Abstract
Background
Chronic hepatitis B (CHB) remains a global health dilemma with high morbidity and mortality. Human males absent on the first (hMOF) (a histone acetyltransferase) is responsible for DNA damage repair, tumorigenesis and cell cycle regulation. Persistence of HBV DNA contributes to cirrhosis and hepatocellular carcinoma (HCC) in CHB patients. Histone acetyltransferase enhances HBV replication, however the precise underlying mechanism of hMOF in HBV replication in CHB patients remains to be explored. This study aims to investigate the correlation between hepatic hMOF and HBV DNA replication in CHB patients, and may provide new insights towards the treatment of CHB patients.
Methods
hMOF in liver biopsy (CHB, n = 33 HBeAg+; n = 20 HBeAg−, and three healthy controls) was determined, using immunohistochemistry, qPCR and Western blot. The correlation between hMOF and HBsAg, as well as, HBeAg were determined.
Results
A positive correlation between hMOF and HBV DNA in overall CHB patients was observed. A distinct positive correlation between hMOF and HBsAg and/or HBeAg in HBeAg+ CHB patients was also detected, however not observed between hMOF and HBsAg in HBeAg− CHB patients. No correlation was observed between hMOF and hepatic inflammation severity and fibrotic stage in CHB patients.
Conclusions
Hepatic hMOF might contribute to host HBV clearance in CHB patients and possible pathogenesis.
Electronic supplementary material
The online version of this article (10.1186/s13578-018-0215-5) contains supplementary material, which is available to authorized users.
Keywords: hMOF, Chronicity, Hepatitis B, Viral replication, Epigenetic regulation
Background
Hepatitis B virus (HBV) infection continues to be a global health problem with high morbidity and mortality, despite decades of extensive research into antiviral drugs and vaccines [1]. Chronicity of HBV infection has been attributed to the unique and stable replication system employed by the virus. The episomal viral genome, the covalently closed circular DNA (cccDNA), forms within infected hepatic nuclei, resulting in increased difficulty for host HBV clearance [2]. Chronic hepatitis B infection (CHB) eventuates in irreversible conditions, including cirrhosis, hepatic decompensation, and hepatocellular carcinoma [3]. The precise pathogenesis of CHB is still a major clinical challenge and remains to be explored.
Males absent on the first (MOF) is a member of MYST family of histone acetyltransferases (HATs). Acetylation of H4K16, a component of the lysine group on the N-terminal tail of histone H4, raises the level of X chromosome transcriptional activity in males at twice the level of females, promoting a dosage compensation effect [4]. Human males absent on the first (hMOF) is involved in the transcriptional regulation of genes, DNA damage repair, tumorigenesis, and cell cycle regulation [5]. Abnormal hMOF expression has been detected in many malignancies, including kidney [6], ovarian [7], lung [8], breast [9] and medulloblast [9].
It is well known that HBV DNA is a high-risk factor in the progression of cirrhosis and hepatocellular carcinoma (HCC) in CHB patients [10]. Epigenetic regulation alters the chromatin structure without variation of gene sequences, including gene transcription, recombination, DNA replication, and damage repair [11]. Epigenetic regulation also participates in post-translational modifications of HBV, such as DNA methylation and histone modifications, specifically acetylation of H4K16 [12]. HBV cccDNA, organized into minichromosomes in the nucleus of the host cells by histone and non-histone proteins [13], is the key transcriptional template for HBV RNA. Histone acetyltransferase has been demonstrated to enhance HBV replication, while histone deacetylase has been identified to suppress HBV replication [12]. Furthermore, our previously published data demonstrates that depletion of hMOF represses HBV replication in vitro, leading to a decrease in HBsAg and HBeAg levels [14]. The directly linkage between hMOF and CHB status remains to be explored.
Our current study aims to investigate the correlation between hMOF and HBV DNA replication in the liver of CHB patients. Understanding the correlation between the expression of hMOF and HBV replication may shed light on pharmaceutical development towards the prevention and treatment of CHB patients.
