Abstract
Mature microRNAs (miRNAs) are single-stranded RNAs with approximately 18–25 bases, and their sequences are highly conserved among animals. miRNAs act as posttranscriptional regulators by binding mRNAs, and their main function involves the degradation of their target mRNAs. Recent studies revealed altered expression of miRNAs in the kidneys during the progression of acute kidney injury (AKI) and chronic kidney disease (CKD) in humans and experimental rodent models by using high-throughput screening techniques including microarray and small RNA sequencing. Particularly, miR-21 seems to be strongly associated with renal pathogenesis both in the glomerulus and tubulointerstitium. Furthermore, abundant evidence has been gathered showing the involvement of miRNAs in renal fibrosis. Because of the complex morphofunctional organization of the mammalian kidneys, it is crucial both to determine the exact localization of the kidney cells that express the miRNAs, which has been addressed mainly using in situ hybridization methods, and to identify precisely which mRNAs are bound and degraded by these miRNAs, which has been studied mostly through in vitro analysis. To discover novel biomarker candidates, miRNA levels in urine supernatant, sediment, and exosomal fraction were comprehensively investigated in different types of kidney disease, including drug-induced AKI, ischemia-induced AKI, diabetic nephropathy, lupus nephritis, and IgA nephropathy. Recent studies also demonstrated the therapeutic effect of miRNA and/or anti-miRNA administrations. The intent of this review is to illustrate the state-of-the-art research in the field of miRNAs associated with renal pathogenesis, especially focusing on AKI and CKD in humans and animal models.
Keywords: microRNA, kidney disease, acute kidney injury, chronic kidney disease, biomarker, exosome
Introduction
Mature microRNAs (miRNAs) act as posttranscriptional regulators by binding mRNAs, and their main function involves the degradation of their target mRNAs. Recent studies revealed altered expression of miRNAs in the kidneys during the progression of acute kidney injury (AKI) and chronic kidney disease (CKD) in humans and experimental animals by using high-throughput screening techniques. Because of the complex morphofunctional organization of the mammalian kidneys, it is crucial both to determine the exact localization of the kidney cells that express the miRNAs, which has been addressed mainly using in situ hybridization methods, and to identify precisely which mRNAs are bound and degraded by these miRNAs, which has been studied mostly through in vitro analysis. To discover novel biomarker candidates, miRNA levels in urine supernatant, sediment, and exosomal fraction were comprehensively investigated in different types of kidney disease. The intent of this review is to illustrate the state-of-the-art research in the field of miRNAs associated with the pathogenesis of AKI and CKD in humans and animal models.
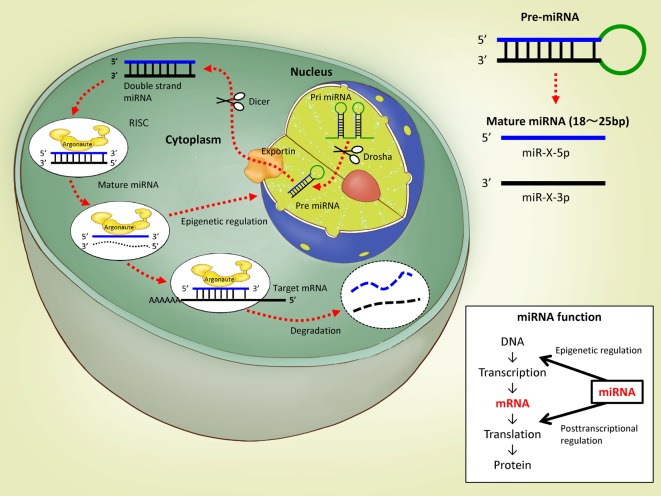
The Biosynthesis of microRNAs (miRNAs)
Non-coding RNAs (ncRNAs) are transcribed from the non-coding region of the genome, which is considered to be a “gene desert,” and they can regulate the expression of genes on the coding region. miRNAs are one type of ncRNAs; their mature form is a single-stranded RNA molecule between 18 and 25 bases long, and their sequences are highly conserved among animal species. Initially, an miRNA molecule is transcribed by RNA polymerase II as a primary miRNA (pri-miRNA) having a stem-loop structure, and then this pri-miRNA is cleaved into a precursor miRNA (pre-miRNA) of approximately 70 bases by an RNase III enzyme, Drosha (Fig. 1). The pre-miRNA is transported into the cytoplasm through the nuclear export protein Exportin 5 and is cleaved into double-stranded miRNA mainly by Dicer, another RNase III enzyme. The double-stranded miRNA is incorporated into the RNA-induced silencing complex (RISC) containing Argonaute proteins, and one strand of the duplex (called passenger strand, miR-X*, or miR-X-star) is