Abstract
Nod2 is a cytosolic, innate immune receptor responsible for binding to bacterial cell wall fragments such as muramyl dipeptide (MDP). Upon binding, subsequent downstream activation of the NF-κB pathway leads to an immune response. Nod2 mutations are correlated with an increased susceptibility for Crohn’s disease (CD) and ultimately results in a misregulated immune response. Previous work had demonstrated that Nod2 interacts with and is stabilized by the molecular chaperone Hsp70. In this work it is shown using purified protein and in vitro biochemical assays that the critical Nod2 CD mutations (G908R, R702W, 1007fs) retain the ability to bind bacterial ligands. A limited proteolysis assay and luciferase reporter assay reveal regions of Hsp70 that are capable of stabilizing Nod2 and rescuing CD-mutant activity. A minimal 71 amino acid subset of Hsp70 that stabilizes the CD associated variants of Nod2 and restores a proper immune response upon activation with MDP was identified. This work suggests that CD-associated Nod2 variants could be stabilized in vivo with a molecular chaperone.
Keywords: Nod2, Hsp70, chaperone, innate immunity, muramyl dipeptide, nucleotide cofactors, protein stability, protein-protein interaction
Graphical Abstract
The innate immune system uses a series of receptors to properly respond to both pathogenic and commensal bacteria1–2. Nod2 (nucleotide-binding oligomerization domain-containing protein 2) is an innate immune receptor that is correlated with an increased susceptibility for developing Crohn’s disease (CD), a debilitating, incurable gastrointestinal disease, when mutated3–4. In the presence of muramyl dipeptide (MDP, Figure 1A), a small peptidoglycan (PG) fragment, Nod2 initiates the NF-κB pathway5. It is hypothesized that this is a result of direct binding to MDP and the recruitment of signaling proteins6–10; ultimately leading to an immune response which is misregulated in individuals with CD3–4. The misregulated response of the Nod2 CD variants could stem from a variety of sources: inability to bind PG fragments, failure to initiate an immune response, and/or a general protein stability effect11.
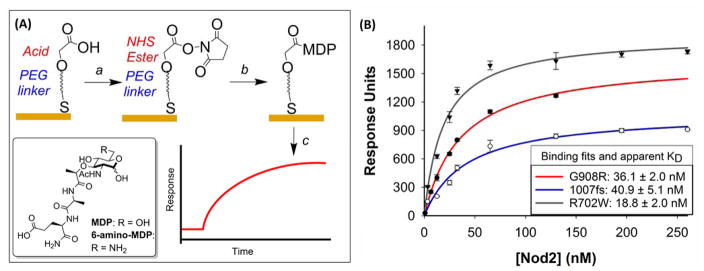
Figure 1.
(A) Schematic of SPR binding assay: (a) activate carboxylic acids using NHS/EDC chemistry, (b) immobilize ligand of interest (6-amino-MDP) to activated chip surface and (c) apply purified protein and generate sensorgram. (B) Binding curves of purified mutant Nod2 to MDP analyzed by SPR. WT Nod2 has previously been reported to bind ligand (10 – 50 nM)6, 21. Details can be found in Supporting Information.
We have previously shown that the chaperone protein, heat shock protein 70 (Hsp70), binds and stabilizes Nod212. Hsp70 is responsible for a diverse range of functions, which when bound to the client protein aids the protein in adopting a more stable conformation, protects vulnerable regions from disruptive interactions, or inhibits self-aggregation13–14. Overexpressed Hsp70 increases MDP induced Nod2-dependent NF-κB activity and lengthens its half-life in cell-based assays for both the wild type (WT) Nod2 and the three major CD variants (accounting for approximately thirty percent of CD cases): G908R, R702W, 1007fs12, 15. These results suggest that the CD variants are able to sense their ligands. Recently two groups reported that the binding site for MDP exists in the LRR domain of Nod216–17. Recent work by Park and coworkers evaluated the ability of two major CD mutations in the LRR domain, G908R and 1007fs, to bind MDP17. Here we investigate a potential biochemical cause for misregulation by the CD-associated NOD2 variants.
