Abstract
The current Ebola virus disease outbreak in West Africa has revealed serious shortcomings in national and international capacity to detect, monitor, and respond to infectious disease outbreaks as they occur. Recent advances in diagnostics, risk mapping, mathematical modelling, pathogen genome sequencing, phylogenetics, and phylogeography have the potential to improve substantially the quantity and quality of information available to guide the public health response to outbreaks of all kinds.
Introduction
The Ebola virus disease (EVD) epidemic in West Africa exemplifies how gaps in capacity for early detection and effective management of an infectious disease outbreak can contribute to a public health crisis. Overcoming these gaps is a global public good with benefits that accrue beyond the boundaries of the country first affected (1). Surveillance—defined in the 2005 International Health regulations (2) as “the systematic, ongoing collection, collation and analysis of data for public health purposes and the timely dissemination of public health information for assessment and public health response as necessary”—is a critical component of outbreak management. Technological advances in diagnostic tools, genome sequencing, computing power and communications devices can augment traditional surveillance methods to accrue and disseminate information in real time, offering the possibility of better outbreak management and thereby saving lives. Lessons from Ebola as well as other infectious diseases, such as influenza and Middle East respiratory syndrome (MERS), may guide the integration of these technologies for successful disease surveillance.
Detection and Monitoring
Most infectious disease outbreaks are first detected through clinical investigation by vigilant frontline healthcare workers. However, clinical surveillance can be an unreliable tool for outbreak detection and monitoring for a number of reasons. Inadequate surveillance and/or reporting systems, a major issue for EVD in West Africa (3), may lead to delayed detection and substantial under-reporting. Misdiagnosis, such as the misdiagnosis of sleeping sickness as malaria, may have fatal consequences for the patients concerned (4). Also, mild or subclinical may not be detected and/or reported to the health services. Such cases accounted for the great majority of infections with pandemic H1N1 influenza in 2009. Indeed, an entirely clinically-orientated view can massively underestimate the burden of infection, leading to inaccurate empirical estimates of the scale and trajectory of an outbreak and compromising outbreak management.
One solution to these problems is the development and deployment of rapid, point-of-care (POC) diagnostic tests, linked to modern information technology (5). For acute infections, improving detection times by as little as 24 hrs, or even less, can make a critical difference to our ability to contain an outbreak (6). The focus of POC testing is to generate rapid results that meet WHO “ASSURED” criteria (Affordable, Sensitive, Specific, User-friendly, Rapid & Robust, Equipment-free, and Delivered – to which we would add ‘Connected’). Suitable platforms are already available for application to a range of viral, bacterial and protozoal infections. These include nucleic acid amplification techniques (NAATs) that encompass thermal, PCR-based tests, isothermal methods (perhaps more suitable for field epidemiology), enzyme immunoassays and immuno-chromatographic tests (5). Although rapid POC testing has yet to play a substantial role during any major infectious disease epidemic, it is currently being evaluated for dengue and influenza virus and is likely to be increasingly important in the future.
There are also technologies that are of limited use for clinical care but of great value for epidemiological surveillance, providing estimates of cumulative exposure at the population level. One such approach is sero-surveillance, the use of serological tests for screening an at-risk population. Sero-surveillance has been used to estimate levels of exposure to H5N1 influenza A (7) and MERS-CoV (8). Sero-surveillance during the H1N1 influenza pandemic gave estimates of 30-40% population exposure in many countries (9), far higher than clinical surveillance indicated. Protocols for the rapid development and deployment of serological tests have been proposed for influenza (10) and could, in principle, be designed for other infections.
Monitoring indirect markers of disease activity, such as internet use and activity on social media may also contribute to epidemiological surveillance. However, an early warning system to detect influenza outbreaks (Google Flu Trends) did not detect the arrival of pandemic H1N1 in the USA in 2009, and the challenge for internet-based and social media-based surveillance systems is to develop methods good enough to be used as surrogates for clinical data (11). However, other new technologies, such as real-time sequencing and mathematical modelling, may be ready for integrating into surveillance systems.
Real-Time Sequence Data
Probably the most important addition to the arsenal of tools for outbreak investigation and guiding public health interventions is the production and use of time-resolved and geo-located pathogen genome data. Over the last decade not only has a deep understanding and a detailed evolutionary framework been developed for, in particular, virus genetics (12), but powerful computational tools and high throughput methods for producing virus genomes are now available.
Large-scale sequencing has been used extensively as a research tool, especially in the fields of HIV and influenza. HIV sequences for parts of the virus genome conferring drug resistance have been routinely determined as part of clinical patient management for nearly two decades, with peripheral blood samples being taken for virus genome load also being used for determining HIV protease and reverse transcriptase sequences for prediction of likely drug sensitivity or resistance (13). When organised nationally, such sequences can be linked under appropriate data governance and ethics to other clinical and demographic data. From this, the sequences can inform transmission network analysis (14) and HIV infection dynamics (15). For influenza viruses, large-scale sequencing of virus isolates, linked to geo-location, provides a rich and detailed insight into global influenza virus transmission both in humans and in animal species (16).
HIV and influenza virus both illustrate that access to and analysis of large numbers of samples (typically hundreds or thousands) is essential. These samples need to be collected without additional sampling of the patient or specialist processing of samples where they are obtained. Fortunately, clinical samples are processed into virus nucleic acid either manually or on robot systems with as little as 20% of the virus nucleic acid used in the diagnostic PCR. It is at this point, when all the costs and logistics associated with diagnosis have been met, that virus genomes can be retrieved from the sample. In short, residual clinical diagnostic nucleic acid should never be discarded before the option of converting to a pathogen genome has been considered.
