Abstract
Infertility and early embryo miscarriage is linked to inadequate endometrial decidualization. Although transcriptional reprogramming is known to drive decidualization in response to progesterone, the key signaling effectors that directly mediate this hormone response are not fully known. This knowledge gap is clinically significant because identifying the early signals that directly mediate progesterone-driven decidualization will address some of the current limitations in diagnosing and therapeutically treating patients at most risk for early pregnancy loss. We recently revealed that the promyelocytic leukemia zinc finger (PLZF) is a direct target of the progesterone receptor and is essential for decidualization of human endometrial stromal cells (hESCs). The purpose of this current work was to identify the genome-wide transcriptional program that is controlled by PLZF during hESC decidualization using an established in vitro hESC culture model, siRNA-mediated knockdown methods, and RNA-sequencing technology followed by bioinformatic analysis and validation. We discovered that PLZF is critical in the regulation of genes that are involved in cellular processes that are essential for the archetypal morphological and functional changes that occur when hESCs transform into epithelioid decidual cells such as proliferation and cell motility. We predict that the transcriptome datasets identified in this study will not only contribute to a broader understanding of PLZF-dependent endometrial decidualization at the molecular level but may advance the development of more effective molecular diagnostics and therapeutics for the clinical management of female infertility and subfertility that is based on a dysfunctional endometrium.
Keywords: decidua, endometrium, female reproductive tract, implantation, progesterone/progesterone receptor, transcriptional regulation, uterus
Summary Sentence
PLZF drives progesterone-dependent transcriptional reprogramming of the human endometrial stromal cells to enable decidualization.
Introduction
Embryo implantation into the maternal uterus requires endometrial stromal cell (ESC) decidualization, a cellular transformation process in which quiescent ESCs proliferate and differentiate into epithelioid decidual cells [1,2]. Apart from providing histotrophic nutrition, protection against physiological stressors, and an immunotolerant microenvironment for embryo development, decidual cells are thought to modulate invasion of the embryonic trophoblast to a sufficient depth to establish the hemochorial placenta [3]. Accordingly, aberrant decidualization causes not only implantation failure or early fetal miscarriage, but is implicated in adverse outcomes that become symptomatic in subsequent pregnancy trimesters [3]. Therefore, an improved understanding of the key molecular underpinnings of progesterone-driven decidualization is essential for development of more effective diagnostics, prognostics, and/or therapeutics not only for peri-implantation failure but also for a broad range of later pregnancy complications that arise from early defects in endometrial decidualization.
Progesterone, through its nuclear receptor (the progesterone receptor (PGR)), transcriptionally reprograms the ESC to a decidual cell. Recent genome-wide analyses have disclosed complex hierarchal transcriptional outputs that underscore PGR-dependent decidualization [4]. Through integrative analysis of genome-wide datasets in conjunction with studies on primary human ESCs (hESCs) in culture, we recently revealed that the promyelocytic leukemia zinc finger (PLZF) transcription factor is rapidly induced from basal levels by progesterone and that the induction of this transcription factor is critical for progesterone-dependent hESC decidualization [5]. Moreover, follow-up chromatin immunoprecipitation (ChIP) studies implicate PLZF as a direct target of the PGR during progesterone-dependent decidualization [5], which is in keeping with the immediate-early transcriptional response of PLZF to progesterone exposure.
Containing an N-terminal BTB/POZ domain and nine C-terminal C2-H2 zinc finger motifs, PLZF (also known as Zinc Finger And BTB Domain Containing 16 (ZBTB16) or Zinc Finger Protein 145(ZNF45)) belongs to the POK (POZ and Kruppel) family of pleiotropic transcription factors [6]. The BTB/POZ domain is required for both homo- and heterodimerization, modulation of chromatin remodeling and transcriptional activity, formation of high-molecular weight DNA–protein complexes, and nuclear sublocalization; the nine C2-H2 zinc finger motifs enable direct sequence-specific DNA binding to target genes [6]. The PLZF transcription factor is required for a broad spectrum of developmental processes and physiological responses, from hematopoiesis and immunomodulation to spermatogenesis and limb skeletal patterning [6]. For many of these physiologies, PLZF regulates stem cell self-renewal and function, cell–cell communication, cell cycle progression, differentiation and programmed cell death [6–8].
Because of PLZF’s pivotal role in other physiological systems along with the high-level position occupied by this transcription factor in the hierarchy of PGR transcriptional targets in hESCs, we performed RNA sequencing (RNA-seq) and follow-up bioinformatic analysis to disclose the PLZF-dependent transcriptomic changes that occur on a genome-wide scale as cultured primary hESCs transform into decidual cells. As with our recent PGR transcriptional profiling in primary hESCs in culture [4], we believe such information will furnish significant molecular insights into the involvement of PLZF in peri-implantation biology.
Materials and methods
Human endometrial stromal cell isolation and culture
Study participants provided signed informed consent in advance of the endometrial biopsy procedure [5]. Endometrial biopsies were obtained during the proliferative phase of the menstrual cycle (cycle day: 7–12) from healthy women of reproductive age (27–38 years) without a history of gynecological abnormalities. Biopsy of the uterine fundus using a pipelle catheter was conducted under sterile conditions following a protocol prospectively approved by the Institutional Review Board at Baylor College of Medicine and in accordance with the guidelines of the Declaration of Helsinki [9]. An established enzymatic/size separation protocol was used to prepare primary hESCs from endometrial biopsies [5]. Following isolation, hESCs were cultured in Dulbecco's modified eagle medium nutrient mixture F-12 (DMEM/F-12) containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, 0.1 mg/ml streptomycin, 250 ng/ml amphotericin B, 10 mM Hepes, and 0.1% sodium bicarbonate (termed hESC medium); only hESCs of low passage number (≤4) were used in these studies.
Decidualization and RNA interference
In a six-well culture plate at 60%–70% cell confluence, hESCs were transfected with nontargeting (NT or control) small interfering (si) RNAs (D-001810-10-05) or siRNAs specifically targeted to PLZF (L-005196-00-0005; GE Dharmacon, Lafayette, CO). Using Lipofectamine RNAi Max reagent in 1XOpti-MEM 1 reduced-serum medium (Thermo Fisher Scientific, Waltham, MA), hESCs in each well were transfected with siRNAs (60 pmoles); each transfection was performed in triplicate. Forty-eight hours posttransfection, hESC decidualization was induced by culturing in 1XOpti-MEM I reduced serum medium containing 2% charcoal-stripped FBS, 10 nM 17β-estradiol (E2), 1 μM medroxyprogesterone acetate (MPA), and 50 μM cAMP (Sigma-Aldrich, St. Louis, MO); this decidualizing medium is termed EPC medium from hereon. The EPC medium was changed every 48 hours of hESC culture; cells were harvested at specified time points as explained in the experimental design. Transcript levels of prolactin (PRL) and insulin like growth factor binding protein-1 (IGFBP-1) were used as positive output signals for the hESC decidual response at the molecular level [5].
