Abstract
B-Raf proto-oncogene, serine/threonine kinase (BRAF) has previously been identified as a candidate target gene in endometriosis. Wild-type and mutated BRAF serve important roles in different diseases. The aim of the present study was to explore BRAF mutation, the mRNA and protein expression of wild-type BRAF (wtBRAF) in endometriosis, and the association between the expression levels of wtBRAF and the predicted transcription factor cAMP responsive element binding protein 1 (CREB1). In the present study, BRAF mutation was detected using Sanger sequencing among 30 ectopic and matched eutopic endometrium samples of patients with endometriosis as well as 25 normal endometrium samples, and no BRAF mutation was detected in exons 11 or 15. A region of ~2,000 bp upstream of the BRAF gene was then screened using NCBI and UCSC databases, and CREB1 was identified as a potential transcription factor of BRAF by analysis with the JASPAR and the TRANSFAC databases. Quantitative polymerase chain reaction was used to analysis the mRNA expression levels of wtBRAF and CREB1, and the corresponding protein expression levels were evaluated using immunohistochemistry and western blot analysis. The results revealed that the mRNA and protein expression levels of wtBRAF and CREB1 were significantly upregulated in the eutopic endometrial tissues of patients with endometriosis compared with normal endometrial tissues (P<0.05) and no significant difference in wtBRAF and CREB1 levels was detected between the ectopic and eutopic endometrium (P>0.05). In addition, correlation analysis revealed that the protein expression of CREB1 was positively correlated with the transcript level and protein expression of wtBRAF. It is reasonable to speculate that CREB1 may activate the transcription of wtBRAF through directly binding to its promoter, increasing BRAF expression and regulating the cell proliferation, migration and invasion of endometriosis.
Keywords: endometriosis, endometrium, B-Raf proto-oncogene, serine/threonine kinase, cAMP responsive element binding protein 1, transcription factors
Introduction
Endometriosis is an estrogen-dependent chronic gynecological disease that is difficult to cure. The main clinical characteristics include pelvic masses, chronic pelvic pain and infertility. Although it is a benign disorder, endometriosis exhibits invasive growth potential, which is similar to that of malignant tumors (1). The incidence of endometriosis is increasing year by year, but its etiology and pathogenesis remain unclear (2). Although the classical theory of endometriosis is Sampson's 'retrograde menstruation theory', which suggests that endometrial fragments undergo retrograde menstruation through the fallopian tubes and implant in the peritoneal cavity, this does not explain why the prevalence of endometriosis is only ~10% in fertile women, the majority of whom experience retrograde menstruation (3). Investigations have shown that cell adhesion, invasion, angiogenesis (4) and apoptosis (5) in the eutopic endometrium in endometriosis are different from that of the normal endometrium, particularly for the secretory endometrium. There is also evidence to suggest that abnormal molecular aberrations in the eutopic endometrium promote the development of endometriosis (6). Thus, an evaluation of specific genes and their associated molecular mechanisms in the eutopic endometrium may provide a new theoretical basis for the pathogenesis of endometriosis. The present study team previously identified 10 upregulated genes in the eutopic endometrium using cDNA representational difference analysis (7). Among them, the abnormal expression of the cofilin 1, methionine adenosyltransferase 2A and LIM domain kinase 1 genes in the eutopic endometrium has been reported (8). Additionally, the present study team also detected that B-Raf proto-oncogene, serine/threonine kinase (BRAF) was overexpressed in the eutopic endometrium of endometriosis (7).
BRAF is a component of the RAS-rapidly accelerated fibrosarcoma (RAF)-mitogen-activated protein kinase kinase (MEK)-extracellular signal-regulated kinase (ERK) signaling pathway. Mutations in BRAF are associated with the development of malignant tumors (9). BRAF mutations have been detected in exons 11 and 15 in numerous tumors, and the most common mutation is the V600EBRAF mutation (10). BRAF mutations may activate the mitogen-activated protein kinase (MAPK) signal pathway constitutively, leading to abnormal cell proliferation, differentiation and tumorigenicity (11). Other studies have shown that the overexpression of wild-type BRAF (wtBRAF) promotes the activation of the RAS-BRAF-MAPK signaling pathway (12,26). Notably, BRAF has been reported to promote cell proliferation through regulation of the Ras/Raf/MAPK signaling pathway in the eutopic endometrial stromal cells of patients with endometriosis (13). Previous studies by the present research team have demonstrated that the mRNA and protein levels of BRAF are markedly overexpressed in the eutopic endometrium tissues of endometriosis compared with normal endometrial tissues (7,14). This suggests that BRAF may regulate the occurrence and development of endometriosis. However, BRAF mutations and the mechanism associated with the upregulation of wtBRAF expression in endometriosis remain unclear.
In the present study, BRAF mutations were detected and the potential transcription factors binding to the region upstream of the wtBRAF transcription start site (TSS) were predicted. The correlation of mRNA and protein expression between wtBRAF and the predicted transcription factor was then analyzed. The results may provide a novel insight into the molecular mechanisms of BRAF in the regulation of the occurrence and development of endometriosis.