In practice, full-length virus genomes are not always required. Partial virus genomes that are not ‘finished’ may provide all the information required for molecular epidemiology. A range of genome criteria should be considered in producing high value or Actionable Virus Genomes (AVGs) (17). The important addition here is a set of criteria for assessing the quality of the assembled genomes and the desire to limit these criteria to the majority/consensus pathogen genome, rather than the requiring accurate reporting of minority sequence variants in the sample. This strategy has been shown to work in practice for both MERS-CoV and ebolavirus (Box 1).
Box 1. Success rates for obtaining ‘actionable’ virus genomes (AVGs) from clinical samples by next generation sequencing (NGS).
One crucial aspect of using NGS during an outbreak or during epidemic surveillance is the estimation of the number of samples that are required for sequencing. The number of genomes required per number of cases and per time unit in an epidemic trajectory is a topic of on-going research but the drive for larger datasets as a buffer against incomplete knowledge of what is required to inform an analysis is clear. The only case study for the use of NGS in an outbreak is provided for MERS-CoV (46). At the time of writing, the genome-to-case ratio is approximately 1:10, and this appears just adequate in that context. In order to achieve this number of genomes it is necessary to understand the success rate of achieving a high quality genome, here defined as a genome with greater that 50% total genome coverage. For MERS-CoV ~40 of 112 (~36%) clinical samples yielded useful genomes (47). Tracheal aspirates and bronchoalveolar lavage specimens yielded significantly higher MERS-CoV genome loads and genome sequenced fractions than sputum and nasopharyngeal swab samples. When stratified by the most productive sample type 22/33 samples yielded useful genomes. For comparison, for an outbreak the size of Ebola (~20 000 cases), to sample at the same 1:10 ratio level would require ~2000 genomes, which at a 67% success rate this would require the processing and sequencing of over 3000 samples, a number that should be attainable. The issue, however, is speed: obtaining 2000 genomes over the first 10 months of the outbreak would require a steady 200 genomes from 300 samples per month, a small amount of sequencing but a major logistical and political undertaking.
Analysis of Sequence Data
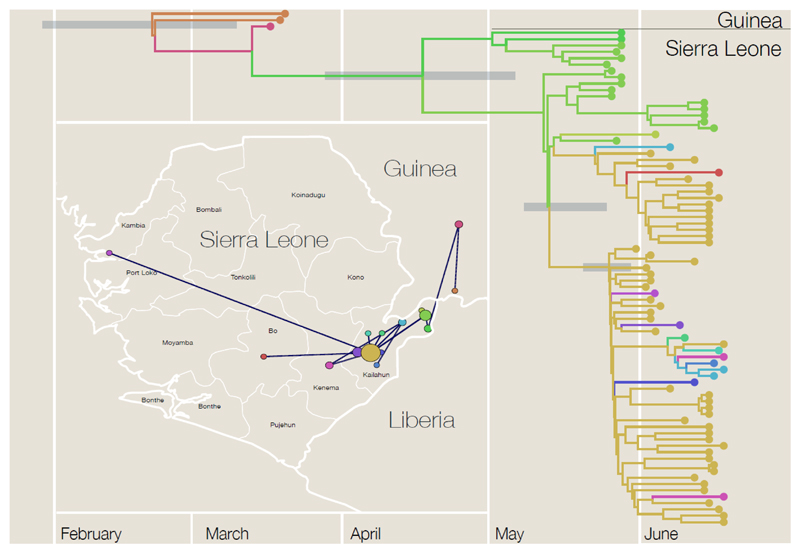
In recent years there has been a profusion of methods that link virus gene sequences with other information to reveal the evolutionary and epidemiological dynamics of the virus. The critical data are dates of sampling of the virus, which transform a phylogeny from a classification procedure into an epidemiological tool (Figure 1). With a time axis the branching events represent transmissions between hosts and thus the times between these events can be used, in a mathematical model, to learn about the key parameters for the outbreak.
Figure 1.
Time-scaled phylogenetic tree based on ebolavirus sequences from ref. (25) from Kenema Government Hospital, Sierra Leone, May and June 2014 plus early samples from Guinea from ref. (23). Branch colours represent probable location of infection with the corresponding locations shown in the inset map. In the map, the radius of the circles denotes the number of sampled sequences and the lines represent the phylogenetic tree projected onto the map.
For many infectious disease outbreaks, estimates of the sampling proportion (the proportion of the epidemic the sample viruses represent), may be the most crucial inferences to be made, revealing the extent of the hidden epidemic due to sub-clinical cases or otherwise unreported cases. Another key motivation for the collection of virus sequence data is to understand the relationship between human cases and an animal reservoir. An important example is MERS Co-V (18), where phlyogenetic analysis of virus sequences obtained from camels, particularly camels with no link to a human case, suggests the directionality of transmission from camel to human (19). Virus sequence analysis can also model how the virus spreads through space and time, using either individual locations of sampled individuals, at the level of map coordinates and assuming that movement of the virus is through a process of diffusion (20) or by treating geography as a limited set of discrete locations (e.g. cities) and interpreting movements as jumps between them that occur at particular rates (21). These approaches can be equally used to investigate the evolution of phenotypic traits of viruses, such as host switching (19) or virulence, resistance, or the antigenic evolution of influenza (22).
