Abstract
Microglia, the primary immune cells in the brain, are the predominant cells regulating inflammation-mediated neuronal damage. In response to immunological challenges, such as lipopolysaccharide (LPS), microglia are activated and the inflammatory process is subsequently initiated. The aim of the present study was to determine whether LPS induces interactions between the Toll-like receptor 4 (TLR4) and the ionotropic glutamate receptor N-methyl-D-aspartate subunit 1 (GluN1) in N9 and EOC 20 microglial cells. Immunocytochemistry demonstrated co-localization of TLR4 and GluN1 in response to LPS, and the direct binding of TLR4 and GluN1 was further validated by antibody-based Fluorescence Resonance Energy Transfer technology. Inhibition of the group I metabotropic glutamate receptor 5 with its selective antagonist, MTEP, abolished LPS-induced direct binding of TLR4 to GluN1. Therefore, these data demonstrated that GluN1 and TLR4 act reciprocally in response to LPS in N9 and EOC 20 microglial cells.
Keywords: lipopolysaccharide, microglia, Toll-like receptor 4, N-methyl-D-aspartate receptor, group I metabotropic glutamate receptor 5
Introduction
It generally known that the central nervous system (CNS) and the immune system are isolated with little interaction, except during disease and/or trauma. The blood-brain barrier prevents infiltration of immune cells and molecules into the CNS (1,2). However, recent studies have reported clear and convincing evidence of bidirectional communication between these two systems. Neuronal transmitters and immune cytokines are considered to mediate communication between CNS and the immune system; however, since both immune receptors and neural receptors may be co-expressed in neurons or innate immune cells, such as microglia, whether the two types of receptors can interact directly by protein-protein interactions has not been reported to date.
Microglial cells have been described as the resident macrophages of the brain that perform surveillance functions to maintain the integrity of the CNS (3). In response to a wide range of invading pathogens, microglia initiate innate immune responses characterized by the production of cytokines and chemokines, upregulation of cell surface molecules and expansion of local immune responses (4,5). Neuron-microglia interactions play a pivotal role in controlling microglial functions and restraining their activation. Neurons are able to affect microglia through the release of neurotransmitters, as well as through direct cell-to-cell interactions mediated by membrane-bound antigens and their cognate receptors. Similar to neurons, microglia express a number of neurotransmitter receptors that enable microglia to respond to the same signals acting on neurons.
Microglia endogenously express both neurotransmitter and immune receptors, such as N-methyl-D-aspartate (NMDA) receptors (6) and Toll-like receptor 4 (TLR4). NMDA receptors are glutamate-gated ion channels, which play a key role in CNS function. NMDA receptor dysfunction is involved in various neurological disorders, including stroke, pathological pain, neurodegenerative diseases and neural inflammation. TLR4 is considered to be a receptor essential for proper response to the Gram-negative bacterial endotoxin lipopolysaccharide (LPS) (7).
The interaction between neurotransmitter NMDA receptor subunit 1 (GluN1) and immune receptor TLR4 in N9 and EOC 20 microglial cells was examined in the present study. LPS was found to trigger direct binding of TLR4 to GluN1 in microglia, whereas inhibition of the metabotropic glutamate receptor 5 (mGluR5) with its selective antagonist, MTEP, abolished LPS-induced binding of TLR4 with GluN1. These results indicate that GluN1 directly interacts with TLR4 in response to LPS in N9 and EOC 20 microglial cells.
Materials and methods
Materials
The EOC 20 microglial cell line was purchased from American Type Culture Collection (Manassas, VA, USA) and the N9 cell line was provided by Professor Yun Bai (Department of Medical Genetics, Third Military Medical University, Chongqing, China). DMEM/F12 medium, fetal bovine serum and trypsin were purchased from HyClone (Logan, UT, USA). Goat anti-TLR4 antibody (cat. no. sc-16240) was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). β-actin antibody (cat. no. A2066), goat IgG (purified immunogloblin) and LPS from Escherichia coli 0111:B4 were purchased from Sigma-Aldrich; Merck KGaA (St. Louis, MO, USA). Pierce™ ECL Western Blotting substrate, goat anti-rabbit IgG (cat. no. 31460) and rabbit anti-goat IgG (cat. no. A27014) conjugated HRP, and immobilized protein A/G agarose were purchased from Pierce (Rockford, IL, USA). Polyvinylidene fluoride (PVDF) membranes were purchased from Millipore (Billerica, MA, USA). Anti-GluN1 antibody (cat. no. 5704) was purchased from Cell Signaling Technologies (Danvers, MA, USA). The rabbit anti-GluN1 antibody (cat. no. LS-B13901) used in immunochemistry was purchased from LifeSpan BioSciences (Seattle, WA, USA). Donkey anti-goat IgG Alexa Fluor 555 (cat. no. A-21432) and donkey anti-rabbit IgG Alexa Fluor 488 (cat. no. A-21206) were purchased from Invitrogen; Thermo Fisher Scientific (Carlsbad, CA, USA). 3-[(2-Methyl-1,3-thiazol-4-yl)ethynyl]pyridine (MTEP) was purchased from Tocris (Ellisville, MO, USA).
Cell culture
The N9 and EOC 20 microglial cell lines were maintained in DMEM/F12 with 10% fetal calf serum, 2 mM L-glutamine, 1X NEAA, 100 µg/ml penicillin and 100 µg/ml streptomycin. The cultures were maintained at 37°C in 5% CO2 and the medium was changed every other day.