Materials and methods
Tissue collection
Ectopic endometrium and paired eutopic endometrium samples were collected from 30 patients (37.30±6.83 years old) with endometriosis who were undergoing total hysterectomy at the Department of Gynecology, Cancer Hospital of China Medical University (Shenyang, China) from January 2015 to June 2016 as the experimental group. Normal endometrium samples from 25 patients (40.46±5.26 years old) without estrogen-dependent disease were also collected as the control group. All patients had regular menstrual cycles and none had malignant diseases, autoimmune disease, surgical diseases or inflammatory diseases. They also did not receive gonadotrophin-releasing hormone analogs, other hormonal medications or antibiotic therapy in the 6 months prior to the surgery. The tissue samples were all collected in the secretory phase of the menstrual cycle, which was confirmed by pathology. The present study was approved by the China Medical University Research Ethics Committee in accord with the Declaration of Helsinki. Written informed consent was obtained from each patient prior to the surgical procedures. All samples were divided into two groups: One was immersed in 10% formalin solution for immunohistochemistry (IHC), and another was frozen in liquid nitrogen for polymerase chain reaction (PCR) and direct sequencing, quantitative PCR (qPCR) or western blot analysis.
Genomic DNA isolation, PCR and direct sequencing
Genomic DNA was extracted from freshly frozen tissue (100 mg) using the TIANamp Genomic DNA kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer's protocol. Approximately 200 ng genomic DNA was used for PCR in a 20-µl reaction system. The DNA polymerase used for PCR is TaKaRa Ex Taq® (Cat. no. R001A) purchased from Takara Bio, Inc., Otsu, Japan. The primer sequences are shown in Table I. The reaction conditions were as follows: 94°C for 5 min, 1 cycle; 94°C for 30 sec, 56°C (for exon 11)/59°C (for exon 15) for 30 sec, 72°C for 30 sec, 35 cycles, and a final extension at 72°C for 10 min. The integrity of all PCR products was observed by 2% agarose gel electrophoresis. PCR products were analyzed using an ABI 3730xl DNA sequencer following purification (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The positive control, V600EBRAF mutation, was detected using BCPAP thyroid carcinoma cells (provided by the Central Laboratory of Shengjing Hospital of China Medical University).
Table I.
Primer sequences of exons used in the present study.
| Exon | Primer sequences (5′→3′) | Product length (bp) |
|---|---|---|
| Exon 11 | F: ATAAGGTAATGTACTTAGGGTGAAACATAA | 356 |
| R: TTTTGTTAGAAACTTTTGGAGGAGTC | ||
| Exon 15 | F: GCTTGCTCTGATAGGAAAATGAGA | 249 |
| R: AATGACTTTCTAGTAACTCAGCAGCA |
F, forward; R, reverse.
Prediction of transcription factor binding sites upstream of the wtBRAF TSS
A region of ~2,000 bp located upstream of the wtBRAF TSS was screened using National Center for Biotechnology Information (NCBI: http://www.ncbi.nlm.nih.gov/) and University of California, Santa Cruz (http://www.genome.ucsc.edu/) databases. Transcription factors that may be able to bind to the ~2,000-bp region upstream of the wtBRAF TSS were then predicted using JASPAR (http://jaspar.genereg.net/) and Transcription Factor (TRANSFAC: http://www.gene-regulation.com/pub/databases.html) datasets.
RNA extraction, reverse transcription and qPCR
Total RNA was extracted from freshly frozen tissues (100 mg) using RNAiso Plus (Takara Bio, Inc.) according to the manufacturer's protocol. RNA purity and concentration were detected by spectrophotometry, and RNA integrity was observed by 1% agarose gel electrophoresis. Total RNA was synthesized into cDNA using the PrimeScript RT Reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. Primers were designed using PrimerPremier 5.0 (Premier Biosoft International, Palo Alto, CA, Canada), and the sequences used in this study are shown in Table II. qPCR analysis was performed using SYBR Premix Ex Taq (Takara Bio, Inc.) according to the manufacturer's protocol using a LightCycler 480 detection system (Roche Diagnostics International AG, Rotkreuz, Switzerland). The conditions were as follows: One cycle at 95°C for 5 min, followed by 45 cycles at 95°C for 10 sec and 60°C for 30 sec. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. All experiments were performed in triplicate. The relative expression of cAMP responsive element binding protein 1 (CREB1) and BRAF was calculated using the 2−ΔΔCq method (15).
Table II.
Primer sequences used for quantitative polymerase chain reaction in the present study.
| Gene | Primer sequences (5′→3′) | Product length (bp) |
|---|---|---|
| CREB1 | F: GGAGTGCCAAGGATTGAAGAAGA | 333 |
| R: TGCTGTGCGAATCTGGTATGTT | ||
| wtBRAF | F: GGCAGAGTGCCTCAAAAAGAA | 134 |
| R: AACCAGCCCGATTCAAGGA | ||
| GAPDH | F: GCACCGTCAAGGCTGAGAAC | 138 |
| R: TGGTGAAGACGCCAGTGGA |
CREB1, cAMP responsive element binding protein 1; wtBRAF, wild-type B-Raf proto-oncogene, serine/threonine kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; F, forward; R, reverse.
