Abstract
Cell junctions are necessary for spermatogenesis, and there are numerous types of junctions in testis, such as blood–testis barrier, intercellular bridge, and ectoplasmic specialization (ES). The details of their functions and construction are still unknown. To identify a novel protein essential to the function of a cell junction, we enriched testis membrane protein and analyzed it using a proteomics approach. Here, we report a novel ES protein, which is encoded on the X chromosome and an ortholog of hypothetical human protein KIAA1210. KIAA1210 is expressed in testis predominantly, localized to the sex body in spermatocyte, acrosome, and near ES. Moreover, KIAA1210 possesses a topoisomerase 2 (TOP2)-associated protein PAT1 domain, a herpes simplex virus 1 (HSV-1) large tegument protein UL36 hypothetical domain, and a provisional DNA translocase FtsK domain. Using IP-proteomics with specific antibody to KIAA1210, we identified proteins including TOP2 isoforms as components of a complex with KIAA1210, in cell junctions in testis. The interaction between KIAA1210 and TOP2 was confirmed by two different proteomic analyses. Furthermore, immunofluorescence showed that KIAA1210 and TOP2B co-localize around the sex body in spermatocyte, apical ES, and residual bodies in elongated spermatids. Our findings suggest that KIAA1210 may be essential cell junction protein that interacts with TOP2B to regulate the dynamic change of chromatin structures during spermiogenesis.
Keywords: KIAA1210, testis, ectoplasmic specialization, acrosome, topoisomerase II beta
Summary Sentence
KIAA1210, which is a novel X-chromosome-linked protein, is localized to the acrosome and associates with ectoplasmic specialization, and has a direct or indirect interaction with DNA topoisomerase 2.
Introduction
Mammalian males have a pair of distinct sex chromosomes. Abnormal phenomena may be observed by genetic mutations of X or Y. Notably, many X-chromosome-linked genes are predominantly expressed in testis and a mutation in any of these results in abnormal spermatogenesis or severe infertility [1]. Single-copy X-chromosome-linked genes are highly conserved between human and mouse, and account for over 80% of the common X-chromosome-linked genes [2].
Spermatogenesis begins with spermatogonial stem cells, located on the basement membrane, can self-renew and produce differentiated spermatogonia. After several mitotic divisions, spermatogonia enter meiosis to form spermatocytes and after meiotic division form four haploid round spermatids. Subsequently, male germ cells change their morphology to become mature spermatozoa [3]. During this complex process of spermatogenesis, the differentiation of male germ cells is supported not only by Sertoli cells but also by interactions between germ cells and Sertoli cells, which are mediated by three major cell junctions in the seminiferous tubules: intercellular bridge (ICB), blood–testis barrier (BTB), and ectoplasmic specialization (ES).
Intercellular bridge is a stable junction interconnecting daughter cells that are derived from a single spermatogonial stem cell. Intercellular bridges are thought to aid in the sharing of essential factors among interconnected germ cells, synchronization of germ cell divisions, and chromosome dosage compensation in haploid germ cells [4]. Blood–testis barrier (BTB), which is composed of a gap junction formed between adjacent Sertoli cells, prevents large molecules from invading the blood into the seminiferous tubules [5, 6]. Ectoplasmic specialization is an actin-based adherens junction and splits into two types, a connection between adjacent Sertoli cells (basal ES) and a connection between a Sertoli cell and spermatid (apical ES). In contrast to basal ES, which coexists with BTB, an apical ES anchors elongating spermatids to Sertoli cells until they are fully differentiated and regulates cellular polarity of spermatids [7, 8].
The testis expressed gene 14 (TEX14) is an essential molecule for the formation and the maintenance of ICB [9]. TEX14 binds centrosomal protein 55 kDa (CEP55) and inhibits CEP55 from interacting with apoptosis-linked gene 2 interacting protein X (ALIX) or tumor susceptibility gene 101 (TSG101) [10]. Because interaction between CEP55 and ALIX or TSG101 is required for abscission of cell division, an expression of TEX14 results in the failure to complete cytokinesis and supports the formation of stable ICBs. When TEX14 is knocked out, ICBs are never formed and spermatogenesis is arrested at the spermatocyte stage [9]. We also identified other ICB proteins from male mice using proteomic analyses [10, 11, 12]. Because BTB and ES include insoluble junctional proteins like ICB, we performed a similar proteomics strategy with adult mouse testes to identify not only ICB proteins but also BTB and ES proteins. We investigate a novel single-copy X-chromosome-linked protein, which is predominantly expressed in the testis and is highly conserved among species to be expected great significance.
Materials and methods
Proteomics analysis
Enriched membrane proteins were obtained from 8-week-old testes as described [11]. Sixteen micrograms of isolated protein was run on a 4%–20% gradient Bis-Tris gel (Promega; Madison, WI) and stained with Coomassie Brilliant Blue. The lane was cut into 10 pieces, and each piece was subjected to in-gel digestion with trypsin/Lys-C (WAKO; Osaka Japan) and applied to a Finnigan LTQ Orbitrap Velos Pro Mass Spectrometer (Thermo Fisher Scientific; Waltham, MA) for LC-MS/MS analysis. Immunoprecipitation (IP) experiments for proteomics analysis (IP-proteomics) were performed using Dynabeads Protein A (Life Technologies; Carlsbad, CA); the 25 μl 50% slurry beads, which bound 50 μl anti-KIAA1210 antiserum or guinea pig normal serum separately, according to the manufacturer's instructions, were incubated with insoluble or soluble 8-week-old testis lysates, and left on a rotator at 4°C overnight. The gel, containing boiled samples, was stained with silver.
