Abstract
Introduction
Identification of adenocarcinoma (AC) and squamous cell carcinoma (SCC) histology of non-small cell lung cancer (NSCLC) in biopsies is clinically important but can be inaccurate by routine histopathologic examination. We quantify this inaccuracy at a cancer center, and evaluate the utility of a microRNA-based method to histotype AC/SCC in biopsies.
Methods
RNA was extracted from tissue sections with >90% tumor content that were macro- or micro-dissected from formalin-fixed, paraffin-embedded biopsy specimens. MicroRNAs in RNA from the biopsies and from resected tumors were quantified by TaqMan™ RT-PCR assays and normalized against the RNU6B housekeeping RNA. Publicly available microRNA expression data-sets were examined.
Results
NSCLC subtyping of small biopsy specimens by routine histopathologic examination either failed or mistyped the histology of 21% of 190 cases. Using 77 resectates, an RT-PCR-based assay of microRNAs miR-21, miR-205 and miR-375 was developed to identify AC and SCC subtypes of NSCLC. This method identified the AC/SCC histotypes of 25 biopsies with an accuracy of 96%, and correctly histotyped all 12 cases for which the histology had been mistyped by routine histopathologic examination of the biopsy. Examination of publicly available data-sets identified miR-205 and miR-375 as microRNAs with the best ability to histotype AC and SCC, and that levels of the two microRNAs in AC or SCC are unaffected by the pathologic stage of the tumor or the age or race of the patient.
Conclusions
Histotypic microRNA assays can aid the subtyping of NSCLC biopsies as AC or SCC by standard histopathologic methods.
Introduction
Subtyping of non-small cell lung cancer (NSCLC) to identify adenocarcinoma (AC) and squamous cell carcinoma (SCC) histotypes which constitute ~85% of NSCLC is clinically important. AC is more susceptible to the anti-folate drug pemetrexed whereas the SCC subtype is largely unresponsive to it1, and SCC but not AC patients treated with the anti-angiogenic agent bevacizumab are significantly likely to suffer pulmonary hemorrhage2. Identification of the correct histology also prompts the appropriate evaluation of the tumor for genetic changes that predict response to targeted agents; e.g., for EGFR mutations and ALK gene rearrangement in case of AC but not SCC, and for FGFR1 gene amplification and DDR2 mutation in SCC but not AC3.
Examination of morphology and protein expression is the standard histopathologic method to identify AC and SCC subtypes in small biopsy specimens. While it can be highly accurate with the right protein markers and expertise4–8, AC/SCC subtyping by histopathologic examination of biopsy material in routine clinical settings can be incorrect for 15%–35% cases9–13. A number of studies have demonstrated the histotyping utility of microRNAs for NSCLC14–21, a class of approximately 2500 ultra-short, non-coding cellular RNAs that are also remarkably preserved after formalin fixation of biospecimens22–24. In particular, a reverse transcription (RT)-PCR-based assay of microRNAs miR-205 and miR-21, and the RNU6-2 (U6B) small nucleolar RNA developed by Lebanony and colleagues14 has been successfully used by other groups to identify AC and SCC subtypes of resected specimens with accuracy rates of 90% to 100%21, 25, 26. This method had an accuracy rate of 95% with biopsy material in the one study in which this was examined27.
In this work, we note the inaccuracy of routine histopathologic examination at a cancer hospital for subtyping NSCLC in biopsies by examining the discordance of the histological diagnoses with those made for the resected tumor specimens. We evaluate the performance of the microRNA assay of Lebanony et al. for AC/SCC histotyping of resected tumors. By analyzing publicly available data on microRNA expression in NSCLC tumors, we identify miR-375 as a histotypic microRNA whose measurement can be used to improvise the performance of the Lebanony assay, and we test this improved method with biopsied tumor tissue.
Materials and methods
Statement on ethics
This study was conducted in accordance with the amended Declaration of Helsinki under protocols I129008 and NHR015110 of the Institutional Review Board of Roswell Park Cancer Institute (RPCI).
Analyses of external lung squamous cell carcinoma and adenocarcinoma microRNA expression data-sets
Four studies28–31 were identified to have publicly available, high-throughput expression data for ≥24 each of SCC and AC cases for microRNAs, including miR-21 and miR-205 that are assessed in the Lebanony method14 for AC/SCC subtyping. This identification, and the analyses of microRNA expression data from these studies is described in Appendix 1 and Table S1 in Supplemental Digital Content. Clinical and microRNA expression data generated on Genome Analyzer or HiSeq™ RNA sequencing platforms (Illumina®, San Diego, CA) for AC/SCC cases of the Cancer Genome Atlas (TCGA) project were obtained from the Broad Institute Firehose (data standardization run of 22 February 2013). The edgeR Bioconductor package32 was used in R to normalize microRNA count data for analyzing differential expression with Fisher’s exact test. Characteristics of the TCGA patient cohorts, and processing and analyses of the data are described in Appendix 2, and Tables S2 and S3 in Supplemental Digital Content.
Identification of cohort to evaluate the accuracy of subtyping non-small cell lung cancer by histopathologic examination of biopsies
The tumor registry of Roswell Park Cancer Institute (RPCI) was queried to identify 190 cases of primary non-small cell cancer (NSCLC) of lung parenchyma that were treated by resective surgery at RPCI during 2008–2010, and for whom histopathologic examinations had been performed for both pre-surgery biopsy and surgical resectate tissue specimens, with a diagnosis of NSCLC obtained with the latter. Relevant characteristics of this cohort are noted in Table S4 in Supplemental Digital Content. All resectate specimens were histopathologically examined at RPCI. For about two-thirds of the cases, biopsies had been performed outside RPCI, reflecting the stature of RPCI as a tertiary referral center. When multiple biopsies had been performed for a case, only the result of the biopsy used for final diagnosis was considered. Slides of biopsy specimens were re-examined at RPCI if histopathologic examination of biopsy specimens had been conducted elsewhere, and RPCI’s evaluation of histology was considered for analysis, but for the rare cases for which slides of biopsy specimens were unavailable, the non-RPCI pathologists’ evaluations of histology were used. Histology of a resectate, determined from tissue morphology, and immunohistochemical examinations in some cases, was taken as the true NSCLC histology when evaluating the accuracy of histology determined for the biopsy specimens.
