Abstract
DHHC-type protein acyltransferases may regulate the localization, stability, and/or activity of their substrates. In this study, we show that the protein palmitoyltransferase DHHC3 is upregulated in malignant and metastatic human breast cancer. Elevated expression of DHHC3 correlated with diminished patient survival in breast cancer and six other human cancer types. ZDHHC3 ablation in human MDA-MB-231 mammary tumor cell xenografts reduced the sizes of both the primary tumor and metastatic lung colonies. Gene array data and fluorescence dye assays documented increased oxidative stress and senescence in ZDHHC3-ablated cells. ZDHHC3-ablated tumors also showed enhanced recruitment of innate immune cells (antitumor macrophages, natural killer cells) associated with clearance of senescent tumors. These antitumor effects were reversed upon reconstitution with wild-type, but not enzyme-active site-deficient DHHC3. Concomitant ablation of the upregulated oxidative stress protein TXNIP substantially negated the effects of ZDHHC3 depletion on oxidative stress and senescence. Diminished DHHC3-dependent palmitoylation of ERGIC3 protein likely played a key role in TXNIP upregulation. In conclusion, DHHC3-mediated protein palmitoylation supports breast tumor growth by modulating cellular oxidative stress and senescence.
Introduction
Posttranslational palmitoylation affects protein localization, stability, molecular interactions, and functions (1, 2). Protein palmitoylation is typically mediated by protein acyl transferases (PAT), containing conserved DHHC (Asp-His-His-Cys) motifs needed for enzymatic activity (1, 3). Among 23 mammalian DHHC enzymes, only DHHC17 (HIP-14) and DHHC5 were so far suggested to affect tumor xenograft growth (4, 5), but mechanistic details have been lacking. Golgi-resident enzyme DHHC3 (GODZ) is upregulated in breast, prostate, and colon carcinomas (6), but a possible role in cancer has not been previously addressed.
Moderate oxidative stress levels can promote early cancer stages, but excess levels limit tumor growth (7, 8). One major consequence of tumor cell oxidative stress is senescence (9, 10), leading to clearance by the innate immune system (11, 12). Here we show DHHC3 upregulation in human breast cancer, elevated ZDHHC3 expression correlating with reduced survival in multiple human cancers, and DHHC3 actively supporting breast tumor xenograft growth. Furthermore, ZDHHC3 ablation upregulates tumor cell oxidative stress, senescence, and infiltration by innate immune cells, leading to diminished in vivo tumor growth.
Materials and Methods
Cell culture, other reagents, and Western blotting
Human breast cancer (MDA-MB-231, MCF-7, ZR-75, BT549, BT474) and kidney (HEK 293) cell lines were obtained from ATCC without further authentication. After 10–20 passages, frozen cells were newly thawed. Mycoplasma was tested using the MycoAlert kit (Lonza Biologics). Cells were cultured in DMEM and/or RPMI medium (Invitrogen) containing 10% FBS (Sigma), HEPES, and 1% penicillin–streptomycin (Invitrogen). Antibodies to DHHC3 (pAb) and MCP-1 (pAb) were from Abcam and antibodies to p-FAK (pAb), total FAK (pAb), VEGF (pAb), total STAT3 (pAb), and IL8 (pAb) were from Santa Cruz Biotechnology, Inc. Antibodies to p-STAT3 (mAb), p-ERK (pAb), and total ERK (mAb) were from Cell Signaling Technology, anti-TXNIP (mAb) was from MBL International, 4G10 (mAb) was from Millipore, and anti-EGFR (mAb) was from BD Biosciences. Dyes for measuring oxidative stress (CellROX) and cellular senescence (C12FDG) were from Invitrogen. Oxidative stress inhibitors, N-acetyl cysteine (NAC) and α-lipoic acid (α-LA) were from Sigma, and atrovastatin was from Cayman Chemicals. Chemokine array analysis kit was from R&D Systems. Cultured cells were lysed in 1% Triton X-100, and total protein was quantified using BCA protein estimation kit. Western blotting and relative band intensity densitometry were described previously (13).
Gene ablation and reconstitution
Transient and stable, control and ZDHHC3 ablations were performed as described previously (13). TXNIP was ablated using two different siRNAs from OriGene. For ZDHHC3 reconstitution, ZDHHC3 cDNA was mutated to escape shRNA targeting (D3R) and two "DHHC" palmitoylation site mutations were made (D3R+DH/AA & D3R+C/S). These three cDNAs were cloned into lentiviral plasmids downstream of DHHC3 shRNA, and followed by DNA coding for C-terminal 2A linker peptide and GFP tag. These lentiviral plasmids were transfected into HEK293 cells along with pCMV-dR8.91 and VSV-G packaging plasmids, to produce lentiviral particles, which were subsequently used to infect target cells (MDA-MB-231) for stable expression. Plasmid expression in target cells was verified by GFP analysis and GFP-positive cells were sorted by flow cytometry.
Mouse xenograft growth and lung metastasis assays
For in vivo tumor growth experiments, control and ZDHHC3-ablated MDA-MB-231 cells were injected into female nude mice on both flanks either ectopically (1.0 × 106 cells, s.c.) or orthotopically (0.5 × 106 cells, into mammary fat pads), with 5 mice/group. Starting at day 5 postinjection, tumors were measured using calipers and tumor volumes were calculated (length × width2 × 0.5). Mice were sacrificed when tumor size reached 2 cm, and tumors were excised, weighed, and portions were fixed and embedded in paraffin sections for IHC staining, while remaining portions were frozen and used for RNA isolation and DNA array analysis. For lung metastasis, control and ZDHHC3-ablated MDA-MB-231 (1.0 × 106 cells) were injected into tail veins of SCID Beige mice (3 mice/group). After 5 weeks, mice were sacrificed and lungs were perfused with India Ink, excised, and fixed with Fekete solution. Tumor colonies (white) on lung surfaces were counted using a stereomicroscope. Paraffin-embedded lung sections were hematoxylin and eosin (H&E) stained to assess colony size (using light microscope; ×2 magnification). Mouse studies were reviewed and approved by the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee board.
Measurement of oxidative stress and senescence
To assess oxidative stress/reactive oxygen species (ROS), control, and ZDHHC3-ablated tumor cells were trypsinized and suspended in complete medium. After PBS wash, cells were loaded with 2 µmol/L CellROX dye in complete medium (37°C, 30 minutes). Cells were then washed twice with 1× PBS, incubated in complete media (37°C, 30 minutes), washed twice again with 1× PBS, and then analyzed by flow cytometry. Similarly, senescence was measured by loading suspended control and ZDHHC3-ablated cells with C12FDG dye (30 minutes, 37°C) in complete medium. The C12FDG dye is converted by intracellular β-galactosidase (elevated in senescent cells) to produce fluorescence, which was quantitated by flow cytometry.