degraded. The other chain of the duplex (called the guide strand or miR-X) is thus free to bind the 3′ untranslated region (UTR) of the target mRNA. In most cases, the guide strand miR-X showed higher abundance compared with the passenger strand miR-X*. On the other hand, to avoid problems if the abundance of each strand changes between tissues, developmental stages, or species, this previously accepted nomenclature has been largely abandoned, and the latest convention is to name mature miRNAs by the arm of the pre-miRNA from which they are derived, regardless of their abundance: those from the 5′ arm are named miR-X-5p, and those from the 3′ arm are named miR-X-3p. If the miRNA sequence corresponds without any mismatches to the target mRNA sequence and if their binding is therefore perfect, RISC will degrade the target mRNA, but if they do not completely match, the mRNA degradation will be delayed, and the miRNA-mRNA duplex will accumulate in the cytoplasmic processing body (P body). For binding to the target mRNA, 2 to 8 bases (called the seed sequence) of the 5′ region of the miRNA are important. Thus, miRNAs regulate the expression of their target mRNAs at the posttranscriptional level. A recent review by Luo et al.1 pointed out the importance of the regulatory circuit existing between epigenetic modulation and miRNAs, with special regard to cancer, as miRNA genes can be epigenetically regulated by DNA methylation and/or histone modification, and in turn, a subclass of miRNAs named “epi-miRNAs” was recognized to directly target epigenetic regulators such as DNA methyltransferases (DNMTs), HDACs, or components of the polycomb repressor complexes.
Fig. 1.
The biosynthesis and function of miRNAs.
miRNAs in the Kidneys
At present (August 2017), the miRNA database (miRbase; http://www.mirbase.org/) contains 2,588, 690, 793, 411, 453, 765, and 1,915 miRNAs found in humans, horses, bovines, pigs, dogs, rats, and mice, respectively. In 2004, using miRNA microarray analysis, Sun et al.2 reported that miR-192, miR-194, miR-204, miR-215, and miR-216 are abundantly expressed in the human kidneys. In a later study, Tian et al.3 showed that the expression levels of miR-192 and miR-194 are higher in the cortex than in the medulla of the rat kidneys, while miR-27b was highly expressed in the medulla. These basic comprehensive miRNA expression data are quite important to help clarify the function of miRNAs and identify the miRNAs that could be successfully used as biomarkers in several kidney diseases, because the kidney structures are complexly organized with essential contributions from various cell types, such as glomerulus-composing cells including podocytes, mesangial cells, and endothelial cells; tubulointerstitium-composing cells including the epithelial cells of the proximal tubules (PTs), attenuated tubules, distal tubules (DTs), and collecting ducts (CDs); capillary endothelial cells; interstitial cells; and immune cells. Renal pathological events can be mainly divided into two types, glomerular and tubulointerstitial lesions. One of the pathological features of renal lesions, i.e., their localization in the organ, differs among kidney diseases. The glomeruli tend to be primarily damaged in focal segmental glomerulosclerosis (FSGS) and in the various types of glomerulonephritis due to genetic mutations or immunological changes and secondarily damaged in systemic diseases such as numerous infectious diseases or autoimmune diseases including systemic lupus erythematosus (SLE; https://www.kidney.org/). On the other hand, the tubulointerstitium tends to be damaged due to the use of antibiotics or anticancer drugs, which usually involve injuries of PTs4. Furthermore, the localization of lesions in hereditary kidney diseases depends on the kind of mutated genes. Thus, since renal injury features differ among kidney diseases, we have to carefully examine the miRNA expression profile in each kidney disease in relation to the type of cells expressing the miRNA in question, in particular for application of miRNA molecules as disease markers.