It has previously been reported that the CD Nod2 mutants have diminished NF-κB activity8, and this activity can be recovered through the overexpression of Hsp7012. We reasoned that these mutations make the protein inherently unstable and therefore difficult to express and purify. Previous attempts using small affinity tags were unsuccessful for the expression and purification of the CD mutants. Large affinity tags can provide additional stability and solubility to proteins, and to this effect we opted to use an N-terminal glutathione S-transferase (GST)-tag for its known ability to enhance protein stability18. Prior accounts for expressing and purifying regions of Nod2 report success using large affinity tags such as maltose binding protein (MBP) and GST16–17. The addition of the N-terminal GST-tag resulted in the first successful expression and purification of the three CD mutants from Sf21 cells (Figure S1, S2). With purified full-length CD mutants in hand, the ability of full-length protein to bind Nod2’s proposed ligand MDP could be assessed.
Upon cleavage of the GST tag, the CD mutant protein was analyzed in the surface plasmon resonance (SPR) assay previously reported in our lab6, 16, to assess their ability to bind to a biologically active MDP derivative, 6-amino-MDP19. Briefly, the SPR assay monitors binding of Nod2 to a PG fragment that has been covalently attached to a gold surface via a self-assembled monolayer (Figure 1A)6, 20. Purified protein was applied to immobilized 6-amino-MDP over a concentration range and equilibrium dissociation constants were calculated (Figure 1, S4). The three prominent CD mutations of Nod2 maintain the ability to bind 6-amino-MDP with nanomolar affinity on the same order of magnitude as the WT protein (10 – 50 nM)6–7,21. The R702W displayed the smallest change in affinity with an apparent KD of 18.8 ± 2.0 nM. The site of this mutation is far from the proposed binding region for MDP and the small changes in affinity were attributed to slight changes in the fold of that region. The G908R mutation disturbed binding to a higher degree, with an apparent KD of 36.1 ± 2.0 nM, which can be accredited that this mutation occurs next to the proposed binding pocket22. Previous work found that the G908R mutation did not affect binding when evaluating the LRR domain alone, suggesting that other regions of the protein are involved in recognition of the ligand. Lastly, the 1007fs mutation displayed an apparent KD of 40.9 ± 5.1 nM. Previously the LRR domain 1007fs mutation as an MBP fusion indicated no binding to MDP in a fluorescence polarization assay17.
The experimental setup employed here used full-length Nod2 protein devoid of a tag, suggesting that either the full protein is necessary for binding or the addition of a tag to this mutant occluded binding.
Previous experiments have shown that overexpressed Hsp70 increases the half-life and NF-κB signaling of Nod2 and the CD mutants in cell based assays12. In order to further characterize this stabilization effect, an in vitro assay was designed to control for external factors that influence Hsp70’s protective effect on Nod2. These additional factors could then be applied to further enhance the stability of the CD mutants. A limited proteolysis mediated degradation assay was selected to investigate Hsp70’s effect on its client proteins23–24. Briefly, Nod2 was subjected to trypsin-mediated degradation under varying conditions, and Nod2 levels were quantified by Western blot (Figure 2, Figure S5, Scheme S1).
Figure 2.