Chemokine assay
As per array kit instructions (ARY017 Kit, R&D Systems), 500 µL of supernatant was collected from MDA-MB-231 cells (after 30 hours in DMEM media with 1% BSA), and mixed with detection antibody (biotinylated) cocktail. Next nitrocellulose membranes (in duplicate) containing immobilized antibodies to 31 human chemokines were incubated overnight at 4°C with supernatant/antibody mix. Membranes were then incubated with streptavidin-HRP solution followed by Chemi reagent mix and autoradiography detection. Signal intensities for each chemokine were estimated from pixel densities (Image Quant, version 5.2 software, GE Healthcare).
IHC staining
Human malignant and metastatic breast tumor microarray slides and normal controls (BR2082, BR10010a) were from US Biomax Inc., and stained for DHHC3 using alkaline phosphatase immunohistochemistry detection kit (ZYAGEN). Briefly, slides were deparaffinized, rehydrated, boiled in 10 mmol/L citrate buffer (pH 6.0, 15 minutes), followed by blocking in serum (1 hour). Next, slides were incubated with 1:75 dilution of anti-DHHC3 antibody, Sigma at 4°C overnight, and then with biotinylated secondary antibody (1 hour) and streptavidin–alkaline phosphatase (AP) conjugate (30 minutes). Finally, slides were incubated with fast red solution and counterstained for hematoxylin. Staining intensity was analyzed by light microscopy and scoring (from 0 = no staining to 3 = high staining) was performed, in an unbiased and blinded fashion, by a pathology expert at Brigham and Women's Hospital (Boston, MA).
Microarray analysis
RNA was isolated using RNeasy kit (Qiagen) from mammary fat pad–derived xenograft tumors from control and ZDHHC3-ablated MDA-MB-231 cells. RNA was analyzed from two tumors of each group using U133A 2.0 Affymetrix gene chip array at Dana-Farber Cancer Institute, Microarray Core facility. Microarray data were processed using dChip software. Results were submitted to the GEO database and assigned accession number GSE102776.
Patient survival and tissue expression data
Data used for Fig. 1A and Supplementary Table S1 were gathered from publicly available The Cancer Genome Atlas (TCGA) data accessed through the cbioportal.org database (14, 15). Survival curves and log-rank tests were performed using the cBioPortal "survival" tool. Breast cancer patient samples are from nonidentifiable patients, from a publicly available commercial source (US Biomax).
Figure 1.
DHHC3 expression, patient survival, and breast carcinoma growth. A, Kaplan–Meier overall survival plot comparing breast invasive carcinoma patients with elevated or not elevated ZDHHC3 gene expression. Elevated expression is defined as mRNA expression Z score ≥ 2.0 (data from cBioPortal analysis of TCGA data; refs. 14, 15). B, IHC for DHHC3 was done on paraffin-embedded normal, malignant, and metastatic human breast tumor tissue sections (scale bar, 100 µm). C, Human tissue microarray samples included 42 "others" (normal breast tissue, cancer adjacent to normal breast, inflamed tissue, hyperplastic and benign tissue), 104 "malignant" and 31 "metastatic." The percent of each group expressing high (score = 2+, 3+; black bars) or low (score = 0, 1+; white bars) DHHC3 is indicated. D, MDA-MB-231 cells ± ZDHHC3 ablation were injected orthotopically into female nude mice (5 mice per group with two tumors/mouse), and tumor volumes (left) and weights (at 35 days; right) were determined. E, MDA-MB-231 cells ± ZDHHC3 ablation were injected into tail veins of SCID beige mice (three mice per group). After 30 days, metastatic foci (arrows) in lungs were assessed by H&E staining (scale bar, 200 µm) and colony diameters were quantitated (N = 77 control, 86 D3 knockdown colonies).
Differential protein palmitoylation
Using described procedures (16), protein lysates from MDA-MB-231 cells were subjected to TCEP [tris(2-carboxyethyl)phosphine] treatment to selectively reduce all disulfide bonds, followed by irreversible alkylation with NEM (N-ethylmaleimide) to block all free sulfhydryl moieties. Then, proteins were subjected to hydroxylamine treatment to cleave thioester bonds and newly freed SH moieties subsequently were biotinylated using BMCC-biotin reagent. Finally, biotinylated proteins were immunoprecipitated using neutrAvidin agarose beads, and transferred to nylon membrane for blotting.
Statistical analysis
For evaluation of statistical significance, unpaired t tests were used unless otherwise indicated.
Results
DHHC3 upregulation and cancer patient survival
Analysis of publicly available human breast invasive carcinoma patient data, from the cBioPortal TCGA database, indicated that elevated ZDHHC3 gene expression significantly correlated with diminished patient overall survival (Fig. 1A). Furthermore, DHHC3 protein expression was upregulated significantly in malignant human breast cancer and even more in metastatic breast cancer samples, in comparison with noncancerous breast tissue (Fig. 1B and C). DHHC3 protein was also significantly upregulated in 4 of 5 major malignant primary breast cancer subtypes and 5 of 5 major metastatic breast cancer subtypes (Supplementary Fig. S1A and S1B).
Upregulated ZDHHC3 gene expression correlated with significantly diminished overall patient survival in six other cancer types (Supplementary Table S1). Upregulation of ZDHHC7, the protein acyl transferase with most sequence similarity to ZDHHC3, correlated with significantly diminished overall survival in 2 of 7 cancers (Supplementary Table S1). In contrast, upregulation of other DHHC genes (ZDHHC5 and ZDHHC17), although previously linked to tumor xenograft growth, did not correlate with survival of these patients, except for ZDHHC5 in melanoma (Supplementary Table S1).
DHHC3 supports breast cancer xenograft growth
MDA-MB-231 human breast cancer cells stably ablated for ZDHHC3 (D3) yielded significantly reduced xenograft growth in female nude mice, whether in mammary fat pads (Fig. 1D, Supplementary Fig. S1C) or at a subcutaneous site (Supplementary Fig. S1D). Diminished ZDHHC3-ablated tumor growth was independently confirmed using another shRNA targeting sequence (Supplementary Fig. S1E). After injection of ZDHHC3-ablated MDA-MB-231 cells into SCID mice tail veins, H&E-stained lung sections (4 weeks) showed significant reductions in colony size (Fig. 1E) and colony number (Supplementary Fig. S1F), compared with control shRNA–injected cells. ZDHHC3 knockdown efficiency was >90%, as shown by DHHC3 immunoblotting (Supplementary Fig. S2A) and by immunohistochemically stained xenograft tumor sections (Supplementary Fig. S2B).