Acute Kidney Injury and miRNAs
AKI is defined as a sudden renal dysfunction or kidney damage that occurs within 48 hours or 7 days, according to international criteria such as RIFLE, AKIN, and KDIGO criteria5. It includes renal dysfunction as indicated by a decrease in glomerular filtration and increase in serum creatinine and include a wide range of diseases including early renal tubular injuries. It is commonly observed in hospitalized patients, in particular elderly patients. AKI can be induced by decreased blood flow due to shock, bleeding, severe diarrhea, organ failure, and drugs/chemicals5. For the creation of AKI animal models, ischemia reperfusion (IR) models (pre-renal factor), folic acid or adenine administration models (renal factor), or unilateral ureter obstruction (UUO) models (post-renal factor) have been globally adopted to examine the pathogenesis of these various lesions mainly using experimental rodents including mice and rats. Based on several experimental data and clinical evidence, hepatitis A virus cellular receptor 1 (Havcr1, also known as kidney injury molecule-1; KIM-1/TIM-1), lipocalin 2 (LCN2, also known as neutrophil gelatinase-associated lipocalin; NGAL), and liver-type fatty acid binding protein (L-FABP) have been found to be excellent markers to predict tubulointerstitial lesions in AKI6, as they are localized in the PTs.
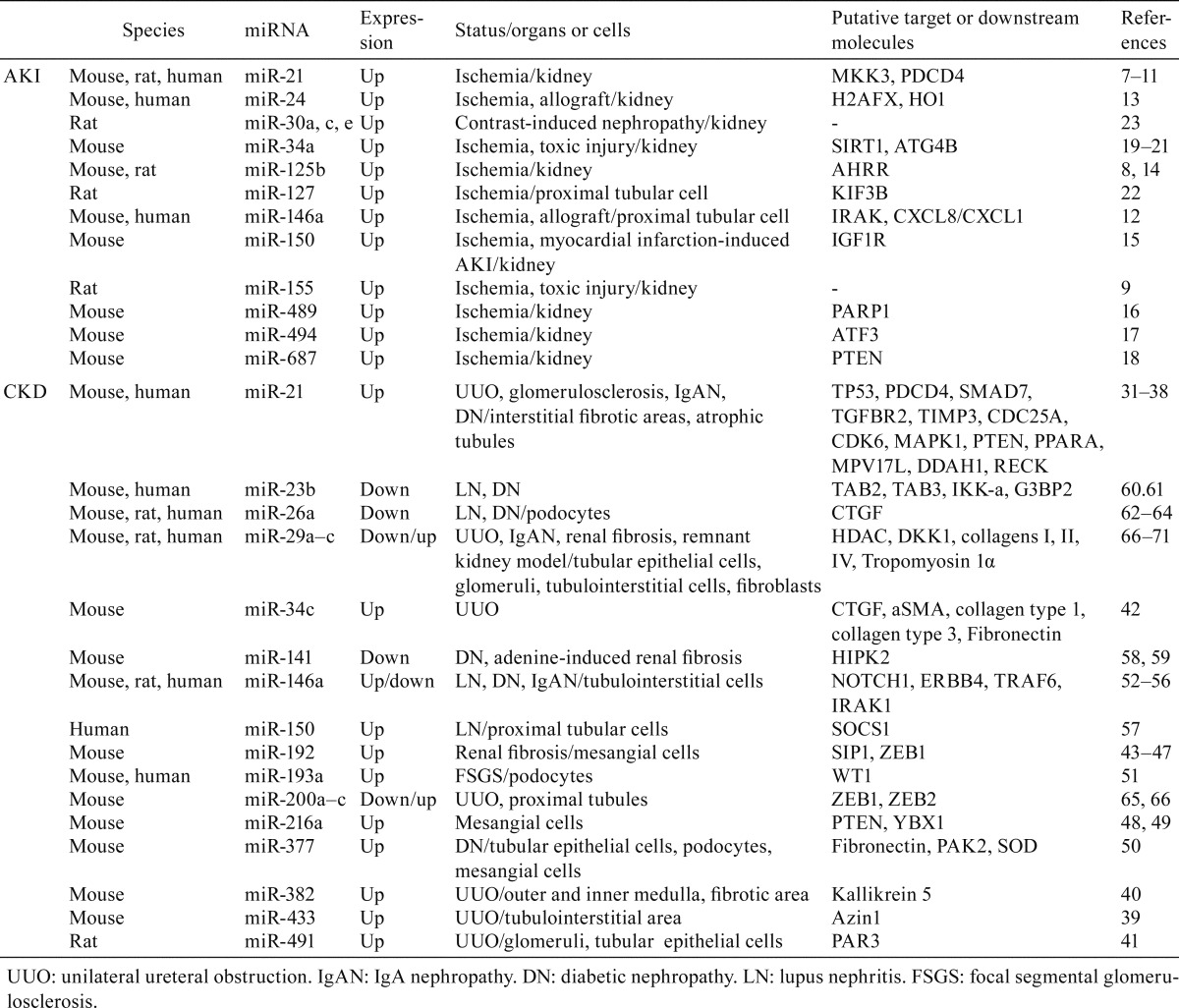
miRNAs associated with the development of AKI have mainly been reported in mouse, rat, and humans showing ischemic status after surgical operations (Table 1). To avoid confusion, we have provided a list of miRNA target molecules with their descriptions (Table 2). The strongest evidence was obtained for miR-217,8,9. In fact, its expression was found to be increased in the mouse IR kidneys and showed a pathological significance by targeting several factors including MKK310, which explains the inhibitory action of miR-21 against interleukin-6 (IL-6) and tumor necrosis factor α (TNF-α). Xu et al.11 reported PDCD4 as a target of miR-21, clarifying the observed protective effect of miR-21 against apoptosis. Similarly, after IR, miR-146a-deficient