RT-PCR
Total RNAs extracted from adult mouse tissues were extracted by RNeasy mini kit (QIAGEN; Hilden, Germany) and reverse transcribed into cDNAs using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific; Waltham, MA) according to the manufacturer's instructions. PCR was performed using the primer sets for kiaa1210 and hypoxanthine-guanine phosphoribosyltransferase (hprt) below:
kiaa1210 primer-forward, 5΄-CAGGAGCAAAGCTCCAAAC-3΄
kiaa1210 primer-reverse, 5΄-AATGAGGACAAATGGCTTGC-3΄
hprt primer-forward, 5΄-CATCACATTGTGGCCCTCTG-3΄
hprt primer-reverse, 5΄- CCTTAACCATTTTGGGGCTGT-3΄
Antibodies and immunoblot analysis
We generated antibodies against KIAA1210 using described methods [9]. Antisera were purified with Dynabeads Protein A (Life Technologies; Carlsbad, CA), following the manufacturer's instructions. Western blot analysis was performed using guinea pig anti-KIAA1210 as described [10].
Other antibodies used were as follows: anti-Zona Pellucida Binding Protein 1 (ZPBP1) [13], anti-N-Cadherin (N-Cad) (No.610920; BD Biosciences; San Jose, CA), anti-TOP2B (ab58442; Abcam; Cambridge, UK), anti-Claudin11 (CLDN11) (sc25711; Santa Cruz Biotechnology; Santa Cruz, CA), anti-TUBA (sc5286; Santa Cruz Biotechnology), anti-Zonula occludeins-1 (ZO1) (No.40-2200; Zymed; South San Francisco, CA), anti-Drebrin (DBN1) (ab11068-500; Abcam), anti- Tyrosine-protein kinase YES (YES) (No.610375; BD Biosciences), anti-Eps8 (No.610144; BD Biosciences), and anti-Claudin3 (CLDN3) (ab15102; Abcam). Phalloidin was used to stain F-actin (R415; Thermo Fisher Scientific).
Table .
| Antibody | Species/Clonality | Source (Catalogue No.) | Usage |
|---|---|---|---|
| Primary antibody (gene name) | |||
| KIAA1210 (mCG1035397) | Guinea Pig/polyclonal | IB, IF, IHC | |
| KIAA1210 (mCG1035397) | Rabbit/polyclonal | IF | |
| ZPBP1 (Zona Pellucida Binding Protein 1) | Goat/polyclonal | IF | |
| N-Cad (N-Cadherin) | Mouse/monoclonal | BD Biosciences (610920) | IF |
| TOP2B (Topoisomerase II beta) | Rabbit/polyclonal | Abcam (ab58442) | IF |
| CLDN11 (Claudin11) | Rabbit/polyclonal | Santa Cruz Biotechnology (sc25711) | IF |
| TUBA (Tubulin-alpha) | Mouse/monoclonal | Santa Cruz Biotechnology (sc5286) | IF |
| ZO1 (Zonula occludeins-1/Tight Junction protein1) | Rabbit/polyclonal | Zymed (40-2200) | IF |
| DBN1 (Drebrin) | Rabbit/polyclonal | Abcam (ab11068-500) | IF |
| YES (Tyrosine-protein kinase YES) | Mouse/monoclonal | BD Biosciences (610375) | IF |
| Eps8 (Epidermal growth factor receptor pathway substrate 8) | Mouse/monoclonal | BD Biosciences (610144) | IF |
| CLDN3 (Claudin3) | Rabbit/polyclonal | Abcam (ab15102) | IF |
| F-ACTN (F-Actin) | Mouse/monoclonal | Thermo Fisher Scientific (ab205) | IF |
| Secondary antibody | |||
| Peroxidase AffiniPure donkey Anti-Guinea Pig IgG (H+L) | Donkey/polyclonal | Jackson Immunoresearch (706-035-148) | IB |
| biotinylated goat anti-guinea pig IgG (H+L) | Goat/polyclonal | Vector (BA-7000) | IHC |
| Alexa 488-conjugated donkey anti-guinea pig IgG | Donkey/polyclonal | Jackson Immunoresearch (706-546-148) | IF |
| Alexa 594-conjugated donkey anti-guinea pig IgG | Donkey/polyclonal | Jackson Immunoresearch (706-586-148) | IF |
| Alexa Fluor 555-conjugated donkey anti-guinea pig IgG | Donkey/polyclonal | Bioss (bs-0358D-A555) | IF |
| Alexa Fluor 555-conjugated donkey anti-rabbit IgG | Donkey/polyclonal | Life (ab150074) | IF |
| Alexa Fluor 647-conjugated donkey anti-guinea pig IgG | Donkey/polyclonal | Bioss (Bs-0358D-A647) | IF |
| Alexa Fluor 488-conjugated donkey anti-goat IgG | Donkey/polyclonal | Thermo Fisher Scientific (A-11055) | IF |
| Alexa Fluor 488-conjugated donkey anti-rabbit IgG | Donkey/polyclonal | Life (406416) | IF |
| Alexa Fluor 488-conjugated donkey anti-mouse IgG | Donkey/polyclonal | Life (A21202) | IF |
IF, Immunofluorescence:IHC, Immunohistochemistry; IB, Immunoblot analysis.
Testis and sperm sample preparation
Three-week-old to 6-month-old mice were euthanized by cervical dislocation under anesthesia; newborn to 2-week-old mice were euthanized by decapitation. Testes were collected, immediately purified RNA/protein for RT-PCR/proteomics, or fixed for histology. Sperm were spread onto glass slides and fixed. All mouse experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine and the Ethics Committee on Animal Experiments at Kyushu University.
