Abstract
MicroRNAs (miRNAs/miRs) serve a role as important regulators in cardiac hypertrophy. The present study aimed to reveal the differential expression profile of miRNAs between young and aging spontaneously hypertensive rats (SHRs) and studied the functional annotation of predicted targets. Briefly, 3-month-old and 12-month-old SHRs (n=3/group) were subjected to echocardiography, histopathological analysis and dihydroethidium staining. Subsequently, small RNA sequencing and data processing was conducted to identify the differentially expressed miRNAs between these two groups. Eight significantly upregulated miRNAs were validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), followed by in silico target gene prediction. Functional annotation analysis of the predicted targets was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. As a result, significantly impaired left ventricular diastolic function was detected in the 12-month-old SHRs, alongside increased myocyte cross-sectional area and percentage area of fibrosis, elevated reactive oxygen species production and reduced microvessel density (P<0.05). Compared with the 3-month-old SHRs, 21 miRNAs were significantly upregulated and five miRNAs were downregulated in 12-month-old rats (P<0.05). Eight upregulated, remodeling-associated miRNAs, including rno-miR-132-3p, rno-miR-182, rno-miR-208b-3p, rno-miR-212-3p, rno-miR-214-3p, rno-miR-218a-5p, rno-miR-221-3p and rno-miR-222-3p, underwent bioinformatics analysis. The target genes were significantly enriched in 688 GO terms and 39 KEGG pathways, including regulation of peptidyl-tyrosine phosphorylation, regulation of protein serine/threonine kinase activity, adrenergic signaling in cardiomyocytes, ErbB signaling pathway, mTOR signaling pathway, FoxO signaling pathway, Ras signaling pathway, insulin secretion, adipocytokine signaling pathway, HIF-1 signaling pathway, Rap1 signaling pathway, VEGF signaling pathway and TNF signaling pathway. Collectively, the present study identified a dysregulated miRNA profile in aging SHRs, which targeted numerous signaling pathways associated with cardiac hypertrophy, autophagy, insulin metabolism, angiogenesis and inflammatory response.
Keywords: microRNA, spontaneously hypertensive rats, cardiac hypertrophy, left ventricular diastolic dysfunction
Introduction
Abnormal left ventricular (LV) diastolic function is closely associated with increased cardiovascular morbidity and mortality, and is a precursor of heart failure (HF) with preserved ejection fraction (HFpEF). HFpEF accounts for 30–50% of cases of HF and is becoming increasingly prevalent; in addition, nearly all patients with HF symptoms, including those with reduced EF, have some component of HFpEF (1). The abnormal deposition of fibrillar collagen, and other extracellular matrix components, may account for this pathological state, which leads to increased stiffness and decreased compliance of the LV wall. In recent years, research has focused on diastolic dysfunction as an early marker of target organ damage in hypertension (2–4). Clinical data and the results of experiments using animal models have indicated that alterations in diastole precede the development of HF in hypertensive heart disease. Spontaneously hypertensive rat (SHR) is a well-established animal model, which resembles numerous aspects of human essential hypertension and has been regarded as a useful tool for studying transitions and the underlying mechanisms of LV diastolic dysfunction and HFpEF. Slama et al (5) detected normal LV systolic function, which was associated with impaired LV relaxation and compliance in SHRs; this was present early in life and progressively increased over the lifetime of SHRs. In order to study the molecular mechanisms underlying the development of LV hypertrophy and its transition to diastolic HF, Rysä et al (6) analyzed global alterations in gene expression in SHRs of different age brackets, and detected increased expression of hypertrophy-associated genes during the transition from LV hypertrophy to HFpEF.
MicroRNAs (miRNAs/miRs) are a group of small non-coding RNAs that regulate gene expression at the post-transcriptional level and serve as key regulators in cardiovascular diseases, including hypertension, cardiac hypertrophy and HF. To the best of our knowledge, no previous study has reported the use of high-throughput small RNA sequencing in identifying miRNA profile alterations between young and old SHRs. Since advanced aging serves an important role in the process of diastolic dysfunction, the determination of alterations in miRNA profiles that may affect LV hypertrophy and to what extent they differ between young and aging SHRs is of great value. The present study aimed to analyze miRNA gene profiles in myocardial tissues between young and aging SHRs, in order to provide novel insights into potential therapeutic targets for the prevention and treatment of HFpEF resulting from primary hypertension.
Materials and methods
Experimental animals
A total of 6 male SHRs weighing about 400 g were purchased from Shanghai Laboratory Animal centre, chinese Academy of Sciences (Shanghai, china), among which 3 rats were 3 months old, and the remaining 3 rats were 12 months old. All rats were housed in a room at 23°C, 60% humidity under a 12-h light/dark cycle. Food and water were supplied ad libitum. All experiments and procedures were performed in compliance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH; publication no. 85–23, revised 1996). The present study was approved by the Animal Care Committee in Zhongshan Hospital, Fudan University (Shanghai, China).
Experimental protocol
Body weight (BW) was determined using an electronic scale. Systolic blood pressure (SBP) was measured according to the tail-cuff method (BP-2000 Blood Pressure Analysis System; Visitech Systems, Apex, NC, USA). Subsequently, rats were subjected to transthoracic high frequency echocardiography. BW, SBP and echocardiography were assessed in one day just before euthanization. Once echocardiography was completed, all rats were euthanized and the LV was rapidly excised and weighed, in order to calculate LV mass index (LVMI), which is expressed as a ratio of LV weight (mg)/total BW (g). In addition, venous blood samples were collected for further study. The middle section of the LV at the papillary level was immediately cut into 2-mm transverse sections, some of which were immersed in 10% neutral buffered paraformaldehyde for histological analysis. Other transverse sections were snap frozen in Tissue-Tek® optimum cutting temperature compound (Sakura Finetek USA, Inc., Torrance, CA, USA) and were stored at −80°C for dihydroethidium (DHE) staining. The remaining tissue was quickly treated with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at −80°C for RNA preparation.
Echocardiography
After anesthesia, the precordial region of the rats was shaved and they were placed in a supine position on the warmed stage of a Vevo 770 High Resolution Echocardiography system (VisualSonics, Inc., Toronto, ON, Canada). Transthoracic echocardiographic measurements were performed using a 17.5 MHz scanhead. Parasternal long- and short-axis two-chamber M-mode views were obtained at midpapillary level, in order to determine LV end-diastolic diameter (LVEDD), LV end-systolic diameter (LVESD), LV posterior wall thickness (LVPW) and interventricular septum thickness in diastole (IVS). LV ejection fraction (LVEF) and fractional shortening (LVFS) were measured directly from the long-axis image. Transmitral flow velocity was assessed from a four-chamber apical view using pulsed-wave Doppler imaging. Peak velocities of early (E) and late (A) mitral inflow waves, isovolumic relaxation time (IVRT) and E-wave deceleration time (DT) were measured, and the E/A ratio was calculated. Lateral mitral annulus velocity was also assessed using tissue Doppler imaging. Peak early (E′) and late (A′) diastolic annular velocities were recorded, and the E′/A′, E/E′ were calculated. All measurements were averaged from four consecutive cardiac cycles and were conducted by an experienced echocardiographer blinded to the protocol.