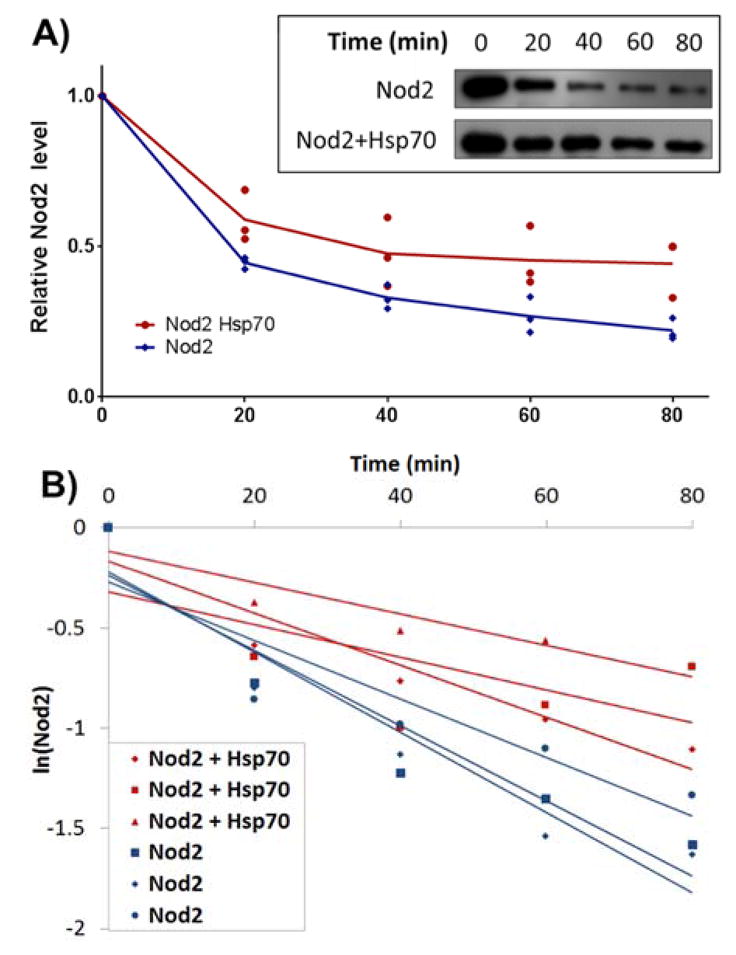
Representative analysis of limited proteolysis data: Western blots (shown as inset) of triplicated limited proteolysis experiments were graphed as shown in A for visualization or B for determination of ½ life. Experimental details, additional blots and graphs for all parameters can be found in Supporting Information.
Nod2 in vitro half-life was calculated for each condition (Table 1, S2). Apo-Nod2 showed a half-life of 39.7 min; upon the addition of excess Hsp70, Nod2’s half-life increases to 75.4 min (Table 1), thus establishing Hsp70 serves a protective role in vitro.
Table 1.
Calculated half-lives for WT Nod2 in the presence of cofactors and/or Hsp70.
| Cofactor | Half-life (min) | |
|---|---|---|
| (−) Hsp70 | (+) Hsp70 | |
| --- | 39.7 ± 1.8 | 75.4 ± 1.0 |
| MDP | 48.4 ± 1.5 | 77.8 ± 1.9 |
| ADP | 201.2 ± 1.0 | 112.3 ± 0.4 |
| ATP | 70.8 ± 3.3 | 200.1 ± 1.9 |
| ATP + MDP | 51.7 ± 3.0 | 282.3 ± 1.7 |
This protective role was further evaluated in the presence of nucleotide cofactors. When ADP is added to the Nod2/Hsp70 complex, the half-life increases to 112.3 min. This result is not surprising as rabbit Nod2 was crystallized in the presence of ADP22, suggesting the closed conformation of Nod2 has an increased stability.
In addition to probing the effects of chaperones and nucleotide substrates on Nod2’s stability, Nod2 was subjected to trypsin-mediated degradation in the presence of Nod2’s PG ligand, MDP. It is known that the addition of PG fragments can stabilize Nod2 in cells25. When MDP is added to a mixture of Hsp70, ATP and Nod2, the half-life of Nod2 increases to 282.3 min (Table 1), revealing that the full complex (peptidoglycan ligand, molecular chaperone, nucleotide cofactor) amplifies the protective effect of Hsp70.
In order to further probe the requirements for Hsp70 to assist Nod2, the role of ATP and ADP in conferred stability was investigated. Two mechanisms that involve ATP and ADP were focused on: hydrolysis of ATP confers conformational changes to the bound client, while ADP increases affinity for its client proteins to protect them from degradation. A mutant form of Hsp70 that forces the chaperone to bind ADP selectively over ATP26 was used to differentiate the two mechanisms (Figure S3). This mutant replaces a key lysine residue with a serine in the nucleotide binding pocket and is commonly referred to as the Hsp70(K71S) mutation26. In the trypsin mediated degradation assay, the addition of Hsp70(K71S) to Nod2 increases the half-life to 403.2 min, approximately seven times higher than the wild type Hsp70 alone (Table 2). These data indicate that ATP hydrolysis is not necessary for Hsp70 to stabilize Nod2.
