Summary
Human melanomas exhibit relatively high somatic mutation burden compared to other malignancies. These somatic mutations may produce neoantigens that are recognized by the immune system, leading to an anti-tumor response. By irradiating a parental mouse melanoma cell line carrying three driver mutations with UVB and expanding a single cell clone, we generated a mutagenized model that exhibits high somatic mutation burden. When inoculated at low cell numbers in immunocompetent C57BL/6J mice, YUMMER1.7 (YUMM Exposed to Radiation) regresses after a brief period of growth. This regression phenotype is dependent on T cells as YUMMER1.7 tumors grow significantly faster in immunodeficient Rag1−/− mice and C57BL/6J mice depleted of CD4 and CD8 T cells. Interestingly, regression can be overcome by injecting higher cell numbers of YUMMER1.7, which results in tumors that grow without effective rejection. Mice that have previously rejected YUMMER1.7 tumors develop immunity against higher doses of YUMMER1.7 tumor challenge. In addition, escaping YUMMER1.7 tumors are sensitive to anti-CTLA-4 and anti-PD-1 therapy, establishing a new model for the evaluation of immune checkpoint inhibition and anti-tumor immune responses.
Keywords: melanoma, immunotherapy, mouse models, Braf, UV radiation
Introduction
Mouse models of melanoma have helped define the factors that regulate tumor formation and progression; however, studying effective anti-tumor immune responses has been more difficult. For example, therapeutic cancer vaccines that showed promise in animal models have rarely translated into successes in clinical trials (Mak et al., 2014). One possible explanation for this observation is that existing syngeneic transplantation models such as the B16 melanoma cell line may not accurately recapitulate select driver mutations combined with the numerous passenger mutations that are present in human melanomas (Castle et al., 2012). B16, derived from spontaneous melanocytic tumors over half a century ago, is also considered poorly immunogenic perhaps due to its low expression of major histocompatibility complex class I molecules and rapid growth (Becker et al., 2010). The development of syngeneic mouse models of melanoma that closely reflect the genetics of human melanoma will be essential for the continued advancement of melanoma immunotherapy.
Somatic mutation burden is variable in human cancers but is relatively high in melanoma. Primary and metastatic melanomas are among the highest mutated tumors analyzed by the Cancer Genome Atlas with a reported mean mutation rate of 16.8 mutations/Mb per exome (Cancer Genome Atlas, 2015). The vast majority of these are UV signature mutations, most of which are thought to be passenger mutations rather than driver gene changes that promote a selective growth advantage (Vogelstein et al., 2013). However, these mutations have the potential to act as neoantigens and elicit anti-tumor immune responses. Indeed, high somatic mutation burden in melanoma has been reported to be associated with improved survival in the setting of immune checkpoint blockade (Snyder et al., 2014; Van Allen et al., 2015). The success of a phase 2 clinical trial using PD-1 blockade to treat mismatch repair-deficient colorectal and non-colorectal cancers further supports this hypothesis (Le et al., 2015).
In melanoma, ultraviolet radiation is significantly involved in the accumulation of somatic mutations and pathogenesis of the malignancy. Recent studies corroborate the role of ultraviolet radiation in all evolutionary stages of melanoma (Martincorena et al., 2015; Shain et al., 2015). The number of mutations correlates with cumulative sun exposure inferred from age and anatomical site and with histological features of sun exposure including solar elastosis (Shain et al., 2015). We have recently reported the generation of the YUMM (Yale University Mouse Melanoma) cell lines, which are derived from various genetically engineered mouse models of melanoma with human relevant driver mutations (Meeth et al., 2016). These lines, however, possess a low number of somatic genetic changes and, in general, are minimally immunogenic as reflected by their growth in immunocompetent C57BL/6J hosts. Here, we describe a derivative of YUMM1.7, which carries three driver mutations: BrafV600E, Pten−/− and Cdkn2a−/−. We hypothesized that UV-induced passenger mutations would enhance the immunogenicity of the cell line and stimulate a functional adaptive immune response in vivo. Accordingly, we developed a mutagenized cell line named YUMMER1.7 (YUMM Exposed to Radiation) by irradiating YUMM1.7 and expanding a single cell-derived clone. When engrafted at low cell numbers into immunocompetent C57BL/6J mice, YUMMER1.7 gives rise to tumors that persist for one to two weeks but eventually regress completely, in contrast to the parental YUMM1.7 tumors that uniformly grow out from the same inoculation cell number. Interestingly, injection of higher numbers of YUMMER1.7 cells allows for escape and growth of the tumors. YUMMER1.7 cells injected into immunocompromised Rag1−/− mice even at low cell numbers also reproducibly form tumors, and antibody-based depletion of CD4, CD8 or both leads to enhanced tumor growth in C57BL/6J mice. These experiments demonstrate that T cells are required for rejection of YUMMER1.7 tumors at low injection cell numbers, but this anti-tumor response can be overcome with greater initial tumor burden. Moreover, tumors resulting from high cell number injections of YUMMER1.7 are responsive to immune checkpoint therapies. YUMMER1.7 lies on the fulcrum between the opposing forces of cancer cell growth and immune-mediated cytotoxicity. Based on these experiments, we propose that this cell line better recapitulates the genomic landscape of human melanoma and provides a model for the evaluation of anti-tumor immune responses to novel immune therapeutics in cancer.