Plasma N-terminal pro-natriuretic peptide (NT-proBNP) assay
Venous blood was collected into EDTA tubes immediately after sacrifice. Samples were centrifuged at 4°c, at3500 × g for 10 min and the plasma samples were collected and stored at −80°C for subsequent analysis. Plasma NT-proBNP concentration was quantified using an ELISA kit (SEA485Ra; Cloud-Clone Corp., Katy, TX, USA) according to the manufacturer's protocol. Briefly, standard or sample, detection reagent, substrate solution and stop solution were sequentially added to the wells and incubated, with repeated washing as appropriate. Absorbance was measured using a microplate reader at a wavelength of 450 nm, and sample values were calculated from the standard curve.
Histopathological analysis and DHE staining
Transverse LV specimens were fixed in 10% buffered formalin for 24 h at room temperature, embedded in paraffin and cut into 4-μm sections. Hematoxylin and eosin staining at room temperature was used to assess cardiomyocyte cross-sectional area; an average of ≥100 myocytes taken from five randomly selected high-powered fields were analyzed (magnification, ×400) for each specimen. Masson trichrome staining was used to evaluate the extent of fibrosis, which was obtained by dividing the sum of the fibrotic area by total tissue area under five randomly selected high-powered fields (magnification, ×200). Microvessel density was assessed by cluster of differentiation (CD)31 immunohistochemical staining. Following deparaffinization by gradient xylene and ethanol, inactivation of endogenous enzyme with hydrogen peroxide, heat-induced antigen retrieval and sealing with goat serum, slides were incubated with rabbit anti-CD31 monoclonal antibody (1:500; ab182981; Abcam, Cambridge, UK) for 12 h at 4°c, followed by incubation with biotinylated goat anti-rabbit immunoglobulin G and streptavidin-biotin-peroxidase complex (1:100; SA2002; Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 20 min each at room temperature. Subsequently, the slides were treated with 3′-diaminobenzidine; brown staining was considered positive. Microvessel density was expressed as the number of CD31+ endothelial cells (including single cells, cluster or tubular structures) in five randomly selected high-powered fields (magnification, ×400) for each section. Digital images were captured under a microscope (Leica Microsystems GmbH, Wetzlar, Germany) and analyzed using a high-resolution digital image analysis system (Qwin V3; Leica Microsystems GmbH). DHE staining was used to quantify reactive oxygen species (ROS) production in heart samples. Unfixed frozen sections (8 μm) of myocardium were incubated with DHE (5 μmol/l, 100 μl/section; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C in a humidified chamber protected from light for 30 min. Digital images were captured using a fluorescence microscope (Leica Microsystems GmbH) and were analyzed by ImageJ software (version 1.44; NIH, Bethesda, MD, USA). Images of five randomly selected fields (magnification, ×200) of each sample were captured with the in situ fluorescence intensity calculated and averaged.
RNA extraction, small RNA sequencing and data processing
Small RNA sequencing was commercially conducted by Guangzhou RiboBio Co., Ltd (Guangzhou, China). Total RNA was isolated from tissues using the TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA purity was assessed using the ND-1000 Nanodrop (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Each RNA sample had an A260:A280 ratio >1.8 and an A260:A230 ratio >2.0. RNA integrity was evaluated using the Agilent 2200 TapeStation (Agilent Technologies, Inc., Santa Clara, CA, USA) and each sample had a RINe >7.0. For cDNA library construction, 1.0 μg RNA was ligated with 3′ RNA adapter (AAGATCGGAAGAGCACACGTCT), followed by 5′ adapter ligation (GUUCAGAGUUCUACAGUCCGACGAUC). Subsequently, the adapter-ligated RNAs were subjected to reverse transcription-polymerase chain reaction (RT-PCR) and were amplified for 12 cycles. Subsequently, the PCR products were size-selected by PAGE, according to the instructions of the TruSeq® Small RNA Sample Prep kit (Illumina, Inc., San Diego, CA, USA). The purified library products were evaluated using the Agilent 2200 TapeStation (Agilent Technologies, Inc.) and were diluted to 10 pM for cluster generation in situ on the HiSeq 2500 single-end flow cell, followed by sequencing (1×50 bp) on HiSeq 2500 (Illumina, Inc.).
After both 3′ and 5′ adaptor sequences were trimmed, reads containing the adapter and poly-N regions, and those of low quality were filtered according to base quality value, including a phred quality mean score >30 and reads between 18 and 40 nt in length. Clean reads were aligned against Rat Nov. 2014 (rn6) assembly, by means of Burrows-Wheeler Aligner (https://sourceforge.net/projects/bio-bwa/), allowing seven mismatches and a gap length of 7 bp. The functional annotations of small RNA variants were then generated. The predicted miRNAs were annotated and identified using miRBase v.21.0 (http://www.mirbase.org/). In each sample, miRNA expression levels were scaled and normalized as transcripts per million of total aligned miRNA reads. Read counts of 3-month-old and 12-month-old groups were compared and log2-transformed. Candidate miRNAs with fold-change >2.0 and P<0.05 were subjected to clustering analysis using Cluster 3.0 and Java Treeview software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm).
Validation of miRNA expression and inflammation-associated mRNA detection
RT-quantitative PCR (RT-qPCR) was conducted to verify eight significantly altered and remodeling-associated miRNAs, which were revealed by small RNA sequencing. In addition, the mRNA expression levels of tumor necrosis factor-α (TNF-α), intercellular cell adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) were detected by RT-qPCR. Firstly, total RNA was isolated from LV tissue samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized in a 20 μl reaction volume using PrimeScript™ RT reagent kit (Takara Bio, Inc. Otsu, Japan) according to the manufacturer's protocol. qPCR was performed using the SYBR® Premix Ex Taq™ kit (Takara Bio, Inc.) in a 10 μl reaction volume containing cDNA template, SYBR® Premix Ex Taq™, PCR forward primer, PCR reverse primer and RNase-free water on a CFX96 Touch™ Real-Time PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). miRNA-specific stem-loop RT and PCR primers were designed and provided by Guangzhou RiboBio Co., Ltd., whereas mRNA primers were provided by Sangon Biotech Co., Ltd. (Shanghai, China) (Table I). PCR thermal cycling involved a denaturing step at 95°C for 30 sec, followed by 40 cycles of annealing at 95°C for 5 sec and extension at 60°C for 34 sec; each sample was analyzed in triplicate. Quantification of relative miRNA expression was normalized to RNU6 as an endogenous control and mRNA expression was normalized to β-actin using the standard 22−ΔΔCq method (7).