The 2009 H1N1 influenza A pandemic was remarkable for being the first serious outbreak to be tracked in real-time by virus genetic data, using data provided by the US Centers for Disease Control and Prevention (CDC) within days of samples being taken from suspected cases. These data were shared as part of the Global Initiative on Sharing All Influenza Data (GISAID) which had been set up a few years before to encourage the exchange of influenza data. However, no similar initiatives exist for other viruses with epidemic potential.
Virus genome sequencing of the earliest EVD cases from Guinea attributed the outbreak in West Africa to the species ebolavirus within weeks of the first cases being diagnosed (23). The genetic similarity to viruses that had previously caused human outbreaks in Central Africa provided an expectation of the epidemiological and pathological properties of the virus: the Zaire species of ebolavirus had caused 14 documented outbreaks of no more than a few hundred cases but with a case fatality rate of up to 90%. However, even though this was a known virus, the outbreak occurred in an unexpected geographical area and in a population that had a very different demography from previous outbreaks (24).
In June 2014, the Broad Institute in collaboration with partners at the Kenema Government Hospital in Sierra Leone shared 78 virus genome sequences from patients that had presented with EVD in the preceding weeks. These provided information on the rate of evolution and revealed no evidence of virus adaptation to humans (25), a major concern at the time. Another important finding was that the epidemic was not being driven by multiple zoonotic transfers from an animal reservoir. These sequences provided crucial insights into the virus just at a time when the outbreak was growing rapidly. The publication of these sequences (25) inspired a series of analytical papers extracting additional inferences about the outbreak including estimates of epidemiological parameters such as the case reproduction rate, infectious period and sampling fraction (26), and the identification of lineages of potential epidemiological significance (27). Estimates of the case reproduction rate (similar to R0 during the early phase of an outbreak) were broadly in line with epidemiological estimates, providing helpful confirmatory evidence given concerns over the reliability of case reporting data. However, given that the epidemic had grown to the point where hundreds of cases per week were being reported from the three affected countries by the time these studies were published (in October and November 2014), the results had limited practical value.
Mathematical Modelling
Mathematical modelling is an established tool in infectious disease epidemiology (28). Real-time projections of case numbers using mathematical models have been provided during many epidemics in the past three decades, including EVD (29,30). At a minimum, actionable projections require: i) an appropriate model framework that captures heterogeneities in risks of infection and rates of transmission; ii) appropriate methods for model parameterisation; iii) rapid access to infection and disease data. Recent applications of mathematical and statistical models to project the course of the 2014 EVD epidemic provide instructive examples. Two studies (29,30) were based on the standard compartment model framework (28), extended to allow for heterogeneous transmission related to clinical disease, hospitalization and funerals, and calibrated against early case data. Another study (31) fitted both regression and branching process models to clinical case data. A variant of the latter approach incorporated separate probability distributions for different transmission routes, resulting in a multi-type branching process model (32) (Figure 2). Together, these make for a set of very different modelling approaches, but all are essentially extrapolations, implicitly assuming near-exponential growth of the epidemic.
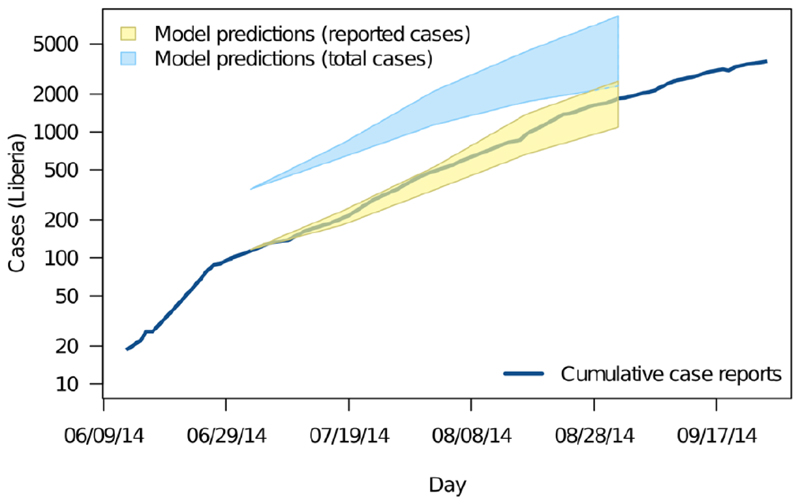
Figure 2.
Projected numbers of cases of EVD in Liberia in 2014 obtained using a branching process model with an ensemble of plausible parameter values. 95% prediction intervals from 07/04/14 (yellow shading) are compared with observed cumulative case numbers (logarithmic scale) over the following two months (blue line). 95% prediction intervals for a model that incorporates estimated levels of under-reporting are also shown (blue shading). Reproduced with authors’ permission from ref. (32).
Accurate projections depend to a large degree on accurate parameter estimation, not least because exponential processes are highly sensitive to exact parameter values. Two key parameters are R0 (the average number of secondary cases generated by a single primary case introduced into a previously unexposed population) and the generation time (the average time between initial infection of a case and of cases it gives rise to) (28). Together, R0 and the generation time determine the doubling time of an outbreak during the early, exponential phase. However, exponential growth during the early stages of an outbreak is not expected in all circumstances (33), such as when R0≈1, a realistic scenario for which large outbreaks (hundreds of cases or more) is entirely possible (34) or when there are multiple introductions separated in time and space, some of which die out due simply to demographic stochasticity. This was the case with pandemic H1N1 influenza A in Scotland in 2009, as indicated by the analysis of virus sequence data (35). The history of ebolavirus in Liberia in 2014 may also have involved multiple introductions (32) but, again, this can only be confirmed from virus genome sequence data.