IHC staining
The IHC SP kit (ZSGB-BIO, Beijing, China) was used to detect the protein expression levels of CREB1 and wtBRAF. Paraffin-embedded specimens were cut into 4-µm sections. The sections were dewaxed with xylene and dehydrated with graded alcohol. Following washing with phosphate-buffered saline (PBS), the sections were incubated in 3% H2O2 for 15 min at room temperature followed by microwave antigen retrieval (oven fire to 100%, for 7 min). The sections were blocked with serum (contained in The IHC SP kit) for 30 min at 37°C and incubated with mouse anti-human BRAF monoclonal antibody (cat. no. sc-5284; 1:50; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and rabbit anti-human CREB1 polyclonal antibody (cat. no. 12208-1-AP; 1:200; ProteinTech Group, Inc., Chicago, IL, USA) overnight at 4°C. The sections were washed with PBS and then incubated with secondary antibody (contained in the IHC SP kit) for 30 min at 37°C. Staining was performed using a diaminobenzidine kit (Beyotime Institute of Biotechnology, Nanjing, China) according to the manufacturer's protocol. Immunostaining results were scored using a light microscope (E100; Nikon, Tokyo, Japan) according to the positive cell percentage and positive staining intensity. First, the extent of the staining was scored according to the positive cell percentage: <5%, 0 points; 5–10%, 1 point; 10–50%, 2 points; >50%, 3 points. Additionally, the intensity of staining was scored as follows: no staining, 0 points; light yellow, 1 point, moderate yellow, 2 points; strong yellow, 3 points. The extent of the staining multiplied by the intensity was the final score; scores of 0–3 and 4–9 were considered negative and positive expression, respectively.
Western blot detection of protein expression levels
To 100 mg frozen endometrial tissue was added lysis buffer (quantity to volume ratio in mg/µl; 1:5) with 1% protease inhibitor, lysed in ice and centrifuged at 12,000 × g and 4°C for 15 min. A bicinchoninic acid reagent kit (Beyotime Institute of Biotechnology) was used to detect the protein concentration of the supernatant. The protein sample (80 µg) was separated by 8% SDS-PAGE, followed by transfer to a polyvinylidene fluoride (EMD Millipore, Billerica, MA, USA) membrane. After blocking with 5% non-fat milk for 2 h at room temperature, the membranes were incubated with mouse anti-human BRAF monoclonal antibody (cat. no. sc-5284; 1:100; Santa Cruz Biotechnology, Inc.), rabbit anti-human CREB1 polyclonal antibody (cat. no. 12208-1-AP; 1:500) and mouse anti-human GAPDH monoclonal antibody (cat. no. 0004-1-Ig; 1:10,000) (both from ProteinTech Group, Inc.) overnight at 4°C. GAPDH was used as a loading control. The membrane was washed with TBS with 0.1% Tween-20 buffer three times, and then the matched secondary antibodies (goat anti-mouse; cat. no. A00001-1; 1:2,000; goat anti-rabbit; cat. no. A00001-2; both ProteinTech Group, Inc.) were added for 2 h at room temperature. The binding was detected using a BeyoECL Plus kit (Beyotime Institute of Biotechnology), and the integrated density was analyzed using ImageJ 1.48v software (National Institutes of Health, Bethesda, MD, USA). The ratio of the integrated density between the target band and GAPDH was considered the relative expression level of each protein.
Statistical analysis
All data are presented as mean ± standard deviation. ANOVA and Chi-squared test were used to analyze differences in the mRNA and protein expression of BRAF and CREB1 among eutopic and ectopic endometrium tissues from patients with endometriosis and normal endometrium. Pearson's coefficient correlation was applied to analyze the correlation between the expression of CREB1 protein and the mRNA and protein expression of wtBRAF. Data analysis was performed with the statistical software SPSS 20.0 (IBM Corp., Armonk, NY, USA), and P<0.05 was considered to indicate a statistically significant result.
Results
BRAF mutation at exons 11 and 15
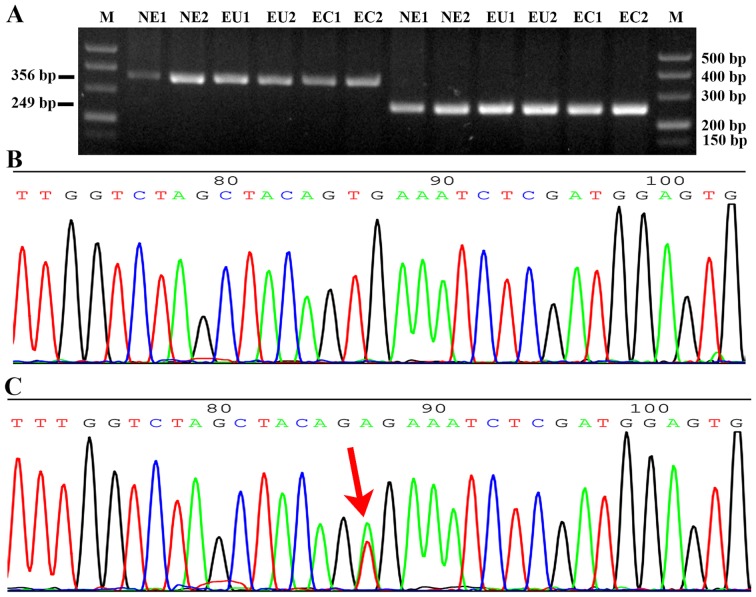
All DNA specimens were amplified using specific primers and detected using gel electrophoresis. The band size of exon 11 was 356 bp and that of exon 15 was 249 bp (Fig. 1A). No BRAF mutations were detected among the 30 cases of ectopic and matched eutopic endometrium samples from patients with endometriosis and the 25 cases of normal endometrium (Fig. 1B), compared with the positive control in which the BRAF mutation (T1799A) was detected at exon 15 in the BCPAP papillary thyroid cancer cell line (Fig. 1C).