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction quantification of differentially expressed miRNAs and inflammation-associated mRNAs.
| miRNA/mRNA | Primer sequence |
|---|---|
| rno-miR-132-3p | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCGACCAT-3 |
| Forward | 5′-GCGTAACAGTCTACAGCCAT-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| rno-miR-182 | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCGGTGTG-3′ |
| Forward | 5′-GGTTTGGCAATGGTAGAACTCA-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| rno-miR-208b-3p | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACCTTTT-3′ |
| Forward | 5′-GGCCGGGATAAGACGAACAA-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| rno-miR-212-3p | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTGGCCGT-3′ |
| Forward | 5′-TAACAGTCTCCAGUCACGGCCA-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| rno-miR-214-3p | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTGCCTG-3′ |
| Forward | 5′-CGACAGCAGGCACAGACA-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| rno-miR-218a-5p | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACATGGT-3′ |
| Forward | 5′-GCGGTTGTGCTTGATCTAAC-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| rno-miR-221-3p | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGAAACCC-3′ |
| Forward | 5′-GGAGCTACATTGTCTGCTGG-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| rno-miR-222-3p | |
| RT primer | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACCCAGT-3′ |
| Forward | 5′-AGCUTACATCTGGCTACTGGGT-3′ |
| Reverse | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| U6 | |
| RT primer | 5′-AACGCTTCACGAATTTGCGT-3′ |
| Forward | 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse | 5′-AACGCTTCACGAATTTGCGT-3′ |
| TNF-α | |
| Forward | 5′-ATGGGCTCCCTCTCATCAGT-3′ |
| Reverse | 5′-GCTTGGTGGTTTGCTACGAC-3′ |
| ICAM-1 | |
| Forward | 5′-ACCTGGACAAGAAGGACTGC-3′ |
| Reverse | 5′-GGTCAGATTAGGGGCTGGAT-3′ |
| VCAM-1 | |
| Forward | 5′-TGGGAAACTGGAAAGAGGAA-3′ |
| Reverse | 5′-CATCAGGAGCCAAACACTTG-3′ |
| β-actin | |
| Forward | 5′-GAAGTGTGACGTTGACATCCG-3′ |
| Reverse | 5′-TGCTGATCCACATCTGCTGGA-3′ |
ICAM-1, intercellular cell adhesion molecule-1; miR/miRNA, microRNA; RT, reverse transcription; VCAM-1, vascular cell adhesion molecule-1.
miRNA targets prediction, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
As aforementioned, for the eight differentially expressed and remodeling-associated miRNAs, target genes were predicted using the miRDB-miRanda algorithm and were then subjected to GO enrichment analysis using the Blast2GO program (https://www.blast2go.com/). All target genes were mapped to each term of the GO database (http://www.geneonology.org) to calculate the number of genes corresponding to each GO term. The significantly enriched GO terms (P<0.05) in biological process, cellular component and molecular function were identified by hypergeometric distribution. As a major public pathway-associated database, KEGG (http://www.genome.jp/kegg/) identifies significantly enriched signaling pathways in target gene candidates compared with the entire reference gene background. Significantly enriched KEGG pathways (P<0.05) of the predicted target genes were identified using KOBAS software version 2.0 (KOBAS, Surrey, UK).
Statistical analysis
Experiments were repeated 3 times and all values are expressed as the means ± standard deviation. Group differences were evaluated using Student's t-test. P<0.05 was considered to indicate a statistically significant difference. All analyses were conducted using SPSS 18.0 for Windows (SPSS Inc., Chicago, IL, USA). Data processing for miRNAs screening and target gene functional annotation was performed by Guangzhou RiboBio Co., Ltd.
Results
SEP, LVMI and plasma NT-proBNP
SBP (187.3±5.5 mmHg vs. 162.3±9.6 mmHg, P<0.05) and LVMI (2.89±0.27 mg/g vs. 2.07±0.26 mg/g, P<0.05) were significantly increased in 12-month-old SHRs compared with in 3-month-old SHRs. Plasma NT-proBNP concentration was also elevated from 259.93±24.74 pg/ml at 3 months to 419.10±28.11 pg/ml at 12 months (P<0.01).
Echocardiographic parameters for 3- and 12-month-old SHRs
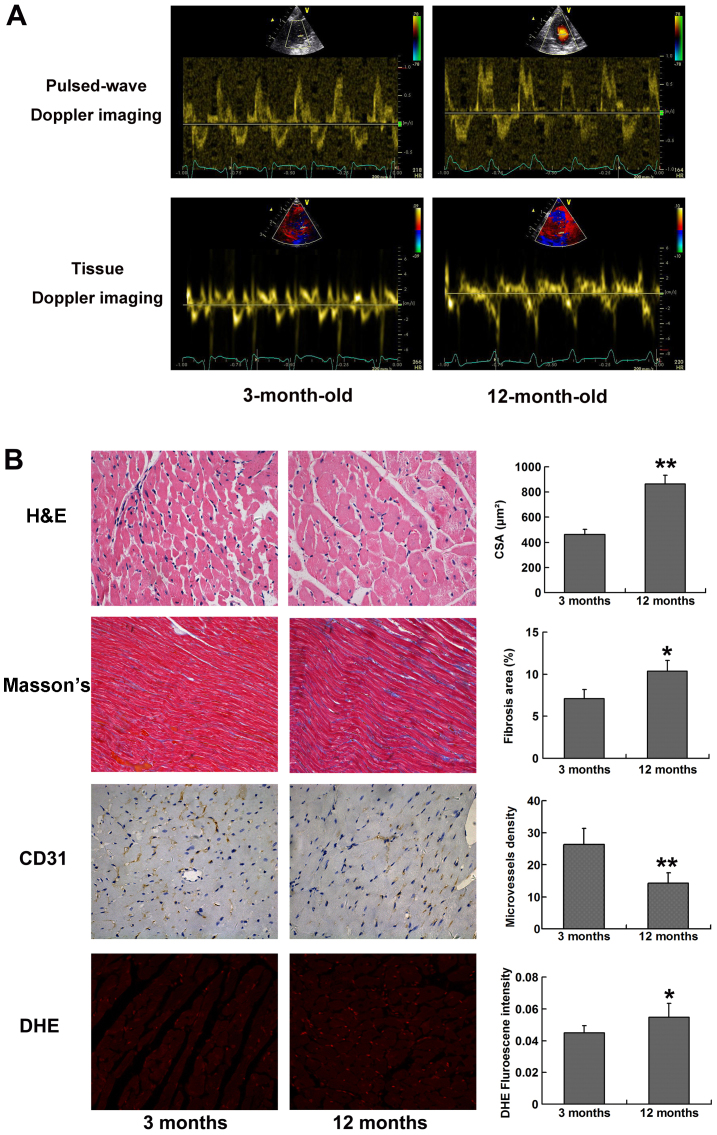
Compared with at 3 months of age, LV geometry of SHRs at 12 months exhibited eccentric hypertrophy and a tendency towards LV dilation, which was associated with increased IVS, LVPW and LVEDD. Conversely, LVESD, LVFS and LVEF remained unchanged between 3 and 12 months of age, thus indicating that LV systolic function was unimpaired in 12-month-old SHRs. Diastolic function was assessed by transmitral pulsed-wave Doppler imaging and tissue Doppler imaging of lateral mitral annulus. E/A ratio and E′/A′ ratio were significantly decreased, whereas E/E′ was increased in 12-month-old SHRs compared with in 3-month-old SHRs (Fig. 1A). In addition, at 12 months of age, SHRs exhibited prolonged IVRT and DT (Table II). Collectively, these data suggested that aging SHRs developed HF with a preserved LVEF, marked LV hypertrophy, and abnormal LV diastolic properties, as characterized by delayed LV relaxation and shortened LV filling time.
Figure 1.