Results
The YUMM cell lines including YUMM1.7 have been previously described, are distributed by the American Type Culture Collection (ATCC), and are widely used (Ho et al., 2015; Homet Moreno et al., 2016; Meeth et al., 2016; Scortegagna et al., 2015). Briefly, the YUMM lines are a collection of C57BL/6J syngeneic melanoma cell lines with driver mutations common in human melanoma and can be injected subcutaneously in immune competent C57BL/6J mice, resulting in tumor formation. Here, YUMM1.7 cells were exposed to three rounds of UVB radiation at 1500J/m2 to generate a version of the cell line with an increased number of somatic mutations, YUMMER1.7 (Figure 1A). After the last round of radiation, a single cell-derived clone was selected and expanded. In order to characterize the number of UV-induced mutations in YUMMER1.7, whole exome sequencing of both the parental YUMM1.7 and mutagenized YUMMER1.7 cell lines were performed and compared to the wild-type C57BL/6J exome. Compared to C57BL/6J, YUMM1.7 exhibited 310 nonsynonymous exonic point mutations (Table 1), which likely reflects incomplete backcrossing of the four alleles in the original genetically engineered mouse model. There were an additional 1446 unique nonsynonymous exonic mutations in YUMMER1.7 relative to YUMM1.7. A large proportion of these single-base changes (81.5%) were also C>T transitions, which is consistent with ultraviolet light treatment and mutagenesis (Figure 1B). To assess the stability of the mutations in culture, four mutations unique to YUMMER1.7 and not detected in YUMM1.7 were evaluated in early and late passages of YUMMER1.7. All four mutations were detected in both passage 15 and 25 of YUMMER1.7 (Suppl. Figure 1). Upon evaluation of DNA content in the cell lines, it was determined that YUMM1.7 contains both diploid and tetraploid clones whereas YUMMER1.7 is tetraploid with twice the DNA content as splenocytes (Suppl. Figure 2A). The allele frequencies of YUMMER1.7 mutations compared to C57BL/6J background center around 0.25, further supporting that YUMMER1.7 is a tetraploid clone (Suppl. Figure 2B).
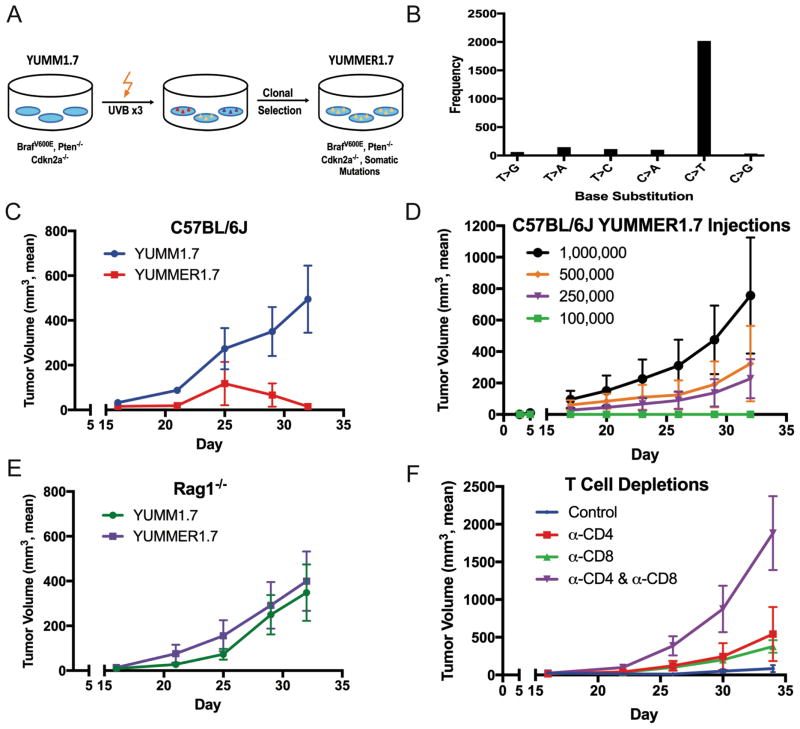
Figure 1. Generation and Characterization of YUMMER1.7.
(A) YUMMER1.7 was generated from YUMM1.7 using three rounds of UVB radiation and a single cell-derived clone was expanded.
(B) Total point mutations in YUMMER1.7 compared to YUMM1.7 categorized based on the type of base substitution.
(C) Growth of 100,000 cell YUMM1.7 and YUMMER1.7 tumors engrafted into wild-type C57BL/6J mice.
(D) Growth of 100,000, 250,000, 500,000, and 1,000,000 cell YUMMER1.7 tumors engrafted into wild-type C57BL/6J mice.
(E) Growth of 100,000 cell YUMM1.7 tumors and YUMMER1.7 tumors engrafted into Rag1−/− C57BL/6J mice.
(F) Growth of 100,000 cell YUMMER1.7 tumors engrafted into wild-type C57BL/6J mice that were treated with depleting antibodies against CD4, CD8 or both.
Tumor growth curves are representative of two independent experiments (mean ± SEM, n = 5 each).
Table 1.