Resectate and biopsy specimens
Paraffin-embedded formalin-fixed specimens for primary AC/SCC tumor biopsies (n = 25) or resectates (n = 77), and clinical data were obtained from RPCI’s Pathology Research and Clinical Data Networks. Relevant characteristics for the specimens are noted in Tables S5 and S6 in Supplemental Digital Content. The 6th American Joint Committee on Cancer tumor staging system was used.
Tissue microdissection and RNA extraction
Areas with >90% cellular tumor content in hematoxylin-eosin-stained 8 μm-thick sections of biopsies were microdissected by laser under a pathologist’s supervision on an LMD6000 system (Leica®, Wetzlar, Germany) as described previously33. Dissected areas averaged 25.6 mm2 (range = 0.9–98.4; standard deviation [SD] = 26.9). Six biopsy specimens with large contiguous areas of tumor were dissected with a scalpel under naked eye. RNA extracted from the dissectates with the FFPE RNA Purification kit (Norgen Biotek®, Thorold, Canada) averaged 0.78 μg (range = 0.03–3.76, SD = 0.97) when quantified using RiboGreen33. RNA extraction from resectates with >70% tumor content using RecoverAll™ (n = 56; Ambion®, Austin, TX) or High Pure™ miRNA (n = 21; Roche®, Indianapolis, IN) has been described29, 34.
MicroRNA reverse transcription (RT)-PCR assays
RNA samples were at −80 °C for a few days to 5 years when assayed (detailed in Tables S5 and S6 in Supplemental Digital Content). TaqMan™ MicroRNA assays35 (Applied Biosystems®, Foster City, CA) 000397, 000509, 000564 and 001093 were respectively used to quantify miR-21-5p, miR-205-5p, miR-375-3p, and RNU6-2 (U6B), a nucleolar housekeeping small RNA, as quantification cycle (Cq) values, which are inversely and logarithmically proportional to analyte concentrations. MiR-21-5p, miR-205-5p and RNU6-2 are the three RNAs that analyzed in the AC-SCC typing method described by Lebanony and colleagues14.
Typically, a tissue RNA sample was simultaneously assayed by RT-PCR for miR-21, miR-205 and miR-375 in the same experiment, and U6B was quantified any time that a microRNA was, with RT reactions performed in one batch and all PCR reactions for an RT batch set up on one 384-well plate. RT batches always included a negative, no RNA control (water), and RT-PCR experiments were repeated if a Cq value was detected and <36 for the negative control. RT-PCR experiments that used tissue RNA also included 1–2 standard RNA samples, in duplicate, for each of the microRNAs, and U6B, that were being quantified in the experiments. The standard samples were dilutions in water of synthetic, 5′ phosphorylated small RNA standards with sequences of the human U6B or mature microRNAs (Life Technologies®) such that an RT reaction had 102–7 molecules of a standard.
For calculation of molarity values from the raw Cq value for a microRNA or U6B for a tissue RNA sample, the Cq value was first adjusted using a calibration factor, and then used in a linear equation derived from Cq values obtained with the synthetic RNA standard at different molarities (Figure S1 in Supplemental Digital Content). The calibration factor was the average deviation of the Cq values seen for an experiment’s standard controls of a certain molarity from the Cq values expected for that molarity from the standard curves. Values of calibration factors are listed in Table S7 in Supplemental Digital Content. Calibration of the raw Cq values for molarity determinations also resulted in inter-experiment calibration of the Cq values. X ΔCqY, measurement of microRNA X relative to that of Y, was calculated by subtracting Cq for Y from that for X. The Lebanony score14 was calculated as miR-205 ΔCqU6B − (miR-21 ΔCqU6B)/2. Cq values and calculations are in Tables S7 and S8 in Supplemental Digital Content.
Other
Unless noted otherwise, Prism™ software (version 6.0b; GraphPad®, La Jolla, CA) was used for data-plotting and statistical analyses, a P value below 0.05 was associated with statistical significance, and groups were compared for categorical and quantitative variables respectively using chi square and standard two-tailed t tests. Areas under receiver operating characteristic (ROC) curves (AUCs) were compared as per DeLong et al.36 using the StAR online tool37.
Results
Inaccurate histotyping of NSCLC biopsies
Among all primary NSCLC cases that were treated during 2008–10 by surgical resection at RPCI, a tertiary referral cancer center, histological subtyping of the cancer by routine pathologic examination of both biopsied and resected tumor tissue specimens was done for 190 (characteristics of the cases are noted in Table S4 in Supplemental Digital Content). Though NSCLC was identified in 184 of the 190 biopsies, its subtype was unidentifiable for 24 (13.0%), and discordant from the subtype diagnosed for the resectate for 15 (9.4%) of the remaining 160 (Table 1). The discordance or inaccuracy rate was higher for cases that had neoadjuvant chemotherapy with or without radiation (33% vs. 17%; P = 0.02), or for which the biopsy method was fine needle aspiration instead of core biopsy (30% vs. 16%; P = 0.02), but was unaffected if cases had sole radiation therapy, bronchoscopy instead of computer tomography for biopsy, or biopsy was performed outside RPCI. Among the 101 AC cases, the AC histotype was missed in biopsy examination of 22 (21.8%), with 4 (18%) of them mistyped as SCC. Mistyping of SCC as AC was 50% among the 12 (17%) cases whose SCC histotype was missed in biopsy examination.