Mechanistic insights into effects of DHHC3 ablation
ZDHHC3 ablation minimally affected primary tumor xenograft angiogenesis (CD31 staining; Supplementary Fig. S2C), cell proliferation (Ki67 staining), or apoptosis (TUNEL assay, caspase-3 cleavage). Furthermore, ZDHHC3 ablation minimally affected in vitro cell proliferation (up to 72 hours) or 3D soft agar growth (14 days; Supplementary Fig. S2D and S2E). However, invasion through Matrigel was significantly reduced (Supplementary Fig. S2F), which may partly explain reduced lung colony numbers (Supplementary Fig. S1F).
For unbiased mechanistic insight, DNA microarray analysis was performed on ZDHHC3-ablated orthotopic xenograft tumor samples. The complete list of results (Supplementary Table S2) included 25 upregulated and 27 downregulated genes (>1.5-fold increase or decrease; P < 0.05; Supplementary Table S3). ZDHHC3 itself topped the list of downregulated genes. Importantly, changes in 29 of 52 genes (55.8%; Supplementary Table S3) are consistent with increased oxidative stress and/or senescence. Altered expression of 21 additional genes typically linked to senescence, but not quite meeting rigorous criteria for inclusion in Supplementary Table S3, are indicated in Supplementary Table S4. Tumor suppressor genes VGLL3 and TXNIP were also upregulated in ZDHHC3-ablated samples (Supplementary Table S3).
DHHC3 ablation promotes oxidative stress
A fluorescent dye conversion assay confirmed that oxidative stress/ROS is significantly elevated in stable ZDHHC3-ablated MDA-MB-231 (Fig. 2A), and in siRNA ZDHHC3-ablated MCF-7 cells (Supplementary Fig. S3A). Oxidative stress inhibitors NAC (N-acetyl cysteine; Supplementary Fig. S3B) and α-LA (α-lipoic acid) substantially prevented ZDHHC3 ablation effects. Increased oxidative stress can decrease tyrosine phosphatase activities (17), which can elevate tyrosine phosphorylation of focal adhesion kinase (FAK) and STAT3 proteins (18, 19). Accordingly, siRNA or shRNA ablation of ZDHHC3 markedly increased tyrosine phosphorylation of FAK and STAT3 in MDA-MB-231 (Fig. 2B), and STAT3 in MCF-7 and ZR-75 mammary cells (Supplementary Fig. S3C). These results are consistent with diminished phosphatase activity. In contrast, tyrosine phosphorylation of EGFR protein, not known to be affected by oxidative stress, was unaltered (Fig. 2B). In addition, serine phosphorylation of ERK (as another negative control) was unchanged (Fig. 2B). As further evidence for diminished phosphatase activity, ZDHHC3 ablation diminished time-dependent FAK dephosphorylation in suspended MDA-MB-231 cells (Supplementary Fig. S3D). Oxidative stress inhibitors NAC, α-LA, and atorvastatin substantially prevented effects of ZDHHC3 ablation on FAK tyrosine phosphorylation (Fig. 2C, top). Together, these results support ZDHHC3-ablation diminishing tyrosine phosphatase activity due to increased oxidative stress.
Figure 2.
DHHC3 ablation promotes oxidative stress. A, MDA-MB-231 cells ± stable ZDHHC3 ablation were loaded with CellROX dye and oxidative stress was assessed using flow cytometry. Bottom, mean percent of cells with staining elevated above cut-off line (± SD; n = 3). B, MDA-MB-231 cells ± ZDHHC3 ablation with siRNA or shRNA were lysed, FAK and EGFR proteins were immunoprecipitated, and resulting samples were blotted with tyrosine phosphorylation antibody, 4G10. Also, cell lysates were directly blotted with anti-phosphotyrosine-STAT3 and anti-phosphoserine ERK antibodies. Control panels show total FAK, EGFR, STAT3, and ERK proteins. Numbers at the bottom of Western blots indicate levels of p-FAK and p-STAT3 normalized against total FAK and STAT3 levels, respectively. Bottom, DHHC3 levels ± siRNA and shRNA ablation. The images shown are representative of each experiment performed multiple times. C, Effects of oxidative stress inhibitors (NAC, α-LA, and atrovastatin) on FAK phosphorylation (p-FAK) and MCP-1 protein secretion in/from control and ZDHHC3-ablated MDA-MB-231 cells.
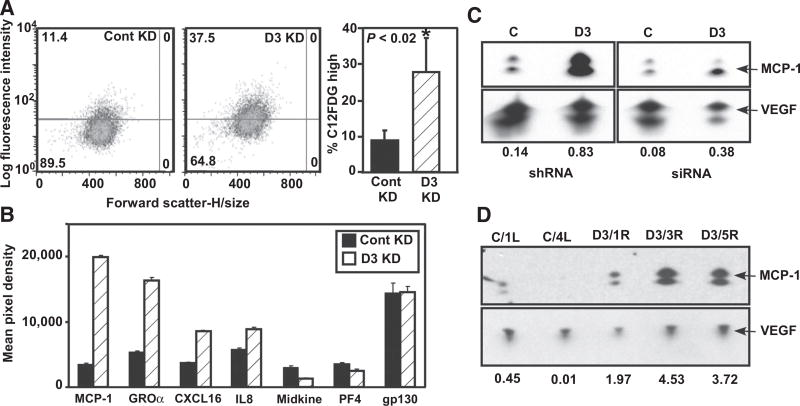
DHHC3 ablation enhances cellular senescence
Increased oxidative stress often triggers senescence (9, 10, 20). Consistent with this (and with results in Supplementary Table S3), ZDHHC3 ablation markedly elevated senescence-associated β-galactosidase (SA-β-Gal) activity in MDA-MB-231 (Fig. 3A) and MCF-7 cells (Supplementary Fig. S3E). Furthermore, ZDHHC3-ablated MDA-MB-231 cells increased secretion (Fig. 3B) of proteins (MCP-1, GROα, CXCL16, and IL8) linked to senescence (21). Secretion of three other proteins was not increased (Fig. 3B), and 24 other chemokines were absent or barely detectable. Enhanced MCP-1 secretion was confirmed by Western blotting (Fig. 3C, top), whereas VEGF control protein was not upregulated (Fig. 3C, bottom). Increased secretion of MCP-1 (but not VEGF) was further validated in 3 of 3 ex vivo tumor cell lines derived from ZDHHC3-ablated xenograft tumors, compared with two control (i.e., nonablated) tumor-derived cell lines (Fig. 3D). Oxidative stress inhibitors (NAC, α-LA, Atorvastatin) markedly diminished MCP-1 upregulation in ZDHHC3-ablated cells (Fig. 2C, bottom), consistent with senescence being triggered by oxidative stress.