Mutations between YUMM1.7 and YUMMER1.7 categorized by mutation type based on whole exome sequencing.
| Synonymous Mutations | Nonsynonymous Mutations | Other Mutations1 | Mutational Load (total # of exonic mutations) | |
|---|---|---|---|---|
|
| ||||
| YUMM1.7 vs C57BL/6J | 554 | 310 | 17 | 881 |
| YUMMER1.7 vs C57BL/6J | 1457 | 1731 | 120 | 3308 |
| YUMMER1.7 vs YUMM1.7 | 924 | 1446 | 111 | 2481 |
Frameshift indels, nonframeshift indels, stoploss, stopgain
In order to understand in vivo growth characteristics of YUMMER1.7 relative to YUMM1.7, we subcutaneously implanted 100,000 cells of each model into the flanks of C57BL/6J mice and measured tumor volume over time. As expected, all YUMM1.7 tumors exhibited rapid and continuous growth (Figure 1C). In contrast, YUMMER1.7 cells initially grew to a palpable tumor of approximately 10–50 mm3 followed by striking tumor regression without regrowth. Regression occurred in approximately 80% of YUMMER1.7 tumors following 100,000 cell injections and was even more consistently observed following 50,000 and 10,000 cell injections (Suppl. Figure 3). Clinical regression of YUMMER1.7 melanomas (tumor volume decrease) was observed as early as 14 days post-injection, and complete regression was durable for at least 120 days.
Next, we implanted higher numbers of YUMMER1.7 cells in a similar fashion. In contrast to the 100,000 cell injections, the inoculations of more than 250,000 YUMMER1.7 cells resulted in tumors that generally did not regress and eventually required euthanasia of the mouse, suggesting that tumor regression could be overcome by increasing the number of cells injected (Figure 1D). Approximately four out of five 250,000 cell YUMMER1.7 tumors and nearly all 500,000 cell YUMMER1.7 tumors exhibited exponential growth. To further investigate why inoculation sizes of 100,000 YUMMER1.7 cells or less leads to tumor regression, we hypothesized that the phenomenon was immune mediated. Thus, we injected 100,000 YUMMER1.7 cells into the flanks of immunodeficient Rag1−/− mice that lack functional T and B cells. Interestingly, all YUMMER1.7 tumors in Rag1−/− mice grew without any cases of regression (Figure 1E). Indeed, the YUMMER1.7 tumors grew at a similar rate compared to the parental model YUMM1.7 tumors. Antibody-mediated depletion of CD4 and CD8 T cells in wild-type C57BL/6J mice also increased tumor growth of YUMMER1.7 when 100,000 cells were injected (Figure 1F). We found that depletion of both CD4 and CD8 T cells significantly accelerated tumor growth compared to the isotype control (p = 0.006). These results suggested tumor regression was the result of an adaptive immune response involving both CD4 and CD8 T cells.
Subsequently, we explored whether mice that had rejected 100,000 cell YUMMER1.7 tumors would develop immunity against higher doses of YUMMER1.7 tumors (Suppl. Figure 4). We injected 100,000 YUMMER1.7 cells into the right flanks of C57BL/6J mice and allowed tumors to completely regress after 30 days. This cohort was then rechallenged with 500,000 cell YUMMER1.7 (I + YUMMER1.7) or 500,000 cell YUMM1.7 (I + YUMM1.7) injections on the left flanks. All five immunized mice rejected YUMMER1.7 tumors, whereas none rejected YUMM1.7 tumors (n = 5). 4/5 tumors grew out in the control naïve C57BL/6J mice that were injected with 500,000 YUMMER1.7 cells. Interestingly, although all YUMM1.7 tumors grew out, they were smaller in the I + YUMM1.7 group than in the N + YUMM1.7 group, albeit without reaching significance (p = 0.19). Finally, antibody-mediated depletion of CD4 and CD8 eliminated the effect of immunization as evidenced by 4/4 tumors failing to regress in the I + YUMMER1.7 + anti-CD4/CD8 group. These observations suggest that mice that rejected tumors following injection of low cell numbers of YUMMER1.7 developed a T cell memory response capable of rejecting a challenge of YUMMER1.7 cells that would otherwise escape immune surveillance in naïve animals.