Transcript profiling
Using the Qiagen RNeasy Plus Mini Kit (Qiagen, Inc., Germantown, MD), total RNA was isolated from primary hESCs derived from endometrial biopsies obtained from three individual patients. For each treatment and time point, samples were prepared in triplicate. Following quality control, technical replicates were pooled for RNA-seq. Total RNA purity (based on absorbance ratios: at 260 and 280 nm and at 260 and 230 nm) and RNA quantity were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Total RNA integrity was evaluated on Agilent RNA Nano 6000 microfluidic chips using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Samples scoring a RNA integrity number above 8 were used for the generation of mRNA-focused libraries for sequencing. Using 250 ng of total RNA as input, the TruSeq RNA library preparation kit v2 (Illumina, Inc., San Diego, CA) was used for mRNA isolation, fragmentation, and subsequent first and second-strand DNA synthesis. The kit also was used for follow-up end repair along with base and adaptor ligation before products were purified and then enriched by PCR (15 cycles) to create the cDNA libraries. The resulting libraries were quantitated using the NanoDrop spectrometer and fragment size assessed using a DNA 1000 chip on the Agilent 2100 Bioanalyzer. A qPCR assay was performed on the libraries to determine the concentration of adapter-ligated fragments using the Applied Biosystems ViiA 7 Real-Time PCR System (Thermo Fisher Scientific) with a KAPA library Quant kit (Kapa Biosystems, Inc., Wilmington, MA). Relevant samples were pooled in equimolar amounts and re-quantitated by qPCR. Library pools (27 pM) were loaded into a high output flow cell for clonal cluster generation by bridge amplification using the cBot 2 system (Illumina, Inc.). Indexed paired-end sequencing (2 × 100 bp read lengths) was performed using the ultrahigh-throughput HiSeq 2500 sequencing system (Illumina, Inc.).
Bioinformatic analysis
Raw reads in fastq file format were mapped to human genome hg19 (Human assembly: GRCh37/hg19; http://genome.ucsc.edu) through use of the ultrafast universal RNA-seq aligner: Spliced Transcripts Alignment to a Reference (STAR) software [10]. To reduce possible PCR bias, read duplicates were removed with Picard Tools (http://broadinstitute.github.io/picard/). The number of reads aligned to known genes was determined by the Python-based software package HTSeq [11] (http://www-huber.embl.de/users/anders/HTSeq). Using raw gene count data, principal component analysis was conducted with R function prcomp (https://cran.r-project.org; Supplemental Figure S1). The Bioconductor package edgeR was applied to the gene expression data to detect differentially expressed genes between different treatment groups [12]. The false discovery rate (FDR) of differentially expressed genes was estimated using the Benjamini and Hochberg method [13]. Gene expression comparisons with an FDR <0.05 and an absolute fold change (|FC|) > 1.5 were considered to be significantly differentially expressed between groups of interest. Integration of datasets was performed using FileMaker Pro 14.0.1 (FileMaker Inc., Santa Clara, CA). All raw data files were deposited in Gene Expression Omnibus repository at the National Center for Biotechnology Information (www.ncbi.nlm.gov/geo). Gene ontology enrichment analysis of different functional databases was performed using the functional annotation and clustering tools within DAVID (Database for Annotation, Visualization, and Integrated Discovery; http://david.abcc.ncifcrf.gov/). To identify established gene sets overrepresented in datasets in this study, Gene Set Enrichment Analysis (GSEA; http://software.broadinstitute.org/gsea/) was used [14]. For this analysis, all identified genes were filtered based on the FDR and |FC| cut-offs described above.
Quantitative PCR
For quantitative PCR experiments, total RNA was isolated from cultured cells using the RNeasy total RNA isolation kit (Qiagen Inc., Germantown, MD). First and second-strand cDNA synthesis was performed using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). Using the Applied Biosystems StepOnePlus Real-Time PCR system (Thermo Fisher Scientific), quantitative real-time PCR was conducted using Applied Biosystems TaqMan Universal Master Mix II without UNG and TaqMan assays. Ribosomal RNA (18S) was used as an internal control for each gene-specific TaqMan assay; a list of TaqMan assays used in this study is shown in Supplemental Table S1.
Chromatin immunoprecipitation-quantitative PCR assay
For ChIP, hESCs were fixed with 1% formaldehyde for 15 min and subsequently quenched with 0.125 M glycine. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. Lysates were sonicated using the EpiShear Probe Sonicator (Active Motif Inc., Carlsbad, CA) with an EpiShear Cooled Sonication Platform (Active Motif Inc.) and the DNA sheared to an average length of 300—500 bp. Genomic DNA (input) was prepared by treating aliquots of chromatin with RNase, proteinase K, and heat for de-crossing, followed by ethanol precipitation. Pellets were resuspended, and the resulting DNA was quantified on a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield. Aliquots of chromatin (30 μg) were precleared with protein G agarose beads (Invitrogen Inc., Carlsbad, CA). Genomic DNA fragments bound by PLZF were isolated using an antibody against PLZF (Santa Cruz Biotechnologies, Dallas, TX; sc-11146). Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase treatment. Crosslinks were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. Quantitative PCR reactions were carried out in triplicate using SYBR Green Supermix (Bio-Rad Inc., Hercules CA; #170-882) on a CFX Connect Real Time PCR system. The ChIP reaction was validated using one positive control site (QRICH1), one negative control (Untr12, #71001; Active Motif Inc.), and the two test sites of interest. The resulting signals were normalized for primer efficiency by carrying out qPCR for each primer pair using input DNA (pooled unprecipitated genomic DNA from each cell line).
Cell proliferation assay
Following siRNA transfection, cells were plated on 96-well plates at a density of 5 × 103 cells per well in EPC medium. Three hours after plating followed by 24 intervals, EPC medium was replaced with the DNA-binding fluorescent dye Cyquant NF reagent (ThermoFisher Scientific); hESCs were incubated for 1 h at 37°C before measurement. The fluorescent signal was measured using a Bio-TEK Synergy HT microplate reader (485 nm excitation; 528 nm emission).
Transwell migration assay
Post-siRNA transfection, cells were plated at 1.2 × 105 cells per well in minimal essential medium without serum on hanging polyethylene terephthalate cell culture inserts with a pore size of 8.0 μm. Cell culture inserts with cells were placed into 12-well plates with hESC medium (which contains 10% FBS). After 48 h in culture, hESCs that had not migrated through the membrane pores were thoroughly removed while cells that passed through were stained with 0.5% crystal violet solution, imaged, and counted using ImageJ’s Cell Counter plugin (https://imagej.nih.gov/ij/plugins/cell-counter.html)
Statistical analyses
Results are presented as means ± standard deviation. Experiments were performed in triplicate. Quantile comparisons were performed to inspect normality of data. Equality of variances was analyzed using the Bartlett test of homogeneity of variances. Statistical analyses were performed with either ANOVA with post hoc analysis performed with Welch two sample test or Tukey range test, in R Studio (R Studio Inc., Boston, MA). Multiple comparisons were adjusted with the Holm method; P > 0.05 were considered as nonsignificant differences, while P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) were considered significant.