Typical images for echocardiography and histological analysis. (A) Transmitral pulsed-wave Doppler imaging and tissue Doppler imaging at the level of lateral mitral annulus obtained from apical four-chamber view. Peak flow velocity of the E/A waves and peak tissue velocity of E′/A′ diastolic waves were assessed. A decreased E/A ratio and E′/A′ ratio could be observed in 12-month-old SHRs compared with in 3-month-old SHRs. (B) H&E staining (magnification, ×400), Masson's trichrome staining (magnification, ×200), CD31 staining (magnification, ×400) and DHE staining (magnification, ×200), for the calculation of cardiomyocyte CSA, relative fibrosis area (%), microvessel density and reactive oxygen species production, respectively. Five randomly selected fields from each left ventricular section were measured. *P<0.05 and **P<0.01 vs. 3-months-old SHRs. A, late; CD31, cluster of differentiation 31; CSA, cross-sectional area; DHE, dihydroethidium; E, early; H&E, hematoxylin and eosin; SHRs, spontaneously hypertensive rats.
Table II.
Comparison of echocardiographic data between 3- and 12-month-old spontaneously hypertensive rats.
| Variable | 3-month-old | 12-month-old | P-value |
|---|---|---|---|
| LVEDD (mm) | 5.23±0.25 | 6.27±0.55 | 0.042 |
| LVESD (mm) | 2.90±0.17 | 3.27±0.25 | 0.106 |
| IVS (mm) | 1.83±0.25 | 2.47±0.17 | 0.033 |
| LVPW (mm) | 1.87±0.15 | 2.93±0.25 | 0.003 |
| LVEF (%) | 82.6±2.2 | 82.1±2.4 | 0.778 |
| LVFS (%) | 44.6±1.4 | 47.7±5.4 | 0.391 |
| DT (ms) | 20.3±1.5 | 24.7±1.5 | 0.025 |
| IVRT (ms) | 34.7±1.5 | 38.3±1.2 | 0.029 |
| E/A | 2.25±0.07 | 1.72±0.23 | 0.018 |
| E′/A′ | 1.64±0.26 | 0.78±0.02 | 0.004 |
| E/E′ | 26.0±2.5 | 38.2±2.0 | 0.003 |
A, late; DT, deceleration time of E wave; E, early; E/A, ratio of peak transmitral flow velocities of E and A waves; E′/A′, ratio of peak tissue velocities of E′ and A′ diastolic waves at the level of lateral mitral annulus; E/E′, ratio of E to E′; IVRT, isovolumic relaxation time; IVS, interventricular septum thickness; LVEDD, left ventricular end-diastolic diameter; LVEF, left ventricular ejection fraction; LVESD, left ventricular end-systolic diameter; LVFS, left ventricular fractional shortening; LVPW, left ventricular posterior wall thickness.
Histological alterations and ROS production in LV myocardial tissues
Histological analysis demonstrated that LV myocardium developed significant hypertrophy and fibrosis in SHRs at 12 months old compared with at 3 months old, as presented by markedly increased myocyte cross-sectional area (P<0.01) and percentage of fibrosis area (P<0.05). Progressive myocardial hypertrophy and interstitial fibrosis may account for LV stiffness and diastolic dysfunction in aging SHRs. The results of CD31 immunohistochemical staining demonstrated that microvessel density was significantly reduced in 12-month-old SHRs (P<0.01), thus indicating that capillary rarefaction is present in the myocardial tissue of aging SHRs. In addition, DHE fluorescence intensity was significantly increased in 12-month-old SHRs compared with in 3-month-old SHRs (P<0.05), thus suggesting that ROS production was elevated in aging SHRs (Fig. 1B).
Deep small RNA sequencing data analysis and verification
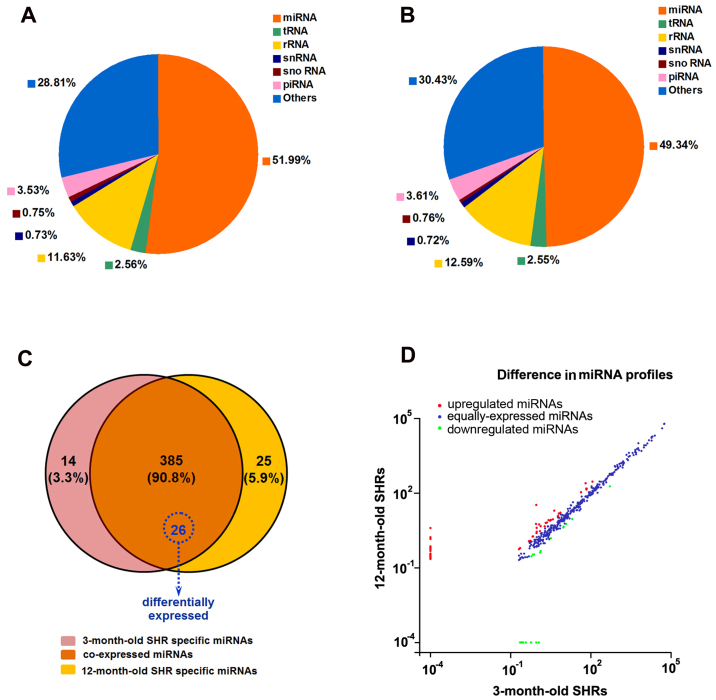
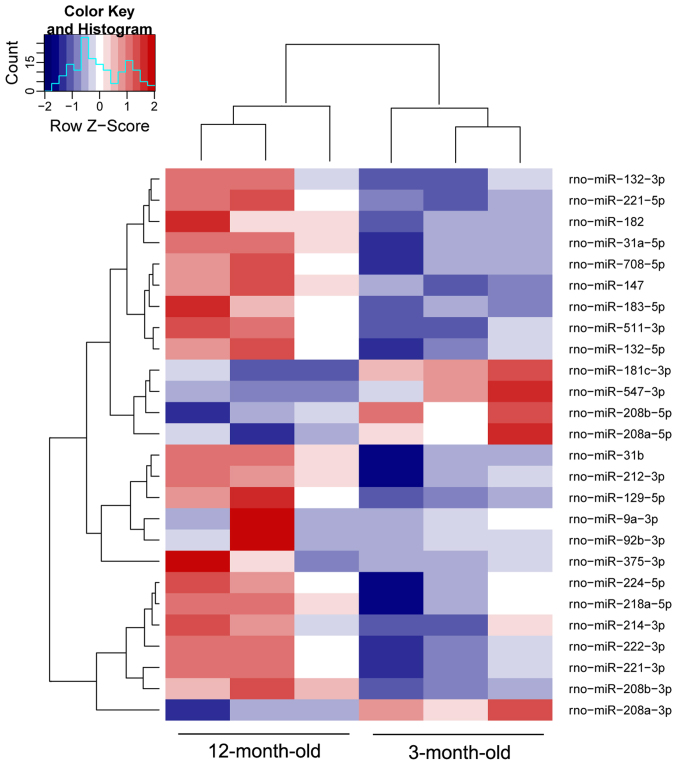
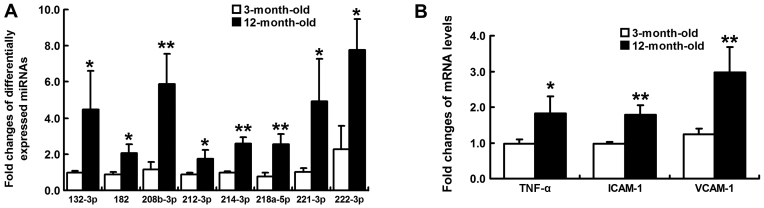
To identity miRNAs associated with the progression of spontaneous hypertension, six small RNA differential expression libraries were constructed and sequenced using the high-throughput HiSeq 2500 platform. After trimming the 3′ and 5′ adapter sequences, and removing low quality reads, as well as reads containing poly N tags and reads <18 nt or >40 nt, an average of 14,295,600 and 13,165,714 clean reads were obtained from the 3-month-old and 12-month-old SHR groups, respectively. The sequenced clean small RNAs included various categories of transfer RNA, ribosomal RNA, small nuclear RNA, small nucleolar RNA, piwi-interacting RNA, miRNA and other unannotated reads, of which miRNA tags accounted for 7,432,397 (51.99%) and 6,495,689 (49.34%) for these two groups, respectively (Fig. 2A and B). The results of high-throughput sequencing indicated that 385 miRNAs were coexpressed in the 3- and 12-month-old SHR groups; whereas 14 and 25 were preferentially expressed in 3- and 12-month-old SHRs, respectively (Fig. 2C). Of the 385 coexpressed miRNAs, 82 miRNAs were up- or downregulated between the 3-month-old and 12-month-old SHRs with a fold-change >2.0 (Fig. 2D), among which 26 mature miRNAs were differentially expressed with a fold-change >2.0 and P<0.05, including those expressed from the 5′-arm and 3′-arm. Compared with 3-month-old SHRs, 21 miRNAs were upregulated and five miRNAs were downregulated in 12-month-old SHRs (Table III). A heat map was constructed to visualize the results of hierarchical clustering of the differentially expressed miRNAs between the 3-month-old and 12-month-old SHR samples (Fig. 3). Notably, the majority of those closely associated with myocardial remodeling were upregulated, including rno-miR-132-3p, rno-miR-182, rno-miR-208b-3p, rno-miR-212-3p, rno-miR-214-3p, rno-miR-218a-5p, rno-miR-221-3p and rno-miR-222-3p. The expression levels of these miRNAs were validated by RT-qPCR, the results of which identified a similar pattern of upregulation consistent with small RNA sequencing data (Fig. 4A).