Western blot and immunoprecipitation analysis
The cells were washed thoroughly with cold phosphate-buffered saline (PBS) and lysed in cold lysis buffer (1% Triton X-100, 10 mM Tris pH 7.6, 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM NaF, 5 mM EDTA and 0.1 mM Na3VO4) with protease inhibitor cocktail tablets on ice for 20 min. The lysates were centrifuged at 4°C at 16,000 × g for 30 min. Supernatant fractions containing equal amounts of total protein were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto PVDF membranes and analyzed by western blotting. Enhanced chemiluminescence detection was performed according to the manufacturer's protocol. For the co-immunoprecipitation assay, total cell lysates were first incubated with protein A agarose with gentle shaking at 4°C for 10 min, then centrifuged at 4°C at 16,000 × g for 15 min to remove non-specific binding. The cleared lysates containing equal amounts of total protein were incubated with anti-TLR4 antibody or goat IgG with gentle shaking at 4°C overnight, then incubated with protein A/G agarose for a further 2 h at 4°C. After centrifugation, the agarose was washed three times in cold lysis buffer. The proteins were eluted with Laemmli buffer and separated by SDS-PAGE.
Cell surface biotinylation
The cells were rinsed thoroughly with ice-cold PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS2+) and incubated twice with 1 ml of 1.0 mg/ml NHS-SS-biotin (Pierce) for 20 min (a total of 40 min) at 4°C. Non-reactive biotin was quenched with 2X 20-min incubations at 4°C in ice-cold PBS2+ containing 0.1 M glycine. After incubation, the cells were washed with PBS2+ without 0.1 M glycine and solubilized for 1 h at 4°C with gentle shaking in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate and 0.1% SDS) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 µg/ml each leupeptin, aprotinin and pepstain). The cell lysate was centrifuged at maximum speed for 20 min at 4°C to remove cell debris. Biotinylated and non-biotinylated proteins were separated from equal amounts of cellular protein (100 µg) by incubation with 100 µl immobilized streptavidin (Pierce) for 12 h at 4°C. The mixture was centrifuged at maximum speed for 2 min and the beads were washed with 1 ml RIPA buffer four times. Proteins bound to streptavidin beads were diluted in Laemmli sample buffer. Biotinylated proteins were analyzed by SDS-PAGE.
Immunocytochemical examination
The cells were plated on poly-D-lysine-coated glass coverslips and cultured for at least 24 h. After being treated with 1 µg/ml of LPS for different intervals, the cells were washed with PBS and fixed with 4% formaldehyde for 15 min at room temperature, then permeabilized in 0.1% Triton X-100 for 20 min, and blocked with 5% goat serum for 1 h at room temperature. Co-immunostaining with anti-TLR4 and anti-GluN1 antibody (1:1,000) was performed at 4°C overnight; donkey anti-rabbit IgG Alexa Fluor 488 (1:1,000) and donkey anti-goat IgG Alex Fluor 555 (1:1,000) were incubated for 1 h at room temperature and the nuclei were then stained with DAPI. Images were acquired under a Leica TCS SP2 confocal microscope with a CCD camera (Leica Microsystems, Wetzlar, Germany). Five to ten fields from each coverslip were randomly selected and analyzed with CoLocalizer Pro software (8) and Protein Proximity Analysis software (9).
Fluorescence resonance energy transfer (FRET)
FRET was measured with the acceptor bleaching method using a Leica confocal laser scanning microscope (Leica Microsystems) according to the method described by König et al (10) and modified by Liu et al (11). Briefly, microglia were seeded onto poly-D-lysine-coated glass coverslips (24 mm in diameter). After a 24-h culture, cells were treated with 1 µg/ml LPS for 30 min, and fixed. Images were captured using a 40X objective and a LUDL filter wheel that allows for rapid exchange of filters (<100 msec). The system was equipped with the following fluorescence filters: Donor filter (excitation, 488 nm; emission, 500–535 nm) and acceptor filter (excitation, 543 nm; emission, 555–620 nm, 75% intensity). The acquisition of the images was performed with Leica confocal software (Leica Microsystems). Background fluorescence was subtracted from all images, and fluorescence intensity was measured in different regions of interest. The change in fluorescence intensity is expressed as FRET efficiency (FRETeff), percentage of fluorescence increase. To calculate FRETeff, the following equation was used: FRETeff = (Ipost−Ipre)/Ipost ×100, where Ipost is the donor intensity after bleaching and Ipre is the donor intensity before bleaching. Negative control was performed with goat IgG and rabbit IgG antibody rather than TLR4 and GluN1, respectively. One cell was selected as the region of interest 1 (ROI1), which was bleached. Four other cells and background were selected as controls, and were not bleached. Five ROI1 regions were analyzed for each coverslip and the experiment was repeated three times independently.
Statistical analysis
All statistical analyses were performed with the SPSS 10.0 software (SPSS Inc., Chicago, IL, USA). The data are expressed as mean ± standard error of the mean. Unpaired, two-tailed Student's t-tests were performed to evaluate the significance of the differences between two groups. For comparison of multiple groups, analysis of variance was used. P<0.05 was considered to indicate statistically significant differences.
Results
LPS triggers interactions of TLR4 and GluN1 in N9 and EOC 20 microglial cells
LPS is the most frequently used model agent to study microglial activation and inflammatory signaling (12). It is generally accepted that LPS activates microglia through TLR4 (7), one of LPS's interaction proteins.