Results
Transcriptomic changes in human endometrial stromal cells in response to EPC exposure
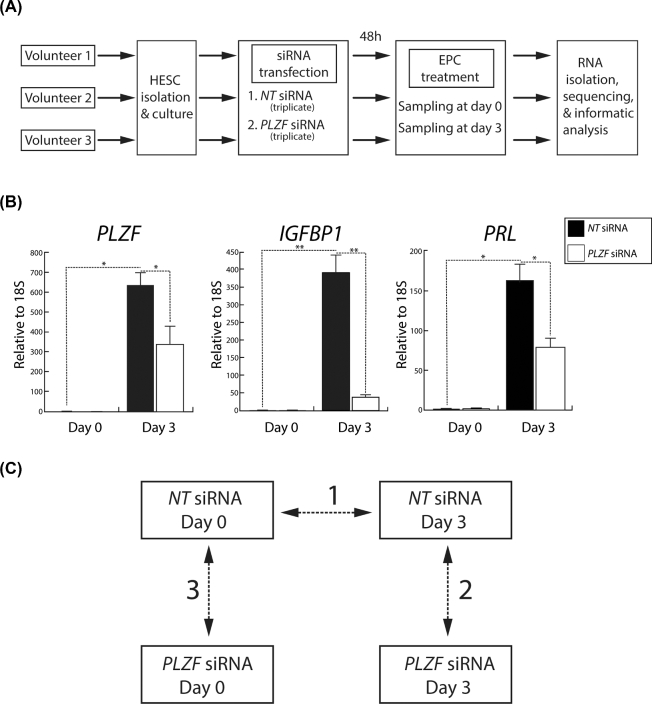
For the RNA-seq experiments, hESCs were isolated from endometrial tissue biopsied from three volunteers (Figure 1A). Note hESCs used in this study were obtained from the same volunteers as described in [5]. Cultured primary hESCs were transfected with either nontargeting (NT (control)) siRNAs or siRNAs targeting PLZF [5]. Forty-eight hours post-transfection, hESCs were cultured in EPC medium to elicit decidualization. For both siRNA transfection groups, total RNA was isolated from hESCs that were harvested on day 0 and day 3 of EPC treatment (Figure 1A). Three days of EPC treatment was chosen as an early sampling time point for transcriptional profiling during hESC decidualization because this time point was used in our recent published hESC RNA-seq [4] and PLZF ChIP-seq studies [5], thereby allowing future integration of all of these datasets. Note the experimental design and samples were specifically formulated and prepared for this study. Furthermore, because PLZF is rapidly induced to high transcript levels following progestin exposure [5], we did not want to compromise the effectiveness of siRNA mediated knockdown of PLZF by extending the EPC exposure time beyond 3 days. Quantitative PCR confirmed that PLZF transcript levels are significantly reduced by PLZF siRNA transfection and that this reduction correlates with a marked decrease in the transcript levels for IGFBP1 and PRL, both biomarkers for decidualization [15] (Figure 1B). For follow-up bioinformatic analysis, three comparisons of the resultant transcriptome datasets were conducted in this study (Figure 1C). The complete list of genes with associated gene expression changes for the three comparisons is reported in Supplemental Table S2.
Figure 1.
Experimental design of the RNA-Seq study to identify the PLZF transcriptome in cultured HESCs. (A) Workflow of the RNA-Seq experiment showing that HESCs were isolated from endometrial biopsies from three healthy volunteers and transfected with either nontargeting (NT) or PLZF-targeting siRNAs. Forty-eight hours post-transfection, HESCs were treated with EPC to induce HESC decidualization. Cells were collected for RNA isolation and sequencing at day 0 and day 3 of EPC treatment. Each siRNA and EPC treatment time point was prepared in triplicate; following quality control, samples were pooled for RNA-sequencing. (B) Prior to RNA-Seq, quantitative real-time PCR analysis of transcript levels for PLZF and two decidual markers (IGFBP1 and PRL) in HESCs (previously transfected with NT or PLZF siRNAs) is shown at the indicated time points following EPC treatment. Expression data from the patient RNA samples to be used for RNA-Seq analysis confirm significant siRNA-mediated knockdown of PLZF at day 3 of EPC treatment, which closely correlates with a marked reduction in the levels of IGFBP1 and PRL; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, (C) Paired quantitative comparisons of the four sequenced RNA samples (NT siRNA (day 0); NT siRNA (day 3); (PLZF siRNA (day 0); and PLZF siRNA (day 3)) in this experiment are shown: comparison 1: Transcriptional changes that occur with EPC treatment for 3 days; comparison 2: Transcriptional changes that occur with PLZF knockdown following EPC treatment for 3 days; and comparison 3: Transcriptional changes that occur with PLZF knockdown at day 0 of EPC treatment.
To first reveal the normal transcriptomic changes that occur in decidualizing hESCs, RNA-seq was conducted on RNA isolated from control hESCs transfected with NT siRNA following day 0 and day 3 of EPC culture. Comparing day 3 with day 0 of EPC treatment, a total of 4,922 expressed genes (2,195 upregulated and 2,727 downregulated) were significantly changed in these control hESCs (Figure 2A). Differentially expressed genes between the two time points were filtered according to their absolute fold change (|FC| > 1.5) and FDR (<0.05). Using the Database for Annotation, Visualization and Integrated Discovery (DAVID) software, functional clustering analysis of genes was performed (top 20 clusters shown; Figure 2B). Importantly, significantly enriched biological processes are associated with cell cycle regulation, cytoskeletal organization, extracellular matrix remodeling, cell migration, and early cellular responses to steroid hormone and inflammatory stimuli. Collectively, these biological responses are in accordance with the known cellular changes that occur when endometrial stromal fibroblasts transform into polygonal epithelioid decidual cells [2]. Our DAVID analysis is further supported by GSEA (Figure 2C), which revealed a representation of gene sets involved in cell cycle modulation, epithelial–mesenchymal transition, and early response to steroid hormone and inflammatory stimuli.
Figure 2.
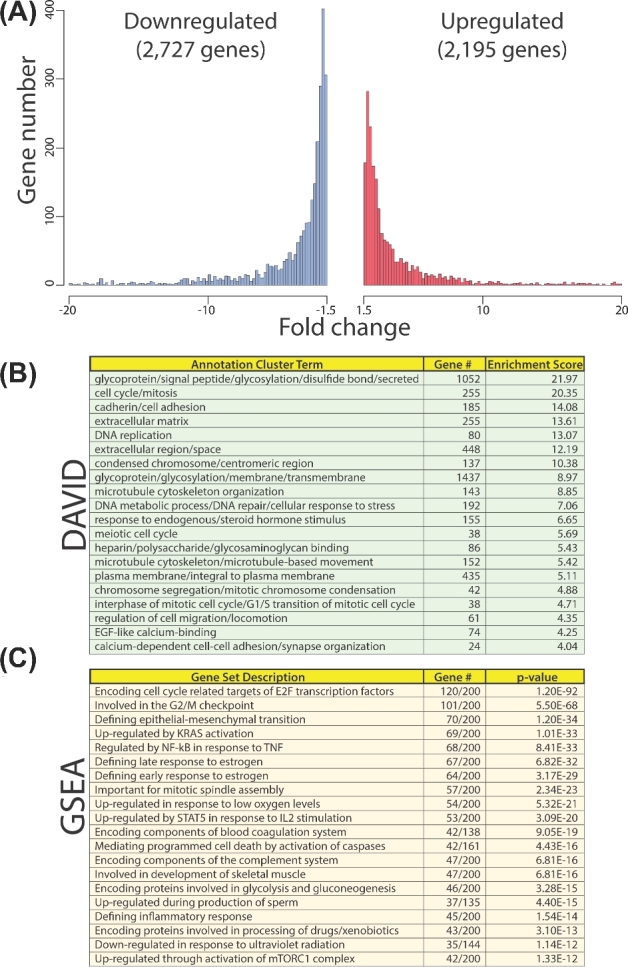
Global gene expression changes during HESC decidualization. (A) Histogram of the number of genes up- and downregulated (red and blue, respectively) during decidualization (NT siRNA day 3 (EPC) vs NT siRNA day 0 (EPC)) with corresponding fold changes. (B) DAVID analysis of gene expression changes during decidualization (as shown in (A)). (C) GSEA of gene expression changes during decidualization (as shown in (A)). Enrichment of genes was surveyed against collection H (the hallmark gene sets collection representing “well-defined biological states or processes”).