Though short-term projections are feasible for many outbreaks, extrapolation methods are much less useful in the longer term. This is partly because the confidence intervals on the projections quickly become very wide (Figure 2) but, more fundamentally, because the exponential growth assumption breaks down as the epidemic progresses (often with the introduction of control measures). This underlines the need for clear communication of how model outputs – particularly “worst case” scenarios – should be interpreted (3).
Several initiatives (e.g. 36) aim to increase the availability of open access tools kits for epidemiological modelling—both for model parameterisation and development. Indeed, formal, robust and rapid model fitting procedures, generally based on maximum likelihood or Markov chain Monte Carlo (McMC) methods (37), are being developed to replace the ad hoc approaches – “calibration” or “tuning” – which are still often utilised in practice (33).
One potentially useful approach is pattern oriented modelling (POM) (38). POM is a technique used originally in ecological modelling both to distinguish between possible model structures and to reduce parameter uncertainty. POM identifies models that reproduce a set of pre-selected patterns observed in the data – whether qualitative or quantitative. The ability to consider multiple patterns and different kinds of data simultaneously greatly increases both discriminatory power and flexibility. It also addresses a legitimate reluctance to apply very precise model fitting procedures to poor quality disease data. POM has only rarely been applied to infectious diseases (38) but a very similar approach has been used to parameterise a model of EVD cases, generating encouragingly precise estimates of a set of seven different parameters (32).
Risk Mapping
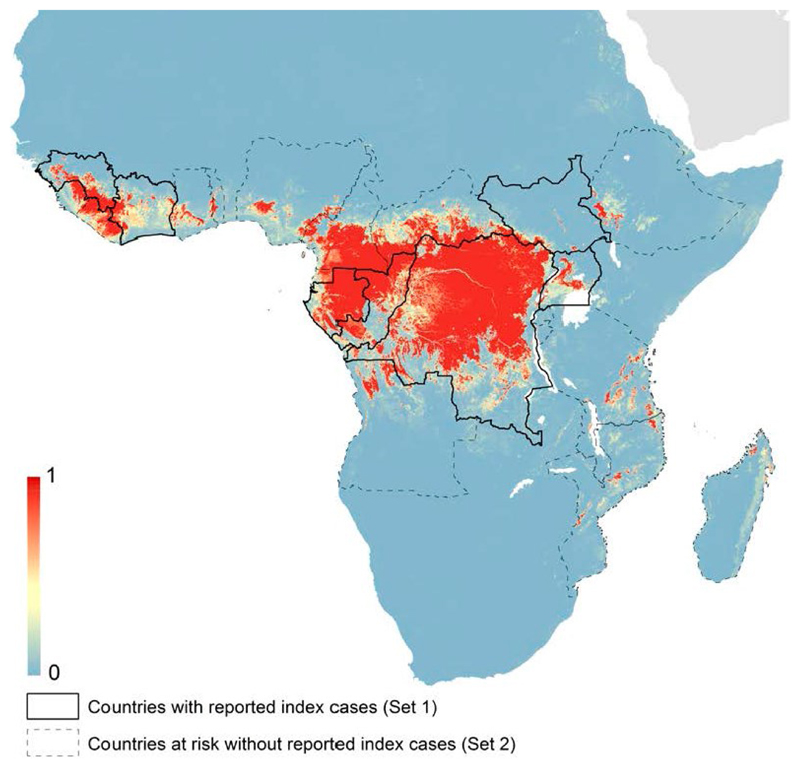
Risk mapping has been applied to a range of diseases, including EVD in Africa. The EVD risk map (24) incorporated a set of predictors including elevation, an index of vegetation cover, other environmental variables, and estimated composite distribution data for three bat species suspected to be reservoirs of Ebola virus. The output (Figure 3) suggests that several countries, notably Nigeria and Cameroon, are at risk of EVD but lie outside its currently reported range.
Figure 3.
Predicted probability distribution (blue=low, red=high) of zoonotic EVD cases in Africa based on a risk mapping analysis and highlighting at-risk countries with and without index cases reported up to 2014. Reproduced with authors’ permission from ref. (24).
Spatial risk analyses are restricted to predictors for which spatial data are available. In resource-poor settings this often equates to data available via remote sensing. Analyses are also limited by the quantity and quality of the disease data used to calibrate the models, in particular the issue of ascertainment bias, i.e. “pseudo-absence” at locations where health reporting is unreliable. The utility of the models is determined to a large extent by how well they deal with this issue. However, even with these limitations, risk maps provide information that helps direct national and international surveillance efforts and contributes to planning and preparedness between outbreaks.
Applications of Modelling
Outbreak size distribution analysis has been used successfully to monitor the epidemiology of measles in the UK following a fall-off in childhood vaccination rates in the late 1990s, charting the approaching loss of herd immunity through shifts in the size and frequency of small outbreaks (39). It has also been applied to monkeypox (40), anticipating a possible increase in monkeypox transmissibility as the fraction of the population immunised against smallpox dwindles. It has recently been applied, using McMC techniques, to EVD (34), confirming that prior to 2013 R0 for ebolavirus in humans was close to, or possibly above, 1, indicative of a high risk of major epidemics.
Modelling can aid in predicting the impact of so-called ‘reactive’ control measures (6) on the course of an infectious disease outbreak. For example, the expected impact of case isolation and/or quarantine of at-risk individuals on the course of an outbreak is determined, inter alia, by the relative timings of a case becoming infectious (and thus potentially transmitting infection to others) and being detected, typically following the appearance of clinical signs (6). Surprisingly, such basic information on the time course of an infection is often lacking, even for well-studied infections such as influenza, and there is a need for greater investment in experimental studies to fill this gap. This example illustrates a wider concern: many public health interventions are designed to reduce pathogen transmission rates and neither their intended nor actual impact can be quantified without reference to changes in transmission rates; however, research on pathogen transmission consumes a miniscule fraction of research effort expended on infectious diseases, the bulk of which is aimed at understanding and preventing infection and pathology.