Figure 1.
Detection of BRAF gene mutation at exons 11 and 15. (A) Gel electrophoresis of the PCR products of exon 11 and 15 of the BRAF gene. The PCR product size of exon 11 is 356 bp, and that of exon 15 is 249 bp. Part of a sequence chromatogram from the Sanger sequencing of BRAF from (B) a patient and (C) the positive control. The red arrow indicates a BRAF mutation (T1799A) of exon 15 in the BCPAP papillary thyroid cancer cell line. BRAF, B-Raf proto-oncogene, serine/threonine kinase; PCR, polymerase chain reaction; M, DNA marker; NE, normal endometrium; EU, eutopic endometrium of endometriosis; EC, ectopic endometrium of endometriosis.
Prediction of the transcription factor binding sites of the BRAF TSS
A region of ~2,000 bp upstream of the BRAF TSS was screened, and transcription factors binding to the ~2,000-bp region were predicted using the JASPAR core transcription factor database. In total, 323 transcription factors (profile score threshold 80%) were identified. Among them, 5 were filtered (relative score >1.000; Table III), including CREB1 (NCBI Gene ID 1385), Spi-B transcription factor (SPIB; NCBI Gene ID 6689), nuclear factor of activated T-cells 2 (NFATC2; NCBI Gene ID 4773), zinc finger protein 354C (ZNF354C; NCBI Gene ID 30832) and SRY-box 10 (SOX10; NCBI Gene ID 6663). Combined analysis with the JASPAR and TRANSFAC databases predicted a cAMP-responsive element (CRE) binding site (TGACGTCA) at −266 to −259 bp upstream of the wtBRAF TSS (Fig. 2). Additionally, in a previous study, it was detected that CREB1 mRNA expression was significantly higher in the eutopic endometrium of patients with endometriosis compared with that in normal endometrium (16). These findings suggest that CREB1 may activate BRAF gene transcription by directly binding to the CRE sequence of the BRAF promoter region.
Table III.
Transcription factor binding sites predicted by the JASPAR database.
| Model ID | Model name | Score | Relative score | Start | End | Strand | Predicted site sequence |
|---|---|---|---|---|---|---|---|
| MA0018.2 | CREB1 | 11.569 | 1.0000161 | 1735 | 1742 | −1 | TGACGTCA |
| MA0018.2 | CREB1 | 11.569 | 1.0000161 | 1735 | 1742 | 1 | TGACGTCA |
| MA0081.1 | SPIB | 10.470 | 1.0000147 | 1599 | 1605 | 1 | AGAGGAA |
| MA0152.1 | NFATC2 | 11.360 | 1.0000115 | 938 | 944 | 1 | TTTTCCA |
| MA0152.1 | NFATC2 | 11.360 | 1.0000115 | 1243 | 1249 | −1 | TTTTCCA |
| MA0130.1 | ZNF354C | 8.916 | 1.0000095 | 823 | 828 | 1 | ATCCAC |
| MA0442.1 | SOX10 | 8.910 | 1.0000092 | 252 | 257 | −1 | CTTTGT |
| MA0442.1 | SOX10 | 8.910 | 1.0000092 | 1098 | 1103 | −1 | CTTTGT |
CREB1, cAMP responsive element binding protein 1; SPIB, Spi-B transcription factor; NFATC2, nuclear factor of activated T-cells 2; SOX10, SRY-box 10.
Figure 2.
The CRE sequence site predicted by the TRANSFAC database. The arrow represents the CRE binding site (TGACGTCA) from −266 to −259 bp upstream of the wild-type B-Raf proto-oncogene, serine/threonine kinase transcription start site. CRE, cAMP-responsive element.