Table 1.
Histology determined by pathologic examination of biopsies of 190 cases of resected non-small cell lung cancera
| Resectate histology
|
|||||||
|---|---|---|---|---|---|---|---|
| Biopsy histology | Adenocarcinoma (101) | Adenosquamous carcinoma (3) | Large cell carcinoma (1) | Neuroendocrine tumor (9) | Squamous cell carcinoma (69) | Unspecified malignant tumor (1) | Unspecified non-small cell carcinoma (6) |
| Adenocarcinoma (86) | 79 | 6 | 1 | ||||
| Neuroendocrine tumor (9) | 9 | ||||||
| Small cell carcinoma (2) | 1 | 1 | |||||
| Squamous cell carcinoma (65) | 4 | 3 | 57 | 1 | |||
| Unspecified malignant tumor (4) | 3 | 1 | |||||
| Unspecified non-small cell carcinoma (24) | 15 | 1 | 5 | 3 | |||
Sums along columns and rows are noted in parentheses
Identification of miR-375 as a microRNA that may improve the performance of the Lebanony method for AC/SCC subtyping
The Lebanony method is based on the relatively higher expression of miR-205 in SCC compared to AC to distinguish these subtypes of lung cancer. When the work described here was initiated, four studies28–31 with publicly available, high-throughput microRNA expression data-sets for ≥24 each of lung AC and SCC tumors had been published (Appendix 1 in Supplemental Digital Content). ROC analyses of these data-sets for association of microRNA levels with AC/SCC histology showed that, besides miR-205, microRNA miR-375 had the best AUC values among all microRNAs, with miR-375 levels being higher in AC compared to SCC (Table S1 in Supplemental Digital Content). This suggests that measurement of miR-375 may be useful for improving the accuracy of the Lebanony method for AC/SCC subtyping.
To check for microRNAs that may have a better AC/SCC histotyping ability than miR-205 and miR-375, tumor microRNA expression data from the Cancer Genome Atlas project for all 334 primary lung AC and 300 primary lung SCC cases of the project for which data was available was analyzed for differential expression and receiver operating characteristics (Appendix 2 and Table S2 in Supplemental Digital Content). Expression of 214 and 216 microRNAs in SCC was, respectively, significantly higher and lower in SCC compared to AC. Average fold-change and ROC AUC values were respectively ≥2 and ≥0.8 for 12 of the 430 differentially expressed microRNAs (Table S3 in Supplemental Digital Content). These 12 microRNAs, thus deemed to have significant histotyping potential, included miR-205 and miR-375 whose AUC values 0.96 and 0.88 respectively were highest among the sub-sets of upregulated and downregulated microRNAs. MiR-205 or miR-375 levels in either AC or SCC tumors did not significantly vary by tumor stage (pathologic stage I/II vs. III/IV), or patient age (below vs. above group mean) or race (Caucasian vs. other). The TCGA data was also examined for expression of the histotypic microRNAs in tumor-adjacent normal lung tissue (Figure 1) because biopsy specimens contain normal tissue to a varying degree and expression of a microRNA in normal lung has to be considered when evaluating the microRNA’s histotypic utility. While miR-375 is overexpressed in AC, its expression was noted as unchanged in SCC compared to normal tissue (P = 0.79), and miR-205 expression, higher in SCC, was unchanged in AC compared to normal tissue (P = 0.08; Figure 1). These observations from the largest AC/SCC microRNA expression data-set therefore support the use of miR-205 and miR-375 for AC/SCC subtyping.
Figure 1. Histotypic microRNAs to distinguish lung squamous cell carcinoma (SCC) and adenocarcinoma (AC) identified in analyses of data from the Cancer Genome Atlas.
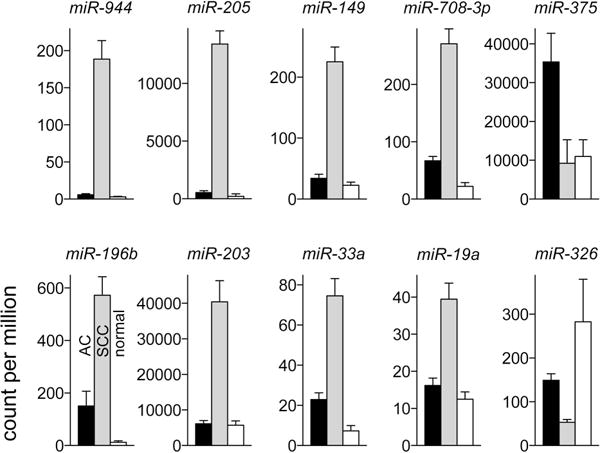
Count per million microRNAs in primary AC (n = 334, black) and SCC (n = 300, gray) tumor or tumor-adjacent normal lung tissues (n = 79, white) for 10 of the 12 microRNAs identified in the analyses are depicted as mean and 95% confidence interval values.
MicroRNA assays of resected tumors
To test the Lebanony method14 for AC/SCC subtyping, TaqMan™ RT-PCR was used to quantify miR-21, miR-205, and U6B in 5 ng RNA of 49 AC and 28 SCC stage I tumor resectates (characteristics of the cases are noted in Table S5 in Supplemental Digital Content). MicroRNA miR-375 was also quantified because, as noted earlier, it had the highest AUC value besides miR-205 in ROC analyses of microRNA levels and AC/SCC histology, and we wished to test the performance of an independent miR-375 assay to histotype AC and SCC as well as check if miR-375 assessment could improve the performance of the Lebanony method. Two RNA samples were assayed in 2–5 different experiments to assess replicability of the RT-PCR assays.