Figure 2.
High-throughput small RNA sequencing analysis in 3- and 12-month-old SHRs. Pie-charts representing the distribution of different classes of small RNA reads in (A) 3- and (B) 12-month-old SHR groups, in which miRNAs constituted 51.99 and 49.34% of all small RNA reads. Data were averaged from three samples from each group. (C) Venn diagram indicating the distribution of 424 unique miRNAs between 3- and 12-month-old SHRs. The overlapping section contains 385 coexpressed miRNAs, among which 26 miRNAs were significantly differentially expressed (P<0.05). (D) Differences in the miRNA profiles from 3- and 12-month old SHRs, presented as a scatter plot with normalized values (number of reads per million clean tags). A total of 57 miRNAs were upregulated (red) and 25 miRNAs were downregulated (green) with a fold-change >2 irrespective of P-value. The remaining 303 miRNAs were equally expressed (blue) with a fold-change <2. miRNA, microRNA; piRNA, piwi-interacting RNA; rRNA, ribosomal RNA; SHRs, spontaneously hypertensive rats; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; tRNA, transfer RNA.
Table III.
Significantly up- and downregulated miRNAs between 3- and 12-month-old spontaneously hypertensive rats (including those expressed from the 5′-arm and 3′-arm).
| miRNA | Normalized read no.
|
Fold (log2) | P-value | |
|---|---|---|---|---|
| 3 m | 12 m | |||
| Upregulated | ||||
| rno-miR-132-3p | 7.31 | 16.31 | 1.16 | <0.001 |
| rno-miR-132-5p | 2.07 | 6.37 | 1.62 | 0.03 |
| rno-miR-182 | 6.43 | 15.16 | 1.24 | 0.01 |
| rno-miR-208b-3p | 64.66 | 250.22 | 1.95 | <0.001 |
| rno-miR-212-3p | 0.95 | 4.03 | 2.08 | 0.04 |
| rno-miR-214-3p | 68.72 | 153.49 | 1.16 | <0.001 |
| rno-miR-218a-5p | 69.29 | 140.69 | 1.02 | <0.001 |
| rno-miR-221-3p | 116.83 | 290.16 | 1.31 | <0.001 |
| rno-miR-221-5p | 5.99 | 12.20 | 1.03 | 0.03 |
| rno-miR-222-3p | 43.55 | 109.15 | 1.33 | <0.001 |
| rno-miR-375-3p | 0.92 | 33.60 | 5.19 | <0.001 |
| rno-miR-129-5p | 1.18 | 5.56 | 2.24 | 0.01 |
| rno-miR-9a-3p | 2.65 | 8.30 | 1.65 | 0.04 |
| rno-miR-31a-5p | 4.30 | 19.85 | 2.21 | <0.001 |
| rno-miR-183-5p | 3.01 | 7.91 | 1.39 | 0.02 |
| rno-miR-224-5p | 70.84 | 152.60 | 1.11 | <0.001 |
| rno-miR-31b | 0.90 | 5.81 | 2.70 | <0.001 |
| rno-miR-147 | 3.23 | 8.40 | 1.38 | 0.03 |
| rno-miR-708-5p | 3.86 | 8.89 | 1.20 | 0.04 |
| rno-miR-92b-3p | 1.34 | 4.73 | 1.82 | 0.05 |
| rno-miR-511-3p | 2.30 | 6.63 | 1.53 | 0.03 |
| Downregulated | ||||
| rno-miR-208a-5p | 20.33 | 9.16 | −1.15 | <0.001 |
| rno-miR-208a-3p | 521.89 | 187.50 | −1.48 | <0.001 |
| rno-miR-181c-3p | 12.05 | 5.79 | −1.06 | 0.01 |
| rno-miR-547-3p | 9.81 | 4.5236 | −1.12 | 0.02 |
| rno-miR-208b-5p | 9.38 | 4.1846 | −1.17 | 0.03 |
m, month; miR, microRNA.
Figure 3.
Hierarchical cluster analysis of the 26 significantly differentially expressed miRs in 3-month-old and 12-month-old spontaneously hypertensive rats (including those expressed from the 5′-arm and 3′-arm). Red represents increased expression and blue represents reduced expression. miR, microRNA.
Figure 4.
(A) Validation of eight upregulated miRNAs, according to reverse transcription-quantitative PCR. RNU6 served as a housekeeping control. PCR results were consistent with sequencing data. (B) Quantification of TNF-α, ICAM-1 and VCAM-1 mRNA levels normalized to β-actin. *P<0.05 and **P<0.01 vs. 3-month-old spontaneously hypertensive rats. ICAM-1, intercellular cell adhesion molecule-1; miRNA, microRNA; PCR, polymerase chain reaction; TNF-α, tumor necrosis factor-α; VCAM-1, vascular cell adhesion molecule-1.
Inflammation-associated mRNA expression
In order to investigate myocardial inflammatory reaction, the mRNA expression levels of TNF-α, ICAM-1 and VCAM-1 were compared between young and aging SHRs. As a result, these proinflammatory cytokines were significantly upregulated in 12-month-old SHRs (P<0.05; Fig. 4B).