Figure 3.
DHHC3 ablation enhances cellular senescence. A, MDA-MB-231 cells ± stable ZDHHC3 ablation were loaded with senescence indicator dye C12FDG, which was measured using flow cytometry. Mean percent elevated staining (± SD; n = 3) is shown. B, MDA-MB-231 cells, ± stable ZDHHC3 ablation, were cultured for 30 hours, and cell supernatants were analyzed. Results for 7 chemokines are shown (mean ± SD; n = 2 replicates). Twenty-four other chemokines were not detected and/or not different. C, Enhanced secretion of MCP-1 chemokine from ZDHHC3-ablated MDA-MB-231 cells was validated by Western blotting (representative of multiple experiments), with VEGF secretion used as a control. D, Enhanced MCP-1 secretion was also assessed in supernatants from cells obtained from xenograft tumors. C/1L and C/4R cell samples were cultured from control xenograft tumors; D3/1R, D3/3R, and D3/5R samples are from ZDHHC3-ablated tumor-derived cells. Numbers at the bottom represent MCP-1 levels normalized against VEGF levels.
Recruitment of innate immune cells—further evidence for senescence
The upregulated chemokine pattern (Fig. 3B) emulates a senescence-associated secretory phenotype (SASP) response, which triggers innate immune cell recruitment, and facilitates tumor clearance (11). Indeed, ZDHHC3-ablated xenografts showed enhanced iNOS staining, indicative of antitumor "M1-like" macrophages (Fig. 4A) and moderate upregulation of panmacrophage marker CD68 (Fig. 4B). Conversely, diminished expression of Arginase 1 indicates fewer protumor "M2-like" macrophages (Fig. 4C). Furthermore, elevated granzyme F levels (Fig. 4D) indicate increased NK cell recruitment into tumors of reduced size (as in Fig. 1D, Supplementary Fig. S1D). Notably, of five ZDHHC3-ablated tumors, the one with least granzyme F elevation had the largest tumor volume. Another NK-cell marker, NKTR (natural killer cell triggering receptor), also showed significant upregulation in ZDHHC3-ablated tumors (Fig. 4E). These results are consistent with the innate immune system contributing to reduced tumor appearance.
Figure 4.
DHHC3 ablation affects tumor recruitment of immune cells. A, IHC for iNOS (marker for antitumor "M1-like" macrophages) is shown on representative paraffin sections of control and ZDHHC3-ablated orthotopic tumors (at 35 days, as in Fig. 1D; Supplementary Fig. S2A). Right, percent iNOS positive area from multiple sections (three tumors per group, quantitated using Image J software; mean ± SD). B, Expression of pan-macrophage marker CD68 was assessed, using RT-PCR, from two tumor sections each from control and DHHC3 ablated xenografts. C and D, qPCR was used to assess expression of Arginase 1 (marker for protumor "M2-like" macrophages) and granzyme F (secreted by NK cells). qPCR results are shown as mean ± SD; n = 6 in each group. E, RT-PCR was used to assess expression of NK cell marker NKTR in control and ZDHHC3-ablated xenograft tumors. Shown is mean ± SD; n = 2 tumors per group).
In an in vitro model system, THP1 cells were stimulated by LPS and IFNγ to differentiate into iNOS-positive "M1-like" macrophages (Supplementary Fig. S4A). Conditioned media was then collected from control and ZDHHC3-ablated MDA-MB-231 breast cancer cells, to use in a Transwell migration assay as a chemoattractant. As indicated (Supplementary Fig. S4B), THP1-derived "M1-like" macrophages showed significant preferential migration toward conditioned media from ZDHHC3-ablated breast cancer cells. ZDHHC3 ablation increased secretion of chemoattractant proteins MCP-1 and IL8 (Supplementary Fig. S4C), consistent with enhanced "M1-like" macrophage chemoattraction. In another control assay, undifferentiated THP1 cells showed essentially no Transwell migration.
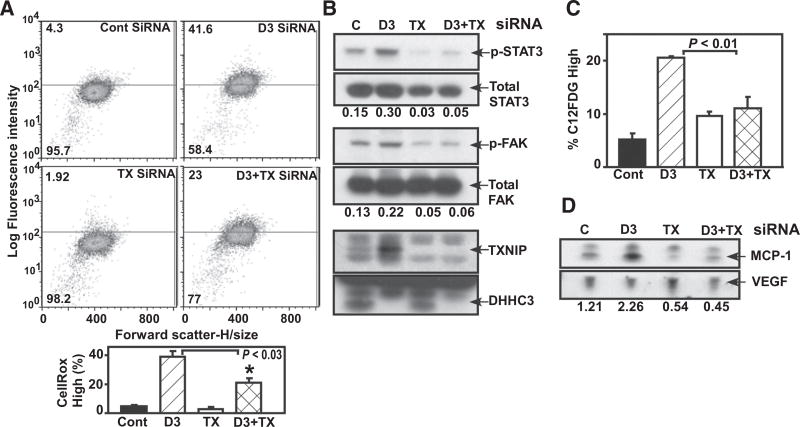
TXNIP contributes to the DHHC3 ablation phenotype
Among genes with expression altered by ZDHHC3 ablation, we focused on TXNIP (#5 in Supplementary Table S3), which is linked to oxidative stress, senescence, and tumor suppression (10, 22, 23). Enhanced TXNIP protein expression was validated in multiple breast cancer lines (Supplementary Fig. S4D) and in tumor xenograft–derived cells (Supplementary Fig. S4E). Notably, TXNIP removal from MDA-MB-231 cells partially, but significantly, reversed oxidative stress (Fig. 5A), and almost completely reversed ZDHHC3 ablation effects on oxidative stress–dependent STAT3 and FAK tyrosine phosphorylation (Fig. 5B). DHHC3 and TXNIP protein knockdowns were >90% (Fig. 5B, bottom). TXNIP removal also largely reversed senescence upregulation (colorimetric senescence assay, Fig. 5C; senescence-associated MCP-1, Fig. 5D). TXNIP or ZDHHC3 ablation did not upregulate control VEGF protein (Fig. 5D).
Figure 5.