Another consideration, all-too-often ignored until an outbreak occurs, is the logistic capacity of the affected health system to respond. For the West African EVD epidemic a key issue was the capacity to roll out isolation units fast enough to ‘catch up’ the epidemic curve (3). However, similar arguments apply more generally to the capacity to administer drugs, vaccines or any other reactive measures that contribute to reducing the net rate of transmission. In this context, models can help quantify an “effective” response. For EVD, models indicated that hospital capacity and individual behaviour (particularly social distancing) were particularly important (32).
Parameterising the variables that capture both the intended and the actual impact of interventions can be extremely difficult (33). There is a need firstly to monitor the implementation of interventions (noting that targets set by policy makers do not always correspond to events on the ground) and secondly to analyse these data in real time in order to evaluate their impact. These activities require resources and are often neglected. Moreover, many of the measures that may be taken have effects, particularly on the rate of transmission, that are difficult to quantify. Examples include the wearing of face masks and social distancing (i.e. reducing the risk of infection by changing patterns of contact with the rest of the population, whether in response to public health warnings or through individual initiative).
An important general principle that emerges from the infectious disease modelling literature is that there are substantial benefits arising from the implementation of reactive control measures as early as possible (6). This is a straightforward consequence of the expectation that absolute numbers of cases will increase exponentially during the early stages of an outbreak. Indeed, during this phase the costs of delay also increase over time; for an acute infection such as ebolavirus, each week’s delay permits a greater number of extra cases than the previous week (3).
Practical Steps
The call for better surveillance systems has been made repeatedly during the past decade (41), but there has been too little effective change on the ground (42). One of the most important barriers to the modernisation of infectious disease surveillance systems is that non-traditional approaches are all-too-often seen as an unnecessary distraction from immediate health needs, particularly during an emergency when resources are likely to be severely stretched. This can be exacerbated by real or perceived gaps in technical capacity and expertise (a health emergency is not the best time to be learning new techniques), and by those involved in collecting samples and data (sometimes in extremely challenging circumstances) being disconnected from the subsequent work that depends on their efforts. The best way to remove such barriers to adoption may be to promote a wider appreciation of what is possible, how it can be achieved and the immediate benefit to public health.
It has been argued that improving global surveillance for emerging infectious diseases is feasible and cost-effective (2) but substantial investment in infrastructure, technology, training and organization is required. Ultimately, improved global surveillance will emerge from strengthening and connecting national surveillance systems. Similar kinds of investment are needed to strengthen national and international capacity to respond effectively to infectious disease events, and there is an ongoing discussion in the light of the current EVD epidemic as to whether that should include an international rapid response force (1). In addition, there is a need for a greater investment in health policy and systems research, an underfunded and unappreciated field that has a central role to play in meeting the challenge of achieving effective infectious disease surveillance and outbreak management on a global scale.
Any response to an infectious disease outbreak, and especially a coordinated international effort, is contingent not just on the presence of functional national surveillance systems but on the rapid sharing of information between countries and with international agencies. The revolution in information and communications technology that has occurred over the past 20-30 years has removed virtually all technological barriers to this process, even in remote, resource-poor settings. Moreover, as several of the above examples illustrate, it is now routine to integrate and analyse data from multiple sources, such as public health, demographic, location (e.g. global positioning system), movement, geographic, animal distribution, remote sensing and genome sequence data.
Arguably the biggest remaining barrier to real-time data sharing is cultural, reflecting a reluctance to report disease events. This can be for a number of reasons, not least fear of the imposition of restrictions on freedom of movement or trade, or of adverse effects on tourism and investment (2). The 2005 International Health Regulations provide a framework for disease reporting, but do not directly address the question of disincentives, and their implementation has been very patchy to date (42). An obvious solution is to balance the negative consequences of reporting with the promise of effective assistance.
For maximum benefit, data sharing should be as rapid and as open as possible. Again, there are few if any technological barriers to this: data and information sharing platforms such as GenBank, Dryad and ArXiv have been available for many years. However, although lines of reporting from front-line health official to international agencies are fairly well set out (2), there is no agreement on responsibility for data sharing and all too often this is left to individual or institutional preference. One approach is to penalise countries that do not implement and report from an adequate surveillance system, as was required for participation in the international cattle trade during the BSE epidemic.
One possible consequence of data sharing is a proliferation of analyses of those data, as was seen during the 2009 H1N1 influenza A pandemic and during the current EVD epidemic. While we regard this as a positive development, it can have perceived disadvantages, notably a loss of control by national or international agencies, and as creating uncertainty over which analyses should be trusted. These issues are not insurmountable and should not be regarded as obstacles to data sharing. In other fields, notably climate change, an ensemble approach to data analysis, interpretation and projection has been the norm for many years (43). Although this is challenging for many infectious diseases, if only because of the much shorter timescales involved, suitable systems are already in place and there has been, for example, real-time evaluation of multiple models of pandemic influenza (44).