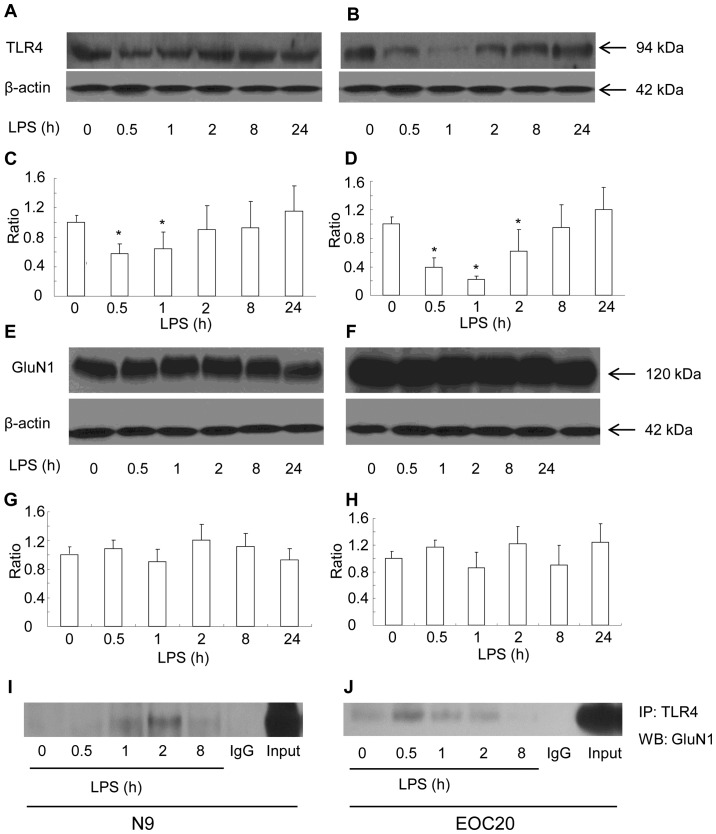
First, the surface expression of TLR4 was measured with the biotinylation method in response to LPS in N9 microglial cells. Briefly, cell surface proteins were biotinylated, and cleared total cell lysates were incubated with streptavidin beads to pull down biotinylated proteins originally localized on the cell surface. The surface expression of TLR4 was found to be gradually reduced in the N9 microglial cells when treated with 1 µg/ml of LPS within the first 2 h. The reduction at 30 min and 1 h after exposure to LPS was significantly lower compared to that prior to LPS treatment (Fig. 1A and C) (P<0.05 vs. prior to LPS treatment). At 2 h after exposure to LPS, the surface expression of TLR4 had recovered to the level before treatment (Fig. 1A and C). Thereafter and until 24 h after exposure to LPS, the surface expression of TLR4 remained at a similar level. The total TLR4 expression was not altered at all the tested timepoints in response to LPS (data not shown). The expression of GluN1 was also observed in N9 microglial cell. The surface expression of GluN1 (data not shown) and total GluN1 expression were not affected by LPS treatment (Fig. 1E and G). The cellular level of β-actin did not differ among the LPS-treated groups. We tested whether TLR4 was bound to GluN1. The total cell lysates of the N9 microglia following LPS treatment for various intervals were immunoprecipitated with the TLR4 antibody. Anti-GluN1 western blot analysis of the TLR4 immunoprecipitates revealed that GluN1 was co-immunoprecipitated with TLR4 (Fig. 1I). The co-immunoprecipitates of TLR4 with GluN1 appeared at 1 h, reached a maximum level at 2 h, and declined 8 h after LPS treatment. The N9 microglial cell line is a type of primary mouse microglial cells immortalized with the v-myc and v-mil oncogenes of the avian retrovirus MH2, which share many phenotypical characteristics with primary mouse microglia and express functional TLR4 receptor (13,14). To elucidate whether LPS-induced co-immunoprecipitation of GluN1 and TLR4 is dependent on TLR4 receptor, TLR4-mutant EOC 20 microglial cells were utilized (15). EOC 20 microglial cells were derived from the C3H/HeJ mouse brain, which carries a missense mutation in the TLR4 gene. This destructive mutation in the TLR4 receptor leads to a defective response to LPS in EOC 20 microglia (15,16). As shown in Fig. 1B and D, LPS treatment significantly reduced the surface expression of TLR4 in EOC 20 microglia within the first 8 h (P<0.05 vs. before LPS treatment). The surface expression of TLR4 reached a minimum at 1 h and gradually returned back to the level before LPS treatment at 8 h after LPS treatment. Compared with N9 microglia, the recovery of surface TLR4 was slow in EOC 20 microglia. However, similar to N9 microglia, the total TLR4 expression at all tested timepoints was at a similar level (data not shown). The surface and total expression of GluN1 in EOC 20 microglia were not affected by LPS treatment, and the total expression of GluN1 is shown in Fig. 1F and H. Similar to N9 microglial cells, co-immunoprecipitates of TLR4 and GluN1 were detected in EOC 20 microglial cells. The co-immunoprecipitates of TLR4 and GluN1 reached a maximum level at 0.5 h after LPS treatment, were gradually reduced, and diminished at 8 h after LPS treatment. These results demonstrated that LPS induced co-immunoprecipitation of TLR4 and GluN1 in both N9 and EOC 20 microglial cells, which is not affected by TLR4-destructive missense mutations.
Figure 1.