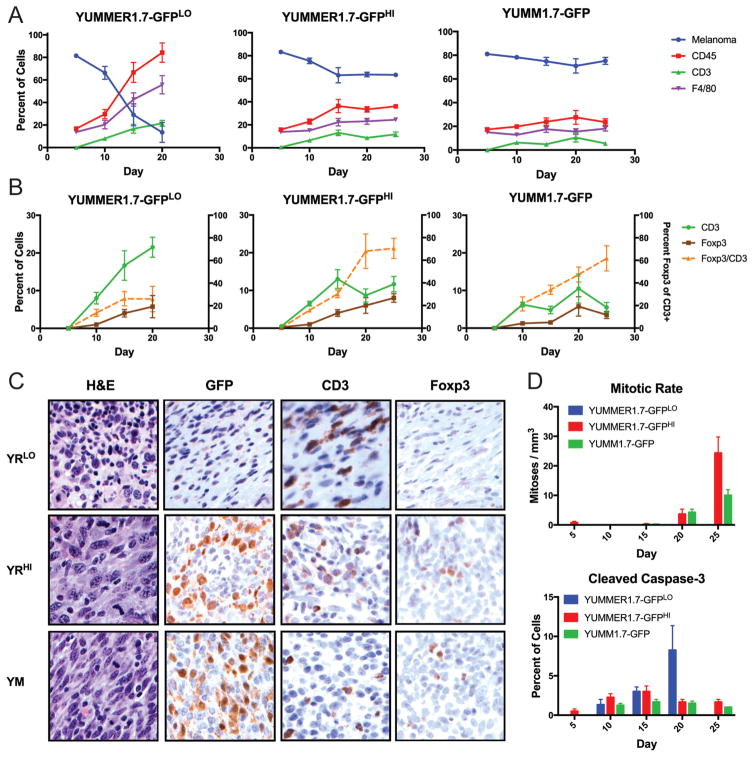
Additionally, we sought to compare the intratumoral immune infiltration, mitotic rate and apoptotic cell death between YUMM1.7 and YUMMER1.7 tumors as a function of time. In order to track very early tumors, YUMM1.7 and YUMMER1.7 cell lines were labeled with GFP, implanted and harvested at different time points—day 5, 10, 15, 20, 25 post-implantation for analysis by immunohistochemistry. Similar to the unlabeled lines, 100,000 cell YUMM1.7-GFP and 500,000 cell YUMMER1.7-GFP (YUMMER1.7-GFPHI) tumors grew out whereas 100,000 cell YUMMER1.7-GFP (YUMMER1.7-GFPLO) tumors regressed over time as shown by GFP staining as a percentage of nucleated cells (Figure 2A). As a result of regression before day 25, staining for YUMMER1.7-GFPLO tumors at day 25 is not available. The infiltration of CD45 and CD3 positive nucleated cells increased over time in YUMMER1.7-GFPLO tumors whereas the percentage of these cells remained relatively constant or decreased in YUMMER1.7-GFPHI and YUMM1.7-GFP tumors (Figure 2A, B). At day 20, a period when YUMMER1.7-GFPLO is regressing and YUMMER1.7-GFPHI and YUMM1.7-GFP are escaping in growth, the fraction of CD45 positive nucleated cells were significantly greater in YUMMER1.7-GFPLO than YUMMER1.7-GFPHI (p = 0.0045) and YUMM1.7-GFP (p = 0.0016) tumors. Similarly, CD3 staining was increased in YUMMER1.7-GFPLO tumors compared to YUMMER1.7-GFPHI (p = 0.0141) and YUMM1.7-GFP (p = 0.0574) tumors. In YUMMER1.7-GFPLO tumors, although CD3 infiltration increased over time, the amount of Foxp3 positive staining (a marker of immunosuppressive regulatory T cells) remained relatively low. This contrasts with increased Foxp3:CD3 ratio within YUMMER1.7-GFPHI and YUMM1.7-GFP tumors (Figure 2B). Representative images of each cell line and condition are shown for the day 20 time point (Figure 2C). Additionally, the mitotic rate was low for all three tumor types until day 20, when mitotic rate markedly increased in YUMMER1.7-GFPHI and YUMM1.7-GFP tumors only (Figure 2D). In contrast, cleaved caspase-3 staining, a marker of apoptosis, progressively increased only in YUMMER1.7-GFPLO tumors over time and remained low in both the escaping tumors.
Figure 2. Immune infiltration, mitotic rate and apoptotic cell death within YUMM1.7-GFP and YUMMER1.7-GFP tumors.
(A) GFP (melanoma), CD45, CD3, F4/80-positive cells as a percent of nucleated cells. Data are averages of counts from 3–4 independent tumors (mean ± SEM).
(B) CD3, Foxp3 as a percent of nucleated cells and Foxp3 as a percentage of CD3 positive cells (orange). Data are averages of counts from 3–4 independent tumors (mean ± SEM).
(C) Representative immunohistochemical images of YUMM1.7 and YUMMER1.7 tumors on day 20 with the indicated staining at 40X magnification. YRLO = YUMMER1.7-GFPLO, YRHI = YUMMER1.7-GFP HI, YM = YUMM1.7-GFP.
(D) Mitotic rate and cleaved caspase-3 staining for YUMM1.7-GFP and YUMMER1.7-GFP tumors over time (mean ± SEM, n = 3–4).
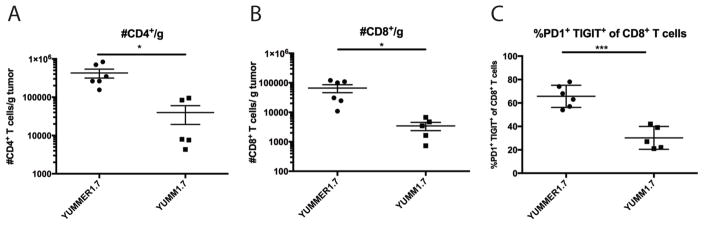
To further characterize the T cell infiltrate, tumors were harvested from wild-type C57BL/6J mice injected with either 500,000 YUMMER1.7 or 500,000 YUMM1.7 cells. Tumors were evaluated upon reaching 500–1000 mm3 in volume for flow cytometry analysis (between day 30–40). Both CD4 (p = 0.017) and CD8 T cells (p = 0.020) were present at significantly higher numbers per gram of tumor in YUMMER1.7 implants compared to YUMM1.7 implants (Figure 3). Moreover, tumor-associated YUMMER1.7 CD8 T cells expressed increased activation/exhaustion markers such as PD-1 and TIGIT (p = 0.0002).