Many of the practical aspects of preparedness for an infectious disease outbreak can and should be addressed in advance of a crisis. These include: contingency planning and coordination; developing and stockpiling diagnostics, drugs, and vaccines; setting up sequencing pipelines; designing data-sharing protocols; constructing, verifying and validating mathematical models; agreeing reporting and communication pathways; and anticipating public engagement and ethical issues. One approach to this is to set up sentinel cohorts. This ensures that data collection and reporting (including self-reporting) systems are all in place and tested in advance of an outbreak. Importantly, it would also cover ethical requirements. Ethical considerations both delayed and limited surveillance in the UK during the 2009 H1N1 influenza A pandemic (45) and can be difficult to deal with rapidly even during a major emergency, as recent experience with trials for ebolavirus vaccines illustrates.
To facilitate the provision of virus genomes, we would propose an approach similar to the WHO’s Pandemic Influenza Preparedness Framework for the sharing of influenza viruses and access to vaccines and other benefits (or PIP Framework), an international arrangement that brings together key stakeholders to strengthen preparedness for the next influenza pandemic. This has now been extended to address sequencing data though a Technical Expert Working Group, with the overall PIP framework encouraging collaborative, transnational working under a framework of a more structured, efficient and equitable system.
We also need to recognize that managing infectious disease of all kinds is a multi-disciplinary problem and, if it is to be done as effectively as possible, requires input from beyond traditional clinical medicine and public health. An integrated, global infectious disease surveillance system needs to take a One Health approach and embrace livestock and wildlife health, as well as geography and environmental sciences, sociology, economics and anthropology, informatics, communications science and health technology.
The surveillance systems that are set up also need to be flexible and responsive. The infectious disease threat is diverse and dynamic, and periodically presents “out-of-the-blue” challenges such as BSE/vCJD in the 1980s or SARS in the 2000s.
Conclusions
We can readily identify the components of a surveillance system that would enable the collection of infectious disease surveillance data from multiple sources for use as inputs into state-of-the-art epidemiological analysis (Figure 4). Advances in diagnostics, sequencing platforms, communications technology, and computing and informatics over the past 5-10 years mean that such analyses can now make an effective contribution to outbreak management in real time. This is a highly significant new capability that we should exploit fully in order to improve the public health response to future infectious disease outbreaks. A cultural shift is required among health care workers such that these activities come to be regarded as a valuable compliment to the clinical care of individual patients, and not as unwelcome competition for resources, time and effort.
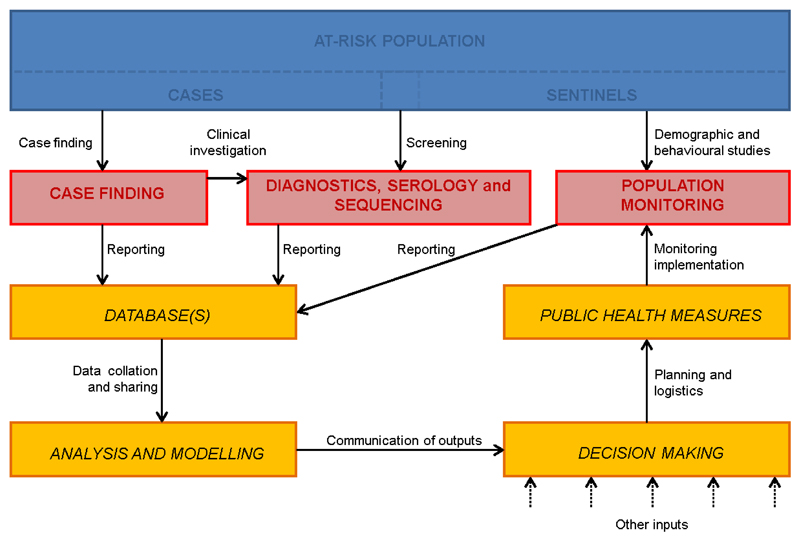
Figure 4.
Key elements of data capture and information flows for real-time quantitative analysis to inform outbreak management. The at-risk population encompasses cases and, where available, a sentinel subpopulation (blue boxes). Three types of data capture activities are identified (red boxes): case finding (including associated epidemiological investigations such as contact tracing); diagnostic information on individual patients, including serological testing and pathogen sequencing; and so-called ‘denominator’ studies on the population at risk, including demography, behaviour, e.g. social media activity, and the impact of health measures. Information flows (yellow boxes) involve communication between data gatherers, data analysts and modellers, policy makers and public health authorities. We note, however, that decision making never relies solely on the outputs of real-time epidemiological analyses.
Strengthening surveillance and response capacity around the world would require investment estimated at tens of billions of dollars per annum, but is likely to be cost-effective. Moreover, capacity strengthening should not be the sole responsibility of individual countries; we emphasize that infectious disease surveillance is a global good and should be financed on that basis. We suggest that not all elements of a state-of-the-art surveillance system need to be replicated at a national level; it will often be much more efficient to integrate local activities into an international network. However, this would require considerably more proactive leadership of global surveillance efforts than exists at present. Ultimately, there will be little progress without strong and trusted international governance systems.
Summary.
A maximally effective public health response to infectious disease outbreaks requires the collection, communication and analysis of multiple data types within the framework of an integrated surveillance system.
Acknowledgments
We thank colleagues involved with VIZIONS, COMPARE and Edinburgh Infectious Diseases for helpful discussions and Catriona Waugh for assistance with manuscript preparation.
Funding: This work was supported by a Wellcome Trust Strategic Award (VIZIONS), with additional support from an EU Horizon 2020 grant (COMPARE), the UK Department of Health, the Wellcome Trust (grant no. WT098608) and the Health Innovation Challenge Fund (HICF-T5-344).