Lipopolysaccharide (LPS) triggers interaction of Toll-like receptor 4 (TLR4) and glutamate receptor N-methyl-D-aspartate subunit 1 (GluN1) in N9 and EOC 20 microglial cells. N9 (A and E) and EOC 20 (B and F) microglial cells were treated with LPS (1 µg/ml) for various durations. Cell surface expression of TLR4 was blotted with anti-TLR4 (A and B, top panel). Total cell lysates were blotted with anti-GluN1 (E and F, top panel) or anti-β-actin (A, B, E and F, bottom panel). β-actin blots exhibited equal loading. Scanning densitometry values of western blotting for TLR4 (C and D) and GluN1 (G and H) were normalized by densitometry of corresponding β-actin bands and expressed as mean ± standard error of the mean. The mean was calculated from four independent experiments (n=4). *P<0.05 vs. prior to LPS treatment. Co-immunoprecipitation of TLR4 and GluN1 in N9 (I) and EOC 20 (J) microglial cells treated with LPS (1 µg/ml) for various durations. Anti-TLR4 immunoprecipitates were blotted with anti-GluN1. Similar results were obtained from three independent experiments (n=3).
Immunocytochemistry shows co-localization of TLR4 and GluN1 in response to LPS in N9 and EOC 20 microglial cells
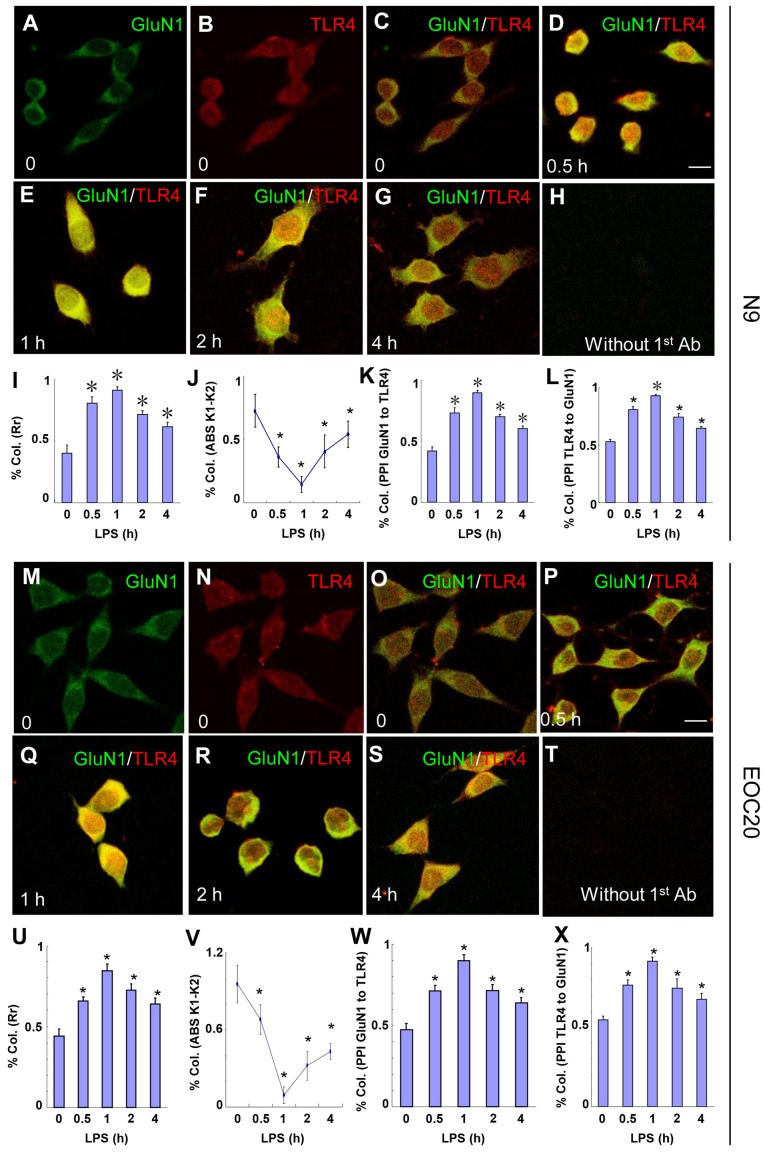
The LPS-induced co-immunoprecipitation of TLR4 and GluN1 in microglia suggests the possibility of co-localization of the two proteins. The co-localization of TLR4 and GluN1 was examined with immunocytochemistry. N9 microglia were treated with 1 µg/ml LPS, and then cells were fixed at various times. As shown in Fig. 2A–L, the co-localization of TLR4 and GluN1 was observed at 30 min (Fig. 2D) after LPS treatment. These images were analyzed with CoLocalizer Pro software (8) and protein proximity index (PPI) (9) and the results are presented in Fig. 2I–L. Pearson's correlation coefficient (Rr) in Fig. 2I describes the correlation of the intensity distributions between two channels, TLR4 (red) and GluN1 (green). The 1.0 value indicates complete co-localization. Overlap coefficients K1 and K2 (AbsK1-K2) in Fig. 2J split the value of co-localization into a pair of separate parameters. The lower the absolute value of K1-K2, the stronger the co-localization. PPI analysis provides a separate value for each channel to characterize the contribution of each protein to the overall co-localization. TLR4 and GluN1 contribute equally to the co-localization based on the PPI analysis (Fig. 2K and L). As shown in Fig. 2D–G, I and J, the co-localization of TLR4 and GluN1 appeared at 30 min, reached a maximum at 1 h, and decreased gradually until 4 h after LPS treatment. Without LPS treatment, co-localization of TLR4 and GluN1 was not observed (Fig. 2C). When TLR4 and GluN1 antibodies were replaced with goat IgG and rabbit IgG, respectively, no specific staining was observed (Fig. 2H). Similar results were obtained when EOC 20 microglia were treated with LPS (Fig. 2M–X). These results suggest that TLR4 and GluN1 are located in the same organelles in microglia following exposure to LPS, and the co-localization of TLR4 and GluN1 was not affected by destructive TLR4 missense mutations.