Methods
Human subjects
A total of 53 CHB patients were recruited, including 33 HBeAg+, 20 HBeAg− CHB patients, and three healthy controls, from December 2012 to December 2014 in the Department of Infectious Diseases, Ruijin Hospital, Shanghai, China. CHB patients aged 18–65 years, were identified as HBV mono-infected with HBsAg+ for at least 6 months and were naïve to antiviral treatment and immunotherapy prior to the initiation of the current study [15]. HBeAg+ chronic hepatitis B patents are characterized by the presence of serum HBeAg with high levels of HBV DNA, while chronic hepatitis B patients are characterized by the presence of serum antibodies to HBeAg (anti-HBe), and persistent or fluctuating moderate to high levels of serum HBV DNA (often lower than in HBeAg+ patients) [16]. Exclusion criteria were: (1) co-infection with HAV, HCV, HDV or HEV; (2) autoimmune liver diseases; (3) non-alcoholic fatty liver diseases; (4) alcoholic liver diseases; (5) congenital metabolic liver disease; (6) evidence of hepatocellular carcinoma (suspicious foci on hepatic ultrasonic examination or CT, or a rising serum level of α-fetoprotein). Liver biopsy (n = 53) was obtained for histological examination.
Laboratory assay
Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were tested routinely, using an automated chemistry analysis system (Beckman Coulter, Fullerton, CA, USA). Serum HBsAg and HBeAg were determined, using commercial enzyme immunoassay kits (AXSYM System; Abbott, Wiesbaden, Germany). Serum HBV DNA level was quantified, using Applied Biosystems PCR system (Prism 7500; Applied Biosystems, Inc., USA), with a lower limit of quantification at 200 IU/mL.
Western blot
Western blot was performed as described previously [17]. Briefly, liver tissues were lysed in RIPA buffer (Beyotime, China), and extracted proteins were quantified using a BCA assay (Beyotime, China). Proteins (10 μg) were transferred to PVDF membranes which were then blocked with 20 mL 5% fat-free milk in 1× TBS at room temperature for 1 h. Membranes were incubated with either rabbit anti-human hMOF, 1:2000, (Abcam, Cambridge, UK); or mouse anti-human GAPDH, 1:2000, (Abcam, Cambridge, UK) at 4 °C overnight, followed by secondary antibody (goat anti-rabbit-HRP or goat anti-mouse–HRP, 1:5000 each; Beyotime, China) for 1 h and visualized, using ImageQuant™ LAS 4000 (Fujifilm, Tokyo, Japan).
qRT-PCR
Trizol reagent (Invitrogen, USA) was used to extract RNA from liver tissues, with cDNA synthesized, using a reverse transcriptase kit (TaKaRa). qPCR was subsequently conducted in in three independent assays according to the manufacturer’s instruction, using SYBR Green PCR Master Mix (TaKaRa) in duplicates. Relative hMOF expression levels were quantified after normalization to GAPDH as an internal control. The primers used were listed in Table 1.
Table 1.
Related primer sequences of qRT-PCR
| Gene | Related primer sequences |
|---|---|
| hMOF-forward primer | GAAGGAGCATGAGGCGATCA |
| hMOF-reverse primer | TTTCGTAGTTCCCGATGTGGAT |
| GAPDH-forward primer | ATCACTGCCACCCAGAAGAC |
| GAPDH-reverse primer | ATGAGGTCCACCACCCTGTT |
Immunohistochemistry
Liver biopsies from 53 CHB patients and three healthy liver transplant donors were collected for immunohistochemistry. Immunohistochemistry was performed as previously described [18], using rabbit anti-human hMOF primary antibody (Abcam, Cambridge, MA, USA). Every test was coupled with a negative control in which the antibody was substituted by the primary rabbit negative control. Immunohistochemical assay and computer-assisted genuine color image analysis system (ImagePro-plus 7.0) was used to quantify objectively the integrated optional density of hepatic hMOF [18, 19].
Liver biopsy
Percutaneous liver biopsy was performed under the guidance of ultrasound. All puncture samples were more than 1 cm in length, and at least six portal tracts were contained for evaluation. Liver histopathology was graded by a pathologist independently in a double-blind fashion. Modified Ishak’s histological activity index (HAI) for necroinflammation and the Ishak fibrosis score for fibrosis were used [20]. Remaining liver biopsy samples were stored at − 80 °C for further use.