mice exhibited more severe tubulointerstitial lesions than wild-type mice, and overexpression of miR-146a reduced both IRAK1 and CXCL8/CXCL1 expression in injured tubular cells12. In vivo, more severe renal ischemia-reperfusion injury (IRI) was also found to be associated with increased miR-146a expression in both allografts and urine of human kidney transplant recipients12. miR-24 was upregulated in mouse IR kidneys and in human ischemic kidneys, especially in renal endothelial and tubular epithelial cells, where it was shown to exert a deleterious pro-apoptotic effect by targeting the H2A.X and HO113. Other reports also showed increased levels of specific miRNAs related to AKI, such as miR-125b, which regulates the AHRR in cisplatin-treated mice14 and IR rats8; miR-150, which modulates the expression of IGF1R in myocardial infarction-induced AKI mice15; miR-489, which binds the PARP-1 mRNA in IR mice16; miR-494, which downregulates ATF3 in IR mice17; and miR-687, which targets PTEN in IR mice18. In cisplatin-induced AKI mice, miR-34a was found to be induced via P5319 and to control the expression of SIRT120. Another study showed that, in IR mice, miR-34a regulates the autophagic activity in tubular epithelial cells by targeting ATG4B21. In the IR rat kidneys, increased miR-127 expression appeared to be positively associated with cell-matrix and cell-cell adhesion maintenance through downregulation of KIF3B22. Saikumar et al.9 reported that miR-21 as well as miR-155 showed increased levels in a rat model of AKI induced by IR or gentamicin administration. Contrast media also induces AKI and is associated with an increased risk of cardiovascular events. Gutiérrez-Escolan et al.23 revealed increased levels of miR-30 family members (miR-30a, c, e) in the rat kidney showing contrast-induced nephropathy. Thus, several reports showed increased expression for various miRNAs in kidney tissues affected by AKI in the species examined, while similar reports showing decreased miRNA expression levels were relatively scarce.
Table 1. miRNAs Showing Altered Expression in the Kidneys in AKI Or CKD.
Table 2. miRNA Targets Referred to in this Review.