Figure 2.
Toll-like receptor 4 (TLR4) and glutamate receptor N-methyl-D-aspartate subunit 1 (GluN1) co-localize in N9 and EOC 20 microglial cells. (A–L) N9 and (M–X) EOC 20 microglial cells were treated with 1 µg/ml LPS for (D and P) 30 min, (E and Q) 1 h, (F and R) 2 h or (G and S) 4 h, and stained for GluN1 (green) and TLR4 (red). Without LPS treatment, (C and O) no co-localization of TLR4 and GluN1 was observed. Single GluN1 and TLR4 images are shown in (A and M) and (B and N), respectively, in the group without LPS treatment. No signal was observed if TLR4 and GluN1 antibodies were replaced with goat IgG and rabbit IgG, respectively. Five to ten fields from each condition were randomly selected and analyzed with CoLocalizer Pro software and Protein Proximity Analysis software. The results are shown in (I–L) for N9 and (U–X) for EOC 20 microglial cells. *P<0.05 vs. 0 (without LPS treatment). Scale bar, 20 µM. The experiments were repeated at least three times with similar results, and two coverslips were examined for each condition in one test (n=6). Col., colocalization; Rr, Pearson's correlation coefficient; LPS, lipopolysaccharide.
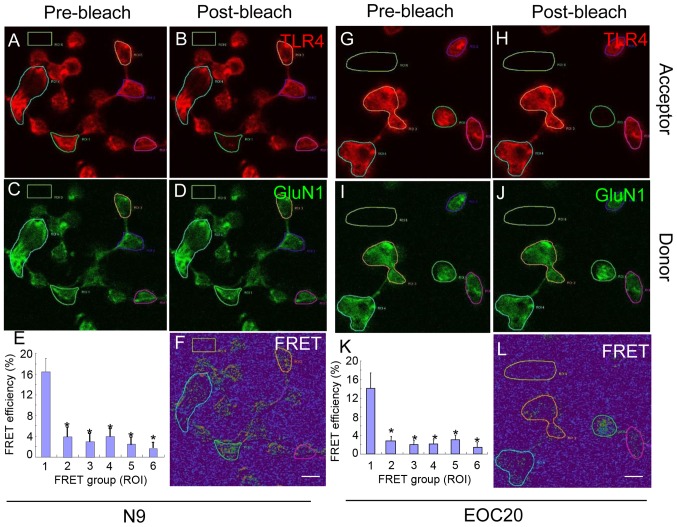
To further validate our findings, antibody-based FRET technology was utilized. N9 microglia were treated with 1 µg/ml LPS for 30 min and co-stained with antibodies against TLR4 (red) and GluN1 (green). Images pre- and post-bleach are shown for TLR4 in Fig. 3A and B, and for GluN1 in Fig. 3C and D. As shown in Fig. 3E and F, the average FRET efficiency in ROI1 was 16.38±2.63%, which was significantly higher compared with that in ROI2-ROI6 (P<0.05 vs. ROI1). Cells at ROI2-5 were not bleached and ROI6 was background control. When the experiment was performed with EOC 20 microglia, similar results were obtained (Fig. 3G–L). Thus, the FRET analysis provided further evidence that LPS triggers direct binding of TLR4 to GluN1 in both N9 and EOC 20 microglial cells, and that the binding of TLR4 with GluN1 was not affected by TLR4 missense mutations.
Figure 3.
The binding of Toll-like receptor 4 (TLR4) and glutamate receptor N-methyl-D-aspartate subunit 1 (GluN1) was investigated with antibody-based fluorescence resonance energy transfer (FRET) in N9 and EOC 20 microglial cells. (A–F) N9 and (G–L) EOC 20 microglial cells were treated with 1 µg̸ml lipopolysaccharide (LPS) for 30 min and stained for TLR4 (red) and GluN1 (green). (A and G) Pre-bleach and (B and H) post-bleach images for TLR4. (C and I) Pre-bleach and (D and J) post-bleach images for GluN1. (F and L) FRET efficiency images. When one cell was randomly selected as the region of interest 1 (ROI1), four other cells and background were selected as controls, which were not bleached. (E and K) Five cells as ROI1 were analyzed from each coverslip and FRET efficiency was calculated. *P<0.05 vs. ROI1. Similar results were obtained from at least three independent experiments and two coverslips were examined for each condition in one test (n=6). Scale bar, 20 µM.
LPS-triggered binding of TLR4 and GluN1 is dependent on mGluR5 in N9 and EOC 20 microglial cells. We have previously demonstrated that LPS triggers calcium [Ca2+]i oscillation through directly binding to mGluR5 in microglial cells (11). A large number of studies have indicated a regulatory role of mGluR5 in the GluN1 responses to its ligand activation (17–19). We tested whether mGluR5 is involved in the direct binding of TLR4 to GluN1 in response to LPS.