Immunofluorescence and immunohistochemistry analyses
Immunofluorescence (IF) and immunohistochemistry (IHC) were performed using primary antibodies listed above and Alexa Fluor 488, 555, 594 (Thermo Fisher Scientific), and 647 (Bioss; Woburn, MA) conjugated and biotinylated goat anti-guinea pig secondary antibodies for IF and IHC, respectively, as described [12]. Sections were examined under a fluorescence microscope (Carl Zeiss MicroImaging; Jena, Germany) or BZ-X700 and its analyzer (Keyence; Osaka, Japan).
Results
Identification of novel protein KIAA1210 and its expression pattern
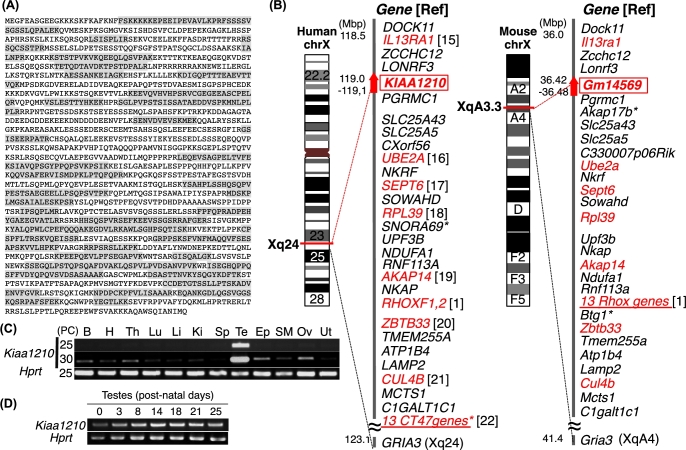
The insoluble proteins containing membranes, cell junctions, and cytoskeletons enriched from 8-week-old mouse testis were analyzed by LC-MS/MS, and 2565 proteins were identified. The top 15 proteins in this more comprehensive collection were A-kinase anchor protein 4 (AKAP4), tubulin-beta (TUBB), A-kinase anchor protein 3 (AKAP110), outer dense fiber protein 2 (ODF2), fibrous sheath-interacting protein 2 (FSIP2), tubulin-alpha (TUBA), an unnamed protein, mCG117026, TOP2, another unnamed protein, Piwi-like protein 1 (PIWIL1), KIAA1210 (mCG1035397), heat shock-related 70kDa (HSP70), PC4 and SFRS1-interacting protein (PSIP1), and hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (HADHA) (Supplemental Table S1). We focused on hypothetical protein KIAA1210, the 12th most abundant protein (Figure 1A). Of 208 peptides matched to KIAA1210, 141 peptides were nonduplicated or partially duplicated and 67 peptides were perfectly duplicated. The nucleotide sequence corresponding to the amino-acid sequence in Figure 1A is identical to Gm14569 on the X chromosome (XqA3.3; 36,423,871–36,484,339) and is located between Lonrf3 and Pgrmc1 (Figure 1B, right), whereas KIAA1210 is between LONRF3 and PGRMC1 on human X chromosome (Xq24; 119,236,245-119,244,466) (Figure 1B, left). Gm14569 was identified as a Kiaa1210 mouse ortholog. The spatiotemporal expression of Kiaa1210 was investigated by RT-PCR using cDNA prepared from adult mouse multiple tissues and developing testes. Kiaa1210 was highly expressed in testis (Figure 1C), and expression was observed from newborn testes (Figure 1D).
Figure 1.
Identification of KIAA1210 as a novel mouse testis protein. (A) KIAA1210. Peptide sequences identified by proteomic analysis are highlighted in gray. (B) The location of Kiaa1210 with neighboring genes on mouse (right) and human (left) X chromosome. Red, testis expressed gene; underline, multi-copy gene; asterisk; species-specific gene. (C, D) Semiquantitative RT-PCR of Kiaa1210 in multiple tissues (C) and during developing testes (D). PC, PCR cycles; B, brain; H, heart; Th, thymus; Lu, lung; Li, liver; Ki, kidney; Sp, spleen; Te, testis; Ep, epididymis; SM, skeletal muscle; Ov, ovary; Ut, uterus. Hprt, internal control.
Localization of KIAA1210 in testis
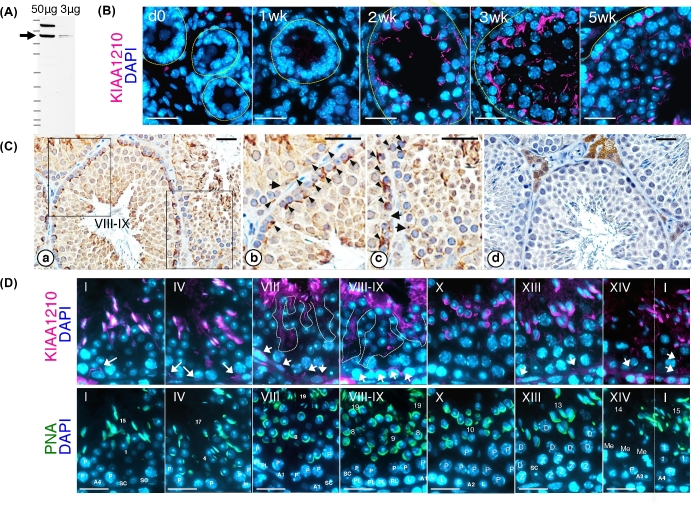
Anti-KIAA1210 antibodies were produced in two guinea pigs and a rabbit. Western blot analysis using affinity purified anti-KIAA1210 antibody was performed and revealed KIAA1210 as a 170kDa protein (Figure 2A).
Figure 2.