TXNIP contributions to DHHC3 ablation phenotype. A–C, MDA-MB-231 cells were transiently transfected with control, DHHC3, TXNIP siRNA, and DHHC3+TXNIP siRNA. After 60 hours, cells were analyzed for oxidative stress via CellRox dye (A); tyrosine phosphorylation of STAT3 and FAK proteins (B), and senescence via C12FDG assay (C). D, Secreted MCP-1 chemokine. Values at the bottom of Western blots (B and D) are normalized levels of p-STAT3, p-FAK, and MCP-1 proteins. Data shown are representative of multiple experiments, and bar graphs in panels A and C show mean ± SD; n = 3.
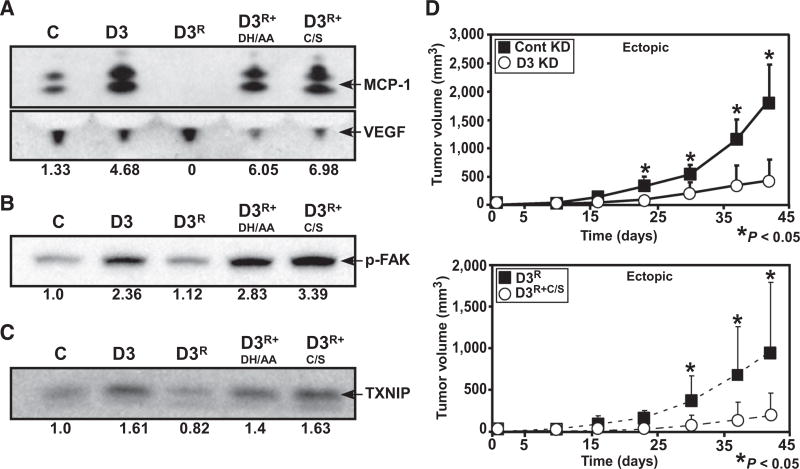
DHHC3 ablation effects are specific and require the enzyme-active site
To confirm ZDHHC3 ablation specificity, we designed rescue vector D3R, containing ZDHHC3 knockdown shRNA, ZDHHC3 cDNA resistant to the shRNA, 2A peptide linker, and GFP tag (Supplementary Fig. S5A). Reconstitution vectors D3R+DH/AA and D3R+C/S contained DHHC3 active site mutations (DHHC→AAHC; DHHC→DHHS) that abolish palmitoylation activity (1, 3). By GFP analysis (Supplementary Fig. S5B), all three vectors are well expressed in MDA-MB-231 cells, with D3R+DH/AA and D3R+C/S present at somewhat higher levels than D3R. Importantly, D3R expression completely reversed ZDHHC3 ablation effects, with respect to increased MCP-1 (indicative of senescence), FAK tyrosine phosphorylation (indicative of oxidative stress), and TXNIP (indicative of oxidative stress and senescence; Fig. 6A–C). Protein levels for D3R are approximately 2-fold greater than endogenous DHHC3 in nonablated control cells. This helps to explain why MCP-1 (Fig. 6A) and TXNIP (Fig. 6C) drop below control levels (compare control "C" and "D3R" results). In contrast, reconstitution with active site mutants (D3R+DH/AA and D3R+C/S) failed to reverse ZDHHC3 knockdown effects on oxidative stress and senescence (Fig. 6A–C, compare lanes 4 and 5 with lane 2 in each of the three panels).
Figure 6.
Reversal of DHHC3 ablation effects by wild-type and mutant DHHC3 re-expression. MDA-MB-231 cells stably expressing control shRNA (C), DHHC3 shRNA (D3), DHHC3 rescue vector (D3R) unmutated or with palmitoylation site mutations (D3R+DH/AA and D3R+C/S) were analyzed by Western blotting for MCP-1 secretion (A), FAK protein tyrosine phosphorylation (p-FAK; B), and TXNIP expression (C). Blots shown are representative of multiple experiments, and values at the bottom represent normalized protein levels. D, Ectopic xenograft growth in female nude mice (four mice in each group with two tumors/mouse) was carried out simultaneously for control and ZDHHC3-ablated cells (top graph) and reconstituted (D3R, D3R+C/S) cells (bottom graph).
Recapitulating Fig. 1D results, stable ZDHHC3 ablation significantly reduced in vivo MDA-MB-231 tumor xenograft growth (Fig. 6D, top). In the same experiment, tumor xenograft growth was almost completely restored upon ZDHHC3 reconstitution, using rescue vector D3R (Fig. 6D, bottom). As expected, reconstitution with the D3R+C/S negative control mutant did not reverse the diminished tumor xenograft growth that was caused by ZDHHC3 ablation (Fig. 6D, bottom).
Disruption of ERGIC3 upregulates TXNIP
To understand how ZDHHC3-ablation might upregulate TXNIP, we focused attention on disruption of ERGIC3 (endoplasmic reticulum Golgi intermediate compartment-3) protein, which is known to cause ER stress (24), which then upregulates TXNIP (25). First, we confirmed again that ZDHHC3 ablation in MDA-MB-231 cells upregulates TXNIP (Fig. 7A, top, lane 2) and then showed that ERGIC3 ablation upregulates TXNIP to an even greater extent (lane 3). Second, we established that ZDHHC3 ablation causes a marked decrease in ERGIC3 palmitoylation (Fig. 7B), accompanied by ERGIC3 subcellular distribution becoming considerably less punctate (Fig. 7C). In a control experiment, DHHC3 ablation did not alter the amount of total ERGIC3 protein in the cell lysate (Fig. 7B).
Figure 7.
DHHC3 ablation effects on ERGIC3. A, After MDA-MB-231 cells were transfected with control (C), DHHC3 (D3), and ERGIC3 (E3) siRNAs (60 hour), expression of TXNIP, ERGIC3, and DHHC3 proteins were assessed by Western blotting. B, After reduction and alkylation steps (see Materials and Methods), protein lysates from control (C) and DHHC3 (D3) siRNA–ablated MDA-MB-231 cells were treated with hydroxylamine (HA) to cleave thioester bonds from palmitoylated proteins. Newly exposed SH moieties (which should be less abundant in putative substrates for DHHC3) were biotinylated. After immunoprecipitation using neutrAvidin agarose beads and transfer to nylon membrane, blotting with ERGIC3 antibody was carried out. Numbers indicate relative amounts in "D3" versus "C" lanes. C, Distribution of ERGIC3 in control and DHHC3-ablated cells was analyzed in permeabilized MDA-MB-231 cells by IHC staining using anti-ERGIC3 antibody (green). Nuclei were stained with DAPI (blue). Results similar to those in panels A–C were obtained in multiple independent experiments.