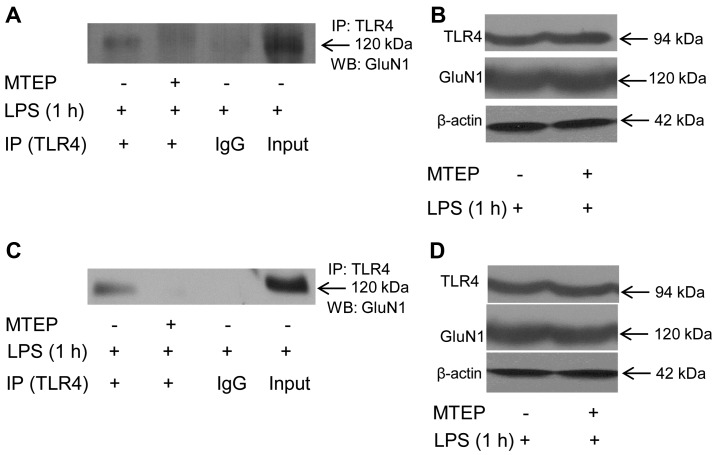
As shown in Fig. 4A, when N9 microglia were first treated with the mGluR5 selective antagonist MTEP (0.1 mM) for 30 min, and then treated with 1 µg/ml LPS for another 1 h, the co-immunoprecipitates of TLR4 and GluN1 was significantly reduced. The cellular level of β-actin did not differ between groups with and without MTEP treatment (Fig. 4B), and the total cellular levels of TLR4 and GluN1 were also similar (Fig. 4B), suggesting that the decreased co-immunoprecipitation of TLR4 and GluN1 by MTEP pretreatment was not due to the change of turnover of TLR4 or GluN1. Similarly, MTEP abolished LPS-triggered co-immunoprecipitation of TLR4 and GluN1 in TLR4-mutant EOC 20 microglia (Fig. 4C and D). Co-localization of TLR4 and GluN1 was not observed in N9 or EOC 20 microglial cells when MTEP was applied prior to LPS treatment (data not shown). These results indicate that inhibition of mGluR5 abolished LPS-induced direct binding of TLR4 to GluN1 in N9 and EOC 20 microglial cells.
Figure 4.
Metabotropic glutamate receptor 5 (mGluR5) antagonist attenuates the binding of Toll-like receptor 4 (TLR4) and glutamate receptor N-methyl-D-aspartate subunit 1 (GluN1) induced by lipopolysaccharide (LPS) in N9 and EOC 20 microglial cells. (A and B) N9 and (C and D) EOC 20 microglial cells were pretreated with 0.1 mM 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]pyridine (MTEP) for 30 min, and then treated with 1 µg/ml LPS for 1 h. Anti-TLR4 immunoprecipitates from (A) N9 or (C) EOC 20 microglial cells were blotted with anti-GluN1. Total cell lysates from (B) N9 or (D) EOC 20 microglial cells were blotted with anti-TLR4 (B and D, top panel), anti-GluN1 (B and D, middle panel), or anti-β-actin (B and D, bottom panel). Similar results were obtained from four independent experiments (n=4).
Discussion
Microglia, the primary immune cells in the brain, have been implicated as the predominant cells regulating inflammation-mediated neuronal damage. In response to immunological challenges, such as LPS, microglia are activated and subsequently the inflammatory process is initiated as evidenced by the release of pro-inflammatory chemokines and cytokines. Activated microglia accumulate around brain lesions that evident in neurodegenerative disorders, such as Alzheimer's disease and Parkinson's disease, and likely play a role in the neuronal damage that occurs in these diseases (20,21). The effective function of microglial cells is critical for controlling neuroinflammation and alleviating neuropathogenesis. In the present study, it was observed that LPS may trigger interactions between TLR4 and GluN1 in N9 and EOC 20 microglial cells. Immunocytochemistry demonstrated co-localization of TLR4 and GluN1 in response to LPS, and the direct binding of TLR4 and GluN1 was further validated by antibody-based FRET technology. Inhibition of mGluR5 with its selective antagonist, MTEP, abolished LPS-induced direct binding of TLR4 with GluN1.
TLR4, a member of the Toll-like receptor family, has been shown to serve as the main upstream sensor for response to LPS (7). TLR4 is a type I transmembrane protein characterized by an extracellular domain containing leucine-rich repeats (LRRs) and a cytoplasmic tail that contains a conserved region referred to as the Toll/IL-1 receptor (TIR) domain. After TLR4 encounters LPS, the first signaling pathway mediated by a pair of proteins, namely TIRAP and MyD88, is activated from the plasma member (22,23). Subsequently, TLR4 undergoes internalization into the endosomal network where the second signaling pathway is triggered through the adaptors TRAM and TRIF (24,25). Thus, LPS-induced endocytosis of TLR4 is essential for its signaling functions. We observed that the surface expression of TLR4 was significantly reduced at 30 min after exposure to 1 µg/ml LPS in both N9 and EOC 20 microglial cells. At 2 h after LPS treatment, the surface expression of TLR4 returned back to the level prior to treatment. However, the total TLR4 expression was not altered by LPS treatment. These results indicate that LPS treatment accelerates TLR4 internalization and reduces its surface expression, but does not disturb TLR4 turnover, which is in agreement with previous studies in murine macrophages (24,26), suggesting that TLR4 undergoes internalization in response to LPS in microglial cells.