Normal human endometrial stromal cell transcriptional responses to EPC exposure require PLZF
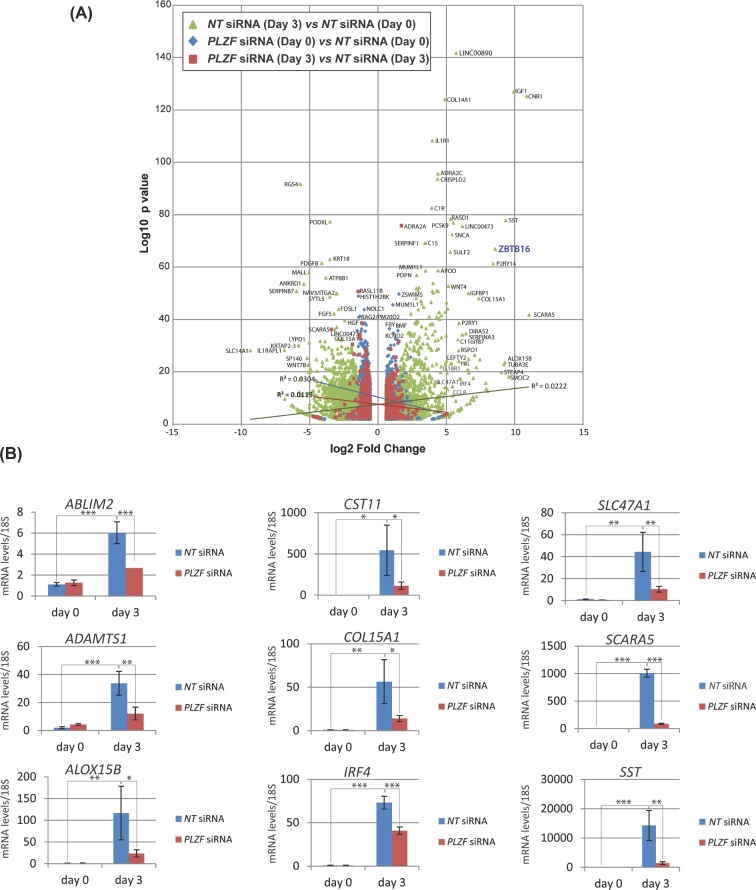
In Figure 3A, the volcano plot provides a global perspective of the genome-wide transcriptional changes that result during normal decidualization (green triangles) and changes in transcription in response to knockdown of PLZF at day 0 (blue diamonds) and day 3 (red squares) of decidualization. A Venn diagram displaying the numbers of genes in the three datasets and the overlaps between them (described in Figure 1C) is shown in Supplemental Figure S2. As an example, Figure 3B shows nine genes differentially expressed with PLZF knockdown that were validated by quantitative real-time PCR. In aggregate, these genes are involved in a broad range of biological processes, such as controlling intra- and extracellular structure (actin binding LIM protein 2 (ABLIM2) and collagen 15A1 (Col15A1)); exhibiting critical enzymatic activities (arachidonate 15-lipoxygenase type B (ALOX15B) and a disintegrin and metalloproteinase with thrombospondin motifs 1 (ADAMTS1)) or inhibiting enzymatic activity (cystatin 11 (CST11)); modulating immune regulatory responses (interferon regulatory factor 4 (IRF4)); transmembrane trafficking (multidrug and toxin extrusion 1 (MATE1/SLC47A1); and scavenger receptor A5 (SCARA 5); and regulating the endocrine system and tissue and cell growth (somatostatin (SST)). A number of these genes have been shown to be expressed in the endometrium [16–18] and function in endometrial decidualization [19].
Figure 3.
Global gene expression alterations which occur with HESC decidualization and which require PLZF. (A) Volcano plot of the number of genes for which expression levels were significantly altered by at least a |FC| ≥ 1.5 with FDR of less than 0.05 in comparisons: 1 to 3 shown in Figure 1C. (B) Quantitative real-time PCR analysis confirms significant changes in transcript levels for a number of diverse genes following PLZF knockdown; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
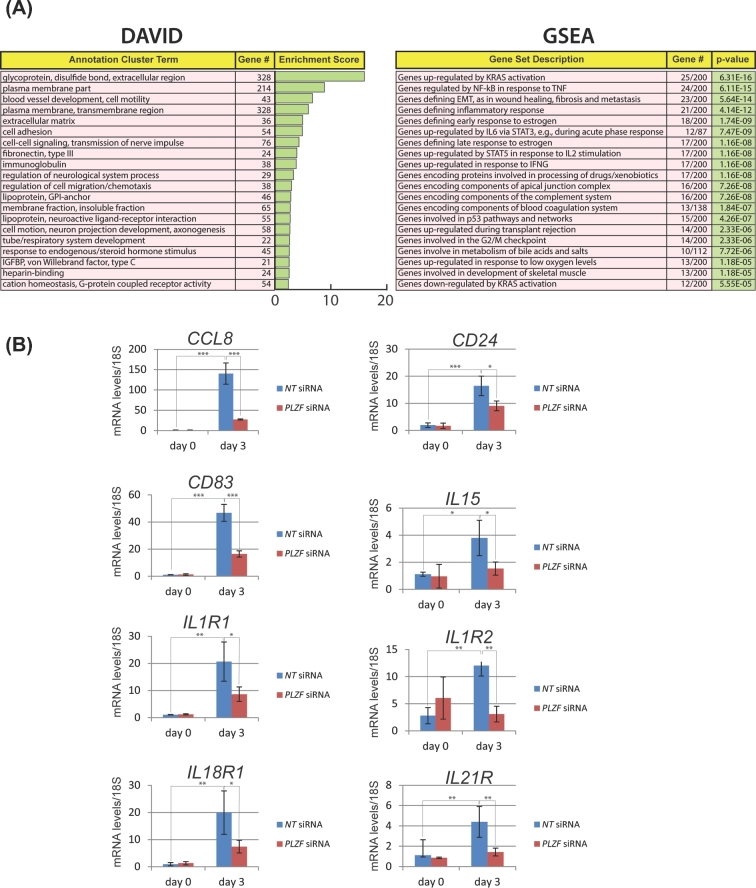
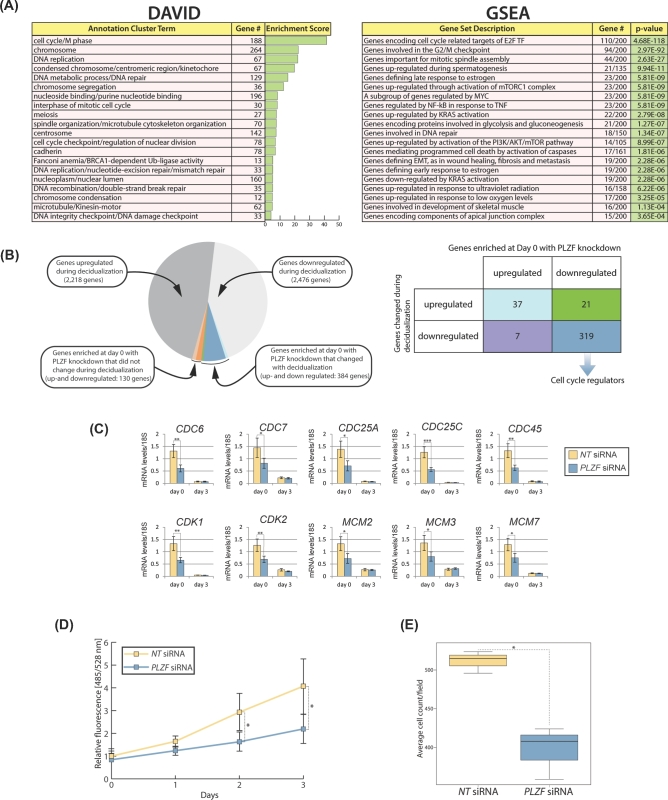
Through DAVID analysis, we revealed a wide spectrum of biological processes that are significantly affected in hESCs with PLZF knockdown following 3 days of EPC exposure (Figure 4A). Many of these biological responses are predicted to be essential for hESC decidualization, such as cell adhesion, extracellular matrix remodeling, regulation of cell migration and chemotaxis, and response to endogenous/steroid hormone stimuli. DAVID analysis is further supported by GSEA, which reveals overlaps between the dataset resulting from PLZF knockdown at day 3 of decidualization and gene sets associated with epithelial–mesenchymal transition, chemotactic and inflammatory responses, cell cycle progression, and cell–cell signaling cues (Figure 4A). For example, our data demonstrate that normal induction of genes associated with immunomodulation is remarkably sensitive to PLZF levels as hESCs decidualize from day 0 to day 3 following EPC exposure (Figure 4B). These genes include chemokine (c-c motif) ligand 8 (CCL8 [20]), also known as monocyte chemoattractant protein 2 (MCP2), which is chemotactic for and activates many types of immune cells (i.e. mast cells, eosinophils, basophils, monocytes, T cells, and natural killer (NK) cells). Cluster of Differentiation 83 and 24 (CD83 and CD24 respectively [21,22]) have