Statistics
Data are presented as mean ± SD. Statistical analysis was performed using SPSS 17.0 statistical software (SPSS Inc, Chicago, IL, USA) and GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA). For normally distributed data, independent-sample t test was used for comparisons between two groups. For abnormally distributed data, non-parametric statistics was performed, and Mann–Whitney U test was used for comparisons between two groups. Chi square tests were performed in comparisons between categorical factors. Spearman rank correlation analyses were performed to analyze the association between measured parameters and ranked data, otherwise Pearson correlation analyses were used. All p values mentioned were two-sided. All values p < 0.05 were considered to be statistically significant.
Results
Clinical characteristics
All 53 CHB patients were classified by HBeAg status. Among 33 HBeAg+ CHB patients, the level of serum HBsAg, HBeAg, ALT or AST was 20,157 ± 29,156 IU/mL, 689 ± 623 S/CO, 46.06 ± 21.45 IU/L or 35.38 ± 13.69 IU/L, respectively. Conversely, among the 20 HBeAg− CHB patients the level of serum HBsAg, ALT or AST was 1047 ± 1443 IU/mL, 43.10 ± 27.99 IU/L or 34.70 ± 17.52 IU/L, respectively (Table 2).
Table 2.
Clinical characteristics of CHB patients
| Total (n = 53) | HBeAg+ (n = 33) | HBeAg− (n = 20) | |
|---|---|---|---|
| Male, n (%) | 34 (64.15%) | 21 (63.63%) | 13 (65.00%) |
| Age (years) | 37.62 ± 9.38 | 35.12 ± 8.94 | 41.75 ± 8.79 |
| ALT (IU/L) | 44.94 ± 23.90 | 46.06 ± 21.45 | 43.10 ± 27.99 |
| AST (IU/L) | 35.06 ± 15.11 | 35.38 ± 13.69 | 34.70 ± 17.52 |
| HBV DNA (log10IU/mL) | 5.63 ± 1.97 | 6.54 ± 1.74 | 4.14 ± 1.36 |
| HBsAg (IU/mL) | 12,838 ± 24,624 | 20,157 ± 29,156 | 1047 ± 1443 |
| HBeAg (S/CO) | / | 689 ± 623 | / |
Data was expressed as mean ± standard deviation
Correlation between hepatic hMOF and serum HBV DNA, HBsAg and/or HBeAg levels
Among all CHB patients, significant correlation was observed between hepatic hMOF and serum HBV DNA (p < 0.001, r = 0.7762, Fig. 1a), or between hepatic hMOF and serum HBsAg (p = 0.0013, r = 0.5025, Fig. 1b). Similarly, within 33 HBeAg+ CHB patients, a significant correlation was identified between hepatic hMOF and serum HBV DNA (p < 0.0001, r = 0.7725, Fig. 1e), hepatic hMOF and serum HBsAg (p = 0.0010, r = 0.5878, Fig. 1f), and hepatic hMOF and serum HBeAg (p = 0.0087, r = 0.4861, Fig. 1g). Combining HBsAg and HBeAg together, we also found a significant correlation between hepatic hMOF and HBsAg–HBeAg in HBeAg+ CHB patients (adjusted p = 0.0015, r = 0.6007, Fig. 1h). Furthermore, correlation between hepatic hMOF and serum HBV DNA levels was observed (p < 0.0001, r = 0.8169, Fig. 1c) in HBeAg− CHB patients, however no correlation was detected between hepatic hMOF and serum HBsAg levels (p = 0.4755, Fig. 1d).
Fig. 1.