Discussion
Here we show initial evidence for DHHC3 having a key role in human breast cancer. Also, data are provided suggesting that DHHC3 enables tumor expansion in vivo, by a mechanism requiring an active enzyme palmitoylation site, and involving downmodulation of oxidative stress and senescence in cancer cells. DHHC3 was not previously known to function in breast cancer or other cancers. Furthermore, neither DHHC3, nor other mammalian DHHC enzymes, were known to control oxidative stress or senescence. Hence, these studies represent new research findings.
DHHC3 and tumor growth
Elevated ZDHHC3 gene expression correlates with significantly reduced human breast cancer patient survival. Furthermore, DHHC3 protein levels were elevated in malignant breast cancer, and even more in metastatic breast cancer. These results, combined with ZDHHC3 ablation effects on ectopic and orthotopic tumor xenograft size and metastatic lung colony size, strongly suggest a novel pro-breast tumor growth role for DHHC3.
Notably, ZDHHC3 gene upregulation correlated with reduced patient survival in six other human cancers. Also, DHHC3 protein expression was elevated in most breast cancer subtypes (our results), and in prostate and colon cancers (6). In addition, ZDHHC3 ablation reduced prostate cancer xenograft growth (to be published elsewhere). Hence, DHHC3 contributes to growth of multiple cancer types. Upregulation of ZDHHC7, closest homolog to ZDHHC3, correlated with reduced patient survival in 2 of 7 human cancers. In contrast, upregulation of other potentially oncogenic DHHC enzymes [i.e., ZDHHC5 (4) and ZDHHC17 (5)] was, with one exception, not significantly correlated with reduced human survival in the same seven cancers. Other DHHC-type enzymes have been suggested to show cancer expression correlations, or palmitoylate cancer–related substrates (26, 27), but definitive cancer links remain to be established.
Mechanistic insights: a role for oxidative stress
ZDHHC3 ablation minimally affected cell proliferation or soft agar growth in vitro, or cell proliferation, angiogenesis, or apoptosis in vivo. However, unbiased DNA array analysis results (for 12/52 genes with significantly altered expression) strongly suggested oxidative stress upregulation in ZDHHC3-ablated tumors. Upregulation of six genes (GTF2i, TXNIP, AVIL, FKBP11, SETD6, and SETX) and downregulation of six other genes (S100A4, PDE4B, HNMT, NUDT2, AKR1C1, and GSTZ1) is consistent with increased oxidative stress in ZDHHC3-ablated MDA-MB-231 xenograft tumor cells. Upregulation of TXNIP is particularly notable. TXNIP (thioredoxin inhibitory protein) binds to, and inhibits antioxidant function of thioredoxin protein TRX-1, thus enhancing oxidative stress (10, 28).
Increased oxidative stress in ZDHHC3-ablated cells was confirmed, in multiple cell types, by measuring increased ROS levels. Both direct and indirect effects of oxidative stress were substantially reversed by oxidative stress inhibitors (NAC, α-LA, atorvastatin) or by knockdown of TXNIP, a major oxidative stress-inducing gene upregulated in ZDHHC3-ablated cells. Increased oxidative stress was further confirmed by typical downstream oxidative stress–dependent events, such as induced senescence (next section), and diminished phosphatase activity (17). The latter is manifested as (i) increased tyrosine phosphorylation (of FAK and STAT3), and (ii) diminished time-dependent loss of tyrosine phosphorylation (of FAK).
Elevated FAK and STAT3 tyrosine phosphorylation may also contribute to deficiencies seen in ZDHHC3-ablated tumor cells. FAK hyperphosphorylation caused by diminished tyrosine phosphatase activity may interfere with FAK turnover, resulting in diminished FAK function (29, 30), which would adversely affect tumor cell invasion and dissemination. Regarding STAT3, elevated tyrosine phosphorylation is not only oncogenic, but also may be suppressive by multiple potential mechanisms (ref. 31 and references within), including induction of cell senescence (32, 33).
Although the transcription factor NRF2 has been termed the "master regulator" of antioxidant responses (8), NRF2 expression was only slightly upregulated (1.2-fold) in ZDHHC3-ablated cells. Furthermore, expression levels of 22 different redox-related genes known to be controlled by NRF2 (8) were only minimally altered in ZDHHC3-ablated cells. Hence, NRF2 does not appear to play a role in DHHC3-regulated redox-related events.
Elevated oxidative stress leads to senescence
Increased oxidative stress, coupled with other tumor microenvironment stresses, can limit tumor survival by triggering apoptosis (34). However, instead of increased apoptosis in DHHC3-ablated tumors, we observed oxidative stress-dependent induction of senescence (20, 35). Evidence for increased senescence includes 10 upregulated senescence-linked genes (PIP5K1b, COL6A3, TXNIP, AMY1A, RSRP1, ITSN2, KLHL28, ATM, TRIM38, and COL13A1) and downregulation of 8 genes typically diminished during senescence (BCL2A1, ANAPC15, PF4, HePTP, CDKN3, CENPN, LIPA, HCLS1). Also significantly altered were CDKN2A, PAI-1, HLA-DRB4, TJP1, RBBP6, IGFBP7, Smurf2, Ask1, THBS1, CBX1, GRP170, CDKN2C, and CCNA2 genes (Supplementary Table S4), which either support senescence or are senescence markers. Additional genes listed in Supplementary Table S4 (Esm1, PDGFB, MAPKAPK2, PDGFC, TP53, and FGF5) showed altered expression, not quite meeting criteria for inclusion in Supplementary Table S3, but nonetheless consistent with enhanced senescence.
Multiple ZDHHC3-ablated cell lines showed increased β-galactosidase activity, indicative of senescence (36). We also demonstrated selective upregulation of specific chemokine proteins (MCP-1, GROα, CXCL16, IL8), characteristic of SASP (senescence-activated secreted protein) responses (21). Increased senescence, measured by β-galactosidase activity and/or SASP marker MCP-1, was substantially reversed by oxidative stress inhibitors (NAC, α-LA, atorvastatin). Senescence in ZDHHC3-ablated cells was also substantially reversed upon knockdown of TXNIP, which supports both oxidative stress and senescence (10, 22). These results further reinforce a mechanism of ZDHHC3 ablation→diminished palmitoylation of key substrates→TXNIP upregulation→oxidative stress→senescence (see summary scheme in Supplementary Fig. S5C).
Consistent with an SASP response (37), we observed increased recruitment of anti-tumor "M1-like" macrophages and NK cells. Furthermore, we confirmed that conditioned media from ZDHHC3-ablated MDA-MB-231 cells (containing SASP elements MCP-1 and IL8) indeed can recruit antitumor "M1-like" macrophages in an in vitro model system. These results are consistent with SASP-dependent clearance of ZDHHC3-ablated tumors by innate immune cells. Diminished recruitment of protumor "M2-like" macrophages (38) may further reduce in vivo growth of DHHC3-ablated tumors.