MiR-21, miR-205, and miR-375 in 53 of the 77 resectate RNA samples had previously been measured using Affymetrix® GeneChip™ miRNA oligonucleotide microarrays34. Comparison of the microRNA quantifications obtained by the RT-PCR and microarray methods revealed Pearson correlation coefficient values of 0.76–0.94 and indicated validity of the RT-PCR assays (Figure S1 in Supplemental Digital Content). Coefficients of variation (CV) for raw Cq values of the microRNAs were 0.002–0.006. CV for microRNA Cq values normalized to U6B (ΔCqU6B) were 0.064–0.088, and was 0.037 for miR-205 ΔCqU6B − (miR-21 ΔCqU6B)/2, the Lebanony score. Use of 25-fold less RNA (0.2 ng) in the assays did not significantly affect the Lebanony score or the miR-205 ΔCqU6B value (Pearson correlation coefficients ≥0.95); coefficients for raw Cq values were 0.74–0.98 (Figure 2).
Figure 2. Correlation between small RNA quantifications obtained by reverse transcription (RT)-PCR using two amounts of tissue RNA.
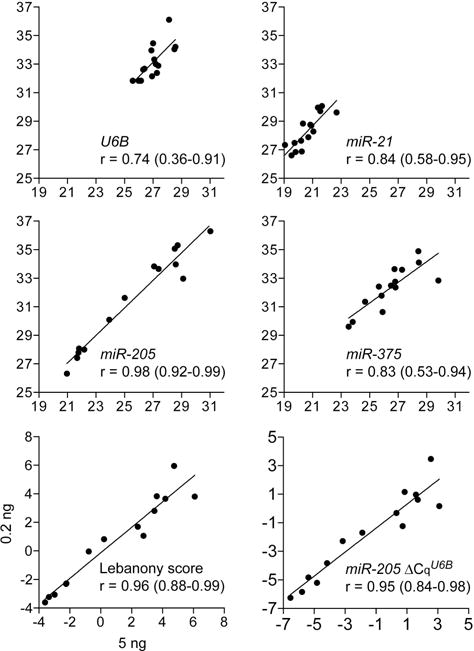
Measurements obtained using 0.2 ng RNA of 14 resected lung cancers of U6B and miR-21, -205 and -375 as quantification cycle (Cq) values, and of miR-205 relative to U6B and miR-21 (Lebanony score), or only U6B (miR-205 ΔCqU6B) are plotted against those obtained with 5 ng RNA. The Pearson correlation coefficient (r) with 95% confidence interval, and the linear regression line generated with the least squares fitting technique are also shown.
The average Lebanony score of the 28 SCC cases was −1.0 (range = −4.1–5.7; SD = 2.6) compared to 5.2 for the 49 AC cases (−2.2–13.2; 3.1), indicating an approximately 26-fold higher miR-205 expression in SCC (Figure 3A). In ROC analysis, the scores had an AUC of 0.93 (Figure 3B). With the suggested cut-off value of 2.5 to classify AC and SCC14, the Lebanony method was found to have an accuracy of 83% (64/77), with misclassification seen for 9 (18%) and 4 (14%) of the AC and SCC cases, respectively. Because synthetic microRNA standards were used in the RT-PCR assays, the molar equivalents of the Lebanony scores could be determined (Figure S2 in Supplemental Digital Content, and Figure 3C) and the cut-off value of 2.5 was calculated to be 10−2.3 (one molecule of miR-205 per 194 of U6B and miR-21).
Figure 3. Expression of miR-205 in lung squamous cell carcinoma (SCC) and adenocarcinoma (AC) resectates.
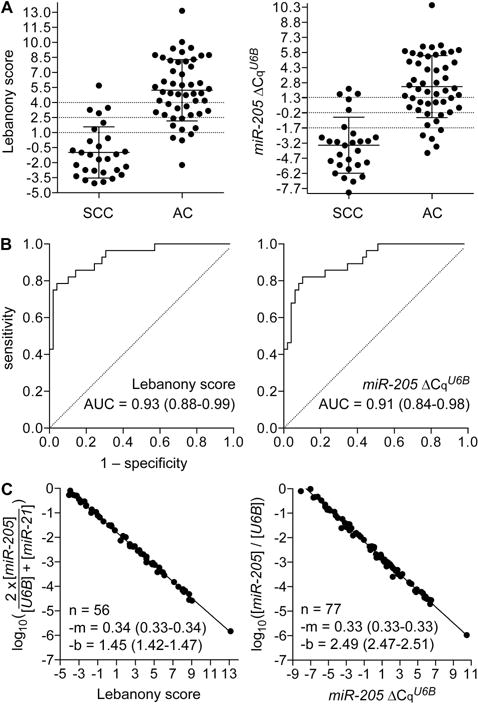
Reverse transcription (RT)-PCR was used to quantify miR-205 relative to U6B and miR-21 (Lebanony score), or only U6B (miR-205 ΔCqU6B) in 5 ng RNA for 49 AC and 28 SCC cases. For each type of measurement, panel A shows dot-plots of individual values along with histology-specific group means and standard deviations; panel B shows receiver operating characteristic curves, along the area under curve (AUC) with 95% confidence interval, for the miR-205 measurements to discriminate SCC from AC; and, panel C shows the measurement values in terms of molarity along with linear regression lines and their slopes (m) and Y intercepts (b) determined by the least squares fitting technique. Molarity equivalent of the Lebanony score could be determined for only 56 of the total 77 cases.