Table 2.
Calculated half-life values for Nod2 variants in the presence of Hsp70 constructs
| Nod2 Variant | (−) | (+) Hsp70 K71S | (+) SBD | (+) S-71 |
|---|---|---|---|---|
| WT | 39.7 ± 1.8 | 403.2 ± 1.7 | 120.1 ± 1.1 | 151.5 ± 0.5 |
| R702W | 10.7 ± 3.5 | 150.8 ± 0.9 | --- | 228.8 ± 0.7 |
| G908R | 20.9 ± 2.5 | 144.3 ± 1.2 | --- | 685.1 ± 0.3 |
| 1007fs | 21.5 ± 2.2 | 185.1 ± 1.4 | --- | 102.1 ± 3.4 |
To confirm ATP hydrolysis is not necessary for NOD2 stabilization, we tested the ability of a truncated HSP70 to stabilize NOD2. To this end, the ATPase domain of Hsp70 (Figure S3) was truncated and the potential effect of the remaining substrate binding domain27 (SBD) of Hsp70 on Nod2 was tested. The SBD-Hsp70 increased Nod2 half-life from 39.7 min to 120.1 min (Table 2). This increase was slightly higher in comparison to the effect Hsp70 has alone (Table 1). This result led to the hypothesis that the SBD could be further narrowed down to a mini-mal fragment for enhancing the stability of Nod2 and the CD-associated mutants.
A small 71 amino acid, evolutionarily conserved region (S-71) of Hsp70 (403 AA – 474 AA) was identified to further assess for a minimal Hsp70 portion that stabilizes Nod2. It is well documented that the hydrophobic portions of peptides or proteins bind to the hydrophobic SBD of Hsp70 and other Hsp70-like chaperones28–30. More specifically, peptides have been shown to directly interact with the loops of the β-sandwich conserved in the SBD29–31, and the S-71 peptide includes these key loops of the relevant β-sandwich. The S-71 fragment, which is only 10% of the full Hsp70 protein was over-expressed and purified from E. coli cells (Figure S3) and analyzed by circular dichroism and mass spectrometry (Figure S9 – S11). Using the limited proteolysis assay, the S-71 minimal fragment of Hsp70 stabilized WT Nod2, increasing the in vitro half-life from 39.1 min to 151.5 min (Table 2). With the isolation of two Hsp70 constructs, the constructs were assayed for the ability to stabilize the CD-associated mutants of Nod2. The CD mutants of Nod2 were subjected to the limited proteolysis assay in the presence of K71S or the S-71 peptide. The data show that recombinant CD Nod2 mutants are significantly stabilized by K71S or the S-71 peptide (Table 2, Figure S5–7). These results point to a minimal region of Hsp70 that is sufficient to stabilize Nod2 using only protein-protein interactions. It is worth noting that these in vitro results correlate with previously published cell based stability assay results12, which indicate the CD mutants are unstable in comparison to WT.
To test whether increased stability of Nod2 in vitro translates to rescued activity of Nod2, the effect of overexpression of the Hsp70 constructs was assayed in a NF-κB luciferase reporter assay8. As shown previously, Hsp70 rescues MDP-induced NF-κB activity in all three CD-associated mutants (Figure 3, S8)12. The addition of conformationally static Hsp70 (K71S), the SBD, or S-71 also increases MDP-induced NF-κB activity of CD Nod2 variants to a similar level of apo-WT Nod2 signaling (Figure 3, S8).
Figure 3.