Figure 3. Immune infiltration of escaping YUMM1.7 and YUMMER1.7 tumors.
(A) Density of CD4+ T cells per gram of tumor.
(B) Density of CD8+ T cells per gram of tumor.
(C) Percent of CD8+ T cells that are PD1+ TIGIT+.
Data are from two independent experiments (mean ± SEM, n=2–3 each).
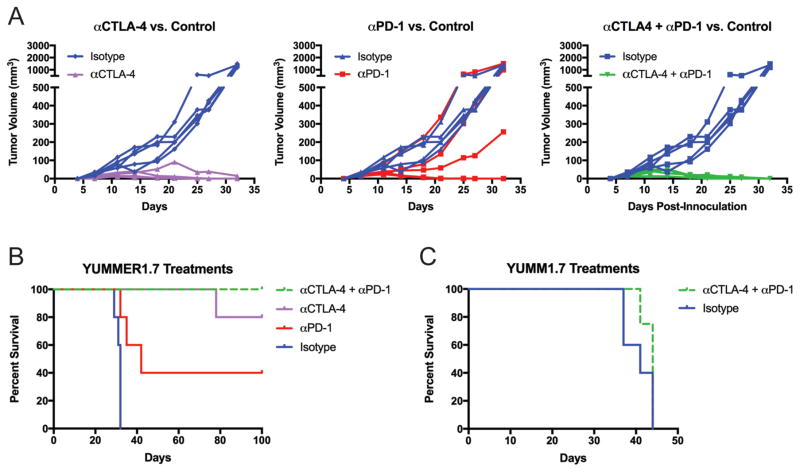
Poor immunogenicity of existing syngeneic tumor models has hindered the study and evaluation of immunotherapies. We tested the effect of PD-1 and CTLA-4 blockade in the 500,000 cell YUMMER1.7 model, which would otherwise result in lethal tumor formation. All 5 mice in the isotype treated control group progressed to endpoint (tumor volume >1,000 mm3) by day 32, whereas 4/5 anti-CTLA-4 treated tumors, 2/5 anti-PD-1 treated tumors and 5/5 combination treated tumors regressed completely (Figure 4A). When regression was complete, no tumors regrew for up to 180 days. The one tumor in the anti-CTLA-4 treatment group that grew out was slowed by the therapy such that it did not reach endpoint until day 78 (Figure 4B). Both anti-CTLA-4 (p = 0.0034) and anti-PD-1 (p = 0.0119) treatments significantly delayed the progression of YUMMER tumors from 500,000 cell inoculations compared to the isotype control. Similarly, the combination of the two checkpoint inhibitors inhibited tumor growth versus isotype control (p = 0.0034). When single and combination treatments were compared against each other, only the PD-1 versus combination treatment comparison was borderline significant (p = 0.0494). These effects were not seen in 500,000 cell YUMM1.7 tumors as combination treated tumors (n = 4) did not significantly differ from isotype antibody control treated tumors (n = 5) (Figure 4C).
Figure 4. YUMMER1.7 but not YUMM1.7, is sensitive to checkpoint inhibitors anti-CTLA-4 and anti-PD-1.
(A) Individual tumor growth curves for YUMMER1.7 tumors treated with anti-CTLA-4 (n = 5), anti-PD-1 (n = 5) or both (n = 4) compared to isotype control (n = 5). Data are representative of two independent experiments.
(B) Kaplan-Meier survival curves for treated YUMMER1.7 and (C) YUMM1.7 tumors with an endpoint of tumor size > 1000 mm3 (n = 4–5).
Discussion
Mouse models of cancer have been a tremendous resource for studying the pathophysiology of human malignancies and have provided preclinical rationale for novel therapies (Day et al., 2015). However, low somatic mutation burden in many mouse models can limit tumor immune responses and the study of immunotherapy, demonstrating a need for immunogenic models (Ward et al., 2016). As reported previously, YUMM1.7 exhibits similar growth in the presence or absence of a functional adaptive immune system likely due to a low number of mutations relative to the host C57BL/6J background (Meeth et al., 2016). Here, we use UVB irradiation and clonal selection of YUMM1.7 to generate the derivative YUMMER1.7 melanoma cell line. We have shown that this modified cell line elicits a robust immune response as evidenced by the quantity and quality of immune infiltration, particularly with respect to T cells. More importantly, normal tumor growth in Rag1−/− and enhanced growth in T cell-depleted C57BL/6J mice suggest that the adaptive immune system may be responsible for YUMMER1.7 tumor regression when 100,000 or fewer cells are injected. Such an immune response also confers protection against future rechallenge of YUMMER1.7 tumor engraftment, likely a manifestation of T cell memory.