ROC analyses of publicly available microRNA expression data obtained in the four identifiable published studies for ≥24 each of lung AC and SCC tumors (Appendix 1 in Supplemental Digital Content) suggested that the histotypic value of miR-205 was not affected by disregarding miR-21 measurements (Figure S3 in Supplemental Digital Content). If indeed miR-205 relative to just U6B, instead of both U6B and miR-21, could be used for histotyping the resectates was therefore tested by examining the RT-PCR data. The average miR-205 ΔCqU6B value of −3.4 for SCC (range = −8.1–2.2; SD = 2.8) was significantly lower than the value of 2.4 for AC (−4.2–10.5; 3.1; Figure 3A). The miR-205 ΔCqU6B equivalent of the Lebanony score cut-off value of 2.5 was identified by linear regression analysis as −0.17 (Figure S4 in Supplemental Digital Content), 10−2.4 in molar terms (one molecule of miR-205 per 272 U6B; Figure 3C). Using the cut-off of −0.17, miR-205 ΔCqU6B values could be used to identify AC/SCC subtypes with an accuracy of 82% (63/77), with misclassification seen for 9 (18%) and 5 (17%) of the AC and SCC cases, respectively. The AUC for miR-205 ΔCqU6B values was 0.91, not significantly different from that for Lebanony scores (P = 0.07; Figure 3B).
Though miR-375 expression was higher in AC than SCC, AUCs in ROC analyses of miR-375 ΔCqU6B or ΔCqmiR-205 respectively were 0.72 and 0.90 (Figure S5 in Supplemental Digital Content), less than those seen for the Lebanony score or miR-205 ΔCqU6B values. However, miR-375 ΔCqU6B values appeared to have the potential to reduce misclassification of AC with a <2.5 Lebanony score as SCC by the Lebanony method (Figure 4). The median of miR-375 ΔCqU6B and SD of Lebanony score values for the AC cases with Lebanony score >2.5 were −1.25 and 2.44, respectively, and we modified the Lebanony classification system to classify a case with Lebanony score between 2.5 and 2.5–2.44 (0.06; 10−1.5 in molar terms) as AC instead of SCC if its miR-375 ΔCqU6B value was below −1.25 (10−4.2 in molar terms). This improved the AC/SCC subtyping accuracy of the Lebanony method from 83% (64/77) to 92% (71/77) by reducing AC misclassification from 18% (9/49) to 4% (2/49) (Figure 4). The improvement in accuracy from 83% to 92% was statistically significant (McNemar’s test P = 0.023).
Figure 4. Expression of miR-205 and miR-375 in lung squamous cell carcinoma (SCC) and adenocarcinoma (AC) resectates.
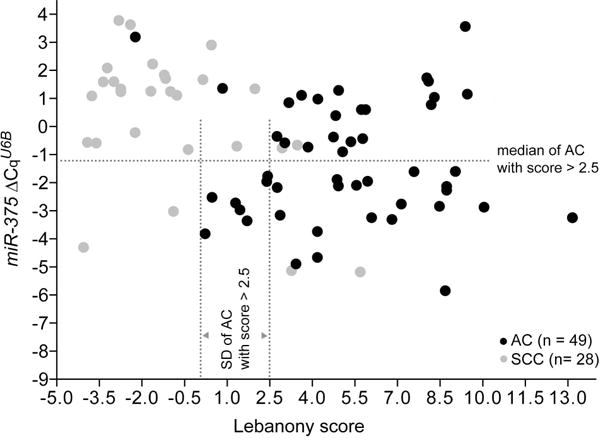
Reverse transcription (RT)-PCR was used to quantify miR-205 relative to U6B and miR-21 (Lebanony score), and of miR-375 relative to U6B (miR-375 ΔCqU6B) in 5 ng RNA for 49 AC (gray) and 28 SCC (black) cases. Dotted lines indicate the median and standard deviation (SD) values of miR-375 ΔCqU6B and Lebanony scores of the AC cases with scores >2.5.
Histotyping biopsies by microRNA measurements
Having evaluated and refined the Lebanony method using resected AC/SCC tumors, the modified method was tested against biopsy specimens of tumors later resected and identified as AC (n = 8) or SCC (n = 17). Macro- or laser-assisted microdissection was used to isolate specimen sections with >90% tumor content. Histopathologic examination of the biopsies had failed at identifying or misidentified the NSCLC subtype for 12 (48%) of the cases (Table 2). The modified Lebanony method using 0.75 ng RNA input, on the other hand, correctly identified the AC/SCC subtype of NSCLC in biopsy tissue for 24 (96%) of the 25 cases; the accuracy with the original Lebanony method was 88% (Table 2). Unlike for the set of 77 resectate samples (Figure 4), the improvement in accuracy of the biopsy histotyping with the modified Lebanony method from 88% to 96% was not statistically significant (McNemar’s test P = 0.48), though this may have been due to the small sample-size of 25. The single case that was mistyped, an SCC, was the only one of this study with an undetectable miR-205 Cq in RT-PCR assays and this may be because of a variation in sequence of miR-205 in the individual.
Table 2.