Reconstitution and a requirement for the DHHC3-active site
Reconstitution with wild-type ZDHHC3 reversed ZDHHC3 knockdown effects on TXNIP protein levels, oxidative stress, senescence and in vivo tumor growth. These results, plus similar results obtained using multiple RNAi-targeting sequences, effectively rule out off-target shRNA/siRNA effects. A conserved "DHHC" motif is required for palmitoyl transferase activity of DHHC3 and other DHHC enzymes (1). Hence, we reconstituted ZDHHC3-ablated cells with two DHHC active site disabled mutants. Both mutants failed to reverse ZDHHC3 ablation effects on oxidative stress, senescence, TXNIP expression, or related events. Furthermore, our D3C/S mutant failed to restore tumor growth in vivo. Hence, DHHC3 palmitoylation activity is needed for reconstituted functions. Although overexpressed DHHC3/GODZ was claimed previously to mediate Ca2+ transport (39), ZDHHC3 ablation did not alter calcium levels in our cells.
DHHC3 palmitoylation activity
The requirement for the DHHC3 palmitoylation active site focused attention on potentially important DHHC3 substrates. Among approximately 50 putative protein substrates palmitoylated by DHHC3 (manuscript in preparation), we focused on ERGIC3, because disruption of ERGIC3 is a known trigger of ER stress (24), which leads to upregulation of TXNIP (25). We confirmed that ERGIC3 ablation upregulates TXNIP, and that ZDHHC3 ablation markedly diminishes ERGIC3 palmitoylation, stimulates ER stress (unpublished result) and considerably alters ERGIC3 subcellular distribution. Hence, diminished ERGIC3 palmitoylation and altered ERGIC3 distribution (and presumably also function) appear to be key mechanistic consequences of DHHC3 ablation (Fig. 7). Loss of DHHC3 also caused reduced palmitoylation of several other proteins, including a few antioxidant-type proteins (manuscript in preparation). We suggest that this additionally contributes to oxidative stress, which can further enhance ER stress (40), and could then further amplify TXNIP upregulation (Step 3, Supplementary Fig. S5C).
Although GABA(A) receptor (41), integrin α6 and β4 subunits (13), G protein α subunit (42), regulator of G-protein signaling 4 (RGS4) (43), and phosphatidylinositol 4-kinase IIα (PI4KIIα; ref. 44) are reported to be DHHC3 substrates, it is unclear that diminished palmitoylation of these few proteins would cause diminished in vivo tumor growth, oxidative stress and/or senescence. Ablation of ZDHHC3 diminished α6β4 integrin palmitoylation and partially reduced integrin expression (13). However, this may not be relevant to current results because α6β4 ablation in MDA-MB-231 cells did not diminish oxidative stress, senescence, or TXNIP expression.
Conclusions
In conclusion, DHHC3 promotes in vivo breast tumor growth, by a mechanism involving palmitoylation of key substrate proteins such as ERGIC3. ZDHHC3 ablation not only diminishes in vivo breast tumor growth, but also promotes oxidative stress and senescence. This may explain reduced tumor growth because oxidative stress can diminish tumor growth and/or metastasis (7, 8, 45) and senescence can lead to tumor clearance by immune cells (11, 12, 46). As a key supporter of breast tumor growth, DHHC3 may be a useful cancer target. Furthermore, we predict that targeting of DHHC3, which enhances oxidative stress, should also markedly improve sensitivity to a variety of oxidative stress-dependent anticancer drug types (7, 47).
Supplementary Material
Acknowledgments
C. Sharma, H-W. Wang, Q. Li, K. Knoblich, and M.E. Hemler received support from NIH grant CA42368. A.L. Richardson received support from Breast Cancer Research Foundation.
We acknowledge Fenghui Xu and Viviana Cremasco for help in mouse studies and cell sorting, respectively.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: M.E. Hemler, C. Sharma, H.-X. Wang
Development of methodology: M.E. Hemler, C. Sharma, H.-X. Wang
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M.E. Hemler, C. Sharma, H.-X. Wang, K. Knoblich, E.S. Reisenbichler, A.L. Richardson
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M.E. Hemler, C. Sharma, H.-X. Wang, Q. Li, K. Knoblich, A.L. Richardson
Writing, review, and/or revision of the manuscript: M.E. Hemler, C. Sharma
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M.E. Hemler, C. Sharma
Study supervision: M.E. Hemler, C. Sharma
References
- 1.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–27. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 3.Politis EG, Roth AF, Davis NG. Transmembrane topology of the protein palmitoyl transferase Akr1. J Biol Chem. 2005;280:10156–63. doi: 10.1074/jbc.M411946200. [DOI] [PubMed] [Google Scholar]
- 4.Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD. Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene. 2004;23:9230–7. doi: 10.1038/sj.onc.1208171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian H, Lu JY, Shao C, Huffman KE, Carstens RM, Larsen JE, et al. Systematic siRNA screen unmasks NSCLC growth dependence by palmitoyltransferase DHHC5. Mol Cancer Res. 2015;13:784–94. doi: 10.1158/1541-7786.MCR-14-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, et al. A global map of human gene expression. Nat Biotechnol. 2010;28:322–4. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–91. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 8.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–47. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 9.Hwang JW, Yao H, Caito S, Sundar IK, Rahman I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic Biol Med. 2013;61:95–110. doi: 10.1016/j.freeradbiomed.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahmood DF, Abderrazak A, El HK, Simmet T, Rouis M. The thioredoxin system as a therapeutic target in human health and disease. Antioxid Redox Signal. 2013;19:1266–303. doi: 10.1089/ars.2012.4757. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Mancera PA, Young AR, Narita M. Inside and out: the activities of senescence in cancer. Nat Rev Cancer. 2014;14:547–58. doi: 10.1038/nrc3773. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–76. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Sharma C, Rabinovitz I, Hemler ME. Palmitoylation by DHHC3 is critical for the function, expression and stability of integrin α6β4. Cell Mol Life Sci. 2012;69:2233–44. doi: 10.1007/s00018-012-0924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanner JJ, Parsons ZD, Cummings AH, Zhou H, Gates KS. Redox regulation of protein tyrosine phosphatases: structural and chemical aspects. Antioxid Redox Signal. 2011;15:77–97. doi: 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- 18.Carballo M, Conde M, El BR, Martin-Nieto J, Camacho MJ, Monteseirin J, et al. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J Biol Chem. 1999;274:17580–6. doi: 10.1074/jbc.274.25.17580. [DOI] [PubMed] [Google Scholar]