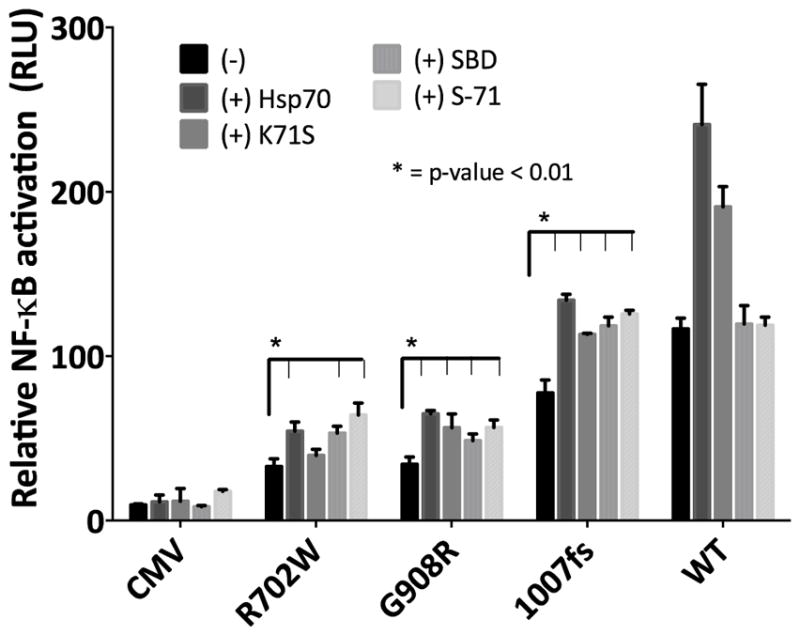
Relative MDP-induced (20 μM) NF-κB activation of Nod2 variants with no chaperones or in the presence (+) of Hsp70 constructs. Experimental details can be found in Supporting Information.
These data suggest that CD mutants do in fact suffer from a stability issue that affects immune signaling. Upon stabilization immune signaling is restored, and these mutants function similarly to that of the WT Nod2 (Figure 3). Interestingly Nature may have already harnessed the ability to stabilize Nod2 and other proteins in a controlled way by varying the expression of Hsp70. The modulation of the innate immune response through variation of Hsp70 levels in cells has previously proven to be an effective tool in reducing inflammation32–35.
Nod2 stabilization becomes an ideal target for the development of a pharmacological chaperone. Pharmacological chaperones have successfully been applied to a number of proteins that contribute to disease due to inherent instability36–37. The CD-associated mutants contribute to pathogenesis in turn because their instability leads to a host of misregulated signaling events. Current therapeutics for CD target broad effects of the disease through immune suppressors and anti-inflammatory agents. An ideal approach to the disease would be a molecular strategy that corrects the response of the CD-associated mutants, as the Hsp70 constructs do (Figure 3). The biochemical assessment of the CD-associated Nod2 variants has identified a potential therapeutic approach that could target the instability that is associated with mutation.
In conclusion, biochemical analysis of the CD-associated Nod2 variants revealed two critical pieces of information about the mutated-receptors: they bind to the PG-fragment with nanomolar affinity and are stabilized by a minimal region of the molecular chaperone, Hsp70. The data presented here suggest that CD caused via a Nod2 mutation might be best characterized as a protein misfolding disease, and therefore future studies could benefit by considering protein stabilization techniques.
Supplementary Material
Acknowledgments
Funding Sources
Delaware COBRE program, supported by the National Institute of General Medical Sciences (NIGMS 1 P30 GM1 10758-02 and 1 P20 GM104316-01A1) of the National Institutes of Health. CLG is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. HCW was supported by the Plastino Scholar Program. AKS and MLL were supported by the NIH CBI grant (NIH T32CM008550).
We thank our colleagues at the University of Delaware: the Koh group for use of the cell culture facility.
ABBREVIATIONS
- CD
Crohn’s disease
- PG
peptidoglycan
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interests.