Histotyping of 25 cases of lung squamous cell carcinoma and adenocarcinoma by microRNA assays of biopsies
| Biopsy | Resectate | Discordanced | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Sample | Typea | Histologyb (Bp) | Protein markers | Histotype by microRNA assayc (Bm) | Histology (Rp) | Protein markers | Bp–Rp | Bm–Rp |
| A | CC | Carcinoma, poorly diff. | CK−, CK7−, TTF1− | SCC/AC | AC | + | +/− | |
| B | CC | AC with focal SCC, poorly diff. | CK+, CK7+, MUC1+, TTF1− | SCC/SCC | SCC, poorly diff. | CK5/6−, mucin−, TTF1−, | + | −/− |
| C | CF | SCC, poorly diff. | AC/AC | AC, poorly diff. | + | −/− | ||
| D | CF | SCC, moderately diff. | SCC/SCC | SCC, poorly diff. | − | −/− | ||
| E | CC | SCC, moderately diff. | SCC/SCC | SCC, moderately diff. | − | −/− | ||
| F | CF | NSCLC, poorly diff. | CK+, CK7+, TTF1− | SCC/AC | AC, moderately diff. | CK−, CK7−, TTF1− | + | +/− |
| G | BC | SCC | SCC/SCC | SCC, moderately diff. | − | −/− | ||
| H | CF | NSCLC, poorly diff. | AC/AC | AC, moderately diff. | + | −/− | ||
| I | BC | SCC | SCC/SCC | SCC, poorly diff. | − | −/− | ||
| J | BC | SCC | SCC/SCC | SCC, poorly diff. | − | −/− | ||
| K | BF | NSCLC, poorly diff. | SCC/SCC | SCC, moderately diff. | + | −/− | ||
| L | CF | AC, moderately diff. | AC/AC | AC, well diff. | − | −/− | ||
| M | CF | NSCLC | AC/AC | AC, poorly diff. | + | −/− | ||
| N | CF | SCC, poorly diff. | CK+, CK7− | SCC/SCC | SCC, poorly diff. | − | −/− | |
| O | BC | NSCLC, poorly diff. | SCC/SCC | SCC, poorly diff. | + | −/− | ||
| P | BC | NSCLC, poorly diff. | AC/AC | AC, poorly diff. | + | −/− | ||
| Q | BC | SCC | SCC/SCC | SCC, moderately diff. | − | −/− | ||
| R | CF | SCC, moderately diff. | SCC/SCC | SCC, moderately diff. | − | −/− | ||
| S | CF | SCC, moderately diff. | AC/AC | SCC, moderately diff. | − | +/+ | ||
| T | CF | SCC, poorly diff. | SCC/SCC | SCC, moderately diff. | − | −/− | ||
| U | CF | SCC, poorly diff. | SCC/SCC | SCC, moderately diff. | − | −/− | ||
| V | CC | AC, poorly diff. | CK+, CK7−, TTF1+ | SCC/SCC | SCC, moderately diff. | CK5/6+, CK7+, mucin−, P63+, TTF1+ | + | −/− |
| W | BC | SCC, poorly diff. | CK5/6+, P63+ | SCC/SCC | SCC, moderately diff. | CK7+, P63+, TTF1− | − | −/− |
| X | CC | AC, poorly diff. | SCC/SCC | SCC, moderately diff. | + | −/− | ||
| Y | CF | NSCLC, poorly diff. | AC/AC | AC, poorly diff. | + | −/− | ||
BC, bronchoscopic core biopsy; BF, bronchoscopic fine needle aspirate; CC, computerized tomography (CT)-guided core biopsy; CF, CT-guided fine needle aspirate
AC, adenocarcinoma; NSCLC, non–small cell lung cancer; SCC, squamous cell carcinoma. Histology including the grade of differentiation (diff.) was characterized by morphological examination of tissue sections and, for some cases, by the presence (+) or absence (−) of protein markers as assessed by periodic acid-Schiff (mucin) or immunohistochemical (others) staining. CK, cytokeratins as recognized by the AE1/AE3 mouse antibody mix; CK7, etc., cytokeratin 7, etc.; TTF-1, thyroid transcription factor-1
As per the original or modified Lebanony methods; i.e., with Lebanony scores evaluated without or with consideration of miR-375 levels. RT-PCR assays were performed on 0.75 ng RNA.
Presence or absence of discordance between histology diagnosed with routine pathologic examination of biopsy material (Bp) or resected tumor tissue (Rp) and histology identified with microRNA assays of biopsy material (Bm) is designated with a + or −, respectively. For the Bm–Rp comparison, discordance is noted for histotypes identified with the original and modified Lebanony methods.
Discussion
Our study finds that histological subtyping of NSCLC in small biopsy specimens by routine histopathologic examination either fails or mistypes the histology for 21% of cases (Table 1), a rate similar to that noted in other studies9–13. Such inaccuracy probably arises from scarcity of tumor content, tissue heterogeneity and loss of tissue architecture that are often seen in small biopsy material. Recent studies have suggested that appropriate immunohistochemical assays can be used to histotype AC and SCC in small biopsy samples with very high accuracy (93%–96%)4, 6, 8. Studies with resected tumor specimens have shown that immunohistochemistry can be used with 100% accuracy to identify AC or SCC tumors5. Highly specific protein markers, such as CD141 (thrombomodulin), desmocollin 3 and Sox2 for SCC38, 39, and napsin A for AC8, 40, 41 have been identified, and the accuracy of AC-SCC histotyping by immunohistochemistry in routine pathologic examinations will improve through the use of such markers and optimal algorithms.