mRNA expression of wtBRAF and CREB1 in endometrial tissues
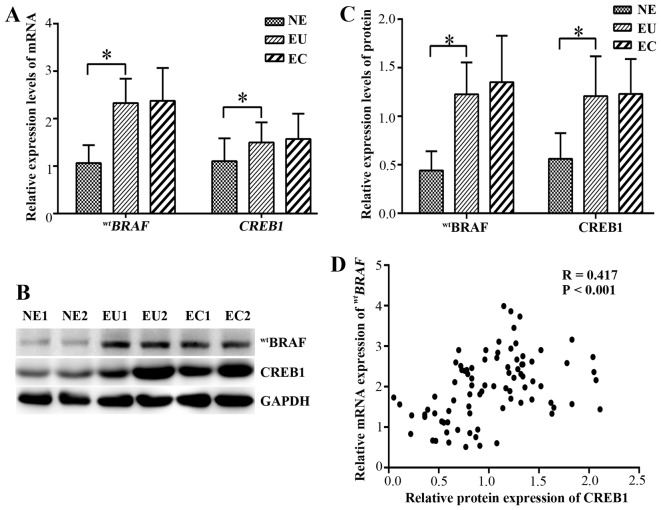
qPCR was used to evaluate the mRNA expression levels of wtBRAF and CREB1 in the ectopic and eutopic endometrium of endometriosis patients. The mRNA and protein levels are shown in (Fig. 3). In the eutopic endometrial tissues, the relative mRNA expression levels of wtBRAF and CREB1 were significantly higher than those in the normal endometrial tissues (P<0.001). However, no significant difference in wtBRAF and CREB1 mRNAs was detected between the ectopic and eutopic endometrium (P=0.989 and P=0.548, respectively; Fig. 3A).
Figure 3.
Expression of the mRNA and protein of wtBRAF and CREB1. (A) Relative mRNA expression levels of wtBRAF and CREB1. (B) Western blotting of wtBRAF and CREB1 proteins. GAPDH was used as a control. (C) Relative protein expression levels of wtBRAF and CREB1 in different endometrial tissues. (D) Correlation analysis between CREB1 protein and wtBRAF transcript levels in all tissues. *P<0.001 as indicated. wtBRAF, wild-type B-Raf proto-oncogene, serine/threonine kinase; CREB1, cAMP responsive element binding protein 1; NE, normal endometrium; EU, eutopic endometrium of endometriosis; EC, ectopic endometrium of endometriosis.
Protein expression of wtBRAF and CREB1 in endometrial tissues by IHC
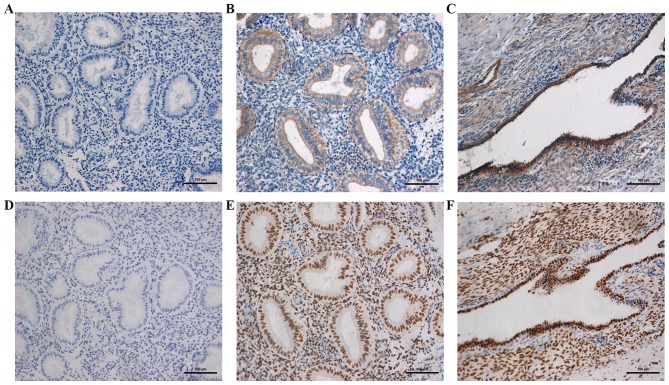
The location and expression of wtBRAF and CREB1 in the ectopic and eutopic endometrial tissues of patients with endometriosis and the normal endometrial tissues of the control group were detected using IHC (Fig. 4). The results revealed that the wtBRAF protein was located in the cytoplasm of the epithelial and stromal cells, while CREB1 was located in the nuclei of these cells. In the normal, eutopic and ectopic endometrial tissues, the positive expression rates of wtBRAF were 24, 70 and 63.3% and those of CREB1 were 36, 80 and 76%, respectively. The positive rates of wtBRAF and CREB1 in the eutopic endometrial tissue were significantly higher than those in normal tissues (P<0.05), while no significant difference was detected between the eutopic and ectopic endometrial tissues (P=0.584 and P=0.766, respectively; Table IV).
Figure 4.
Immunostaining of wtBRAF and CREB1 in different endometrial tissues. Representative images of (A–C) wtBRAF and (D–F) CREB1 staining. (A and D) Normal endometrium from the control group, (B and E) eutopic endometrium from patients with endometriosis and (C and F) ectopic endometrium from patients with endometriosis (magnification, ×200). wtBRAF, wild-type B-Raf proto-oncogene, serine/threonine kinase; CREB1, cAMP responsive element binding protein 1.
Table IV.
Protein expression of wtBRAF and CREB1 in different endometrial tissues determined by immunohistochemistry.
| Protein | Tissue samples, n (%)
|
P-value | ||
|---|---|---|---|---|
| EC (n=30) | EU (n=30) | NE (n=25) | ||
| wtBRAF | ||||
| Positive | 19 (63.3) | 21 (70) | 6 (24) | 0.584a |
| Negative | 11 (36.7) | 9 (30) | 19 (76) | <0.001b |
| CREB1 | ||||
| Positive | 22 (76) | 23 (80) | 9 (36) | 0.766a |
| Negative | 8 (24) | 7 (20) | 16 (64) | 0.002b |
EC vs. EU;
EU vs. NE. EC, ectopic endometrium; EU, eutopic endometrium; NE, normal endometrium; CREB1, cAMP responsive element binding protein 1; wtBRAF, wild-type B-Raf proto-oncogene, serine/threonine kinase.
Protein expression of wtBRAF and CREB1 in endometrial tissues by western blot analysis
The protein expression of wtBRAF and CREB1 in the endometriosal tissues was further detected using western blot analysis (Fig. 3B and C). In the eutopic endometrium tissues, the wtBRAF and CREB1 expression levels were significantly upregulated compared with those in the normal endometrial tissue (P<0.001), while no significant differences in expression were observed between the eutopic and ectopic endometrium (P=0.566 and P=0.811, respectively).