Footnotes
Author contributions: MW, AR and PK co-wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Woolhouse MEJ, Drury P, Dye C. Zero infection. Science. 2014;346:1271. doi: 10.1126/science.aaa4117. [DOI] [PubMed] [Google Scholar]
- 2.Davies SE, Youde JR. In: The politics of surveillance and response to disease outbreaks: The new frontier for states and non-state actors. Davies SE, Youde JR, editors. Ashgate Publishing; Surrey: 2015. pp. 9–23. [Google Scholar]
- 3.Whitty CJM, Farrar J, Ferguson N, Edmunds WJ, Piot P, Leach M, Davies SC. Infectious disease: Tough choices to reduce Ebola transmission. Nature. 2014;515:192–194. doi: 10.1038/515192a. [DOI] [PubMed] [Google Scholar]
- 4.Odiit M, Coleman PG, Liu W-C, McDermott JJ, Fèvre EM, Welburn SC, Woolhouse MEJ. Quantifying the level of under-detection of Trypanosoma brucei rhodesiense sleeping sickness cases. Trop Med Int Health. 2005;10:840–849. doi: 10.1111/j.1365-3156.2005.01470.x. [DOI] [PubMed] [Google Scholar]
- 5.Clerc O, Greub G. Routine use of point-of-care tests: Usefulness and application in clinical microbiology. Clin Microbiol Infect. 2010;16:1054–1061. doi: 10.1111/j.1469-0691.2010.03281.x. [DOI] [PubMed] [Google Scholar]
- 6.Charleston B, Bankowski BM, Gubbins S, Chase-Topping ME, Schley D, Howey R, Barnett PV, Gibson D, Juleff ND, Woolhouse MEJ. Relationship between clinical signs and transmission of an infectious disease and the implications for control. Science. 2011;332:726–729. doi: 10.1126/science.1199884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TT, Parides MK, Palese P. Seroevidence for H5N1 influenza infections in humans: Meta-analysis. Science. 2012;335:1463. doi: 10.1126/science.1218888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, Bagato O, Siu LY, Shehata MM, Kayed AS, Moatasim Y, Li M, et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574. doi: 10.2807/1560-7917.es2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 9.Kelly H, Peck HA, Laurie KL, Wu P, Nishiura H, Cowling BJ. The age-specific cumulative incidence of infection with pandemic influenza H1N1 2009 was similar in various countries prior to vaccination. PLoS One. 2011;6:e21828. doi: 10.1371/journal.pone.0021828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurie KL, Huston P, Riley S, Katz JM, Willison DJ, Tam JS, Mounts AW, Hoschler K, Miller E, Vandemaele K, Broberb E, et al. Influenza serological studies to inform public health action: Best practices to optimise timing, quality and reporting. Influenza Other Resp Viruses. 2013;7:211–224. doi: 10.1111/j.1750-2659.2012.0370a.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson DR, Konty KJ, Paladini M, Viboud C, Simonsen L. Reassessing Google Flu Trends data for detection of seasonal and pandemic influenza: A comparative epidemiological study at three geographic scales. PLoS Comput Biol. 2013;9:e1003256. doi: 10.1371/journal.pcbi.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes EC, Grenfell BT. Discovering the phylodynamics of RNA viruses. PLoS Comput Biol. 2009;5:e1000505. doi: 10.1371/journal.pcbi.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee S-Y, Gonzales M, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hue S, Brown AE, Ragonnet-Cronin M, Lycett SJ, Dunn DT, Fearnhill E, Dolling DI, Pozniak A, Delpech VC, Leigh Brown AJ. Phylogenetic analyses reveal HIV-1 infections between men misclassified as heterosexual transmissions. AIDS. 2014;28:1967–1975. doi: 10.1097/QAD.0000000000000383. [DOI] [PubMed] [Google Scholar]
- 15.Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis. 2011;204:1463–1469. doi: 10.1093/infdis/jir550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung C-L, Raghwani J, Bhatt S, Peiris JSM, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 17.Ladner JT, Beitzel B, Chain PSG, Davenport MG, Donaldson E, Frieman M, Kugelman J, Kuhn JH, O’Rear J, Sabeti PC, Wentworth DE, et al. Standards for sequencing viral genomes in the era of high-throughput sequencing. mBio. 2014;5:e01360–14. doi: 10.1128/mBio.01360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah AA, Corman VM, Sieberg A, Makhdoom HQ, Assiri A, Masri MA, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faria NR, Suchard MA, Rambaut A, Streicker DG, Lemey P. Simultaneously reconstructing viral cross-species transmission history and identifying the underlying constraints. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120196. doi: 10.1098/rstb.2012.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemey P, Rambaut A, Welch JJ, Suchard MA. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol. 2010;27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, Russell CA, Smith DJ, Pybus OG, Brockmann D, Suchard MA. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 2014;10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, McCauley JW, Russell CA, Smith DJ, Rambaut A. Integrating influenza antigenic dynamics with molecular evolution. eLife. 2014;3:e01914. doi: 10.7554/eLife.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, De Clerc H, Tiffany A, et al. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 24.Pigott DM, Golding N, Mylne A, Huang Z, Henry AJ, Weiss DJ, Brady OJ, Kraemer MUG, Smith DL, Moyes CL, Bhatt S, et al. Mapping the zoonotic niche of Ebola virus disease in Africa. eLife. 2014;3:e04395. doi: 10.7554/eLife.04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gire SK, Goba A, Andersen KG, Sealfon RSG, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stadler T, Kühnert D, Rasmussen DA, du Plessis L. Insights into the early epidemic spread of Ebola in Sierra Leone provided by viral sequence data. PLoS Curr. 2014;6 doi: 10.1371/currents.outbreaks.02bc6d927ecee7bbd33532ec8ba6a25f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Łuksza M, Bedford T, Lässig M. Epidemiology and evolutionary analysis of the 2014 Ebola virus outbreak. arXiv preprint. 2014 arXiv: 1411.1722. [Google Scholar]