Localization of KIAA1210 during spermatogenesis. (A) Detection of KIAA1210 in the testis by western blot using anti-KIAA1210 antibody. Arrow, 170 kDa KIAA1210. (B) The spatiotemporal localization of KIAA1210 (magenta) with DAPI (blue) during postnatal development of the testis (dO, newborn; 1–5 wk, postnatal weeks). Dotted lines indicate basal membrane of seminiferous tubule. Bar, 25 μm. (C) The localization of KIAA1210 in 3-month-old testis. Panels b and c are higher magnification of insets in panel a. Panel d is a negative control. Arrow, nucleus of Sertoli cell; arrowhead, KIAA1210 signals; roman number, seminiferous tubule stage. Bar, 25 μm. (D) Immunofluorescence of KIAA1210 in seminiferous epithelial cycles in 6-month-old testis. Distributions of KIAA1210 (magenta), PNA (green), and DAPI (blue) are shown. Arrows, KIAA1210 near basal membrane; dotted line, disassembled KIAA1210 signals, roman numbers, seminiferous tubule stage; numbers, differentiation step of spermatids; A1–4, type A1–4 spermatogonia; In, intermediate spermatogonia; P, pachytene; PL, pre-leptotene; L, leptotene; Z, zygotene; D, diplotene spermatocyte; Me, first meiotic division of spermatocyte. Bar, 25 μm.
Expression of KIAA1210 was examined using testes sections from newborn to adult mice by immunostaining (Figure 2B–D). Although KIAA1210 protein was not observed in newborn testis (“0d” in Figure 2B), weak signals were observed in seminiferous tubule of 1-week-old testis (“1wk” in Figure 2B). In 2-week-old testis, the expression was increased (“2wk” in Figure 2B). Furthermore, in 3-week-old testis, the signal intensity of KIAA1210 became stronger and condensed around germ-cell nuclei near the basal membrane (“3wk” in Figure 2B). In 5-week-old testis, KIAA1210 was expressed and localized specifically around the nuclei of elongating spermatids in addition to being localized near the basal membrane (“5wk” in Figure 2B). Immunohistochemistry using 3-month-old testis showed similar localization of KIAA1210 as 5-week-old testis (Figure 2Ca–c). There are strong signals near basal membrane (arrowhead) and around elongating spermatids. KIAA1210 localizes strongly around the basal membrane in stage VIII–IX seminiferous tubules rather than at other stages (Figure 2Ca–c). KIAA1210 localized along the changing shape of the nuclear membrane with the progression of spermatogenesis. Therefore, we analyzed the seminiferous tubule stage-specific localizations of KIAA1210 by co-staining with peanut agglutinin (PNA), an acrosome marker [14]. Thus, we were able to determine seminiferous tubule stages by the shape of PNA signals and by tracking the locomotion of KIAA1210 (lower panels of Figure 2D). According to the change of the shape of elongating spermatids, the signals of KIAA1210 also changed in similar way with PNA around only elongated spermatids (upper panels of Figure 2D). The exposure time was reduced because of the strong signal of KIAA1210 in elongated spermatids; consequently, KIAA1210 expression near the basal membrane appears weak (arrows in Figure 2D).
Identification of the structure that includes KIAA1210
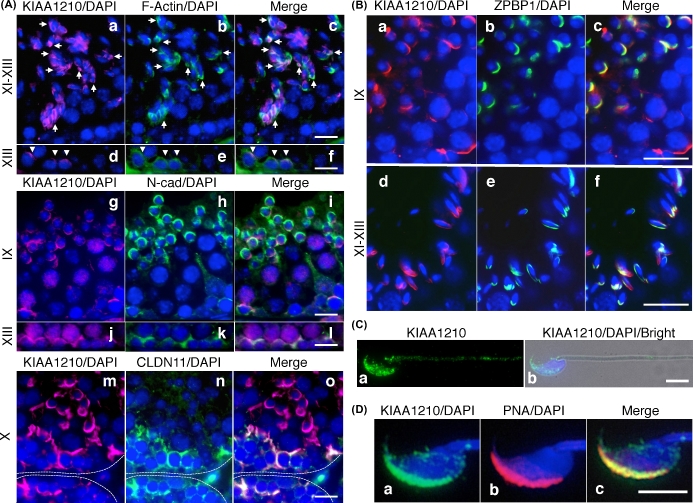
Localization of KIAA1210 was defined near elongated spermatids and in the proximity of the basal membrane. Thus, KIAA1210 was predicted to be a protein involved in ES. To identify the structure including KIAA1210, we performed co-immunofluorescence with anti-KIAA1210 antibody using antibodies specific for ES and/or BTB proteins such as F-actin, N-cadherin, and claudin11 (Figure 3A). F-Actin was mostly localized in BTB/basal ES and rarely in apical ES. Although both KIAA1210 and F-actin were localized around the nuclei of the same elongated spermatids, these proteins typically did not co-localize exactly (Figure 3Aa–c). Some signals show co-localization between F-actin and KIAA1210 in the stage XI–XIII seminiferous tubules (arrow in Figure 3Ac). KIAA1210 was co-localized with F-actin in basal ES in stage XIII seminiferous tubule (arrowhead in Figure 3Ad–f). Likewise, N-cadherin (BTB protein) and KIAA1210 were localized at different sites around the same nuclei of elongating spermatids (Figure 3Ag–i). However, they were co-localized strongly on most apical ES in stages I–III (Supplemental Figure S2G–I). N-Cadherin and KIAA1210 were more abundantly co-localized than F-actin and KIAA1210 on basal ES/BTB (Figure 3Aj–l). Localization of CLDN11 (BTB protein) and TUBA (ES protein) are similar to that of F-actin so that KIAA1210 was localized on BTB/basal ES with them (Figure 3Am–o and Supplemental Figure S1). KIAA1210 was also detected around apical ES but not co-localized with CLDN11 and TUBA (Figure 3Am–o and Supplemental Figure S1). Moreover, KIAA1210 was co-localized at basal ES/BTB with DBN1, YES, and EPS8 (Supplemental Figure S1). To further investigate the distribution of KIAA1210 around elongated spermatids, co-immunofluorescence was performed with an anti-ZPBP1 antibody, a sperm acrosome marker (Figure 3B). In testis, KIAA1210 in apical ES was co-localized with ZPBP1 (Figure 3B). In sperm, KIAA1210 localized to the sperm head with PNA as well as to the tail in a punctate pattern (Figure 3C and D).