CREB1 positively correlates with wtBRAF expression
The correlation between wtBRAF and CREB1 protein expression as determined by IHC was evaluated (Table V). The results indicated that wtBRAF immunostaining was positively correlated with that of CREB1 (correlation coefficient R=0.529, P<0.001). Furthermore, correlation of the western blotting and qPCR data suggested that there was a positive correlation between CREB1 protein and the wtBRAF transcript level in all tissues (correlation coefficient R=0.417, P<0.001; Fig. 3D).
Table V.
Correlation between wtBRAF and CREB1 protein expression in endometriosis.
| CREB1 (cases) |
wtBRAF (cases)
|
P-value | ||
|---|---|---|---|---|
| Positive | Negative | R | ||
| Positive | 40 | 14 | 0.529 | P<0.001 |
| Negative | 6 | 25 | ||
CREB1, cAMP responsive element binding protein 1; wtBRAF, wild-type B-Raf proto-oncogene, serine/threonine kinase.
Discussion
In the present study, no BRAF mutations were detected in exons 11 or 15 in the ectopic and eutopic endometrium samples of patients with endometriosis. However, significant overexpression of wtBRAF and CREB1 mRNA and protein was detected in the eutopic endometrium of these patients compared with normal endometrium. However, no significant difference was identified between the ectopic and eutopic endometrium. In addition, analysis of the protein expression of CREB1 indicated that it was positively correlated with the transcript level and protein expression of wtBRAF.
BRAF is a proto-oncogene that is also known as v-raf murine sarcoma viral homolog B1. It belongs to the Raf kinase family and was first discovered in 1988 (17). BRAF mutations are associated with numerous types of malignant tumor, where they activates the MAPK signaling pathway constitutively, resulting in uncontrolled cellular proliferation and survival (18). The most common BRAF point mutation, V600EBRAF, accounts for ~90% of all BRAF mutations, and derives from a point mutation that results in an amino acid change from valine to glutamic acid (19). However, other malignant tumors, including primary uveal melanoma and uveal melanoma demonstrate a lack of BRAF mutations (20,21). Zannoni et al (22) found no mutations in the hotspot regions of BRAF (i.e. exon 15) in primary clear cell ovarian carcinoma. However, several studies have shown the overexpression of the wtBRAF gene in other cancer types. For example, one study found that the overexpression of BRAF activated the RAS-BRAF-MAPK signaling pathway (12). Furthermore, BRAF has been demonstrated to be overexpressed in advanced hepatocellular carcinoma (23). The overexpression of BRAF may participate in the molecular mechanisms of thyroid papillary carcinoma, and detection of the expression of the BRAF gene has been shown to predict the cell invasion ability of papillary thyroid carcinoma (24). In previous studies, the high expression of BRAF increased the activity of ERK in the Rat-1 cell line (25), and also stimulated the growth of malignant melanoma cells (26). It has been suggested that BRAF serves an important role in tumor development by binding to specific molecular signaling molecules. Previous studies by the present research team demonstrated that BRAF is a candidate gene in the development of endometriosis (7), and preliminarily verified that BRAF mRNA and protein levels are significantly upregulated in the eutopic endometrium of patients with endometriosis compared with normal endometrium (14). In the present study, no BRAF mutation was detected in exons 11 or 15 among the 30 pairs of ectopic and matched eutopic endometrium samples from patients with endometriosis. This is consistent with another study that screened for BRAF mutations in ectopic endometrial tissue (27). Additionally, it was observed in the present study that the mRNA and protein expression levels of wtBRAF in the eutopic endometrial tissues from patients with endometriosis were significantly higher than those in normal endometrium, which further suggests a role for BRAF in endometriosis. However, no significant difference was detected between the eutopic and matched ectopic endometrial tissues from patients with endometriosis. There are two possible reasons for this observation: i) The BRAF gene promotes the occurrence of endometriosis, but does not serve a role in its development; ii) heterogeneity exists in the ectopic endometrium of endometriosis cases, and the quantity of ectopic endometrium tissues obtained from ectopic focus is too little to influence the research results. Further study is required to investigate these hypotheses.
To explore the mechanism of wtBRAF overexpression, a region of ~2,000 bp upstream of the wtBRAF TSS was selected. Using the JASPAR and TRANSFAC databases, a CRE binding site (TGACGTCA) at −266 to −259 bp upstream of the wtBRAF TSS was predicted. In addition, a previous study by the present research team found that 1,216 mRNAs were expressed differentially between eutopic and normal endometrium by long non-coding RNA microarray, among which the function of cyclin-dependent kinase 6 in endometriosis has been preliminarily validated, and CREB1 was an overexpressed mRNA (16). These findings suggest that CREB1 may function as a transcription factor of wtBRAF, and is involved in the regulation of the development of endometriosis.