Chronic Kidney Disease and miRNA
CKD is defined as abnormalities of kidney structure or function continuing for more than 3 months24. Thirteen million Japanese25 and 30 million American adults and have CKD, and millions of others are at increased risk (National Kidney Foundation, https://www.kidney.org/). Complex factors can lead to CKD, such as diabetes, hypertension, autoimmune diseases, systemic infections, urinary tract infections, urinary stones, neoplasia, hereditary mutations, AKI, or drugs/chemicals. Spontaneous animal models for CKD are scarce, but several mouse strains showing autoimmune diseases have been adopted as CKD models26. In addition, experimental rodents showing chronic stages due to UUO or diabetes develop CKD associated with renal fibrosis27,28,29,30.
Several reports helped to clarify the relation between CKD and miRNA (Table 1). Similar to what was observed for miR-21 in AKI, an elevated level of miR-21 was reported in the kidneys of mice suffering either from glomerulosclerosis31 or from UUO32 and in humans affected either by IgA nephropathy33 or diabetic nephropathy34. In particular, glomerular cells34, 35, interstitial fibrotic areas36, 37, and atrophic tubules35 showed elevated miR-21 expression. miR-21 targets several molecules including P53, PDCD4, SMAD7, TGFBR2, TIMP331, CDC25A, CDK634, ERK/MAPK, PTEN, PPARA, MPV17L, DDAH1, and RECK35, 38. In UUO-treated mice or rats, elevated expression levels were reported for miR-43339, miR-38240, and miR-49141 in the tubulointerstitial area, inner and outer medulla, and glomerulus or tubular epithelial cells, respectively. miR-34c showed increased levels in mouse UUO kidneys, and it was reported to have CTGF, α-SMA, collagen type 1, collagen type 3, and fibronectin as downstream factors42. An elevated level of miR-192 was reported in kidneys showing CKD due to UUO43 or diabetes44, 45 and in the mouse or rat model of nephrectomy46. miR-192 seemed to be targeting SIP144 and ZEB147. As for diabetic nephropathy, miR-216a in mouse mesangial cells appears to be important for the development of sclerosis via PTEN48 or YBX149. On the other hand, Wang et al. showed that miR-377 is upregulated in tubular epithelial cells, podocytes, and mesangial cells and can lead to increased fibronectin production in diabetic mice50. Gebeshuber et al.51 showed clearly that FSGS can be induced by miR-193a and that WT1 is an miR-193a target and enforces transgenic expression of miR-193a in mice. Besides, elevated levels of miR-146a in the kidneys are reported in individuals developing lupus nephritis52 or IgA nephropathy53 and in the mouse model for lupus nephritis54 as well as in the rat model for diabetes55. However, the miR-146a level was decreased in diabetic mice55. miR-146a appears to be targeting TRAF6 and IRAK155 or NOTCH1 and ERBB456. In addition, regarding human lupus nephritis, Zhou et al.57 showed increased miR-150 in the kidneys and found SOCS1 as its target.
It should, however be noted that decreased levels of miR-141 were reported in the kidneys of mice developing diabetic nephropathy and adenine-induced renal fibrosis58, and miR-141 was also shown to regulate the epithelial-mesenchymal transition of the tubular epithelium by targeting HIPK259. Furthermore, miR-23b also showed decreased expression in humans, in the mouse model of lupus nephritis60, and in the mouse model diabetic nephropathy61. miR-23b was demonstrated to be targeting TAB2, TAB3, and IKK-a60 in one report and be targeting G3BP2 in another61. In our previous study, we clarified the role of decreased miR-26a levels in the glomerulus of mice and individuals developing lupus nephritis and in the glomerulus of individuals developing IgA nephropathy, and we suggested this decrease was related to podocyte injury62. Other studies also showed decreased miR-26a expression in the kidneys of mice63 and rats64 developing diabetic nephropathy, and the authors of those studies explained the correlation between low miR-26a and nephropathy with findings showing that miR-26a negatively regulates CTGF. Decreased expression of miR-200a was detected in the proximal tubules of UUO-treated rodents, and miR-200a seemed to suppress the expression of ZEB1 and ZEB265. However, another report showed that expression of the miR-200 family, particularly of miR-200b, was increased in a time-dependent manner in the kidneys of UUO mice, that expression of ZEB1 and ZEB2 was also increased after UUO, and that administration of the miR-200b precursor suppressed these increases66. The expression of miR-29a, b, and c was also found to be decreased in the mouse kidneys by UUO67 or diabetic condition68. Importantly, miR-29a, b, and c were found to be expressed in glomeruli and renal tubules68. In later studies, miR-29a expression was confirmed in the mouse glomerulus and renal tubules69, 70 and, in particular, in podocytes69. Besides, decreased levels of miR-29c were reported in the kidneys of individuals developing IgA nephropathy with renal fibrosis and in the 5⁄6 nephrectomy rat71. The miR-29 family has among its targets collagens I and IV68, HDAC69 and DKK170, which are downregulated by miR-29a, and collagen II and tropomyosin 1α, which are under the control of miR-29c71.