Experimental procedures and materials: cloning details, SPR details and sensorgrams, half-life protocols, western blots half-life graphs, NF-κB assay conditions, and protein characterization. The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Janeway CAJ, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Ogura Y, et al. Nature. 2001:411. [Google Scholar]
- 4.Hugot JP, et al. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 5.Girardin SE, et al. J Biol Chem. 2003;278:8869–72. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 6.Grimes CL, Ariyananda LDZ, Melnyk JE, O’Shea EK. J Am Chem Soc. 2012;134:13535–13537. doi: 10.1021/ja303883c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mo J, Boyle JP, Howard CB, Monie TP, Davis BK, Duncan JA. J Biol Chem. 2012;287:23057–67. doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inohara N, et al. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 9.Abbott DW, Wilkins A, Asara JM, Cantley LC. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Yin C, Pandey A, Abbott DW, Sassetti CM, Kelliher MA. J Biol Chem. 2007;282:36223–36229. doi: 10.1074/jbc.M703079200. [DOI] [PubMed] [Google Scholar]
- 11.Valastyan JS, Lindquist S. Dis Model Mech. 2014;7:9–14. doi: 10.1242/dmm.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohanan V, Grimes CL. J Biol Chem. 2014;289:18987–98. doi: 10.1074/jbc.M114.557686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meimaridou E, Gooljar SB, Chapple JP. J Mol Endocrinol. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 14.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. J Mol Biol. 2015;427:1575–88. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buning C, et al. Aliment Pharmacol Ther. 2004:19. doi: 10.1111/j.1365-2036.2004.01967.x. [DOI] [PubMed] [Google Scholar]
- 16.Lauro ML, D’Ambrosio EA, Bahnson BJ, Grimes CL. ACS Infectious Diseases. 2017;3:264–270. doi: 10.1021/acsinfecdis.6b00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann JK, Shen J, Park S. J Cell Biochem. 2017;118:1227–1238. doi: 10.1002/jcb.25778. [DOI] [PubMed] [Google Scholar]
- 18.Harper S, Speicher DW. Methods Mol Biol. 2011;681:259–280. doi: 10.1007/978-1-60761-913-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimes CL, Podolsky DK, O’Shea EK. Bioorg Med Chem Lett. 2010;20:6061–3. doi: 10.1016/j.bmcl.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahiri J, Isaacs L, Tien J, Whitesides GM. Anal Chem. 1999;71:777–790. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer AK, Melnyk JE, Baksh M, Lazor KM, Finn MG, Grimes CL. ACS Chem Biol. 2017 doi: 10.1021/acschembio.7b00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maekawa S, Ohto U, Shibata T, Miyake K, Shimizu T. Nature Communications. 2016:7. doi: 10.1038/ncomms11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heiring C, Muller YA. Protein Eng. 2001;14:183–188. doi: 10.1093/protein/14.3.183. [DOI] [PubMed] [Google Scholar]
- 24.Ivey RAr, Subramanian C, Bruce BD. Plant Physiol. 2000;122:1289–1299. doi: 10.1104/pp.122.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnyk JE, Mohanan V, Schaefer AK, Hou C-W, Grimes CL. J Am Chem Soc. 2015;137:6987–6990. doi: 10.1021/jacs.5b01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klucken J, Shin Y, Hyman BT, McLean PJ. Biochem Biophys Res Commun. 2004;325:367–373. doi: 10.1016/j.bbrc.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 27.Aprile FA, et al. PLoS One. 2013;8:e67961. doi: 10.1371/journal.pone.0067961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma D, Masison DC. Protein Peptide Letters. 2009;16:571–581. doi: 10.2174/092986609788490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer MP, Rudiger S, Bukau B. Biol Chem. 2000;381:877–885. doi: 10.1515/BC.2000.109. [DOI] [PubMed] [Google Scholar]
- 30.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudiger S, Buchberger A, Bukau B. Nat Struct Biol. 1997;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 32.Kim N, Kim JY, Yenari MA. Inflammopharmacology. 2012;20:177–185. doi: 10.1007/s10787-011-0115-3. [DOI] [PubMed] [Google Scholar]
- 33.Kacimi R, Yenari MA. Glia. 2015;63:1200–1212. doi: 10.1002/glia.22811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malago JJ, Koninkx JFJG, Tooten PCJ, van Liere EA, van Dijk JE. Clin Exp Immunol. 2005;141:62–71. doi: 10.1111/j.1365-2249.2005.02810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Bermejo L, Vilaboa NE, Perez C, Galan A, De Blas E, Aller P. Exp Cell Res. 1997;236:268–274. doi: 10.1006/excr.1997.3725. [DOI] [PubMed] [Google Scholar]
- 36.Makley LN, Gestwicki JE. Chem Biol Drug Des. 2013;81:22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.