CREB1 is a proto-oncogenic transcription factor that is involved in oncogenesis in numerous organs. CREB1 increases the expression of its target genes, which are involved in various cell functions, including metabolism, the cell cycle, cell survival and DNA repair (28). As a potent oncogene, CREB1 promotes tumorigenesis by significantly impacting the growth, proliferation, survival, metastasis and invasion of tumor cells. The overexpression of CREB1 promotes these functions, and is also closely associated with the recurrence and poor prognosis of diseases (23). For example, it has been reported that CREB1 is overexpressed in gastric cancer and increases gastric adenocarcinoma cell growth (29). In addition, Yang et al (30) demonstrated that the expression of CREB1 was associated with the migration of hepatocellular carcinoma cells. Furthermore, the overexpression of CREB1 has been reported to be associated with poor prognosis in non-smokers with non-small cell lung cancer and in patients with breast cancer (31,32). In the present study, the expression of CREB1 mRNA and protein in the eutopic endometrium of patients with endometriosis was significantly higher than that in normal endometrium. In addition, it has been suggested that the CREB protein may act as a transcription regulator of aromatase in breast cancer cells to increase the synthesis of estrogen (31). Breast cancer and endometriosis are typical estrogen-dependent diseases. The present study found that the expression of CREB1 was significantly higher in eutopic endometrium than in normal endometrium, which suggests that CREB1 may be a candidate gene for endometriosis intervention and treatment. However, no significant difference in CREB1 expression was detected between the ectopic and eutopic endometrium; this difference remains to be confirmed by microdissection or cell sorting. The correlation analysis conducted in the present study revealed that the protein expression of CREB1 was positively correlated with the transcript level and protein expression of wtBRAF. It is reasonable to speculate that CREB1 may activate the transcription of wtBRAF through directly binding to its promoter, increasing BRAF expression and regulating the cell proliferation, migration and invasion of endometriosis. However, additional studies are required to further verify the exact molecular mechanism by which CREB1 regulates the expression of wtBRAF.
Acknowledgments
The present study was supported as a project of the National Natural Science Foundation of China (grant no. 81270675).
References
- 1.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y, Li Y, Liu K, Chen P, Wang D. Expression and significance of WNT4 in ectopic and eutopic endometrium of human endometriosis. Reprod Sci. 2016;23:379–385. doi: 10.1177/1933719115602763. [DOI] [PubMed] [Google Scholar]
- 3.Gupta D, Hull ML, Fraser I, Miller L, Bossuyt PM, Johnson N, Nisenblat V. Endometrial biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst Rev. 2016;4:CD012165. doi: 10.1002/14651858.CD012165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Chen P, Guo C, Meng X, Wang D. Effect of LIM kinase 1 overexpression on behaviour of endometriosis-derived stromal cells. Cell Tissue Res. 2015;359:885–893. doi: 10.1007/s00441-014-2068-5. [DOI] [PubMed] [Google Scholar]
- 5.Harada T, Kaponis A, Iwabe T, Taniguchi F, Makrydimas G, Sofikitis N, Paschopoulos M, Paraskevaidis E, Terakawa N. Apoptosis in human endometrium and endometriosis. Hum Reprod Update. 2004;10:29–38. doi: 10.1093/humupd/dmh007. [DOI] [PubMed] [Google Scholar]
- 6.Ulukus M, Cakmak H, Arici A. The role of endometrium in endometriosis. J Soc Gynecol Investig. 2006;13:467–476. doi: 10.1016/j.jsgi.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Chen Q, Zhang C, Chen Y, Lou J, Wang D. Identification of endometriosis-related genes by representational difference analysis of cDNA. Aust N Z J Obstet Gynaecol. 2012;52:140–145. doi: 10.1111/j.1479-828X.2011.01405.x. [DOI] [PubMed] [Google Scholar]
- 8.Xu YL, Wang DB, Liu QF, Chen YH, Yang Z. Silencing of cofilin-1 gene attenuates biological behaviours of stromal cells derived from eutopic endometria of women with endometriosis. Hum Reprod. 2010;25:2480–2488. doi: 10.1093/humrep/deq197. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell B, Dhingra JK, Mahalingam M. BRAF and epithelial-mesenchymal transition: Lessons from papillary thyroid carcinoma and primary cutaneous melanoma. Adv Anat Pathol. 2016;23:244–271. doi: 10.1097/PAP.0000000000000113. [DOI] [PubMed] [Google Scholar]
- 10.Rahman MA, Salajegheh A, Smith RA, Lam AK. B-Raf mutation: a key player in molecular biology of cancer. Exp Mol Pathol. 2013;95:336–342. doi: 10.1016/j.yexmp.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Lopes JP, Fonseca E. BRAF gene mutation in the natural history of papillary thyroid carcinoma: diagnostic andprognostic implications. Acta Med Port. 2011;24(Suppl 4):855–868. [PubMed] [Google Scholar]