The striking dependence of tumor phenotype—regression versus progressive growth—on the number of YUMMER1.7 cells injected requires further exploration. In contrast to inoculation cell numbers of 100,000 or less, when 250,000 or more cells are implanted, tumors reproducibly form in immunocompetent host mice. The increased inoculation cell number may allow the mitotic rate of cancer cells to outpace or otherwise evade the developing immune response. Another possibility is that persistently high antigen levels in the context of sub-optimal co-stimulation may lead to an anergic or exhausted T cell phenotype analogous to what has been reported in mice infected with chronic lymphocytic choriomeningitis virus (Utzschneider et al., 2016). Furthermore, regulatory T cells may play a functional role as we observed that regressing tumors are characterized by lower Foxp3:CD3 ratios by IHC compared to tumors that grow out (Figure 2B).
Despite the escape from regression, the 500,000 YUMMER1.7 tumors continue to be infiltrated by significantly larger numbers of CD4 and CD8 T cells when compared to YUMM1.7 tumors. One possible explanation for this difference is that YUMMER1.7 neoantigens are more immunogenic than those present in YUMM1.7 tumors. Indeed, the identification of neoantigens that elicit this immune response in this model is of great interest but beyond the scope of this study. Collectively, these findings point toward a highly immunogenic infiltrate within YUMMER1.7 tumors. The presence of a functional, activated T cell population in tumors that still grow exponentially make YUMMER1.7 a valuable tool for studying modifiers of the immune system such as immune checkpoint inhibitors and other therapeutics.
Immune checkpoint blockade with ipilimumab, nivolumab and pembrolizumab has led to tumor regression and prolonged overall survival in patients with advanced melanoma and other malignancies (Sharma and Allison, 2015; Wolchok et al., 2013). To validate YUMMER1.7 as a model for evaluating immune therapies, we treated 500,000 cell YUMMER1.7 tumors with anti-CTLA-4 and anti-PD-1 therapy. When administered individually or as a combination, these treatments not only inhibited tumor growth but also induced regression and cure of melanoma in a proportion of mice. The combination of anti-CTLA-4 and anti-PD-1 did not significantly delay tumor growth in the less immunogenic YUMM1.7 model. CTLA-4 is believed to regulate immune responses by recruiting phosphatases such as SHP-2 that offset tyrosine and serine/threonine kinase signals induced by the T cell receptor (Topalian et al., 2015). In addition, it also outcompetes CD28 for its ligands, CD80 and CD86, preventing adequate costimulation of naïve T cells (Pardoll, 2012). Because CD4 and CD8 T cells are able to infiltrate YUMMER1.7 tumors to a greater extent than in YUMM1.7 tumors, blockade of CTLA-4 may have a stronger effect in the former model. In fact, the density of tumor infiltrating CD8 T cells is one of the best predictors of response to anti-CTLA-4 (Ji et al., 2012). If the neoantigens in YUMM1.7 do not bind MHC complexes with high affinity, enhanced costimulation may be insufficient to produce an antitumor response. CTLA-4 blockade may also impair immunosuppressive effects of Foxp3+ regulatory T cells in YUMMER1.7 tumors since CTLA-4 has been shown to be constitutively expressed on regulatory T cells and critical for their function (Takahashi et al., 2000; Wing et al., 2008). Moreover, we observed increased PD1+ TIGIT+ CD8 T cells in YUMMER1.7, reflecting either an activated or exhausted state of cytotoxic T cells. PD-1 blockade may exert its effect by reversing the exhausted subset by inhibiting downstream signaling of the PD-1 receptor and recruitment of the phosphatase SHP-2 as well. YUMMER1.7 demonstrates how immune therapies may shift the balance between immune surveillance and cancer cell growth.
We anticipate that YUMMER1.7 will serve as an immunogenic mouse melanoma model for the scientific community and as a valuable addition to the YUMM series. It illustrates the role that antigen burden plays in dictating the extent of the anti-tumor immune response and offers the opportunity to study the immune microenvironment with the goal of evaluating drugs in preclinical stages and developing novel immune therapeutics. Finally, the described approach used to enhance immunogenicity of the YUMM1.7 line could also be used for other syngeneic mouse models of cancer.
Methods
Cell lines and tissue culture
YUMMER1.7 was derived from YUMM1.7, which was generated from a cutaneous mouse melanoma containing the alleles BrafV600E, Pten−/−, Cdkn2a−/− (Meeth et al., 2016). Irradiation of YUMM1.7 included three rounds of 1500J/m2 UVB (3W for 500s) when cells were 50–70% confluent. Cells were given time to recover and proliferate before being re-plated and proceeding to the next UV treatment. After the final UV treatment, a single cell was clonally expanded. YUMM1.7 and YUMMER1.7 DNA content were assessed using a Propidium Iodide Flow Cytometry Kit according to manufacturer instructions (Abcam, Cambridge, UK). YUMMER1.7-GFP and YUMM1.7-GFP were generated using a P-YUK-GFP plasmid with PiggyBac Transposase Expression Vector, a gift from Tian Xu, Department of Genetics, Yale University. Transfection was done with Lipofectamine 2000 (Invitrogen, Carlsbad, California) and cells were selected using Blastomycin resistance. All cell lines were maintained in DMEM/F12 media containing 10% FBS and with 1% nonessential amino acids and 1% penicillin-streptomycin.