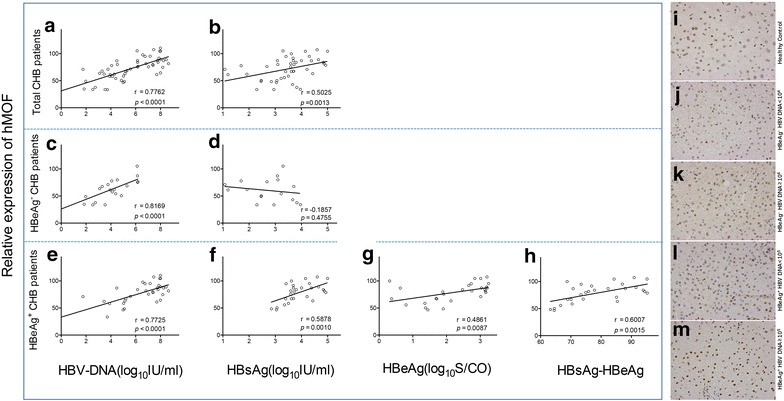
Correlation between hepatic hMOF and serum HBV DNA, HBsAg, and HBeAg levels. Correlation between hepatic hMOF and serum HBV DNA, HBsAg and/or HBeAg levels in CHB patients (a, b), and HBeAg− CHB patients (c, d), and HBeAg+ CHB patients (e–g), and correlation between hepatic hMOF and combination of HBsAg–HBeAg in HBeAg+ CHB patients (h). Representative immunohistochemical micrographs of hepatic hMOF in different liver tissues: Healthy control (i); HBeAg− CHB patients with low HBV DNA load (j) and high HBV DNA load (k); HBeAg+ CHB patients with low HBV DNA load (l) and high HBV DNA load (m)
Consistent with the results above, our imunohistochemical analysis demonstrated that hepatic hMOF was markedly upregulated in HBeAg+ CHB patients compared with healthy controls (Fig. 1i), both in high HBV DNA load groups (HBV DNA ≥ 1 × 105 IU/mL) (Fig. 1m) and in low HBV DNA load groups (HBV DNA < 1 × 105 IU/mL) (Fig. 1l). Moreover, among HBeAg+ CHB patients with high HBV DNA load, hepatic hMOF was 1.4-fold higher than those patients with low HBV DNA load. Similarly, hepatic hMOF was markedly upregulated in high HBV DNA load groups (HBV DNA ≥ 1 × 104 IU/mL) (Fig. 1k) in HBeAg− CHB patients compared with healthy controls. Hepatic hMOF in HBeAg− CHB patients with high HBV DNA load were 1.7-fold higher than those with low HBV DNA load (HBV DNA < 1 × 104 IU/mL) (Fig. 1j). Hepatic hMOF in HBeAg− CHB patients with high HBV DNA load were 2.2-fold higher than healthy controls. However, there was no significant difference of hepatic hMOF between low HBV DNA load of HBeAg− CHB patients and healthy controls.
Hepatic hMOF in HBeAg+ and/or HBeAg− CHB patients
Western blot analysis and qRT-PCR were conducted to illustrate hepatic hMOF expression both in HBeAg+ CHB patients (Fig. 2a) and HBeAg− CHB patients (Fig. 2b). Hepatic hMOF expression was observed to be significantly higher with both high HBV DNA load (HBV DNA ≥ 1 × 105 IU/mL, 10 cases) (2.4-fold) and low HBV DNA load (HBV DNA < 1 × 105 IU/mL, 10 cases) (1.7-fold) in HBeAg+ CHB patients compared to healthy controls (three cases). Furthermore, hepatic hMOF was 1.7-fold higher with high HBV DNA load compared with those with low HBV DNA load (p < 0.01) in HBeAg+ CHB patients. Similarly, hepatic hMOF expression was markedly higher in HBeAg− CHB patients with high HBV DNA load (HBV DNA ≥ 1 × 104 IU/mL, 10 cases) than these with low HBV DNA load (HBV DNA < 1 × 104 IU/mL, 10 cases) and healthy controls (three cases) (2.0-fold and 1.2-fold, respectively). There is no significant difference of the hepatic hMOF expression between HBeAg− CHB patients with low HBV DNA load and healthy controls.
Fig. 2.