- 12.Ewing I, Pedder-Smith S, Franchi G, Ruscica M, Emery M, Vax V, Garcia E, Czirják S, Hanzély Z, Kola B, et al. A mutation and expression analysis of the oncogene BRAF in pituitary adenomas. Clin Endocrinol (Oxf) 2007;66:348–352. doi: 10.1111/j.1365-2265.2006.02735.x. [DOI] [PubMed] [Google Scholar]
- 13.Yotova IY, Quan P, Leditznig N, Beer U, Wenzl R, Tschugguel W. Abnormal activation of Ras/Raf/MAPK and RhoA/ROCKII signalling pathways in eutopic endometrial stromal cells of patients with endometriosis. Hum Reprod. 2011;26:885–897. doi: 10.1093/humrep/der010. [DOI] [PubMed] [Google Scholar]
- 14.Chu DM, Wang DB, Li Y, Liu KR. Expression and significance of BRAF gene in ectopic endometrium of women with endometriosis. Zhongguo Yike Daxue Xuebao. 2014;43:790–793. In Chinese. [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Li Y, Yang Z, Liu K, Wang D. Genome-wide microarray analysis of long non-coding RNAs in eutopic secretory endometrium with endometriosis. Cell Physiol Biochem. 2015;37:2231–2245. doi: 10.1159/000438579. [DOI] [PubMed] [Google Scholar]
- 17.Ikawa S, Fukui M, Ueyama Y, Tamaoki N, Yamamoto T, Toyoshima K. B-raf, a new member of the raf family, is activated by DNA rearrangement. Mol Cell Biol. 1988;8:2651–2654. doi: 10.1128/MCB.8.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sclafani F, Gullo G, Sheahan K, Crown J. BRAF mutations in melanoma and colorectal cancer: a single oncogenic mutation with different tumour phenotypes and clinical implications. Crit Rev Oncol Hematol. 2013;87:55–68. doi: 10.1016/j.critrevonc.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Roskoski R., Jr RAF protein-serine/threonine kinases: Structure and regulation. Biochem Biophys Res Commun. 2010;399:313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- 20.Cohen Y, Goldenberg-Cohen N, Parrella P, Chowers I, Merbs SL, Pe'er J, Sidransky D. Lack of BRAF mutation in primary uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44:2876–2878. doi: 10.1167/iovs.02-1329. [DOI] [PubMed] [Google Scholar]
- 21.Rimoldi D, Salvi S, Liénard D, Lejeune FJ, Speiser D, Zografos L, Cerottini JC. Lack of BRAF mutations in uveal melanoma. Cancer Res. 2003;63:5712–5715. [PubMed] [Google Scholar]
- 22.Zannoni GF, Improta G, Chiarello G, Pettinato A, Petrillo M, Scollo P, Scambia G, Fraggetta F. Mutational status of KRAS, NRAS, and BRAF in primary clear cell ovarian carcinoma. Virchows Arch. 2014;465:193–198. doi: 10.1007/s00428-014-1599-1. [DOI] [PubMed] [Google Scholar]
- 23.Newell P, Toffanin S, Villanueva A, Chiang DY, Minguez B, Cabellos L, Savic R, Hoshida Y, Lim KH, Melgar-Lesmes P, et al. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J Hepatol. 2009;51:725–733. doi: 10.1016/j.jhep.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng L, Li M, Zhang QP, Piao ZA, Wang ZH, Lv S. Utility of BRAF protein overexpression in predicting the metastasis potential of papillary thyroid carcinoma. Oncol Lett. 2011;2:59–63. doi: 10.3892/ol.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erhardt P, Schremser EJ, Cooper GM. B-Raf inhibits programmed cell death downstream of cytochrome c release from mitochondria by activating the MEK/Erk pathway. Mol Cell Biol. 1999;19:5308–5315. doi: 10.1128/MCB.19.8.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanami H, Imoto I, Hirasawa A, Yuki Y, Sonoda I, Inoue J, Yasui K, Misawa-Furihata A, Kawakami Y, Inazawa J. Involvement of overexpressed wild-type BRAF in the growth of malignant melanoma cell lines. Oncogene. 2004;23:8796–8804. doi: 10.1038/sj.onc.1208152. [DOI] [PubMed] [Google Scholar]
- 27.Vestergaard AL, Thorup K, Knudsen UB, Munk T, Rosbach H, Poulsen JB, Guldberg P, Martensen PM. Oncogenic events associated with endometrial and ovarian cancers are rare in endometriosis. Mol Hum Reprod. 2011;17:758–761. doi: 10.1093/molehr/gar049. [DOI] [PubMed] [Google Scholar]
- 28.Tan X, Wang S, Zhu L, Wu C, Yin B, Zhao J, Yuan J, Qiang B, Peng X. cAMP response element-binding protein promotes gliomagenesis by modulating the expression of oncogenic microRNA-23a. Proc Natl Acad Sci USA. 2012;109:15805–15810. doi: 10.1073/pnas.1207787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X, Tang H. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279:1252–1260. doi: 10.1111/j.1742-4658.2012.08519.x. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;288:28893–28899. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chhabra A, Fernando H, Watkins G, Mansel RE, Jiang WG. Expression of transcription factor CREB1 in human breast cancer and its correlation with prognosis. Oncol Rep. 2007;18:953–958. [PubMed] [Google Scholar]
- 32.Seo HS, Liu DD, Bekele BN, Kim MK, Pisters K, Lippman SM, Wistuba II, Koo JS. Cyclic AMP response element-binding protein overexpression: A feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res. 2008;68:6065–6073. doi: 10.1158/0008-5472.CAN-07-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]