In vivo mouse studies
Four to six week old C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and allowed to acclimate for one week prior to use. C57BL/6J Rag1−/− mice were also obtained from the Jackson Laboratory and maintained in our mouse colony. All animal experiment protocols were followed according to the Yale Office of Animal Research Support Committee guidelines. For tumor inoculation, YUMM1.7 and YUMMER1.7 cells were harvested at approximately 60–85% confluence on the day of injection. Cells were trypsinized with 0.25% trypsin for approximately 2–3 minutes before deactivation with media containing 10% serum. They were then washed twice with sterile 1x PBS and counted with an Invitrogen Countess or a hemocytometer. Cells in 100 μL of sterile PBS were injected subcutaneously into a shaved rear flank using a 27G needle. Mice were monitored for the appearance of tumor after injection to begin digital caliper measurements. Three dimensions were taken for calculation of tumor volume, which was calculated using the equation: 0.5233*l*w*h. For depletion experiments, antibodies for CD4 (GK1.5) and CD8 (TIB210) were made in-house using hydridomas. Mice were injected with 10 mg/kg of each antibody on day −1 and then twice per week for the course of treatment. Loss of CD4 and CD8 T cells was verified by flow cytometry. For immunotherapy treatments, anti-CTLA-4 (9H10) and anti-PD-1 (RMP1-14) antibodies were purchased from Bio X Cell (West Lebanon, NH) along with the corresponding isotype controls, Syrian Hamster IgG2 and Rat IgG2a, respectively. Treatments were started with palpable tumors at 6 days after initial cell line injections. Mice were given 10 mg/kg anti-CTLA-4 and 10 mg/kg anti-PD-1 three times per week for four weeks.
Histological analysis
At least three tumors were fixed in 10% formalin for each condition—100,000 cell YUMM1.7-GFP, 100,000 cell YUMMER1.7-GFP and 500,000 cell YUMMER1.7-GFP tumors at every time point—and embedded in paraffin. Cut sections were stained using GFP (Abcam 10558, Cambridge, UK), CD45 (Abcam 290, Cambridge, UK), F4/80 (Thermo Fisher 16363, Waltham, MA), CD3 (Biocare Medical 215, Concord, CA), Foxp3 (eBioscience 145773, San Diego, CA), Cleaved Caspase-3 (Abcam 4051, Cambridge, UK) antibodies. Images of representative fields were taken of the tumor types and quantified at 40X magnification. The positive cells (brown) were counted and compared to the total nucleated cells in the field. Five fields were taken per tumor section and averaged.
Flow cytometry analysis
Single-cell suspensions from tumors or splenocytes were incubated with anti-Fc receptor antibody (2.4G2) on ice for 15 minutes in FACS buffer (PBS with 1% FBS and sodium azide). The cells were then stained with the appropriate antibodies in 2.4G2-containing FACS buffer on ice for 30 minutes. For intracellular cytokine staining, cells were fixed and permeabilized in buffer and stained with antibodies to detect intracellular cytokines or transcription factors. All samples were acquired on LSRII flow cytometers and analyzed with Flowjo (Flowjo, LLC., Ashland, Or). Antibodies against CD45 (A20), CD8 (53-6.7) and CD3 (145-2C11) were purchased from eBioscience. The antibodies against CD4 (RM4-5), PD-1 (RMP1-14), and TIGIT (1G9) were purchased from Biolegend.
Statistical Analyses
Unpaired two-tailed t tests and Kaplan-Meier statistical analyses were performed using GraphPad Prism (Version 6.0a for Mac OS X, GraphPad Software, La Jolla, CA) using a significance cutoff; ns p >0.05, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.005.
DNA Extraction and Exome Sequencing
DNA was extracted from YUMM1.7 and YUMMER1.7 cell lines as well as wild-type C57BL/6J mouse ears using Qiagen’s DNeasy Blood & Tissue kit (Hilden, Germany). All samples passed quality control and were exome sequenced with 100 bp paired end reads using Agilent’s SureSelectXT Mouse All Exon kit by Macrogen (Cambridge, MA). Reads were aligned to the mm10 reference (ftp://ftp-mouse.sanger.ac.uk/ref/GRCm38_68.fa) using BWA version 0.7.15 with the –M option. Duplicate reads were marked with Picard version 2.6.0 MarkDuplicates. The resulting alignments were subjected to base quality score recalibration in GATK 3.6 using the Mus muscularis C57BL/6J SNP and Indel databases according to GATK best practices. Variants unique to YUMMER1.7 compared to YUMM1.7 or cell lines compared to wild-type C57BL/6J were called using GATK MuTect2 with the above mentioned dbsnp and mm10 references and were selected for downstream analysis if they passed the default MuTect2 filters. Then, variants were annotated using Annovar (2016Feb01 release) with the corresponding mm10 ensGene reference according to the program manual.
Sanger Sequencing
Four genomic loci that exhibited mutations in YUMMER1.7 but not in YUMM1.7 based on exome sequencing were Sanger sequenced from different passages of YUMMER1.7 in vitro. Primers amplifying these loci with 200–500 bp flanks were designed using default parameters in Primer-BLAST (NCBI, https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and checked for specificity to the Mus musculus genome. PCR reactions using these primers and YUMMER1.7 genomic DNA were conducted, and PCR products were purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to manufacturer instructions. Sanger sequencing reactions were performed by Genewiz (South Plainfield, NJ), and chromatograms were visualized with 4Peaks (http://nucleobytes.com/4peaks/).