been shown to regulate growth and differentiation of dendritic, mature granulocytes, and B cells. Playing an important role in innate and adaptive immunity, interleukin 15 (IL15 [23]) is a cytokine that controls T and NK cell activation and proliferation. Knockdown of PLZF also significantly attenuates the induction of numerous interleukin receptors (i.e. interleukin 1 receptor 1 (IL1R1), interleukin 1 receptor type 2 (IL1R2), interleukin 18 receptor 1 (IL18R1), and interleukin 21 receptor (IL21R)). Many of these cytokine receptors mediate the proliferative, differentiative, and prosurvival signals of cytokines in T cells, B cells, and NK cells. The immune gene expression signature that is significantly altered in hESCs by PLZF knockdown is in keeping with PLZF’s role in regulating the development and activity of immune cells in other physiological systems [6,8].
Figure 4.
Analysis of gene expression changes which result from PLZF knockdown reveals gene sets and cellular processes that are important for HESC decidualization. (A) DAVID analysis and GSEA of genes for which transcript levels significantly altered with PLZF knockdown (at day 3 EPC treatment). (B) As an example, quantitative real-time PCR analysis confirms that normal transcriptional induction of cytokine and interleukin gene family members is significantly attenuated with PLZF depletion; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Integration of the human endometrial stromal cell PLZF transcriptome and cistrome
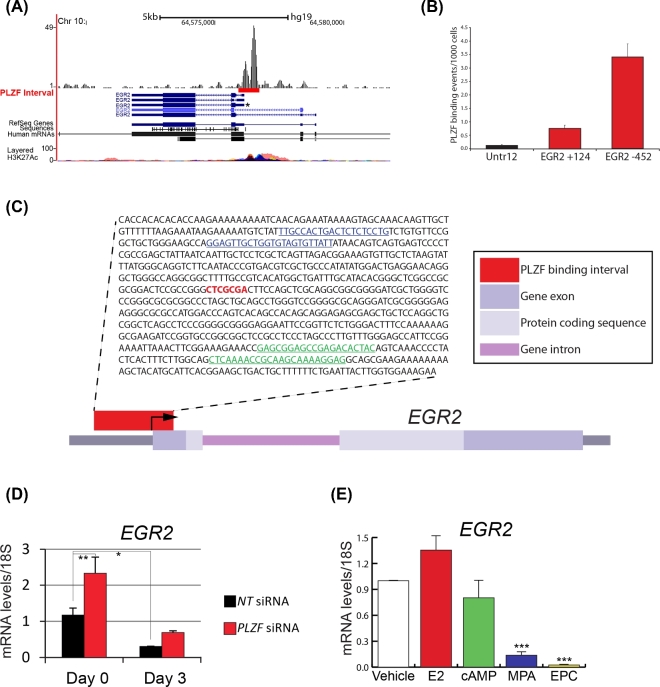
To identify possible transcriptionally responsive genes that are directly bound by PLZF in hESCs, we integrated the above PLZF transcriptome datasets with our recently reported PLZF cistrome data from hESCs treated for 3 days with EPC [5]. A total of 60 genes were found to overlap between the RNA-seq and ChIP-seq datasets (Supplemental Figure S3A). Results from our integrative analysis along with subsequent ChIP-qPCR data indicate that PLZF may directly regulate the transcription of the early growth response 2 (EGR2) gene in hESCs when cultured in EPC medium over a 3-day period (Supplemental Figure S3B). Subsequent ChIP-qPCR data indicate that PLZF may directly regulate the transcription of the (EGR2) gene in hESCs when cultured in EPC medium over a 3-day period (Figure 5A–C; Supplemental Figure S4). This conclusion is further supported by quantitative PCR analysis which demonstrated that EGR2 transcript levels are significantly reduced following PLZF knockdown at day 0 (Figure 5D). Of the three reagents that makeup the EPC cocktail, only the progestin (MPA (which induces PLZF)) significantly reduces EGR2 transcript levels in hESCs (Figure 5E). Together, these results suggest that PLZF may directly downregulate EGR2 transcription during hESC decidualization. Interestingly, we recently demonstrated that the expression levels of EGR1 are also downregulated by PLZF in hESCs with EPC culture and that this downregulation may occur through direct interaction of PLZF with the EGR1 promoter [5]. We previously confirmed by quantitative real-time PCR analysis that EGR1 is significantly downregulated by PLZF during hESC decidualization [5]. Even though our |FC| cutoff of 1.5 in this study excluded EGR1 from our current analysis, EGR1 expression levels were found to be downregulated with an FDR < 0.05 (the FC of EGR1, EGR2, and EGR3 with FDR < 0.05 was equal –1.39, –1.52, and –1.41, respectively).