NMDA receptors (NMDARs) are the major subtype of glutamate-gated ion channels and are crucial for neuronal communication. NMDARs are assembled as heterotetramers composed by multiple subunits, which fall into three subfamilies according to sequence homology, one GluN1 subunit, four distinct GluN2 subunits, and two GluN3 subunits (27,28). NMDARs are typically composed of GluN1 subunits and GluN2 subunits or a mixture of GluN2 and GluN3 subunits. The NMDA receptor is densely distributed in the brain, and the main subunit GluN1 is ubiquitously expressed from embryonic E14 to adulthood (29,30). We observed that GluN1 is expressed in N9 and EOC 20 microglial cells, and its expression is not altered by LPS treatment. NMDARs have been shown to be involved in the LPS-induced innate immune response (31–33). NMDAR antagonists attenuate LPS-induced fever and increase in hypothalamic hydroxyl radicals in rabbits (34) and LPS-enhanced cerebral infarcts in rats (33). Accumulating evidence also indicates that LPS modulates NMDAR subunits, including GluN1 and GluN2 expression (35–37). Our results (Fig. 1I and J) indicate that TLR4 is co-immunoprecipitated with GluN1, the main NMDAR subunit, in response to LPS in N9 and EOC 20 microglial cells. The co-immunoprecipitates of TLR4 and GluN1 emerged at 30 min, reached a maximum at 2 h and diminished at 8 h after LPS application. This time frame is similar to LPS-induced TLR4 internalization, suggesting the possibility that the internalized TLR4 interacts with GluN1. The immunocytochemistry results in the present study (Fig. 2) further verify that TLR4 co-localizes with GluN1 in response to LPS treatment. Similar to co-immunoprecipitates, the co-localization of TLR4 and GluN1 reached a maximum at 1 h after LPS application. Antibody-based FRET analysis (Fig. 3) revealed that TLR4 directly binds to GluN1 in vivo in response to LPS. The binding of TLR4 with GluN1 was not altered by destructive TLR4 missense mutations, as similar binding was observed in both N9 and EOC 20 microglial cells. These results suggest that LPS-induced binding of TLR4 to GluN1 is not mediated by the TLR4 pathway.
The mGluR5 has been shown to be an alternative critical receptor in response to LPS in microglial cells (11). The mGluR5 selective antagonist MTEP abolished LPS-triggered binding of TLR4 with GluN1, suggesting that [Ca2+]i oscillation mediated by mGluR5 in response to LPS may be involved in this process. It has been demonstrated that mGluR5 agonists increase GluN1 phosphorylation in rats (17). It remains unknown whether the phosphorylation status of GluN1 affects its binding with TLR4, which domains of TLR4 and GluN1 are responsible for their binding, and where the binding localizes in the microglial cells. The investigation is still undergoing.
In conclusion, the results of the present study demonstrated that TLR4 directly binds to GluN1 in response to LPS, and mGluR5 modulates LPS-induced binding of TLR4 and GluN1 in N9 and EOC 20 microglial cells. Thus, GluN1 may be a potential target for modulating LPS-induced neuroinflammation.
References
- 1.Goncharova LB, Tarakanov AO. Molecular networks of brain and immunity. Brain Res Brain Res Rev. 2007;55:155–166. doi: 10.1016/j.brainresrev.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Gallowitsch-Puerta M, Pavlov VA. Neuro-immune interactions via the cholinergic anti-inflammatory pathway. Life Sci. 2007;80:2325–2329. doi: 10.1016/j.lfs.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garden GA, Möller T. Microglia biology in health and disease. J Neuroimmune Pharmacol. 2006;1:127–137. doi: 10.1007/s11481-006-9015-5. [DOI] [PubMed] [Google Scholar]
- 4.Aloisi F. The role of microglia and astrocytes in CNS immune surveillance and immunopathology. Adv Exp Med Biol. 1999;468:123–133. doi: 10.1007/978-1-4615-4685-6_10. [DOI] [PubMed] [Google Scholar]
- 5.Kielian T, Mayes P, Kielian M. Characterization of microglial responses to Staphylococcus aureus: Effects on cytokine, costimulatory molecule, and Toll-like receptor expression. J Neuroimmunol. 2002;130:86–99. doi: 10.1016/S0165-5728(02)00216-3. [DOI] [PubMed] [Google Scholar]
- 6.Hirayama M, Kuriyama M. MK-801 is cytotoxic to microglia in vitro and its cytotoxicity is attenuated by glutamate, other excitotoxic agents and atropine. Possible presence of glutamate receptor and muscarinic receptor on microglia. Brain Res. 2001;897:204–206. doi: 10.1016/S0006-8993(01)02114-X. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 8.Zinchuk V, Zinchuk O, Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: Pushing pixels to explore biological phenomena. Acta Histochem Cytochem. 2007;40:101–111. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zinchuk V, Wu Y, Grossenbacher-Zinchuk O, Stefani E. Quantifying spatial correlations of fluorescent markers using enhanced background reduction with protein proximity index and correlation coefficient estimations. Nat Protoc. 2011;6:1554–1567. doi: 10.1038/nprot.2011.384. [DOI] [PubMed] [Google Scholar]
