWE, the sample size of the included studies is too small for rs874945 polymorphism. Fifth, the adjustment of confounders between the original studies is inconsistent, and the study also cannot unify the adjustment of confounding factors (including age, gender, smoking, drinking, etc.) for all included case-control studies due to not obtaining detailed raw data. Finally, due to most of the original publications did not do gender-based stratification analysis, the study failed to perform subgroup analyses of gender. These deficiencies may fail to correctly reveal how HOTAIR polymorphisms affect the risk of cancer. Thus, pooled results of the presented comprehensive analyses should be elucidated cautiously.
Conclusions
The meta-analysis provided that three functional polymorphisms of HOTAIR with rs920778, rs7958904, and rs12826786 might contribute to genetic susceptibility to cancer risk in overall population, whereas rs1899663, rs4759314, and rs874945 had no significant associations. In consequence, well-conducted studies with sufficient sample size are necessarily demanded to further test the association of the abovementioned polymorphisms in HOTAIR and cancer risk, especially in Caucasian and other types of cancers.
Additional files
Table S1. Genotype frequency distributions of HOTAIR polymorphisms and cancer risk. (DOCX 38 kb)
Figure S1. Sensitivity analyses for HOTAIR polymorphisms and cancer risk in heterozygous model. (A) rs920778, (B) rs4759314, (C) rs7958904, (D) rs1899663, (E) rs874945, and (F) rs12826786. (JPEG 754 kb)
Acknowledgements
Not applicable.
Funding
This study was supported by Natural Science Foundation of Liaoning Province (no. 201602870).
Availability of data and materials
The data of the study are available from the corresponding author on reasonable request.
Abbreviations
- ceRNA
Competing endogenous RNAs
- HOTAIR
HOX transcript anti-sense RNA
- PRC2
Polycomb repressive complex 2
Authors’ contributions
JL conceived and wrote the paper. JL and XL collected and analyzed the data. ZC, HL, MG, ZY, BZ, and ZY revised the whole paper. All authors have reviewed the final version of the manuscript and approved to submit to your journal.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors proclaim that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12199-018-0697-0) contains supplementary material, which is available to authorized users.
Contributor Information
Juan Li, Email: 1196349613@qq.com.
Zhigang Cui, Email: zgcui@cmu.edu.cn.
Hang Li, Email: 114870599@qq.com.
Xiaoting Lv, Email: 1564513844@qq.com.
Min Gao, Email: 775532402@qq.com.
Zitai Yang, Email: 424042413@qq.com.
Yanhong Bi, Email: 1757492994@qq.com.
Baosen Zhou, Email: bszhou@cmu.edu.cn.
Zhihua Yin, Phone: 18900910228, Email: zhyin@cmu.edu.cn.
References
- 1.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossi E, Sanchez Y, Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta. 2016;1859:200–208. doi: 10.1016/j.bbagrm.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 8.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 12.Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Li Z. Long non-coding RNA HOTAIR: a novel oncogene (review) Mol Med Rep. 2015;12:5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 15.Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 16.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong W, Wu X, Starnes S, Johnson SK, Haessler J, Wang S, Chen L, Barlogie B, Shaughnessy JD, Jr, Zhan F. An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood. 2008;112:4235–4246. doi: 10.1182/blood-2007-10-119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu H, Wang X, Guo R, Liu Q, Wang Y, Yuan Z, Li J, Shi H. HOTAIR rs920778 polymorphism is associated with ovarian cancer susceptibility and poor prognosis in a Chinese population. Future Oncol. 2017;13:347–355. doi: 10.2217/fon-2016-0290. [DOI] [PubMed] [Google Scholar]