Urinary miRNA and Kidney Diseases
The kidney is a nonregenerative organ, and confirmed diagnosis of kidney disease relies on highly invasive renal biopsy. Identification of biomarkers in body fluids that can help to identify the renal pathology type will be essential for the development of noninvasive diagnostic methods. Urinary albumin, blood urea nitrogen, serum creatinine level, etc., are useful diagnostic indicators, but further information is still needed to be able to grasp the kidney disease type. For the progress of kidney disease research, it will be of crucial importance to evaluate carefully miRNAs as “disease-specific nucleic acid biomarkers.”
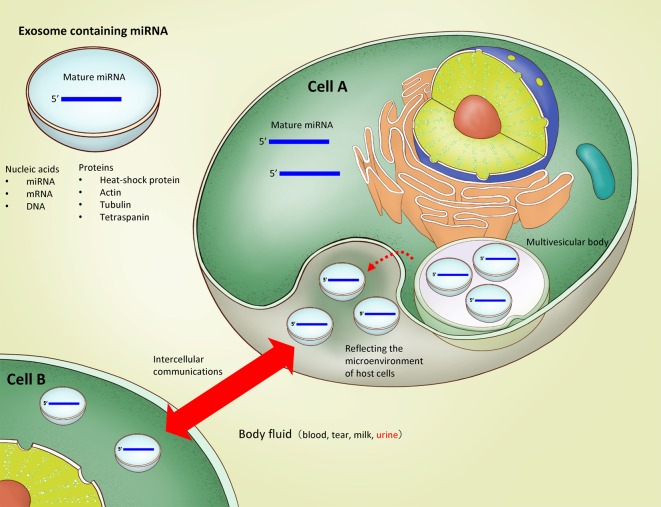
miRNAs are present in body fluids. Weber et al.72 have comprehensively analyzed miRNA extracted from cerebrospinal fluid, tears, pleural effusion, trachea wash, milk, colostrum, ascites, urine, semen, amniotic fluid, and plasma from healthy humans. The number of detected species of miRNA was highest in saliva with 458 miRNAs and lowest in urine with 204 miRNAs. Notably, these authors identified miRNAs that appear specifically in body fluids, such as tear fluid miR-637 and cerebrospinal fluid miR-577, but they were unable to identify any urine-specific miRNAs.
How do miRNAs exist in these body fluids? Johnstone et al.73 discovered vesicles secreted from sheep reticulocytes and named them exosomes (Fig. 2). Exosomes are a type of extracellular vesicles which include microvesicles, apoptotic bodies, and ectosomes74. They are derived from the budding of endosomal membranes, resulting in the formation of multivesicular bodies (MVBs), and are produced by the fusion of MVBs with the plasma membrane. Therefore, exosomal membrane molecules such as CD9, CD63, and CD81 are used as representative exosomal markers75. Valadi et al.76 reported that mRNAs and miRNAs are present in the exosomes, and the miRNAs in body fluids are thought to exist in exosome-encapsulated form. Urinary miRNAs are considered stable because they are also included in exosomes77. In recent years, research has increasingly been conducted to analyze miRNAs in urine in different renal diseases and to compare healthy and disease groups. It is still unknown whether miRNAs could be a good biomarker for kidney disease, but we believe it is worthwhile anyway to summarize the results so far obtained in this field of research.