- 10.König P, Krasteva G, Tag C, König IR, Arens C, Kummer W. FRET-CLSM and double-labeling indirect immunofluorescence to detect close association of proteins in tissue sections. Lab Invest. 2006;86:853–864. doi: 10.1038/labinvest.3700443. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Zhou R, Yan H, Yin H, Wu X, Tan Y, Li L. Metabotropic glutamate receptor 5 modulates calcium oscillation and innate immune response induced by lipopolysaccharide in microglial cell. Neuroscience. 2014;281:24–34. doi: 10.1016/j.neuroscience.2014.09.034. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann A, Kann O, Ohlemeyer C, Hanisch UK, Kettenmann H. Elevation of basal intracellular calcium as a central element in the activation of brain macrophages (microglia): Suppression of receptor-evoked calcium signaling and control of release function. J Neurosci. 2003;23:4410–4419. doi: 10.1523/JNEUROSCI.23-11-04410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Righi M, Mori L, De Libero G, Sironi M, Biondi A, Mantovani A, Donini SD, Ricciardi-Castagnoli P. Monokine production by microglial cell clones. Eur J Immunol. 1989;19:1443–1448. doi: 10.1002/eji.1830190815. [DOI] [PubMed] [Google Scholar]
- 14.Stansley B, Post J, Hensley K. A comparative review of cell culture systems for the study of microglial biology in Alzheimer's disease. J Neuroinflammation. 2012;9:115. doi: 10.1186/1742-2094-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi O, Takeda K, Hoshino K, Adachi O, Ogawa T, Akira S. Cellular responses to bacterial cell wall components are mediated through MyD88-dependent signaling cascades. Int Immunol. 2000;12:113–117. doi: 10.1093/intimm/12.1.113. [DOI] [PubMed] [Google Scholar]
- 17.Choe ES, Shin EH, Wang JQ. Regulation of phosphorylation of NMDA receptor NR1 subunits in the rat neostriatum by group I metabotropic glutamate receptors in vivo. Neurosci Lett. 2006;394:246–251. doi: 10.1016/j.neulet.2005.10.072. [DOI] [PubMed] [Google Scholar]
- 18.Pisani A, Calabresi P, Centonze D, Bernardi G. Enhancement of NMDA responses by group I metabotropic glutamate receptor activation in striatal neurones. Br J Pharmacol. 1997;120:1007–1014. doi: 10.1038/sj.bjp.0700999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Contractor A, Gereau RW, IV, Green T, Heinemann SF. Direct effects of metabotropic glutamate receptor compounds on native and recombinant N-methyl-D-aspartate receptors. Proc Natl Acad Sci USA. 1998;95:8969–8974. doi: 10.1073/pnas.95.15.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Hong JS. Role of microglia in inflammation-mediated neurodegenerative diseases: Mechanisms and strategies for therapeutic intervention. J Pharmacol Exp Ther. 2003;304:1–7. doi: 10.1124/jpet.102.035048. [DOI] [PubMed] [Google Scholar]
- 21.Town T, Nikolic V, Tan J. The microglial 'activation' continuum: From innate to adaptive responses. J Neuroinflammation. 2005;2:24. doi: 10.1186/1742-2094-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J Biol Chem. 2002;277:47834–47843. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 23.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 24.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanimura N, Saitoh S, Matsumoto F, Akashi-Takamura S, Miyake K. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem Biophys Res Commun. 2008;368:94–99. doi: 10.1016/j.bbrc.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 26.Józefowski S, Czerkies M, Sobota A, Kwiatkowska K. Determination of cell surface expression of Toll-like receptor 4 by cellular enzyme-linked immunosorbent assay and radiolabeling. Anal Biochem. 2011;413:185–191. doi: 10.1016/j.ab.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 27.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe M, Inoue Y, Sakimura K, Mishina M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- 30.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 31.Glezer I, Zekki H, Scavone C, Rivest S. Modulation of the innate immune response by NMDA receptors has neuropathological consequences. J Neurosci. 2003;23:11094–11103. doi: 10.1523/JNEUROSCI.23-35-11094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroso M, Balosso S, Ravizza T, Liu J, Aronica E, Iyer AM, Rossetti C, Molteni M, Casalgrandi M, Manfredi AA, et al. Toll-like receptor 4 and high-mobility group box-1 are involved in ictogenesis and can be targeted to reduce seizures. Nat Med. 2010;16:413–419. doi: 10.1038/nm.2127. [DOI] [PubMed] [Google Scholar]
- 33.Cho GS, Lee JC, Ju C, Kim C, Kim WK. N-Methyl-D-aspartate receptor antagonists memantine and MK-801 attenuate the cerebral infarct accelerated by intracorpus callosum injection of lipopolysaccharides. Neurosci Lett. 2013;538:9–14. doi: 10.1016/j.neulet.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 34.Kao CH, Kao TY, Huang WT, Lin MT. Lipopolysaccharide- and glutamate-induced hypothalamic hydroxyl radical elevation and fever can be suppressed by N-methyl-D-aspartate-receptor antagonists. J Pharmacol Sci. 2007;104:130–136. doi: 10.1254/jphs.FP0070272. [DOI] [PubMed] [Google Scholar]
- 35.Weaver-Mikaere L, Gunn AJ, Mitchell MD, Bennet L, Fraser M. LPS and TNF alpha modulate AMPA/NMDA receptor subunit expression and induce PGE2 and glutamate release in preterm fetal ovine mixed glial cultures. J Neuroinflammation. 2013;10:153. doi: 10.1186/1742-2094-10-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harré EM, Galic MA, Mouihate A, Noorbakhsh F, Pittman QJ. Neonatal inflammation produces selective behavioural deficits and alters N-methyl-D-aspartate receptor subunit mRNA in the adult rat brain. Eur J Neurosci. 2008;27:644–653. doi: 10.1111/j.1460-9568.2008.06031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh SH, Hung JJ, Gean PW, Chang WC. Hypoxia-inducible factor-1alpha protects cultured cortical neurons from lipopolysaccharide-induced cell death via regulation of NR1 expression. J Neurosci. 2008;28:14259–14270. doi: 10.1523/JNEUROSCI.4258-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]