- 19.Ben Mahdi MH, Andrieu V, Pasquier C. Focal adhesion kinase regulation by oxidative stress in different cell types. IUBMB Life. 2000;50:291–9. doi: 10.1080/713803721. [DOI] [PubMed] [Google Scholar]
- 20.Panieri E, Gogvadze V, Norberg E, Venkatesh R, Orrenius S, Zhivotovsky B. Reactive oxygen species generated in different compartments induce cell death, survival, or senescence. Free Radic Biol Med. 2013;57:176–87. doi: 10.1016/j.freeradbiomed.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–68. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riahi Y, Kaiser N, Cohen G, Abd-Elrahman I, Blum G, Shapira OM, et al. Foam cell-derived 4-hydroxynonenal induces endothelial cell senescence in a TXNIP-dependent manner. J Cell Mol Med. 2015;19:1887–99. doi: 10.1111/jcmm.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison JA, Pike LA, Sams SB, Sharma V, Zhou Q, Severson JJ, et al. Thioredoxin interacting protein (TXNIP) is a novel tumor suppressor in thyroid cancer. Mol Cancer. 2014;13:62. doi: 10.1186/1476-4598-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong SH, Chang SH, Cho KC, Kim S, Park S, Lee AY, et al. Endoplasmic reticulum-Golgi intermediate compartment protein 3 knockdown suppresses lung cancer through endoplasmic reticulum stress-induced autophagy. Oncotarget. 2016;7:65335–47. doi: 10.18632/oncotarget.11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oslowski CM, Hara T, O'Sullivan-Murphy B, Kanekura K, Lu S, Hara M, et al. Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell Metab. 2012;16:265–73. doi: 10.1016/j.cmet.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaves J, Chamberlain LH. New links between S-acylation and cancer. J Pathol. 2014;233:4–6. doi: 10.1002/path.4339. [DOI] [PubMed] [Google Scholar]
- 27.Yeste-Velasco M, Linder ME, Lu YJ. Protein S-palmitoylation and cancer. Biochim Biophys Acta. 2015;1856:107–20. doi: 10.1016/j.bbcan.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Cadenas C, Franckenstein D, Schmidt M, Gehrmann M, Hermes M, Geppert B, et al. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res. 2010;12:R44. doi: 10.1186/bcr2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu DH, Qu CK, Henegariu O, Lu X, Feng GS. Protein-tyrosine phosphatase Shp-2 regulates cell spreading, migration, and focal adhesion. J Biol Chem. 1998;273:21125–31. doi: 10.1074/jbc.273.33.21125. [DOI] [PubMed] [Google Scholar]
- 30.Manes S, Mira E, Gomez-Mouton C, Zhao ZJ, Lacalle RA, Martinez A. Concerted activity of tyrosine phosphatase SHP-2 and focal adhesion kinase in regulation of cell motility. Mol Cell Biol. 1999;19:3125–35. doi: 10.1128/mcb.19.4.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang HF, Chen Y, Wu C, Wu ZY, Tweardy DJ, Alshareef A, et al. The opposing function of STAT3 as an oncoprotein and tumor suppressor is dictated by the expression status of STAT3beta in esophageal squamous cell carcinoma. Clin Cancer Res. 2016;22:691–703. doi: 10.1158/1078-0432.CCR-15-1253. [DOI] [PubMed] [Google Scholar]
- 32.Leung JY, Wilson HL, Voltzke KJ, Williams LA, Lee HJ, Wobker SE, et al. Sav1 loss induces senescence and Stat3 activation coinciding with tubulointerstitial fibrosis. Mol Cell Biol. 2017;37:e00565. doi: 10.1128/MCB.00565-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JJ, Lee JS, Cui MN, Yun HH, Kim HY, Lee SH, et al. BIS targeting induces cellular senescence through the regulation of 14–3-3 zeta/STAT3/SKP2/p27 in glioblastoma cells. Cell Death Dis. 2014;5:e1537. doi: 10.1038/cddis.2014.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda S, Nakamura H, Masutani H, Sasada T, Yonehara S, Takabayashi A, et al. Redox regulation of caspase-3(-like) protease activity: regulatory roles of thioredoxin and cytochrome c. J Immunol. 1998;161:6689–95. [PubMed] [Google Scholar]
- 35.Dasari A, Bartholomew JN, Volonte D, Galbiati F. Oxidative stress induces premature senescence by stimulating caveolin-1 gene transcription through p38 mitogen-activated protein kinase/Sp1-mediated activation of two GC-rich promoter elements. Cancer Res. 2006;66:10805–14. doi: 10.1158/0008-5472.CAN-06-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 37.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–7. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Hines RM, Kang R, Goytain A, Quamme GA. Golgi-specific DHHC zinc finger protein GODZ mediates membrane Ca2+ transport. J Biol Chem. 2010;285:4621–8. doi: 10.1074/jbc.M109.069849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verfaillie T, Rubio N, Garg AD, Bultynck G, Rizzuto R, Decuypere JP, et al. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19:1880–91. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, et al. The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J Neurosci. 2004;24:5881–91. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutsumi R, Fukata Y, Noritake J, Iwanaga T, Perez F, Fukata M. Identification of G protein alpha subunit-palmitoylating enzyme. Mol Cell Biol. 2009;29:435–47. doi: 10.1128/MCB.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Xie Y, Wolff DW, Abel PW, Tu Y. DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation. FEBS Lett. 2010;584:4570–4. doi: 10.1016/j.febslet.2010.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu D, Sun HQ, Wang H, Barylko B, Fukata Y, Fukata M, et al. Phosphatidylinositol 4-kinase IIalpha is palmitoylated by Golgi-localized palmitoyltransferases in cholesterol-dependent manner. J Biol Chem. 2012;287:21856–65. doi: 10.1074/jbc.M112.348094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woditschka S, Evans L, Duchnowska R, Reed LT, Palmieri D, Qian Y, et al. DNA double-strand break repair genes and oxidative damage in brain metastasis of breast cancer. J Natl Cancer Inst. 2014;106:dju145. doi: 10.1093/jnci/dju145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ewald JA, Desotelle JA, Wilding G, Jarrard DF. Therapy-induced senescence in cancer. J Natl Cancer Inst. 2010;102:1536–46. doi: 10.1093/jnci/djq364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Kim J, Bae J-S. ROS homeostasis and metabolism: a critical liason for cancer therapy. Exp Mol Med. 2016;48:e269. doi: 10.1038/emm.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.