While immunohistochemical markers for AC/SCC that are more accurate than the ones used for the histological diagnoses of samples of our study are known4–8, the findings presented here show that histotypic microRNAs can be used to aid AC/SCC subtyping of NSCLC. Lebanony et al.’s method that measures miR-21, miR-205 and U6B to histotype AC and SCC has been successfully used with 90%-100% accuracy with resectates in at least three studies and with biopsies in one21, 25, 26. The accuracy of the method with resectates in this study was 83%, less than that reported in the previous studies. Thus, as pointed out by the authors of one of these studies25, the Lebanony method misclassifies a significant percentage of cases. However, the accuracy of the Lebanony method could be significantly increased to 92% by incorporating miR-375 in the method (Figure 4). The modified method had an accuracy of 96% against dissected small biopsy specimens with >90% tumor content; 48% of the biopsies had been mistyped in routine histopathologic examination (Table 2). These findings suggest that histotypic microRNA assays can aid the subtyping of NSCLC biopsies as AC or SCC by standard histopathologic methods. In our experience, a specimen that is not formalin-fixed can be evaluated for microRNA expression by RT-PCR within 5–6 hours, including the time taken for RNA extraction; formalin-fixed specimens require an additional, overnight proteinase digestion step. Results of microRNA RT-PCR assays are quantitative, and multiple samples can be subjected to PCR in one batch. Reagents for RNA extraction and RT-PCR assays are inexpensive and readily available from commercial sources. Thus, microRNA assays to histotype AC/SCC can be easily performed in a pathology laboratory equipped with a quantitative PCR equipment, with effort, time and material costs similar to those for immunohistochemical assays.
The utility of miR-375 for AC/SCC subtyping has also been noted in a recent study15 and seems to have a biological basis. Differentiation of alveolar epithelial cells (AEC) from a non-squamous, AEC II phenotype to a squamous AEC I phenotype is accompanied by a decreasing level of miR-375, and artificial overexpression of miR-375 decreases the propensity of cells to undergo squamous differentiation42. Analyses of publicly available microRNA expression data, including that for >600 AC/SCC cases of the TCGA project, shows that miR-205 and miR-375 are likely to be the best microRNA markers to histotype AC and SCC (Tables S1 and S3 in Supplemental Digital Content). Levels of these two microRNAs in tumor tissues don’t seem to be affected by tumor stage or patient age or race.
Our study suggests that the modified Lebanony method may work equally well without measuring miR-21 (Figure 3 and Figure S4 in Supplemental Digital Content). This and the observation on the suitability of the method with only 0.2 ng RNA input (Figure 2) is relevant because the amount of RNA from small biopsy material can be limited. Finally, this study provides the molar equivalents of the cut-off values used for AC/SCC classification by the original and modified Lebanony methods. This should facilitate use of the methods’ principles for AC/SCC subtyping with platforms that do not use specific TaqMan™ RT-PCR assays to quantify microRNAs.
Supplementary Material
Acknowledgments
We thank Zahra Fayazi and Debra Kassay of the Pathology Resource Network, RPCI for tissue microdissection, and Saurabh Chandan of School of Medicine, St. George’s University, Grenada for collation of clinical data.
Funding: Intramural research grant from Roswell Park Cancer Institute to SD and National Cancer Institute (NCI) grant #P30 CA016056
Footnotes
Conflict of interest: None to disclose
Author contributions
SP contributed to study design, data collation and analyses, laboratory work, and project supervision. EK contributed to laboratory work. RM contributed to collation and analyses of TCGA data. RS contributed to collation and analyses of clinical data. WB contributed to collection of clinical specimens and data and supervised the microdissection of the tumor area. SY and SD contributed to study design, project supervision, data analyses, and collection of clinical specimens and data. All authors helped with preparation of the manuscript and approved the final version.
References
- 1.Scagliotti G, Hanna N, Fossella F, et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two Phase III studies. Oncologist. 2009;14:253–263. doi: 10.1634/theoncologist.2008-0232. [DOI] [PubMed] [Google Scholar]
- 2.Gressett SM, Shah SR. Intricacies of bevacizumab-induced toxicities and their management. Ann Pharmacother. 2009;43:490–501. doi: 10.1345/aph.1L426. [DOI] [PubMed] [Google Scholar]
- 3.Travis WD, Brambilla E, Riely GJ. New Pathologic Classification of Lung Cancer: Relevance for Clinical Practice and Clinical Trials. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013 doi: 10.1200/JCO.2012.46.9270. [DOI] [PubMed] [Google Scholar]
- 4.Sigel CS, Moreira AL, Travis WD, et al. Subtyping of non-small cell lung carcinoma: a comparison of small biopsy and cytology specimens. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:1849–1856. doi: 10.1097/JTO.0b013e318227142d. [DOI] [PubMed] [Google Scholar]
- 5.Rekhtman N, Ang DC, Sima CS, et al. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:1348–1359. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 6.Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:451–458. doi: 10.1097/JTO.0b013e31820517a3. [DOI] [PubMed] [Google Scholar]
- 7.Loo PS, Thomas SC, Nicolson MC, et al. Subtyping of undifferentiated non-small cell carcinomas in bronchial biopsy specimens. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2010;5:442–447. doi: 10.1097/JTO.0b013e3181d40fac. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay S, Katzenstein AL. Subclassification of non-small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: Utility of an immunohistochemical panel containing TTF-1, napsin A, p63, and CK5/6. The American journal of surgical pathology. 2011;35:15–25. doi: 10.1097/PAS.0b013e3182036d05. [DOI] [PubMed] [Google Scholar]