Target gene prediction and functional annotation
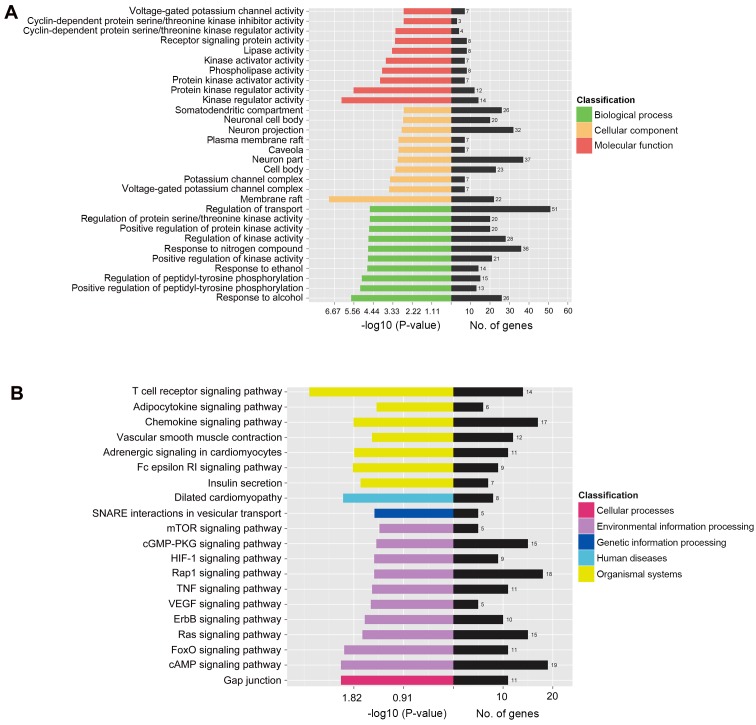
To further elucidate the biological functions and signaling pathways associated with the eight aforementioned miRNAs, target gene prediction was performed using the miRDB-miRanda algorithm; a total of 1,306 genes were identified. Subsequently, the target genes were analyzed using Blast2GO program and KOBAS 2.0 software for GO annotation and KEGG pathway analysis, respectively, in order to understand the molecular interaction and reaction networks. In the GO annotation analysis, 590 biological process terms, 40 cellular component terms and 58 molecular function terms were significantly enriched (P<0.05). Notably, protein kinase regulator activity, regulation of peptidyl-tyrosine phosphorylation, membrane raft, voltage-gated potassium channel complex and activity, and regulation of protein serine/threonine kinase activity were among the most significantly enriched GO terms, thus indicating that the target genes participated largely in cellular signal transduction (Fig. 5A). KEGG pathway analysis identified 39 significantly enriched pathways associated with the target genes. Adrenergic signaling in cardiomyocytes, ErbB signaling pathway, mTOR signaling pathway, FoxO signaling pathway, Ras signaling pathway, insulin secretion, adipocytokine signaling pathway, HIF-1 signaling pathway, Rap1 signaling pathway, VEGF signaling pathway, chemokine signaling pathway and TNF signaling pathway were among the most enriched categories (Fig. 5B).
Figure 5.
GO annotation and KEGG pathway enrichment analysis of predicted miRNA targets. (A) GO enrichment for predicted target genes of the eight upregulated miRNAs. No. of genes enriched and −log10 (P-value) for each term are displayed for the top 10 GO terms in biological process, cellular component and molecular function. (B) Top 20 enriched KEGG pathways of miRNA target genes, as demonstrated by no. of genes and −log10 (P-value). GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA.
Discussion
The present study detected increased LV hypertrophy, fibrosis and stiffness in aging SHRs, leading to diastolic HF; these findings were consistent with those of a previous study (2). Notably, the present study identified a markedly different myocardial miRNA expression profile in 3-month-old SHRs compared with in 12-month-old SHRs, thus suggesting that miRNAs may exert important roles during the transition from hypertension to the onset of diastolic dysfunction and HFpEF in SHRs. A total of 21 miRNAs were upregulated in 12-month-old SHRs, whereas five miRNAs were downregulated. Several of the upregulated miRNAs were involved in cardiac hypertrophy and fibrosis, cardiomyocyte autophagy, angiogenesis, insulin signaling, glucose metabolism and the inflammatory response, and therefore may lead to adverse cardiac remodeling. Bioinformatics analysis revealed that numerous signaling pathways targeting myocardial hypertrophy, autophagy, insulin metabolism, angiogenesis and inflammatory response were involved during the progression of diastolic dysfunction.
Although cardiac hypertrophy is a compensatory mechanism for the maintenance of cardiac output, prolonged pathological hypertrophy has adverse consequences associated with HF and sudden death. Conversely, activation of autophagy during stress conditions is generally considered an adaptive response that compensates for energy loss and rescues spontaneous cardiac hypertrophy. It has been reported that overexpression of the miR-212/132 family directly targets the antihypertrophic and pro-autophagic forkhead box (FoxO)3 transcription factor, which leads to hyperactivation of prohypertrophic calcineurin/nuclear factor of activated T-cells signaling and an attenuated autophagic response (8). In vivo miR-214 overexpression has been reported to induce cardiac hypertrophy by reducing the mRNA expression levels of enhancer of zeste homolog 2 (9). miR-221 has been reported to inhibit autophagy, induce re-expression of fetal genes, and promote cardiac hypertrophy and remodeling by modulating the p27/cyclin-dependent kinase 2/mammalian target of rapamycin (mTOR) axis (10,11). All of these miRNAs were upregulated in 12-month-old SHRs in the present study, and may be associated with the accelerated cardiac hypertrophy detected in aging SHRs. Considering that miR-208b-5p has been defined as the released or destroyed strand, upregulated miR-208b and downregulated miR-208a expression was observed in aging SHRs in the present study. In rodents, miR-208b has been reported to be largely expressed in fetal hearts, whereas its expression is reduced in adult rodents. Conversely, miR-208a exhibits lower expression levels during heart development, becoming predominant in adulthood (12). In addition, it is well known that cardiac hypertrophy is associated with the reappearance of a pattern of gene expression that is similar to that in fetal and newborn rat hearts. The results of the present study are therefore in agreement with the concept of re-expression of fetal genes in cardiac hypertrophy.
Insulin resistance has also been confirmed to be associated with LV diastolic dysfunction, independent of overt diabetes. In addition, adiponectin signaling has been reported to exert protective effects on glucose metabolism and insulin sensitization (13), whereas impaired adiponectin signaling may be associated with increased progression of LV hypertrophy in patients presenting with hypertension (14). Chen et al reported that miR-221 was able to inhibit adiponectin receptor (AdipoR)1 translation by binding to its 3′-untranslated region (15). miR 218 has also been revealed to target AdipoR2 mRNA and inhibit adiponectin-induced AMP-activated protein kinase activation and glucose uptake (16). In addition, overexpression of miR-375 has been reported to suppress glucose-induced insulin secretion, in which myotrophin was predicted to be a target gene and the mechanism was correlated with a direct effect on insulin exocytosis (17). All of these upregulated miRNAs may impair adiponectin signaling, insulin signaling and glucose metabolism, thus promoting deterioration of cardiac diastolic function.