Figure 3.
Identification of the structure involved KIAA1210. (A, B) The co-localization of KIAA1210 with ES or acrosome proteins. Immunofluorescence images of KIAA1210 with ES proteins, F-actin (upper), N-cadherin (middle), and claudin11 (lower) in (A), or acrosome protein, ZPBP1 in (B). Roman numbers, seminiferous tubule stages. Bar, 25 μm (A) and 20 μm (B). Dotted lines, basal membrane of seminiferous tubule. (C) The subcellular localization of KIAA1210 in mature spermatozoa. Immunofluorescence images of KIAA1210 (green) in spermatozoa (a) and merged image with DAPI (blue) and phase (b) are shown. Bar, 5 μm. (D) Localization of KIAA1210 in head of mature sperm. Immunofluorescence image of KIAA1210 (a, green), PNA (b, magenta), and merged (c) are shown. Bar, 5 μm.
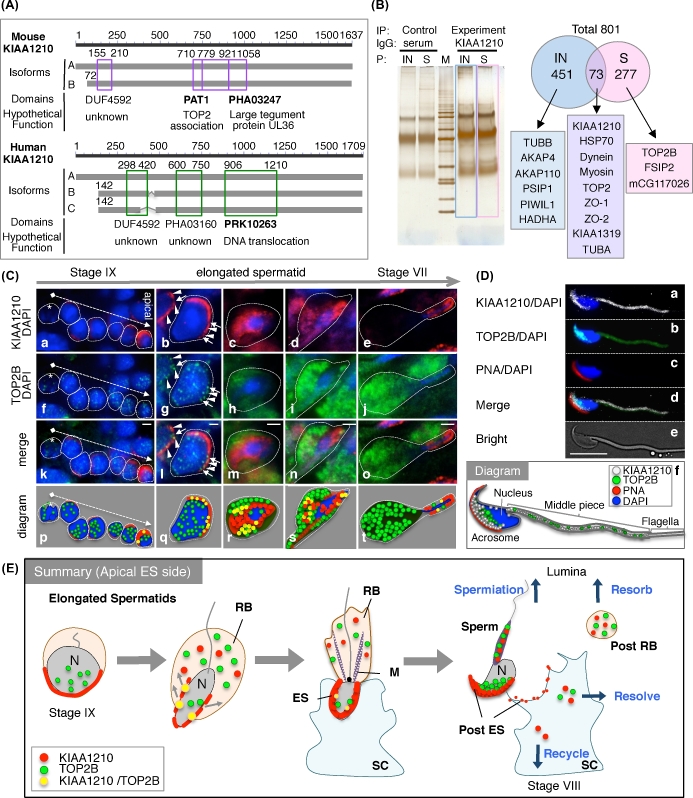
Hypothetical function of KIAA1210 by components that interact with it
KIAA1210 has two hypothetical isoforms in mouse and three in human (Figure 4A and Supplemental Figure S2). There is some variation around the N-terminus and the highly conserved C-terminus among isoforms in human and mouse. There are provisional domains, namely TOP2-associated protein (PAT1), large tegument protein UL36 (PHA03247), a domain of unknown function (DUF4592) in mouse, DNA translocase FtsK (PRK10263) domain, and two additional domains of unknown function (DUF4592 and PHA03160) in human (Figure 4A and Supplemental Table S2). To identify the complex with KIAA1210, IP-proteomics using anti-KIAA1210 antiserum was performed (Figure 4B). The silver staining of SDS-PAGE gel showed a few specific bands in comparison with control sample performed IP using serum (left side of Figure 4B). Proteomic analysis identified total 801 proteins: 350 from soluble part and 524 proteins insoluble part including common 73 proteins; the interested proteins are shown in right panels of Figure 4B. KIAA1210 interacts with 13 proteins including TOP2 family mentioned at first paragraph in result. In addition, cell junction proteins such as dynein family, myosin family, tight junction protein zona occludens protein-1 (ZO-1), -2 (ZO-2), and cingulin (KIAA1319) were identified (right of Figure 4B). Furthermore, to investigate whether the PAT1 domain is functional, we performed co-immunofluorescence assays using antibodies to KIAA1210 and TOP2B (Figure 4C). TOP2B was expressed inside the nuclei of germ cells earlier than KIAA1210 was expressed around the acrosome in stage IX (asterisk of Figure 4Ca, f, k, and p). TOP2B was expressed in only elongated spermatids, specifically from stage IX to stage VII (Figure 4Cf–j, k–o, p–t, and E). Most of the TOP2B signals moved from nuclei to residual body; nevertheless, a few signals were still in the nuclei (Figure 4Cf–j, k–o, p–t, and E). KIAA1210 was located near the acrosome at first around nuclei, then a fraction of the total KIAA1210 transited to the residual body and localized near the acrosome (Figure 4Ca–e, E). The co-localization between KIAA1210 and TOP2B was visible in the merged images (seen as yellow) (Figure 4Ck–o, p–t, and E). To investigate the co-localization of KIAA1210 with TOP2B in sperm, we performed IF using mouse sperm. KIAA1210 and TOP2B were co-localized near the acrosome in the sperm and the middle piece of sperm tail (Figure 4D and E).