Figure 5.
The EGR2 transcription factor is regulated by PLZF in HESCs. (A) Snapshot of the UCSC Genome Browser depicting the PLZF ChIP-seq binding peak on the human EGR2 gene promoter. (B) Direct binding of PLZF at +124 and –452 base pairs from the transcriptional start site (TSS) in the EGR2 promoter was confirmed by ChIP-qPCR. (C) Sequence of PLZF binding interval on the EGR2 promoter with putative PLZF binding motif highlighted in red; primer sequences for +124 and –452 base pair regions used for ChIP-qPCR are highlighted in green and blue respectively. (D) Quantitative real-time PCR confirms that EGR2 transcript levels are markedly reduced with EPC treatment and that this reduction is significantly attenuated with PLZF knockdown. (E) Quantitative real-time PCR confirms that EGR2 transcript levels are significantly reduced by EPC treatment. Of the three EPC components, MPA significantly downregulates EGR2 transcript levels. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Basal levels of PLZF are required for human endometrial stromal cell proliferation and migration
Functional annotation analysis of the genes overlapping between the PLZF transcriptome and cistrome revealed an enrichment of genes with terms involved in the control of the cell cycle and proliferation (Supplemental Figure S3C). The transcriptomic changes that overlapped with the PLZF cistrome occurred primarily at day 0 of EPC treatment when basal levels of PLZF were knocked down (see Figure 1C (comparison 3); Supplemental Figure S5). Further analysis of the transcriptome revealed that predecidual basal levels of PLZF are required for the regulation of gene expression specifically involved in cellular processes driving proliferation such as DNA replication, mitotic spindle assembly, chromosome segregation, and microtubule/cytoskeleton organization (Figure 6A). Examination of genes enriched by DAVID Functional Annotation Clustering (top 20 clusters) at this time point showed that the majority of these genes are also changed during decidualization (Figure 6B). Importantly, most of these overlapping genes are downregulated in both datasets and are involved in cell division processes (Figure 6B). Expression of some of these proliferation regulators was confirmed by qPCR (Figure 6C). In agreement with this finding, cell proliferation assays revealed that PLZF knockdown leads to downregulation of proliferation of hESCs (Figure 6D). Moreover, DAVID analysis and GSEA predict that basal levels of PLZF are also required for the motility potential of hESCs prior to EPC exposure. Results from in vitro migration assays confirm this prediction (Figure 6E).
Figure 6.
Cell cycle progression in HESCs is dependent on PLZF prior to EPC treatment. (A) DAVID analysis and GSEA of genes for which transcript levels significantly changed |FC| ≥ 1.5 with a false discovery rate (FDR) < 0.05 following PLZF knockdown at day 0 EPC treatment. (B) Pie chart shows the number of genes in the indicated datasets and the overlaps between the following datasets: (1) transcriptional changes occurring during decidualization and (2) transcriptional changes that have been enriched by DAVID’s Functional Annotation Clustering at day 0 of decidualization with PLZF knockdown (as described in (A) above). The 384 overlapping genes are further described in the accompanying table. The majority of overlapping genes which are downregulated in both datasets are involved in the regulation of cell cycle progression. (C) Quantitative PCR confirms the significant downregulation in the expression levels of numerous cell cycle regulators following PLZF knockdown at day 0 of EPC treatment. (D) Reduction in HESC proliferation following PLZF knockdown as measured with a DNA content-based cell proliferation assay. (E) Knockdown of PLZF expression in HESC results in a significant reduction in cell migration as measured by a transwell migration assay.
Discussion
Fahnenstich et al were the first to report PLZF expression in the stromal and myometrial cellular compartments of the human uterus, predicting a key role for this progesterone-responsive uterine transcription factor in female fertility [24]. In the case of the endometrium, we recently provided strong in vitro evidence that PLZF is indispensable for progesterone-dependent hESC decidualization by acting as a possible molecular mediator of PGR action [5]. As with any transcription factor, identifying the downstream transcriptome that is regulated by PLZF is predicted to provide significant molecular insights into the role of this transcriptional regulator in a given physiological process. Therefore, we used RNA-seq to disclose the transcriptome that is responsive to PLZF as cultured hESCs undergo decidualization.
Global transcriptomic analyses have previously demonstrated that progesterone-dependent decidualization of hESCs is driven by striking gene expression changes involved in cytoskeletal organization, extracellular matrix remodeling, cell adhesion, immunomodulation, extra- and intracellular signaling, metabolic reprogramming, proliferation, differentiation, and apoptosis [4]. Given PLZF is thought to act as a direct mediator of PGR action in hESCs [5], it was not surprising that experimental reduction of PLZF transcript levels results in alteration of transcriptional responses to EPC. Examined through the context of PLZF’s known cellular and molecular roles in other biological systems [6–8], our first line of analysis focused on the significant expression changes of genes involved in hESCs in either in the regulation of immune cell modulators or cellular proliferation.
Consistent with PLZF’s established role in immune cell development, chemokine induction, and interferon-mediated innate immunity [8], we observed a significant number of immunoregulatory genes that require PLZF for full induction in decidualized hESCs. Of particular interest is the significant dependency of IL15 induction by EPC on PLZF expression; IL15 is a known chemotactic cytokine for uNK recruitment and activation [25]. This observation is significant because the decidua of pregnancy not only comprises decidual cells but also harbors a marked number of uNK cells at the embryo–maternal interface [26]. Concentrated around the maternal spiral arterioles, uNK cells are considered critical innate immune cells that both contribute to an immunotolerant microenvironment for the invading trophoblast and drive remodeling of the spiral arterioles to enable endovascular trophoblast invasion, which eventually establishes the definitive placenta. Noteworthy, previous studies have demonstrated that decidualized hESCs secrete IL-15 along with other cytokines that are implicated in the recruitment and differentiation of uNK cells [23,27]. Furthermore, perturbation in the expression of these cytokines and chemokines has been linked to incomplete implantation or implantation failure as well as placental insufficiency [26]. Therefore, our studies suggest that PLZF may be required for decidualized hESCs to recruit and activate local immune cells, which in turn direct the trophoblast to the maternal vasculature to establish a functional placenta.
Through integration of PLZF RNA-seq and ChIP-seq data from hESCs, we identified EGR2 as one of a few candidate direct targets of PLZF in hESCs. Like EGR1 [5], EGR2 transcription is suppressed in part by PLZF induction as hESCs decidualize. A pleiotropic transcription factor EGR2 is required for a broad range of biological responses from hindbrain ontogenesis to T-cell development and function [28,29]. Interestingly, recent studies have revealed that EGR2 directly binds to the promoter of the PLZF gene to promote invariant (i)NKT cell development and function [30]. Whether a similar transcriptional regulatory control mechanism occurs in hESCs prior to EPC exposure will be an important question to be addressed in the future. Also, whether abrogating the normal downregulation of EGR2 expression adversely affects hESC decidualization—as previously observed for EGR1 [5]—warrants further investigation.