- 19.Bayram S, Sumbul AT, Dadas E. A functional HOTAIR rs12826786 C>T polymorphism is associated with breast cancer susceptibility and poor clinicopathological characteristics in a Turkish population: a hospital-based case-control study. Tumour Biol. 2016;37:5577–5584. doi: 10.1007/s13277-015-4430-y. [DOI] [PubMed] [Google Scholar]
- 20.Gong WJ, Yin JY, Li XP, Fang C, Xiao D, Zhang W, Zhou HH, Li X, Liu ZQ. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37:8349–8358. doi: 10.1007/s13277-015-4497-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062–2067. doi: 10.1093/carcin/bgu103. [DOI] [PubMed] [Google Scholar]
- 22.Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H, Tong N, Chen J, Zhang Z, Wang M. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303–310. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 23.Ulger Y, Dadas E, Yalinbas Kaya B, Sumbul AT, Genc A, Bayram S. The analysis of lncRNA HOTAIR rs12826786 C>T polymorphism and gastric cancer susceptibility in a Turkish population: lack of any association in a hospital-based case-control study. Ir J Med Sci. 2017; 10.1007/s11845-017-1596-x. [DOI] [PubMed]
- 24.Qiu H, Liu Q, Li J, Wang X, Wang Y, Yuan Z, Li J, Pei DS. Analysis of the association of HOTAIR single nucleotide polymorphism (rs920778) and risk of cervical cancer. APMIS. 2016;124:567–573. doi: 10.1111/apm.12550. [DOI] [PubMed] [Google Scholar]
- 25.Guo W, Dong Z, Bai Y, Guo Y, Shen S, Kuang G, Xu J. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumour Biol. 2015;36:2845–2854. doi: 10.1007/s13277-014-2912-y. [DOI] [PubMed] [Google Scholar]
- 26.Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, Zhou L, Yuan Q, Zhou C, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55:90–96. doi: 10.1002/mc.22261. [DOI] [PubMed] [Google Scholar]
- 27.Jin H, Lu X, Ni J, Sun J, Gu B, Ding B, Zhu H, Ma C, Cui M, Xu Y, et al. HOTAIR rs7958904 polymorphism is associated with increased cervical cancer risk in a Chinese population. Sci Rep. 2017;7:3144. doi: 10.1038/s41598-017-03174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayram S, Ulger Y, Sumbul AT, Kaya BY, Rencuzogullari A, Genc A, Sevgiler Y, Bozkurt O, Rencuzogullari E. A functional HOTAIR rs920778 polymorphism does not contributes to gastric cancer in a Turkish population: a case-control study. Familial Cancer. 2015;14:561–567. doi: 10.1007/s10689-015-9813-0. [DOI] [PubMed] [Google Scholar]
- 29.Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J, Ma G, Chu H, Tong N, Zhu H, et al. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6:31255–31262. doi: 10.18632/oncotarget.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, Shang X, Shi Y, Yang Z, Zhao J, Yang M, Li Y, Xu S. Genetic variants of lncRNA HOTAIR and risk of epithelial ovarian cancer among Chinese women. Oncotarget. 2016;7:41047–41052. doi: 10.18632/oncotarget.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan R, Cao J, Song C, Chen Y, Wu Z, Wang K, Dai L. Polymorphisms in lncRNA HOTAIR and susceptibility to breast cancer in a Chinese population. Cancer Epidemiol. 2015;39:978–985. doi: 10.1016/j.canep.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 32.Guo L, Lu X, Zheng L, Liu X, Hu M. Association of long non-Coding RNA HOTAIR polymorphisms with cervical cancer risk in a Chinese population. PLoS One. 2016;11:e0160039. doi: 10.1371/journal.pone.0160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayram S, Sumbul AT, Batmaci CY, Genc A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol. 2015;36:3863–3870. doi: 10.1007/s13277-014-3028-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Lv Z, An C, Shi M, Pan W, Zhou L, Yang W, Yang M. Onco-lncRNA HOTAIR and its functional genetic variants in papillary thyroid carcinoma. Sci Rep. 2016;6:31969. doi: 10.1038/srep31969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xavier-Magalhaes A, Oliveira AI, de Castro JV, Pojo M, Goncalves CS, Lourenco T, Viana-Pereira M, Costa S, Linhares P, Vaz R, et al. Effects of the functional HOTAIR rs920778 and rs12826786 genetic variants in glioma susceptibility and patient prognosis. J Neuro-Oncol. 2017;132:27–34. doi: 10.1007/s11060-016-2345-0. [DOI] [PubMed] [Google Scholar]