Figure 4.
Characterization and localization of KIAA1210 with TOP2B in the testis. (A) The isoforms and domains of KIAA1210, two isoforms (line A, B) in mouse and three isoforms (line A, B, and C) in human were identified. Purple or green box, domain; number, the number of amino acids from N-terminus. (B) Immunoprecipitation experiments using proteomics analysis (IP-proteomics). Immunoprecipitation using anti-KIAA1210 antibody (KIAA1210) and serum as control (serum) to insoluble (IN) or soluble (S) testis lysate was performed, run to SDS-PAGE gel, and suffered silver stain (left). M, protein marker. The proteomics date of blue and pink opened box in left of Figure 4B (right). Arabic numbers, number of identified proteins. (C) Localizations of KIAA1210 (red) and TOP2B (green) with DAPI (blue) in the testis. Immunofluorescence images of KIAA1210 (a–e), TOP2B (f–j), merged (k–o), and diagram (p–t) in stage IX seminiferous tubule (a, f, k, p), surrounding area of elongated spermatids (b–d, g–i, l–n, and q–s), and stage VII seminiferous tubule (e, j, o, t) are shown. Asterisk, nonexpression cell of KIAA1210; dotted arrow, from basal to apical; arrow, strong co-localization of KIAA1210 and TOP2B; arrowhead, TOP2B on weak signal of KIAA1210. Bar, 2 μm. (D) Subcellular localization of KIAA1210 and TOP2B in spermatozoa. Immunofluorescence images of KIAA1210 (a, white), TOP2B (b, green), PNA (c, red), merged (d), and bright field (e) are shown. A diagram of the merged image is shown in panel f. Bar, 10 μm. (E) The summary of dynamic change of KIAA1210 and TOP2B during spermiogenesis. N, nucleus; ES, apical ectoplasmic specialization; RB, residual body; SC, Sertoli cell; M, manchette.
Discussion
In this study, KIAA1210 was identified as a related protein of ES by a proteomics approach, suggesting that our strategy is a useful screening method to identify cell junction proteins in the testis. The reliability was raised with IP-proteomics using antibody against KIAA1210, which we identified as cell junction, ES.
KIAA1210 is a single-copy gene located at position Xq24 on the human X chromosome. Most of genes on Xq24 region are shared with genes on mouse X chromosome XqA3.3 region. Single-copy X-chromosome-linked genes shared between human and mouse are expressed predominantly in the testis with low frequency as well as autosomal genes [2]. However, on the Xq24 and XqA3.3 regions, there are several single-copy genes expressed in the testis [1, 15–22]. Furthermore, multi-copy genes such as CT47 and Rhox families are also predominantly expressed in the testis, although some Rhox genes are homologs of human single-copy RHOXF1 and RHOXF2 [1]. It is expected that testis-expressed multi-copy genes were acquired from a common ancestor onto X chromosome to make it a male germ cell-specific chromosome [2]. Many X-chromosome-linked genes, including KIAA1210, would be essential for spermatogenesis.
We found that KIAA1210 has a TOP2 association protein PAT1 domain. PAT1 controls multiple processes in cell cycle progression as an evolutionarily conserved acetyltransferase homolog [23] and interacts directly with TOP2 for accurate chromosome transmission in Saccharomyces cerevisiae [24]. TOP2 performs DNA double-strand breaks, depending on hyperacetylation of histone H4 at elongating spermatids [25]. Thus, the PAT1 domain of KIAA1210 could be an essential domain for the function of TOP2. TOP2A and TOP2B were ranked ninth together as TOP2 family members out of total 2565 proteins identified in our proteomics analyses. It suggested that TOP2 isoforms could associate with testis membrane proteins including cell junctions. Furthermore, TOP2A and TOP2B, which were identified via IP-proteomics using anti-KIAA1210 antibody, may exist in a complex of ES and/or have a relationship with KIAA1210. The major proteins including TOP2 isoforms, which were identified in first proteomics analysis, were observed in the 801 proteins from second IP-proteomics analysis. We have focused on TOP2B because it is expressed in elongating spermatids and is found with tyrosyl-DNA phosphodiesterase 1 (TDP1), an enzyme that resolves TOP-mediated DNA damage [26]. Interestingly, the expression of KIAA1210 was localized to apical ES with TOP2B immediately after TOP2B expression began in the nucleus of elongating spermatids. Furthermore, some punctate signals of TOP2B co-localized with KIAA1210 at apical ES, suggesting that KIAA1210 is associated with the transition to compact sperm head in elongated spermatids.
Furthermore, KIAA1210 has a large tegument protein UL36 domain found in herpes simplex virus 1 (HSV-1). The inner tegument proteins including UL36 interact with the host motor proteins, dynein, dynactin, kinesin-1, and kinesin-2 to transport capsids on microtubules [27]. Dynein was the eighth protein in our proteomic analysis. KIAA1210 might interact with motor proteins to be transported from nuclei to membrane in basal and apical ES and, possibly, to be transcytosed after ES is disassembled.
In addition, a DNA translocase FtsK domain was identified in human KIAA1210. FtsK is a septum-located DNA translocase found in Escherichia coli, and functions in chromosome segregation [28]. In the near future, more conserved domains will likely be found between human and mouse KIAA1210s (Supplemental Figure S2).
Although our results suggest that KIAA1210 could participate in the exchange of chromatin during spermiogenesis, KIAA1210 was localized not only at apical ES but also at basal ES. Localization of KIAA1210 near basal ES suggests that KIAA1210 might have a function in spermatocytes. Because TOP2B was also localized to the sex bodies of spermatocytes (Supplemental Figure S3), KIAA1210 might be temporarily localized near basal ES to execute its function in spermatocytes. Generation of mutant mice of KIAA1210 will provide clarity toward our molecular understanding of KIAA1210 function.