- 9.Montezuma D, Azevedo R, Lopes P, et al. A panel of four immunohistochemical markers (CK7, CK20, TTF-1, and p63) allows accurate diagnosis of primary and metastatic lung carcinoma on biopsy specimens. Virchows Arch. 2013;463:749–754. doi: 10.1007/s00428-013-1488-z. [DOI] [PubMed] [Google Scholar]
- 10.Edwards SL, Roberts C, McKean ME, et al. Preoperative histological classification of primary lung cancer: accuracy of diagnosis and use of the non-small cell category. J Clin Pathol. 2000;53:537–540. doi: 10.1136/jcp.53.7.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace WA, Rassl DM. Accuracy of cell typing in nonsmall cell lung cancer by EBUS/EUS-FNA cytological samples. Eur Respir J. 2011;38:911–917. doi: 10.1183/09031936.00176410. [DOI] [PubMed] [Google Scholar]
- 12.Thomas JS, Lamb D, Ashcroft T, et al. How reliable is the diagnosis of lung cancer using small biopsy specimens? Report of a UKCCCR Lung Cancer Working Party. Thorax. 1993;48:1135–1139. doi: 10.1136/thx.48.11.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo PS, Thomas SC, Nicolson MC, et al. Subtyping of undifferentiated non-small cell carcinomas in bronchial biopsy specimens. J Thorac Oncol. 2010;5:442–447. doi: 10.1097/JTO.0b013e3181d40fac. [DOI] [PubMed] [Google Scholar]
- 14.Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 15.Hamamoto J, Soejima K, Yoda S, et al. Identification of microRNAs differentially expressed between lung squamous cell carcinoma and lung adenocarcinoma. Mol Med Rep. 2013;8:456–462. doi: 10.3892/mmr.2013.1517. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YK, Zhu WY, He JY, et al. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol. 2012;138:1641–1650. doi: 10.1007/s00432-012-1240-0. [DOI] [PubMed] [Google Scholar]
- 17.Gilad S, Lithwick-Yanai G, Barshack I, et al. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn. 2012;14:510–517. doi: 10.1016/j.jmoldx.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Fassina A, Cappellesso R, Fassan M. Classification of non-small cell lung carcinoma in transthoracic needle specimens using microRNA expression profiling. Chest. 2011;140:1305–1311. doi: 10.1378/chest.11-0708. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, Hu J, Yang DW, et al. Two microRNA panels to discriminate three subtypes of lung carcinoma in bronchial brushing specimens. Am J Respir Crit Care Med. 2012;186:1160–1167. doi: 10.1164/rccm.201203-0534OC. [DOI] [PubMed] [Google Scholar]
- 20.Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solomides CC, Evans BJ, Navenot JM, et al. MicroRNA profiling in lung cancer reveals new molecular markers for diagnosis. Acta Cytol. 2012;56:645–654. doi: 10.1159/000343473. [DOI] [PubMed] [Google Scholar]
- 22.Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. Rna. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Chen J, Radcliffe T, et al. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10:513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weng L, Wu X, Gao H, et al. MicroRNA profiling of clear cell renal cell carcinoma by whole-genome small RNA deep sequencing of paired frozen and formalin-fixed, paraffin-embedded tissue specimens. J Pathol. 2010;222:41–51. doi: 10.1002/path.2736. [DOI] [PubMed] [Google Scholar]
- 25.Del Vescovo V, Cantaloni C, Cucino A, et al. miR-205 Expression levels in nonsmall cell lung cancer do not always distinguish adenocarcinomas from squamous cell carcinomas. The American journal of surgical pathology. 2011;35:268–275. doi: 10.1097/PAS.0b013e3182068171. [DOI] [PubMed] [Google Scholar]
- 26.Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 27.Bishop JA, Benjamin H, Cholakh H, et al. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–619. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 28.Lu Y, Govindan R, Wang L, et al. MicroRNA profiling and prediction of recurrence/relapse-free survival in stage I lung cancer. Carcinogenesis. 2012;33:1046–1054. doi: 10.1093/carcin/bgs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patnaik SK, Kannisto E, Knudsen S, et al. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 30.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 31.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patnaik SK, Kannisto E, Yendamuri S. Factors affecting the yield of microRNAs from laser microdissectates of formalin-fixed tissue sections. BMC Res Notes. 2012;5:40. doi: 10.1186/1756-0500-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahlgaard J, Mazin W, Jensen T, et al. Analytical variables influencing the performance of a miRNA based laboratory assay for prediction of relapse in stage I non-small cell lung cancer (NSCLC) BMC Res Notes. 2011;4:424. doi: 10.1186/1756-0500-4-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 37.Vergara IA, Norambuena T, Ferrada E, et al. StAR: a simple tool for the statistical comparison of ROC curves. BMC Bioinformatics. 2008;9:265. doi: 10.1186/1471-2105-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuta K, Tanabe Y, Yoshida A, et al. Utility of 10 immunohistochemical markers including novel markers (desmocollin-3, glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the Lung. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:1190–1199. doi: 10.1097/JTO.0b013e318219ac78. [DOI] [PubMed] [Google Scholar]
- 39.Kim MJ, Shin HC, Shin KC, et al. Best immunohistochemical panel in distinguishing adenocarcinoma from squamous cell carcinoma of lung: tissue microarray assay in resected lung cancer specimens. Ann Diagn Pathol. 2013;17:85–90. doi: 10.1016/j.anndiagpath.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Whithaus K, Fukuoka J, Prihoda TJ, et al. Evaluation of napsin A, cytokeratin 5/6, p63, and thyroid transcription factor 1 in adenocarcinoma versus squamous cell carcinoma of the lung. Arch Pathol Lab Med. 2012;136:155–162. doi: 10.5858/arpa.2011-0232-OA. [DOI] [PubMed] [Google Scholar]
- 41.Noh S, Shim H. Optimal combination of immunohistochemical markers for subclassification of non-small cell lung carcinomas: A tissue microarray study of poorly differentiated areas. Lung Cancer. 2012;76:51–55. doi: 10.1016/j.lungcan.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Huang C, Reddy Chintagari N, et al. miR-375regulates rat alveolar epithelial cell trans-differentiation by inhibiting Wnt/beta-catenin pathway. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