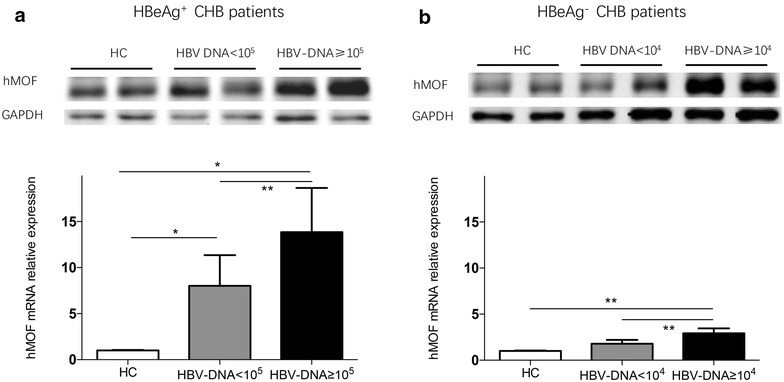
Hepatic hMOF in HBeAg+ and/or HBeAg− CHB patients and healthy controls. Hepatic hMOF production was presented with Western blot and quantitative analysis of hepatic hMOF mRNA expression was measured by qPCR, from HBeAg+ CHB patients (a) and HBeAg− CHB with high HBV DNA load (HBV DNA ≥ 105 IU/mL, 10 cases) and with low HBV DNA load (HBV DNA < 105 IU/mL, 10 cases), and healthy controls (HC) (three cases), and HBeAg− CHB (b) with high HBV DNA load (HBV DNA ≥ 104 IU/mL, 10 cases) and with low HBV DNA load (HBV DNA < 104 IU/mL, 10 cases), and healthy controls (HC) (three cases). The significant difference is expressed as *p < 0.05, **p < 0.01
Correlation between hepatic hMOF and severity of inflammation, or stage of fibrosis of liver
Among CHB patients, no significant correlation was observed between hepatic hMOF and severity of inflammation or hepatic hMOF and stage of fibrosis of liver (Fig. 3a or b). Furthermore, no significant correlation was observed between hepatic hMOF and severity of inflammation or hepatic hMOF and stage of fibrosis of liver in HBeAg+ CHB patients (Fig. 3e, f). Additionally, no significant correlation was observed between hepatic hMOF and severity of inflammation or between hepatic hMOF and severity and stage of fibrosis of liver in HBeAg− CHB patients (Fig. 3c, d).
Fig. 3.
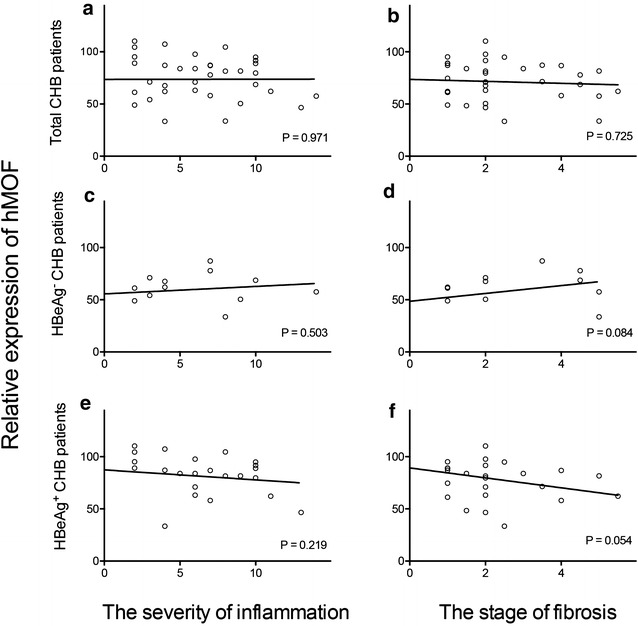
Correlation between hepatic hMOF and severity of inflammation as well as, stage of fibrosis of liver. Correlations between hepatic hMOF and severity of inflammation and stage of fibrosis of liver was described in CHB patients (a, b), in HBeAg− CHB patients (c, d) and in HBeAg+ CHB patients (d, e)
Correlation between serum HBV DNA, HBsAg and/or HBeAg levels and histological damage of liver
In total CHB patients, no significant correlation was observed between serum HBV DNA, HBsAg and/or HBeAg levels and hepatic inflammation severity, or fibrotic stage of liver (Additional file 1: Figure S1A–D). Similarly, there is no significant correlation between serum HBV DNA, HBsAg and/or HBeAg status and hepatic inflammation severity or fibrotic stage of liver neither in HBeAg+ CHB patients (Additional file 1: Figure S1E–H), nor in HBeAg− CHB patients (Additional file 1: Fig S1I–L).
Discussion
HBV infection remains a major clinical challenge worldwide with continued economic burden, as current anti-viral therapy is not completely effective [3]. The precise underlying mechanism of persistent HBV replication in CHB patients, and the associated significant risk of developing liver cirrhosis and HCC, requires further investigation. It is therefore necessary to explore the possible mechanisms involved in the persistence of HBV in CHB patients, and identify potential novel target(s) for the development of pharmaceutical treatments of such a devastating disease.