Supplementary Material
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. Testicular distribution of KIAA1210 and cell junctions. Testicular distribution of KIAA1210 (A–D and M–P) (magenta) with CLDN11 (E), TUBA (F), NCAD (G), ZO1 (H), DBN1 (Q), YES (R), CLDN3 (S), and EPS8 (T)(green) is shown. The nuclei were counterstained with DAPI (blue). Pictures of magenta and blue signals (A–D and M–P), green and blue (E–D and Q–T), and merged signals (I–L and U–X) are shown. Roman numbers, seminiferous tubules stage. Bar, 50 μm.
Supplemental Figure S2. Protein sequence alignment among KIAA1210 isoforms. Isoforms were analyzed by using BLAST (http://blast.ncbi.nlm.nih.gov/) for finding domain and MultAlin (http://multalin.toulouse.inra.fr/multalin/) for alignment. Red, high consensus (90%); blue, low consensus (50%); purple and green box, hypothetical domain; highlighted character, domain name.
Supplemental Figure S3. Subcellular distribution of KIAA1210 and TOP2B in pachytene spermatocyte. Immunofluorescence images of KIAA1210 (red) and TOP2B (green) in the testis are shown. Panels D, E, and F (G, H, and I with instructions) are higher magnification images of insets in A, B, and C, respectively. Arrowhead indicates strong foci of KIAA1210. Dotted line, co-localization; two-way arrow, specific localization of KIAA1210. Bar, 25 μm.
Supplemental Table S1. Information of the proteins identified in proteomics analysis.
Supplemental Table S2. The information of domains on KIAA1210s.
Acknowledgments
We appreciate the support from the Department of Pathology Core Services Laboratory (Baylor College of Medicine, Houston, TX), the Laboratory for Technical Support in Medical Institute of Bioregulation (Kyushu university, Fukuoka, Japan). We thank E. Koba, M. Oda, and K. Motomura for technical assistance, and Dr. John Nelson for critical review of the manuscript. This study was supported in part by the Eunice Kennedy Shriver National Institute of Child Health and Human Development grants R01 HD088412 and U01 HD076508 (to MMM), Grant-in-Aid for Scientific Research (KAKENHI) on Innovative Areas “Mechanisms regulating gamete formation in animals” (#26114506 to TI), and Grant-in-Aid for Young Scientists (B) (#15K21217 to TI).
Supplementary data
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. Testicular distribution of KIAA1210 and cell junctions. Testicular distribution of KIAA1210 (A–D and M–P) (magenta) with CLDN11 (E), TUBA (F), NCAD (G), ZO1 (H), DBN1 (Q), YES (R), CLDN3 (S), and EPS8 (T)(green) is shown. The nuclei were counterstained with DAPI (blue). Pictures of magenta and blue signals (A–D and M–P), green and blue (E–D and Q–T), and merged signals (I–L and U–X) are shown. Roman numbers, seminiferous tubules stage. Bar, 50 μm.
Supplemental Figure S2. Protein sequence alignment among KIAA1210 isoforms. Isoforms were analyzed by using BLAST (http://blast.ncbi.nlm.nih.gov/) for finding domain and MultAlin (http://multalin.toulouse.inra.fr/multalin/) for alignment. Red, high consensus (90%); blue, low consensus (50%); purple and green box, hypothetical domain; highlighted character, domain name.
Supplemental Figure S3. Subcellular distribution of KIAA1210 and TOP2B in pachytene spermatocyte. Immunofluorescence images of KIAA1210 (red) and TOP2B (green) in the testis are shown. Panels D, E, and F (G, H, and I with instructions) are higher magnification images of insets in A, B, and C, respectively. Arrowhead indicates strong foci of KIAA1210. Dotted line, co-localization; two-way arrow, specific localization of KIAA1210. Bar, 25 μm.
Supplemental Table S1. Information of the proteins identified in proteomics analysis.
Supplemental Table S2. The information of domains on KIAA1210s.