- 28.Anderson RM, May RM. Infectious diseases of humans: Dynamics and control. Oxford University Press; New York: 1991. [Google Scholar]
- 29.Pandey A, Atkins KE, Medlock J, Wenzel N, Townsend JP, Childs JE, Nyenswah TG, Ndeffo-Mbah ML, Galvani AP. Strategies for containing Ebola in West Africa. Science. 2014;346:991–995. doi: 10.1126/science.1260612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomes MFC, Pastore y Piontti A, Perra N, Samay N, Zhang Q, Vespignani A, Rossi L, Quaggiotto M, Panisson A, Delfino M, Cattuto C, et al. Assessing the international spreading risk associated with the 2014 West African Ebola outbreak. PLoS Curr. 2014 doi: 10.1371/currents.outbreaks.cd818f63d40e24aef769dda7df9e0da5. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.WHO Ebola Response Team. Ebola virus disease in West Africa — The first 9 months of the epidemic and forward projections. N Engl J Med. 2014;371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drake JM, Kaul RB, Alexander LW, O'Regan SM, Kramer AM, Pulliam JT, Ferrari MJ, Park AW. Ebola cases and health system demand in Liberia. PLoS Biol. 2015;13:e1002056. doi: 10.1371/journal.pbio.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowell G, Nishiura H. Characterizing the transmission dynamics and control of Ebola virus disease. PLoS Biol. 2015;13:e1002057. doi: 10.1371/journal.pbio.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.House T. Epidemiological dynamics of Ebola outbreaks. eLife. 2014;3:e03908. doi: 10.7554/eLife.03908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lycett S, McLeish NJ, Robertson C, Carman W, Baillie G, McMenamin J, Rambaut A, Simmonds P, Woolhouse MEJ, Leigh Brown AJ. Origin and fate of A/H1N1 influenza in Scotland during 2009. J Gen Virol. 2012;93:1253–1260. doi: 10.1099/vir.0.039370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grefenstette JJ, Brown ST, Rosenfeld R, DePasse J, Stone NTB, Cooley PC, Wheaton WD, Fyshe A, Galloway DD, Sriram A, Guclu H, et al. FRED (A Framework for Reconstructing Epidemic Dynamics): An open-source software system for modeling infectious diseases and control strategies using census-based populations. BMC Publ Health. 2013;13:940. doi: 10.1186/1471-2458-13-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinley TJ, Ross JV, Deardon R, Cook AR. Simulation-based Bayesian inference for epidemic models. Comput Stat Data Anal. 2014;71:434–447. [Google Scholar]
- 38.Mitchell KM, Mutapi F, Savill NJ, Woolhouse MEJ. Explaining observed infection and antibody age-profiles in populations with urogenital schistosomiasis. PLoS Comput Biol. 2011;7:e1002237. doi: 10.1371/journal.pcbi.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jansen VAA, Stollenwerk N, Jensen HJ, Ramsay ME, Edmunds WJ, Rhodes CJ. Measles outbreaks in a population with declining vaccine uptake. Science. 2003;301:804. doi: 10.1126/science.1086726. [DOI] [PubMed] [Google Scholar]
- 40.Blumberg S, Lloyd-Smith JO. Inference of R0 and transmission heterogeneity from the size distribution of stuttering chains. PLoS Comput Biol. 2013;9:e1002993. doi: 10.1371/journal.pcbi.1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morse SS, Mazet JAK, Woolhouse MEJ, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braden CR, Dowell SF, Jernigan DB, Hughes JM. Progress in global surveillance and response capacity 10 years after severe acute respiratory syndrome. Emerg Infect Dis. 2013;19:864–869. doi: 10.3201/eid1906.130192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Climate change 2007 synthesis report. Intergovernmental Panel on Climate Change; Geneva, Switzerland: 2007. [Google Scholar]
- 44.van Kerkhove MD, Asikainen T, Becker NG, Bjorge S, Desenclos J-C, dos Santos T, Fraser C, Leung GM, Lipsitch M, Longini IM, Jr, McBryde ES, et al. Studies needed to address public health challenges of the 2009 H1N1 influenza pandemic: insights from modeling. PLoS Med. 2010;7:e1000275. doi: 10.1371/journal.pmed.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLeish NJ, Simmonds P, Robertson C, Handel I, McGilchrist M, Singh BK, Kerr S, Chase-Topping ME, Sinka K, Bronsvoort M, Porteous DJ, et al. Sero-prevalence and incidence of A/H1N1 2009 influenza infection in Scotland in winter 2009–2010. PLoS One. 2011;6:e20358. doi: 10.1371/journal.pone.0020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackay IM, Arden KE. Middle East respiratory syndrome: An emerging coronavirus infection tracked by the crowd. Virus Res. 2015:1–29. doi: 10.1016/j.virusres.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Memish ZA, Al-Tawfiq JA, Makhdoom HQ, Assiri A, Alhakeem RF, Albarrak A, Alsubaie S, Al-Rabeeah AA, Hajomar WH, Hussain R, Kheyami AM, et al. Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome. J Infect Dis. 2014;210:1590–1594. doi: 10.1093/infdis/jiu292. [DOI] [PMC free article] [PubMed] [Google Scholar]