Supplementary Material
Four mutations unique to YUMMER1.7 compared to YUMM1.7 were Sanger sequenced in early and late passages of YUMMER1.7 (passage 15 and 25).
(A) Propidium iodide flow cytometry demonstrates that the vast majority of YUMM1.7 cells (orange) are diploid with similar DNA content as splenocytes (red) whereas YUMMER1.7 (blue) exhibits double the DNA content.
(B) Allele frequencies of the total mutations in the YUMMER1.7 exome compared to the C57BL/6J background.
Tumor growth curves are representative of two independent experiments (mean ± SEM, n = 5 each).
N = naïve C57BL/6J mice, I = C57BL/6J mice injected with 100,000 YUMMER1.7 cells and allowed to regress completely for 30 days before tumor rechallenge with 500,000 cells of YUMM1.7 or YUMMER1.7 on the opposite flank. Tumor growth curves are representative of two independent experiments (mean ± SEM, n = 4–5 each).
Significance.
Mouse models of melanoma that accurately recapitulate the driver mutations as well as the somatic mutation burden present in the human disease can be used to advance our understanding of melanoma and its response to therapy. Here, we describe a novel mouse melanoma model, YUMMER1.7, which reflects the genomic features of human melanoma and elicits a functional anti-tumor immune response in immunocompetent C57BL/6J hosts. Since this model is responsive to immune checkpoint inhibitors including anti-CTLA-4 and anti-PD-1, we anticipate that it will serve as a useful preclinical resource to evaluate new immunotherapies.
Acknowledgments
We would like to thank Tian Xu, Department of Genetics at Yale University for his generous gift of the P-YUK-GFP plasmid. We are grateful to the scientific community for their inspiration and feedback related to our models. This work was supported by R01 CA196660, P01 CA128814, the Melanoma Research Alliance, the Melanoma Research Foundation, and the Hervey Family Foundation.
References
- Becker JC, Houben R, Schrama D, Voigt H, Ugurel S, Reisfeld RA. Mouse models for melanoma: a personal perspective. Exp Dermatol. 2010;19:157–64. doi: 10.1111/j.1600-0625.2009.00986.x. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Kreiter S, Diekmann J, Lower M, Van De Roemer N, De Graaf J, Selmi A, Diken M, Boegel S, Paret C, et al. Exploiting the mutanome for tumor vaccination. Cancer research. 2012;72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- Day CP, Merlino G, Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell. 2015;163:39–53. doi: 10.1016/j.cell.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–28. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homet Moreno B, Zaretsky JM, Garcia-Diaz A, Tsoi J, Parisi G, Robert L, Meeth K, Ndoye A, Bosenberg M, Weeraratna AT, et al. Response to Programmed Cell Death-1 Blockade in a Murine Melanoma Syngeneic Model Requires Costimulation, CD4, and CD8 T Cells. Cancer immunology research. 2016;4:845–857. doi: 10.1158/2326-6066.CIR-16-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–31. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6:114–8. [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, Mclaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–6. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment cell & melanoma research. 2016;29:590–7. doi: 10.1111/pcmr.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortegagna M, Lau E, Zhang T, Feng Y, Sereduk C, Yin H, De SK, Meeth K, Platt JT, Langdon CG, et al. PDK1 and SGK3 Contribute to the Growth of BRAF-Mutant Melanomas and Are Potential Therapeutic Targets. Cancer research. 2015;75:1399–412. doi: 10.1158/0008-5472.CAN-14-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, Dummer R, North J, Pincus L, Ruben B, et al. The Genetic Evolution of Melanoma from Precursor Lesions. The New England journal of medicine. 2015;373:1926–36. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, Mak TW, Sakaguchi S. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192:303–10. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer cell. 2015;27:450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider DT, Alfei F, Roelli P, Barras D, Chennupati V, Darbre S, Delorenzi M, Pinschewer DD, Zehn D. High antigen levels induce an exhausted phenotype in a chronic infection without impairing T cell expansion and survival. J Exp Med. 2016;213:1819–34. doi: 10.1084/jem.20150598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Geukes Foppen MH, Goldinger SM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JP, Gubin MM, Schreiber RD. The Role of Neoantigens in Naturally Occurring and Therapeutically Induced Immune Responses to Cancer. Adv Immunol. 2016;130:25–74. doi: 10.1016/bs.ai.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–5. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, et al. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369:122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Four mutations unique to YUMMER1.7 compared to YUMM1.7 were Sanger sequenced in early and late passages of YUMMER1.7 (passage 15 and 25).
(A) Propidium iodide flow cytometry demonstrates that the vast majority of YUMM1.7 cells (orange) are diploid with similar DNA content as splenocytes (red) whereas YUMMER1.7 (blue) exhibits double the DNA content.
(B) Allele frequencies of the total mutations in the YUMMER1.7 exome compared to the C57BL/6J background.
Tumor growth curves are representative of two independent experiments (mean ± SEM, n = 5 each).
N = naïve C57BL/6J mice, I = C57BL/6J mice injected with 100,000 YUMMER1.7 cells and allowed to regress completely for 30 days before tumor rechallenge with 500,000 cells of YUMM1.7 or YUMMER1.7 on the opposite flank. Tumor growth curves are representative of two independent experiments (mean ± SEM, n = 4–5 each).