- 36.Hu P, Qiao O, Wang J, Li J, Jin H, Li Z, Jin Y. rs1859168 A>C polymorphism regulates HOTTIP expression and reduces risk of pancreatic cancer in a Chinese population. World J Surg Oncol. 2017;15:155. doi: 10.1186/s12957-017-1218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taheri M, Habibi M, Noroozi R, Rakhshan A, Sarrafzadeh S, Sayad A, Omrani MD, Ghafouri-Fard S. HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene. 2017;613:20–24. doi: 10.1016/j.gene.2017.02.031. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Q, Chen F, Fei Z, Zhao J, Liang Y, Pan W, Liu X, Zheng D. Genetic variants of lncRNA HOTAIR contribute to the risk of osteosarcoma. Oncotarget. 2016;7:19928–19934. doi: 10.18632/oncotarget.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge Y, Jiang R, Zhang M, Wang H, Zhang L, Tang J, Liang C. Analyzing 37,900 samples shows significant association between HOTAIR polymorphisms and cancer susceptibility: a meta-analysis. Int J Biol Markers. 2017;32(2):e231–e242. doi: 10.5301/jbm.5000235. [DOI] [PubMed] [Google Scholar]
- 40.Qi Q, Wang J, Huang B, Chen A, Li G, Li X, Wang J. Association of HOTAIR polymorphisms rs4759314 and rs920778 with cancer susceptibility on the basis of ethnicity and cancer type. Oncotarget. 2016;7(25):38775–38784. doi: 10.18632/oncotarget.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Liu X, You LH, Zhou RZ. Significant association between long non-coding RNA HOTAIR polymorphisms and cancer susceptibility: a meta-analysis. OncoTargets and therapy. 2016;9:3335–3343. doi: 10.2147/OTT.S107190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv Z, Xu Q, Yuan Y. A systematic review and meta-analysis of the association between long non-coding RNA polymorphisms and cancer risk. Mutat Res. 2017;771:1–14. doi: 10.1016/j.mrrev.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, Maesawa C. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int J Oncol. 2013;43:63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]
- 45.Cui L, Xie XY, Wang H, Chen XL, Liu SL, Hu LN. Expression of long non-coding RNA HOTAIR mRNA in ovarian cancer. Sichuan Da Xue Xue Bao Yi Xue Ban. 2013;44:57–59. [PubMed] [Google Scholar]
- 46.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 47.Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, Peng C, Xie H, Zhou L, Wu J, Zheng S. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15:4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Wu Z, Mei Q, Li X, Guo M, Fu X, Han W. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 51.Li L, Sun R, Liang Y, Pan X, Li Z, Bai P, Zeng X, Zhang D, Zhang L, Gao L. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J Exp Clin Cancer Res. 2013;32:104. doi: 10.1186/1756-9966-32-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue Y, Wang M, Kang M, Wang Q, Wu B, Chu H, Zhong D, Qin C, Yin C, Zhang Z, Wu D. Association between lncrna PCGEM1 polymorphisms and prostate cancer risk. Prostate Cancer Prostatic Dis. 2013;16:139–144. doi: 10.1038/pcan.2013.6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genotype frequency distributions of HOTAIR polymorphisms and cancer risk. (DOCX 38 kb)
Figure S1. Sensitivity analyses for HOTAIR polymorphisms and cancer risk in heterozygous model. (A) rs920778, (B) rs4759314, (C) rs7958904, (D) rs1899663, (E) rs874945, and (F) rs12826786. (JPEG 754 kb)
Data Availability Statement
The data of the study are available from the corresponding author on reasonable request.