In our present study, we detected the correlation between hepatic hMOF and HBV DNA replication in CHB patients, suggesting that hMOF plays an important role in regulating HBV replication. This is consistent with our previous study that demonstrated HBV replication is markedly repressed in vitro [14]. However, the role of hMOF in HBV replication is still unclear. HBV cccDNA is a key event in the process of HBV replication within the infected hepatocytes post-translational modifications (PTMs) via acetylation [12, 13, 21]. There is no direct linkage between upregulation of hMOF and cccDNA in CHB patients.
One of the key findings of our current study was the significant correlation between hepatic hMOF and serum HBV DNA in total CHB patients. hMOF serves as an enzyme in histone acetylation in the process of post-translational modifications. Serum HBV DNA appears to be a useful marker of HBV replication [22], with cccDNA and relaxed circular (RC) DNA consisting of total intrahepatic HBV DNA [23]. hMOF also participates in cccDNA mediated post-translational modifications (acetylation) [21]. The data from our current study demonstrated that a correlation between hepatic hMOF and circulating HBV DNA in CHB patients in vivo. Our data is supported by our previous study in vitro that depletion of hMOF obviously represses HBV replication, leading to a decrease in HBsAg and HBeAg levels [14]. Our data further suggests that hMOF plays a critical role in enhancing HBV replication, including promoting HBV replication in hepatocytes. The precise underlying mechanism of how hMOF specifically regulates HBV replication remains to be explored.
In our present study, we observed close correlations between hepatic hMOF and serum HBsAg, and HBeAg levels in total CHB patients and in HBeAg+ CHB patients, however not in HBeAg− CHB patients. hMOF may be used as a potential biomarker in predicting HBV replication, particularly in HBeAg+ patients who usually express high loads of HBV due to their low immune status [16]. It is well documented that levels of circulating HBsAg and HBeAg reflect the activity of HBV replication, although not intrahepatic HBV DNA level [24]. Thus, our current data is in line with HBV DNA, suggesting that hepatic hMOF reflects viral replication, particularly in HBeAg+ CHB patients.
The discrepant correlation between serum HBsAg and HBV DNA/cccDNA has been described in previous studies [25–27]. Serum HBsAg levels correlate with cccDNA [25] in HBeAg+ CHB patients [26] but not in HBeAg− CHB patients [27], suggesting that HBsAg may not be a comprehensive predictor of HBV replication in HBeAg− CHB patients. The upregulation of serum HBsAg expression involves the integrated viral genome in addition to the amount of cccDNA in infected hepatocytes [28]. Serum HBsAg levels can therefore reflect cccDNA concentration in HBeAg+ CHB patients during different antiviral therapy phases [25, 29], suggesting that serum HBsAg may be used as a cccDNA predictor when cccDNA levels are high. In HBeAg− CHB patients, the production of cccDNA is reduced by the immune clearance [23], thus the contribution of cccDNA to HBsAg production may be lower than that of HBeAg+ CHB patients. Such reports are in line with our current findings, where no significant correlation was observed between hepatic hMOF and serum HBsAg level in HBeAg− CHB patients.
We hypothesize that hMOF upregulates HBV replication more effectively in HBeAg+ CHB patients than that in HBeAg− CHB patients. We don’t have firm evidence that hMOF is the key molecule determining host immunity against HBV clearance. Nevertheless, our data invites speculation that the interaction between hMOF and the immune system determines host immunity against HBV clearance in HBeAg+ patients, who have relatively low host defense. The precise underlying mechanism is being determined.