References
- 1. Stouffs K, Lissens W. X chromosomal mutations and spermatogenic failure. Biochim Biophys Acta, 2012, 1822:1864–1872. [DOI] [PubMed] [Google Scholar]
- 2. Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet, 2013, 45:1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hess RA, De Franca LR. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol, 2008, 636:1–15. [DOI] [PubMed] [Google Scholar]
- 4. Greenbaum MP, Iwamori T, Buchold GM, Matzuk MM. Germ cell intercellular bridges. Cold Spring Harb Perspect Biol, 2011, 3:a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mruk DD, Cheng CY. The mammalian blood-testis barrier: its biology and regulation. Endocr Rev, 2015, 36:564–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vogl AW, Du M, Wang XY, Young JS. Novel clathrin/actin-based endocytic machinery associated with junction turnover in the seminiferous epithelium. Semin Cell Dev Biol, 2014, 30:55–64. [DOI] [PubMed] [Google Scholar]
- 7. Mruk DD, Cheng CY. Cell–cell interactions at the ectoplasmic specialization in the testis. Trends Endocrinol Metab, 2004, 15:439–447. [DOI] [PubMed] [Google Scholar]
- 8. Mruk DD, Cheng CY. Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev, 2004, 25:747–806. [DOI] [PubMed] [Google Scholar]
- 9. Greenbaum MP, Yan W, Wu M-H, Lin Y-N, Agno JE, Sharma M, Braun RE, Rajkovic A, Matzuk MM. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci USA, 2006, 103:4982–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iwamori T, Iwamori N, Ma L, Edson MA, Greenbaum MP, Matzuk MM. TEX14 interacts with CEP55 to block cell abscission. Mol Cell Biol, 2010, 30:2280–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greenbaum MP, Ma L, Matzuk MM. Conversion of midbodies into germ cell intercellular bridges. Dev Biol, 2007, 305:389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iwamori T, Lin Y-N, Ma L, Iwamori N, Matzuk MM. Identification and characterization of RBM44 as a novel intercellular bridge protein. PLoS One, 2011, 6:e17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin Y-N, Roy A, Yan W, Burns KH, Matzuk MM. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol, 2007, 27:6794–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lybaert P, Danguy A, Leleux F, Meuris S, Lebrun P. Improved methodology for the detection and quantification of the acrosome reaction in mouse spermatozoa. Histol Histopathol, 2009, 24:999–1007. [DOI] [PubMed] [Google Scholar]
- 15. Debinski W, Gibo DM. Molecular expression analysis of restrictive receptor for interleukin 13, a brain tumor-associated cancer/testis antigen. Mol Med, 2000, 6:440–449. [PMC free article] [PubMed] [Google Scholar]
- 16. Koken MHM, Hoogerbrugge JW, Jaspers-Dekker I, de Wit J, Willemsen R, Roest HP, Grootegoed JA, Hoeijmakers JHJ. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev Biol, 1996, 173:119–132. [DOI] [PubMed] [Google Scholar]
- 17. Ihara M, Kinoshita A, Yamada S, Tanaka H, Tanigaki A, Kitano A, Goto M, Okubo K, Nishiyama H, Ogawa O, Takahashi C, Itohara S et al. , Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev Cell, 2005, 8:343–352. [DOI] [PubMed] [Google Scholar]
- 18. Ye Q, Ding S-F, Wang Z-A, Feng J, Tan W-B. Influence of ribosomal protein L39-L in the drug resistance mechanisms of lacrimal gland adenoid cystic carcinoma cells. Asian Pac J Cancer Prev, 2014, 15:4995–5000. [DOI] [PubMed] [Google Scholar]
- 19. Kultgen PL, Byrd SK, Ostrowski LE, Milgram SL. Characterization of an A-kinase anchoring protein in human ciliary axonemes. Mol Biol Cell, 2002, 13:4156–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shumskaya VS, Zhigalova NA, Prokhorchouk AV, Prokhorchouk EB. Distribution of Kaiso protein in mouse tissues. Histochem Cell Biol, 2014, 143:29–43. [DOI] [PubMed] [Google Scholar]
- 21. Lin C-Y, Chen C-Y, Yu C-H, Yu I-S, Lin S-R, Wu J-T, Lin Y-H, Kuo P-L, Wu J-C, Lin S-W. Human X-linked intellectual disability factor CUL4B is required for post-meiotic sperm development and male fertility. Sci Rep, 2016, 6:20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y-T, Iseli C, Venditti CA, Old LJ, Simpson AJG, Jongeneel CV. Identification of a new cancer/testis gene family,CT47, among expressed multicopy genes on the human X chromosome. Genes Chromosomes Cancer, 2006, 45:392–400. [DOI] [PubMed] [Google Scholar]
- 23. Lin R, Allis CD, Elledge SJ. PAT1, an evolutionarily conserved acetyltransferase homologue, is required for multiple steps in the cell cycle. Genes Cells, 1996, 1:923–942. [DOI] [PubMed] [Google Scholar]
- 24. Wang X, Watt PM, Louis EJ, Borts RH, Hickson ID. Pat1: a topoisomerase II-associated protein required for faithful chromosome transmission in Saccharomyces cerevisiae. Nucleic Acids Res, 1996, 24:4791–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laberge R-M, Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol Reprod, 2005, 73:289–296. [DOI] [PubMed] [Google Scholar]
- 26. Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod, 2008, 78:324–332. [DOI] [PubMed] [Google Scholar]
- 27. Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog, 2010, 6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherratt DJ, Arciszewska LK, Crozat E, Graham JE, Grainge I, Diez AA, Farewell A, Nannmark U, Nyström T, Draper GG, McClennan N, Begg K et al. , The Escherichia coli DNA translocase FtsK. Biochem Soc Trans, 2010, 38:395–398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. Testicular distribution of KIAA1210 and cell junctions. Testicular distribution of KIAA1210 (A–D and M–P) (magenta) with CLDN11 (E), TUBA (F), NCAD (G), ZO1 (H), DBN1 (Q), YES (R), CLDN3 (S), and EPS8 (T)(green) is shown. The nuclei were counterstained with DAPI (blue). Pictures of magenta and blue signals (A–D and M–P), green and blue (E–D and Q–T), and merged signals (I–L and U–X) are shown. Roman numbers, seminiferous tubules stage. Bar, 50 μm.
Supplemental Figure S2. Protein sequence alignment among KIAA1210 isoforms. Isoforms were analyzed by using BLAST (http://blast.ncbi.nlm.nih.gov/) for finding domain and MultAlin (http://multalin.toulouse.inra.fr/multalin/) for alignment. Red, high consensus (90%); blue, low consensus (50%); purple and green box, hypothetical domain; highlighted character, domain name.
Supplemental Figure S3. Subcellular distribution of KIAA1210 and TOP2B in pachytene spermatocyte. Immunofluorescence images of KIAA1210 (red) and TOP2B (green) in the testis are shown. Panels D, E, and F (G, H, and I with instructions) are higher magnification images of insets in A, B, and C, respectively. Arrowhead indicates strong foci of KIAA1210. Dotted line, co-localization; two-way arrow, specific localization of KIAA1210. Bar, 25 μm.
Supplemental Table S1. Information of the proteins identified in proteomics analysis.
Supplemental Table S2. The information of domains on KIAA1210s.