Integration of our transcriptome data with our PLZF ChIP-seq dataset followed by DAVID analysis and GSEA predicted that basal levels of PLZF are required for cell cycle progression and migration of hESCs, PLZF-dependent cellular properties that were confirmed by in vitro cell culture assays. A proproliferative role for PLZF was unexpected since the majority of PLZF studies to date have previously characterized this transcription factor as more involved in differentiation rather than proliferation [6], leading some studies to suggest that PLZF acts a tumor suppressor in certain cellular contexts [6,31]. In the broader context of decidualization, however, a role for PLZF—an apex signal for the initiation of progesterone-dependent decidualization—as a mediator of hESC proliferation, fits with the early proliferative responses which hESCs must undergo before these cells morphologically and functionally transform into terminally differentiated epithelioid decidual cells [2]. Moreover, the PLZF transcription factor is known to direct the acquisition of the iNKT effector program during development, which includes not only the cytokine secretory response but also the migratory capacity of iNKT cells [8], supporting a role for PLZF in motile responses of this immune cell. Our data show that PLZF is implicated in hESC motility, which is significant because decidual cell motility and migration are essential cellular properties that allow these specialized cells to localize around the implanting conceptus and subsequently facilitate deep trophoblast invasion into the maternal compartment [32].
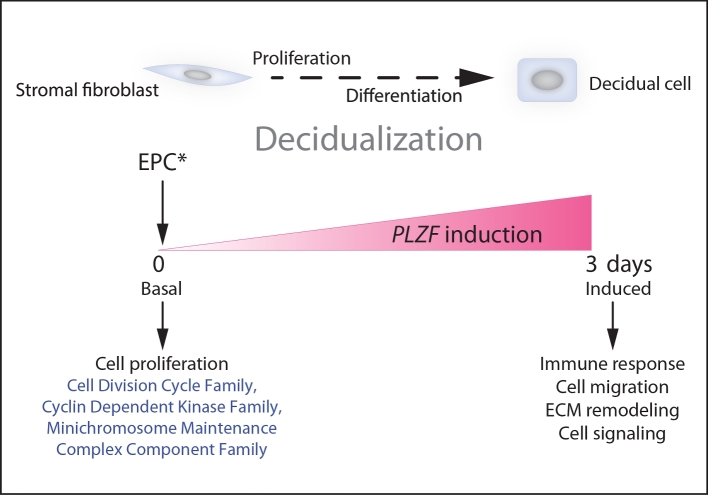
Collectively, genome-wide transcript profiling confirms and significantly extends our current understanding of PLZF’s critical role in hESC decidualization at the molecular level. Our analysis offers the first indication of the possible involvement of this transcription factor in the known cross-communication between decidual cells and resident immune cells as well as in early decidual cell proliferation and migration (Figure 7). Further analysis of this information resource promises to lend a deeper understanding of PLZF’s role in the endometrium, and perhaps its involvement in the juxtaposed uterine compartment, the myometrium [24,33].
Figure 7.
Diverse transcriptional programs are controlled by PLZF as hESCs undergo decidualization in vitro. Prior to hESC decidualization (day 0 of EPC treatment), basal PLZF levels primarily maintain normal transcript levels of a broad spectrum of cell cycle regulators, which are required for hESC proliferation prior to their differentiation into decidual cells. Induction of PLZF by EPC drives hESC decidualization, which entails the transcriptional programming of a plethora of gene families with wide-ranging functions, such as immunomodulation, cell migration, extracellular matrix remodeling, and cell signaling.
Supplementary Material
Acknowledgments
The authors wish to thank Jie Li, Yan Ying, Rong Zhao, and Irene Sheffer for their invaluable technical assistance.
Grant support: This research was supported in part by the Genomic and RNA Profiling Core at Baylor College of Medicine with funding from the NIH NCI grant (P30CA125123 (to LDW)); a Cancer Prevention Research Institute of Texas pre-doctoral fellowship grant (CPRIT: RP101499 (to MMS)); a Baylor College of Medicine Reproductive Endocrinology and Infertility Fellowship (to MCP); National Institutes of Health (NIH)/National Institute of Child Health and Human Development (NICHD) grant (U01: HD-076596 (to DML)); a NIH/NICHD R00 HD080742 to RK; and a NIH/NICHD: R01: HD-042311 grant (to JPL).
Conference presentation: Presented in part at the 50th Annual Meeting of the Society for the Study of Reproduction, 13–16 July 2017, Washington, DC.
Edited by Dr. Peter J. Hansen, PhD, University of Florida.
Supplementary data
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. Principal component analysis of the RNA-seq data. (A) Histogram of percentage of variances of principal components. (B) Distribution of the first and second components (accounting for 66% and 14% of sample variances, respectively). Overall, the first component (PC1) separates samples according to time; the second component (PC2) separates samples according to the patient identification (ID) number.
Supplemental Figure S2. Venn diagram of the number of genes and overlaps of the three gene expression comparisons represented in volcano plot shown in Figure 3A.
Supplemental Figure S3. The integration of HESC RNA-seq and ChIP-seq datasets for PLZF. (A) Overlaps of genes bound by PLZF [5] and genes for which transcription levels are significantly changed by PLZF knockdown at day 0 and day 3 of EPC treatment. (B) Genes bound by PLZF and altered by PLZF knockdown at both day 0 and day 3 of EPC treatment. (C) Genes bound by PLZF and altered by PLZF knockdown at day 0 or day 3 of EPC treatment with a list of associated functional annotation terms.
Supplemental Figure S4. (A) Table of qPCR primers used to confirm PLZF binding to the EGR2 promoter determined by ChIP-qPCR analysis. (B) Histogram of the number of binding events detected/1000 cells for Untr12 (negative control) and QRICH1 (positive control).
Supplemental Figure S5. Knockdown of basal expression levels of PLZF in HESC at day 0 of EPC treatment. (A) Quantitative real-time PCR analysis of PLZF transcript levels in HESCs (at day 0 EPC) 48 h following NT siRNA and PLZF siRNA transfection. (B) Western immunoblotting for PLZF protein expression in HESCs at day 0 of EPC treatment 48 h following NT siRNA and PLZF siRNA transfection. Lanes 1, 2, and 3 represent HESCs without transfection of siRNAs, transfected with NT siRNA, and transfected with PLZF siRNA respectively; β-actin served as a loading control.
Supplemental Table S1.xlsx
Supplemental Table S2.xlsx
Authors’ roles
MMS, RK, and LH performed the experiments and the follow-up analysis. WEG and MCP provided the endometrial tissue biopsies. MMS, LDW, QM, and RBL performed the RNA-seq and ChIP-seq analysis. RBL and MMS conducted advanced bioinformatic and computational analysis of the resultant transcriptome and cistrome datasets. MMS, RK, DML, FJD, and JPL contributed to the design of the study and wrote the manuscript. All authors reviewed the manuscript and approved the revised version for submission.