Angiogenesis serves an important role in cardiac hypertrophy. Hypertrophic stimuli initially promote angiogenesis in the heart, allowing cardiac growth and angiogenesis to be coordinated in the adaptive phase of hypertrophy. It has previously been reported that upregulation of miR-182 is associated with angiogenesis-induced hypertrophic response (18). However, capillary rarefaction, resulting from incoordination between cardiomyocyte growth and angiogenesis in the heart, promotes the transition from adaptive cardiac hypertrophy to HF (19). A previous study indicated that aberrant overexpression of miR-214 may reduce cardiac angiogenesis by targeting X-box binding protein-1 in the maladaptive phase of cardiac hypertrophy (20). The miR-221/222 cluster possesses strictly antiangiogenic properties in mature endothelial cells, and stimulates vascular smooth muscle cell dedifferentiation and a switch from the contractile to synthetic phenotype, thus causing arterial wall thickening (21). Furthermore, increased levels of miR-92a/b in the heart has been suggested to inhibit angiogenesis and contribute to the remodeling process in a previous study (22). In the present study, obvious myocardial capillary rarefaction developed in aging SHRs; such microvascular impairment and cardiac endothelial cell remodeling has a causative role in the onset and progression of HFpEF.
It has been reported that the miR-221/222 cluster has an important role in the regulation of vascular inflammation (15,21). Upregulation of miR-221/222 in senescent endothelial cells may be a consequence of age-associated endothelial dysfunction, and the stimulatory effects of low-grade inflammation commonly persisted in the wall of aged vessels. Furthermore, miR-221 may promote the expression of adhesion molecules and enhance the inflammatory response by inhibiting adiponectin signaling (15). The present study detected significantly higher myocardial mRNA expression levels of proinflammatory cytokines, including TNF-α, ICAM-1 and VCAM-1, in 12-month-old SHRs compared with in 3-month-old SHRs. Furthermore, ROS overproduction was observed in aging SHRs, thus inducing oxidative stress and triggering cardiac remodeling, which serves a key role in the development of hypertension and cardiac hypertrophy (23).
Several of the signaling pathways enriched with the miRNA targets in the present study were associated with cardiac hypertrophy and autophagy pathways (i.e., adrenergic signaling in cardiomyocytes, ErbB signaling pathway, mTOR signaling pathway, FoxO signaling pathway, Ras signaling pathway) (24–30), insulin and glucose metabolism pathways (i.e., insulin secretion, adipocytokine signaling pathway) (31), angiogenesis pathways (i.e., HIF-1 signaling pathway, Rap1 signaling pathway, VEGF signaling pathway) (32), and the inflammatory response (i.e., chemokine signaling pathway, TNF signaling pathway). These results verified the critical role of the aforementioned upregulated miRNAs in regulating cardiac hypertrophy, autophagy, insulin signaling, angiogenesis and inflammation, which are associated with the progressive diastolic dysfunction in aging SHRs. However, biological signaling pathways comprise a complex network and no signaling pathway exerts completely positive or negative effects. For example, ErbB2 overexpression causes activation of prohypertrophic gene pathways, including the phosphoinositide 3-kinase/protein kinase B pathway, whereas disabled ErbB signaling results in a loss of cardioprotection in cardiac hypertrophy and contributes to the development of early failure (25,26). Similarly, basal autophagy serves dual (beneficial and detrimental) roles in cardiac myocyte function; either too little or too much autophagy elicits maladaptive and untoward effects. Therefore, the progression from mere hypertension to HFpEF is associated with a complex spectrum of pathophysiological conditions, including aberrant cardiac homeostasis, instead of simply one-dimensional mechanisms.
Notably, abnormal diastolic filling is closely associated with hypertension and advanced age. Hypertension is much more common in the elderly, and cardiac aging is a specific pathophysiological process, which independently contributes to deterioration of diastolic function. Palao et al (33) reported that the maturation of WKY and SHR vessels exhibited considerable overlap in terms of miRNA expression profiles. Upregulated miR-222 expression was detected in both SHRs and WKY rats when comparing mature rats with young rats, whereas upregulated miR-132 expression was specific for the maturation of SHR vessels. Zhang et al (34) detected increased circulating miR-92a, miR-222 and miR-375 expression during the aging process. In addition, Rippe et al (35) indicated that compared with early-passage arterial endothelial cells, miR-221/222 were upregulated and miR-214 was downregulated in late-passage senescent endothelial cells. Therefore, these findings indicated that the miR-132/212 family may be involved in the development of HFpEF. Since miR-214 and miR-208b expression is usually reduced during the aging process, the upregulation of these miRNAs in aging SHRs in the present study may be associated with HFpEF pathology. However, upregulated miR-92a, miR-221/222 and miR-375 may be associated with the normal aging process. In addition, hypertension and the aging process should not be distinctly separated when referring to the pathogenesis of HFpEF, since hypertension prevalence also increases with aging. The upregulated miRNAs in the present study may elucidate the regulatory mechanisms of HFpEF in aging SHRs.
In conclusion, the present study identified different miRNA expression profiles in aging SHRs compared with in young SHRs, thus indicating the critical role of miRNAs in the process of HFpEF. Notwithstanding lack of transgenic experiments to verify these findings, GO category and KEGG pathway enrichment analyses suggested that the target genes of these differentially expressed miRNAs were involved in signaling pathways associated with cardiac hypertrophy, autophagy, insulin metabolism, angiogenesis and inflammation. Further genetic manipulations are required to identify detailed miRNA-targets for the generation of novel therapeutic strategies for the treatment of HFpEF.
Acknowledgments
The present study was supported by grants from the National Basic Research Program of China (973 Program; grant no. 2012cB518605) and the National Natural Science Foundation of China (grant no. 81370199). The study was presented as an abstract at the 28th Great Wall International Congress of Cardiology.