To determine if hMOF contributes to hepatic damage during the development of CHB, correlation between hMOF and hepatic inflammation/fibrosis was investigated. Interestingly, there was no significant correlation between hMOF and severity of inflammation, nor stage of fibrosis of the liver in CHB patients (with/without HBeAg+). These data suggest hMOF may not be an ideal biomarker for hepatic inflammation or fibrosis. A possible explanation is that hepatic damage is considered to be caused by host cellular immunity against HBV infected hepatocytes, rather than the direct HBV cytopathic effect [30], or that hMOF might not be sensitive at the chronic stage. A close correlation between HBV DNA load and intrahepatic hMOF perhaps can explain that there was not a direct correlation between hMOF and intrahepatic histology, nor serological viral markers, such as HBsAg and HBeAg. Our explanation is consistent with others, showing no significant correlation between serum HBV DNA levels and liver histology in terms of necroinflammation and fibrosis in HBeAg+ CHB patients [31, 32]. Consistently, there was no significant correlations between serum HBV DNA level, HBsAg and/or HBeAg and severity of inflammation, or stage of fibrosis of liver neither in total CHB patients, nor in HBeAg+ CHB patients or in HBeAg− CHB patients.
Conclusion
Hepatic hMOF may be responsible for the persistence of chronicity of HBV infection by promoting HBV replication in CHB patients, particularly in HBeAg+ patients. Such information may provide new insights into pharmaceutical development in prevention and treatment of CHB. Future studies will explore the underlying mechanism hMOF regulates HBV replication in larger cohort.
Additional file
Additional file 1. Correlation between serum HBV DNA level, and HBsAg status and severity of inflammation and stage of fibrosis of liver.
Authors’ contributions
LC and FL performed the experiment. RB revised the manuscript. YW, ZC, XX, LY, GZ and LL collected the samples. CZ, QX, SB and HW designed the experiment and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study complies with the Declaration of Helsinki, and has been approved by the human Ethics Committee, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Written informed consent was obtained from each subject prior to participation.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81570560), Technology Supporting Project of the Science and Technology Commission Shanghai Municipality (16411960300), Project of Shanghai Municipal Health and Family Planning (201440482, 2012Y172), The Shanghai 3-year Plan of the Key Subjects Construction in Public Health-Infectious Disease and Pathogenic Microorganism (15GWZK0102), The Medical Science Research Foundation (YWJKJJHKYJJ-B17503), Baoen Wang for Liver Fibrosis (CFHPC 20131056), and The Suzhou Expert Team of Clinical Medicine (SZYJTD201717).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CHB
chronic hepatitis B subjects without cirrhosis and liver failure
- HBV
hepatitis B virus
- hMOF
human males absent on the first
- HCC
hepatocellular carcinoma
- cccDNA
covalently closed circular DNA
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- HATs
histone acetyltransferases
- PTMs
post-translational modifications
- RC DNA
relaxed circular DNA
Footnotes
Liwen Chen, Chuanwu Zhu and Fengdi Li contributed equally to this work
Electronic supplementary material
The online version of this article (10.1186/s13578-018-0215-5) contains supplementary material, which is available to authorized users.
Contributor Information
Liwen Chen, Email: cliwen0618@163.com.
Chuanwu Zhu, Email: zhuchw@126.com.
Fengdi Li, Email: 531174540@qq.com.
Yun Wang, Email: wangyunruijin@163.com.
Rebecca Bao, Email: rebecca.bao@hotmail.com.
Zhujun Cao, Email: estherlucifer@163.com.
Xiaogang Xiang, Email: shine-xxg@163.com.
Lei Yan, Email: yanlchn@163.com.
Lanyi Lin, Email: lanyilin2002@163.com.
Gangde Zhao, Email: zhaogd1016@foxmail.com.
Qing Xie, Email: xieqingrjh@163.com.
Shisan Bao, Phone: +61 2 9351 6156, Email: bob.bao@sydney.edu.au.
Hui Wang, Phone: +86 2164370045, Email: wanghuirj@163.com.