References
- 1. Carson DD, Bagchi I, Dey SK, Enders AC, Fazleabas AT, Lessey BA, Yoshinaga K. Embryo implantation. Dev Biol 2000; 223:217–237. [DOI] [PubMed] [Google Scholar]
- 2. Gellersen B, Brosens JJ. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev 2014; 35:851–905. [DOI] [PubMed] [Google Scholar]
- 3. Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med 2012; 18:1754–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mazur EC, Vasquez YM, Li X, Kommagani R, Jiang L, Chen R, Lanz RB, Kovanci E, Gibbons WE, DeMayo FJ. Progesterone receptor transcriptome and cistrome in decidualized human endometrial stromal cells. Endocrinology 2015; 156:2239–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kommagani R, Szwarc MM, Vasquez YM, Peavey MC, Mazur EC, Gibbons WE, Lanz RB, DeMayo FJ, Lydon JP. The promyelocytic leukemia zinc finger transcription factor is critical for human endometrial stromal cell decidualization. PLoS Genet 2016; 12:e1005937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suliman BA, Xu D, Williams BR. The promyelocytic leukemia zinc finger protein: two decades of molecular oncology. Front Oncol 2012; 2:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu TM, Lee EH, Lim B, Shyh-Chang N. Concise review: balancing stem cell self-renewal and differentiation with PLZF. Stem Cells 2016; 34:277–287. [DOI] [PubMed] [Google Scholar]
- 8. Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, Bendelac A. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci USA 2016; 113:7602–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. WMA WMA declaration of Helsinki serves as guide to physicians. Calif Med 1966; 105:149–150. [PMC free article] [PubMed] [Google Scholar]
- 10. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 1995; 57:289–300. [Google Scholar]
- 14. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brosens JJ, Hayashi N, White JO. Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells. Endocrinology 1999; 140:4809–4820. [DOI] [PubMed] [Google Scholar]
- 16. Ahmadimoghaddam D, Zemankova L, Nachtigal P, Dolezelova E, Neumanova Z, Cerveny L, Ceckova M, Kacerovsky M, Micuda S, Staud F. Organic Cation Transporter 3 (OCT3/SLC22A3) and Multidrug and Toxin Extrusion 1 (MATE1/SLC47A1) Transporter in the placenta and fetal tissues: expression profile and fetus protective role at different stages of gestation. Biol Reprod 2013; 88:55. [DOI] [PubMed] [Google Scholar]
- 17. Bersinger NA, Wunder DM, Birkhauser MH, Mueller MD. Gene expression in cultured endometrium from women with different outcomes following IVF. Mol Hum Reprod 2008; 14:475–484. [DOI] [PubMed] [Google Scholar]
- 18. Duncan WC, Shaw JL, Burgess S, McDonald SE, Critchley HO, Horne AW. Ectopic pregnancy as a model to identify endometrial genes and signaling pathways important in decidualization and regulated by local trophoblast. PLoS ONE 2011; 6:e23595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vasquez YM, Mazur EC, Li X, Kommagani R, Jiang L, Chen R, Lanz RB, Kovanci E, Gibbons WE, DeMayo FJ. FOXO1 is required for binding of PR on IRF4, novel transcriptional regulator of endometrial stromal decidualization. Mol Endocrinol 2015; 29:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Asselin E, Johnson GA, Spencer TE, Bazer FW. Monocyte chemotactic protein-1 and -2 messenger ribonucleic acids in the ovine uterus: regulation by pregnancy, progesterone, and interferon-tau. Biol Reprod 2001; 64:992–1000. [DOI] [PubMed] [Google Scholar]
- 21. Gilliam DT, Menon V, Bretz NP, Pruszak J. The CD24 surface antigen in neural development and disease. Neurobiol Dis 2017; 99:133–144. [DOI] [PubMed] [Google Scholar]
- 22. Qian ZD, Huang LL, Zhu XM. An immunohistochemical study of CD83- and CD1a-positive dendritic cells in the decidua of women with recurrent spontaneous abortion. Eur J Med Res 2015; 20:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu JJ, Sun HT, Zhang ZF, Shi RX, Liu LB, Shang WQ, Wei CY, Chang KK, Shao J, Wang MY, Li MQ. IL15 promotes growth and invasion of endometrial stromal cells and inhibits killing activity of NK cells in endometriosis. Reproduction 2016; 152:151–160. [DOI] [PubMed] [Google Scholar]
- 24. Fahnenstich J, Nandy A, Milde-Langosch K, Schneider-Merck T, Walther N, Gellersen B. Promyelocytic leukaemia zinc finger protein (PLZF) is a glucocorticoid- and progesterone-induced transcription factor in human endometrial stromal cells and myometrial smooth muscle cells. Mol Hum Reprod 2003; 9:611–623. [DOI] [PubMed] [Google Scholar]
- 25. Patidar M, Yadav N, Dalai SK. Interleukin 15: A key cytokine for immunotherapy. Cytokine Growth Factor Rev 2016; 31:49–59. [DOI] [PubMed] [Google Scholar]
- 26. Hannan NJ, Evans J, Salamonsen LA. Alternate roles for immune regulators: establishing endometrial receptivity for implantation. Expert Rev Clin Immunol 2011; 7:789–802. [DOI] [PubMed] [Google Scholar]
- 27. Germeyer A, Sharkey AM, Prasadajudio M, Sherwin R, Moffett A, Bieback K, Clausmeyer S, Masters L, Popovici RM, Hess AP, Strowitzki T, von Wolff M. Paracrine effects of uterine leucocytes on gene expression of human uterine stromal fibroblasts. Mol Hum Reprod 2009; 15:39–48. [DOI] [PubMed] [Google Scholar]
- 28. De S, Turman JE Jr. Krox-20 gene expression: influencing hindbrain-craniofacial developmental interactions. Arch Histol Cytol 2005; 68:227–234. [DOI] [PubMed] [Google Scholar]
- 29. Kim EY, Lynch L, Brennan PJ, Cohen NR, Brenner MB. The transcriptional programs of iNKT cells. Semin Immunol 2015; 27:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, Singh H, Bendelac A. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol 2012; 13:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsieh CL, Botta G, Gao S, Li T, Van Allen EM, Treacy DJ, Cai C, He HH, Sweeney CJ, Brown M, Balk SP, Nelson PS et al. . PLZF, a tumor suppressor genetically lost in metastatic castration-resistant prostate cancer, is a mediator of resistance to androgen deprivation therapy. Cancer Res 2015; 75:1944–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weimar CH, Macklon NS, Post Uiterweer ED, Brosens JJ, Gellersen B. The motile and invasive capacity of human endometrial stromal cells: implications for normal and impaired reproductive function. Hum Reprod Update 2013; 19:542–557. [DOI] [PubMed] [Google Scholar]
- 33. Momeni M, Kalir T, Farag S, Kinoshita Y, Roman TY, Chuang L, Fishman DA, Burstein DE. Immunohistochemical detection of promyelocytic leukemia zinc finger and histone 1.5 in uterine leiomyosarcoma and leiomyoma. Reprod Sci 2014; 21:1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.