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Owan TE, Hodge DO, Herges RM, Jacobsen SJ, Roger VL, Redfield MM. Trends in prevalence and outcome of heart failure with preserved ejection fraction. N Engl J Med. 2006;355:251–259. doi: 10.1056/NEJMoa052256. [DOI] [PubMed] [Google Scholar]
- 2.Cingolani OH, Yang XP, Cavasin MA, Carretero OA. Increased systolic performance with diastolic dysfunction in adult spontaneously hypertensive rats. Hypertension. 2003;41:249–254. doi: 10.1161/01.HYP.0000052832.96564.0B. [DOI] [PubMed] [Google Scholar]
- 3.Messerli FH, Rimoldi SF, Bangalore S. The transition from hypertension to heart failure: Contemporary update. JACC Heart Fail. 2017;5:543–551. doi: 10.1016/j.jchf.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Gu H, Li Y, Fok H, Simpson J, Kentish JC, Shah AM, Chowienczyk PJ. Reduced first-phase ejection fraction and sustained myocardial wall stress in hyptensive patients with diastolic dysfunction: A manifestation of impaired shortening deactivation that links systolic to diastolic dysfunction and preserves systolic ejection fraction. Hypertesion. 2017;69:633–640. doi: 10.1161/HYPERTENSIONAHA.116.08545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slama M, Ahn J, Varagic J, Susic D, Frohlich ED. Long-term left ventricular echocardiographic follow-up of SHR and WKY rats: Effects of hypertension and age. Am J Physiol Heart Circ Physiol. 2004;286:H181–H185. doi: 10.1152/ajpheart.00642.2003. [DOI] [PubMed] [Google Scholar]
- 6.Rysä J, Leskinen H, Ilves M, Ruskoaho H. Distinct upregulation of extracellular matrix genes in transition from hypertrophy to hypertensive heart failure. Hypertension. 2005;45:927–933. doi: 10.1161/01.HYP.0000161873.27088.4c. [DOI] [PubMed] [Google Scholar]
- 7.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta c(T)) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 8.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang T, Gu H, Chen X, Fu S, Wang C, Xu H, Feng Q, Ni Y. Cardiac hypertrophy and dysfunction induced by overexpression of miR-21. in vivo J Surg Res. 2014;192:317–325. doi: 10.1016/j.jss.2014.06.044. [DOI] [PubMed] [Google Scholar]
- 10.Su M, Wang J, Wang C, Wang X, Dong W, Qiu W, Wang Y, Zhao X, Zou Y, Song L, et al. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 2015;22:986–999. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Wang S, Zhao P, Wang X, Wang J, Wang Y, Song L, Zou Y, Hui R. MiR-221 promotes cardiac hypertrophy in vitro through the modulation of p27 expression. J Cell Biochem. 2012;113:2040–2046. doi: 10.1002/jcb.24075. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira-Carvalho V, Carvalho VO, Bocchi EA. The emerging role of miR-208a in the heart. DNA Cell Biol. 2013;32:8–12. doi: 10.1089/dna.2012.1787. [DOI] [PubMed] [Google Scholar]
- 13.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong SJ, Park CG, Seo HS, Oh DJ, Ro YM. Associations among plasma adiponectin, hypertension, left ventricular diastolic function and left ventricular mass index. Blood Press. 2004;13:236–242. doi: 10.1080/08037050410021397. [DOI] [PubMed] [Google Scholar]
- 15.Chen CF, Huang J, Li H, Zhang C, Huang X, Tong G, Xu YZ. MicroRNA-221 regulates endothelial nitric oxide production and inflammatory response by targeting adiponectin receptor 1. Gene. 2015;565:246–251. doi: 10.1016/j.gene.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Du H, Fu Z, He G, Wang Y, Xia G, Fang M, Zhang T. MicroRNA-218 targets adiponectin receptor 2 to regulate adiponectin signaling. Mol Med Rep. 2015;11:4701–4705. doi: 10.3892/mmr.2015.3282. [DOI] [PubMed] [Google Scholar]
- 17.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Hwangbo C, Jaba IM, Zhang J, Papangeli I, Han J, Mikush N, Larrivée B, Eichmann A, Chun HJ, et al. miR-182 modulates myocardial Hypertrophic response induced by angiogenesis in heart. Sci Rep. 2016;6:21228. doi: 10.1038/srep21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J Mol cell cardiol. 2016;97:245–262. doi: 10.1016/j.yjmcc.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Duan Q, Yang L, Gong W, Chaugai S, Wang F, Chen C, Wang P, Zou MH, Wang DW. MicroRNA-214 is upregulated in heart failure patients and suppresses XBP1-mediated endothelial cells angiogenesis. J Cell Physiol. 2015;230:1964–1973. doi: 10.1002/jcp.24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chistiakov DA, Sobenin IA, Orekhov AN, Bobryshev YV. Human miR-221/222 in physiological and atherosclerotic vascular remodeling. BioMed Res Int. 2015;2015:354517. doi: 10.1155/2015/354517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012;14:147–154. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 23.Bertagnolli M, Schenkel PC, Campos C, Mostarda CT, Casarini DE, Belló-Klein A, Irigoyen MC, Rigatto K. Exercise training reduces sympathetic modulation on cardiovascular system and cardiac oxidative stress in spontaneously hypertensive rats. Am J Hypertens. 2008;21:1188–1193. doi: 10.1038/ajh.2008.270. [DOI] [PubMed] [Google Scholar]
- 24.Barki-Harrington L, Perrino C, Rockman HA. Network integration of the adrenergic system in cardiac hypertrophy. Cardiovasc Res. 2004;63:391–402. doi: 10.1016/j.cardiores.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Sysa-Shah P, Xu Y, Guo X, Belmonte F, Kang B, Bedja D, Pin S, Tsuchiya N, Gabrielson K. Cardiac-specific over-expression of epidermal growth factor receptor 2 (ErbB2) induces pro-survival pathways and hypertrophic cardiomyopathy in mice. PLoS One. 2012;7:e42805. doi: 10.1371/journal.pone.0042805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohrbach S, Yan X, Weinberg EO, Hasan F, Bartunek J, Marchionni MA, Lorell BH. Neuregulin in cardiac hypertrophy in rats with aortic stenosis. Differential expression of erbB2 and erbB4 receptors. Circulation. 1999;100:407–412. doi: 10.1161/01.CIR.100.4.407. [DOI] [PubMed] [Google Scholar]
- 27.Hua Y, Zhang Y, Ceylan-Isik AF, Wold LE, Nunn JM, Ren J. Chronic Akt activation accentuates aging-induced cardiac hypertrophy and myocardial contractile dysfunction: Role of autophagy. Basic Res Cardiol. 2011;106:1173–1191. doi: 10.1007/s00395-011-0222-8. [DOI] [PubMed] [Google Scholar]
- 28.Ni YG, Berenji K, Wang N, Oh M, Sachan N, Dey A, Cheng J, Lu G, Morris DJ, Castrillon DH, et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114:1159–1168. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelb BD, Tartaglia M. RAS signaling pathway mutations and hypertrophic cardiomyopathy: Getting into and out of the thick of it. J Clin Invest. 2011;121:844–847. doi: 10.1172/JCI46399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata R, Ouchi N, Ito M, Kihara S, Shiojima I, Pimentel DR, Kumada M, Sato K, Schiekofer S, Ohashi K, et al. Adiponectin-mediated modulation of hypertrophic signals in the heart. Nat Med. 2004;10:1384–1389. doi: 10.1038/nm1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmona G, Göttig S, Orlandi A, Scheele J, Bäuerle T, Jugold M, Kiessling F, Henschler R, Zeiher AM, Dimmeler S, Chavakis E. Role of the small GTPase Rap1 for integrin activity regulation in endothelial cells and angiogenesis. Blood. 2009;113:488–497. doi: 10.1182/blood-2008-02-138438. [DOI] [PubMed] [Google Scholar]
- 33.Palao T, Swärd K, Jongejan A, Moerland PD, de Vos J, van Weert A, Arribas SM, Groma G, vanBavel E, Bakker EN. Gene expression and microRNA expression analysis in small arteries of spontaneously hypertensive rats. Evidence for ER stress. PLoS One. 2015;10:e0137027. doi: 10.1371/journal.pone.0137027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Yang H, Zhang C, Jing Y, Wang C, Liu C, Zhang R, Wang J, Zhang J, Zen K, et al. Investigation of microRNA expression in human serum during the aging process. J Gerontol A Biol Sci Med Sci. 2015;70:102–109. doi: 10.1093/gerona/glu145. [DOI] [PubMed] [Google Scholar]
- 35.Rippe C, Blimline M, Magerko KA, Lawson BR, LaRocca TJ, Donato AJ, Seals DR. MicroRNA changes in human arterial endothelial cells with senescence: Relation to apoptosis, eNOS and inflammation. Exp Gerontol. 2012;47:45–51. doi: 10.1016/j.exger.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]