References
- 1.Trépo C, Chan HLY, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 2.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 3.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 4.Hilfiker A, Hilfiker‐Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellert HS, McMahon SB. hMOF, a KAT(8) with many lives. Mol Cell. 2009;36:174–175. doi: 10.1016/j.molcel.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Zhang R, Wu D, et al. Epigenetic change in kidney tumor: downregulation of histone acetyltransferase MYST1 in human renal cell carcinoma. J Exp Clin Cancer Res. 2013;32:8. doi: 10.1186/1756-9966-32-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu N, Zhang R, Zhao X, et al. A potential diagnostic marker for ovarian cancer: involvement of the histone acetyltransferase, human males absent on the first. Oncol Lett. 2013;6:393–400. doi: 10.3892/ol.2013.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Ye X, Tang N, et al. The histone acetylranseferase hMOF acetylates Nrf2 and regulates anti-drug responses in human non-small cell lung cancer. Br J Pharmacol. 2014;171:3196–3211. doi: 10.1111/bph.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfister S, Rea S, Taipale M, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer. 2008;122:1207–1213. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- 10.Chen CJ, Yang HI, Su J, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 11.Jin J, Cai Y, Li B, et al. In and out: histone variant exchange in chromatin. Trends Biochem Sci. 2005;30:680–687. doi: 10.1016/j.tibs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Pollicino T, Belloni L, Raffa G, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Bock CT, Schwinn S, Locarnini S, et al. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Liu K, Fang BA, et al. Identification of acetyltransferase genes (HAT1 and KAT8) regulating HBV replication by RNAi screening. Cell Biosci. 2015;5:66. doi: 10.1186/s13578-015-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH. AASLD guidelines for treatment of chronic hepatitis B. Hepatology. 2016;63:261–283. doi: 10.1002/hep.28156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.EASL Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;2017(67):370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Mo R, Wang P, Lai R, et al. Persistently elevated circulating Th22 reversely correlates with prognosis in HBV-related acute-on-chronic liver failure. J Gastroenterol Hepatol. 2016;32(3):677–686. doi: 10.1111/jgh.13537. [DOI] [PubMed] [Google Scholar]
- 18.Lai R, Xiang X, Mo R, et al. Protective effect of Th22 cells and intrahepatic IL-22 in drug induced hepatocellular injury. J Hepatol. 2015;63:148–155. doi: 10.1016/j.jhep.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Yu F, Han Y, et al. SUMO-specific protease 3 is a key regulator for hepatic lipid metabolism in non-alcoholic fatty liver disease. Sci Rep. 2016;6:37351. doi: 10.1038/srep37351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 21.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Bonino F, Hoyer B, Nelson J, Engle R, Verme G, Gerin J. Hepatitis B virus DNA in the sera of HBsAg carriers: a marker of active hepatitis B virus replication in the liver. Hepatology. 1981;1:386–391. doi: 10.1002/hep.1840010503. [DOI] [PubMed] [Google Scholar]
- 23.Volz T, Lutgehetmann M, Wachtler P, et al. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology. 2007;133:843–852. doi: 10.1053/j.gastro.2007.06.057. [DOI] [PubMed] [Google Scholar]
- 24.Ning X, Nguyen D, Mentzer L, et al. Secretion of genome-free hepatitis B virus–single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 2011;7:e1002255. doi: 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werle-Lapostolle B, Bowden S, Locarnini S, et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Tuttleman J, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 27.Lin LY, Wong VW, Zhou HJ, et al. Relationship between serum hepatitis B virus DNA and surface antigen with covalently closed circular DNA in HBeAg-negative patients. J Med Virol. 2010;82:1494–1500. doi: 10.1002/jmv.21863. [DOI] [PubMed] [Google Scholar]
- 28.Zoulim F, Testoni B, Lebosse F. Kinetics of intrahepatic covalently closed circular DNA and serum hepatitis B surface antigen during antiviral therapy for chronic hepatitis B: lessons from experimental and clinical studies. Clin Gastroenterol Hepatol. 2013;11:1011–1013. doi: 10.1016/j.cgh.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Wursthorn K, Lutgehetmann M, Dandri M, et al. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology. 2006;44:675–684. doi: 10.1002/hep.21282. [DOI] [PubMed] [Google Scholar]
- 30.Lee WM. Hepatitis B virus infection. New Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 31.Yuen MF, Ng IO, Fan ST, et al. Significance of HBV DNA levels in liver histology of HBeAg and Anti-HBe positive patients with chronic hepatitis B. Am J Gastroenterol. 2004;99:2032–2037. doi: 10.1111/j.1572-0241.2004.40440.x. [DOI] [PubMed] [Google Scholar]
- 32.Lindh M, Horal P, Dhillon AP, Norkrans G. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J Viral Hepat. 2000;7:258–267. doi: 10.1046/j.1365-2893.2000.00236.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Correlation between serum HBV DNA level, and HBsAg status and severity of inflammation and stage of fibrosis of liver.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