Fig. 2.
The biosynthesis and function of exosomes containing miRNAs.
Urinary miRNAs and AKI
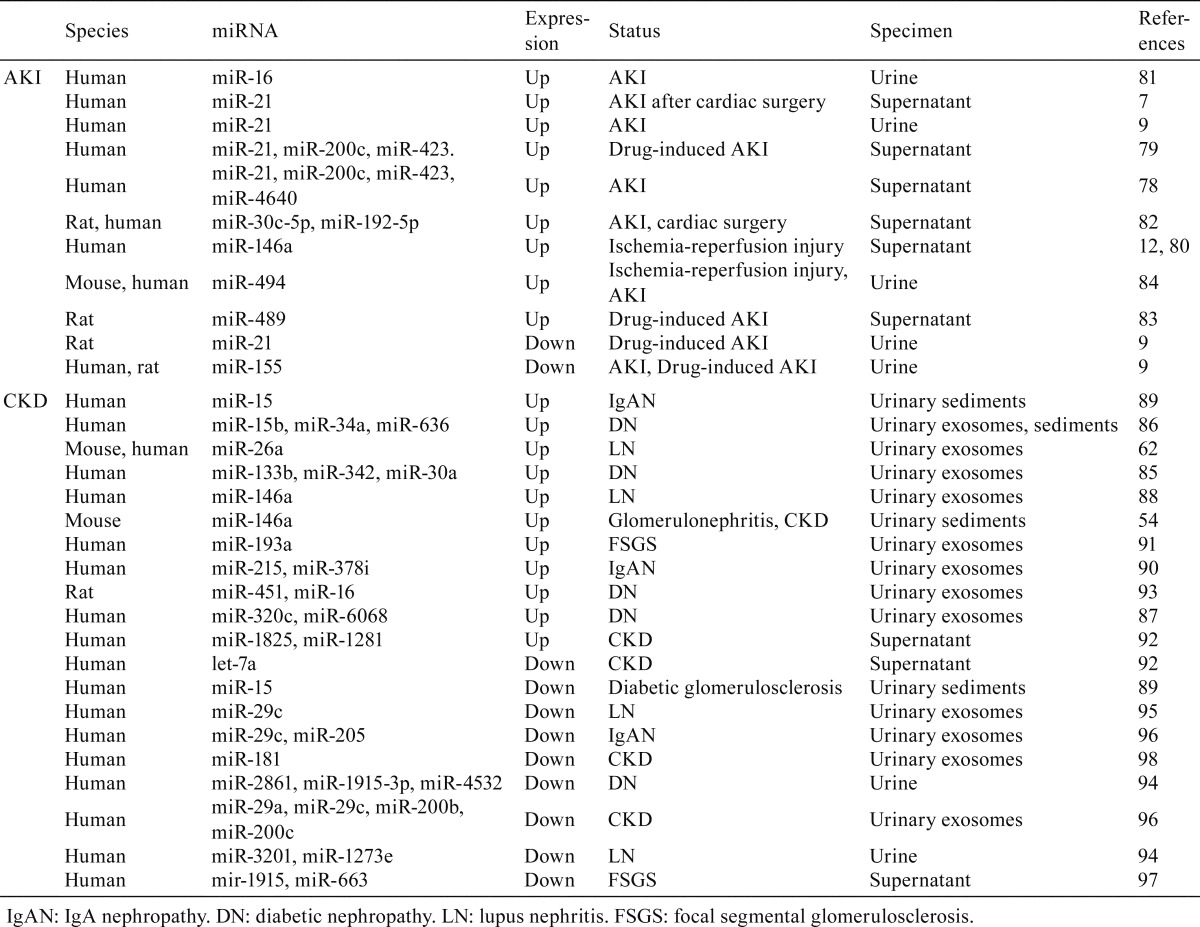
As summarized in Table 3, miR-21, miR-200c, miR-423, and miR-4640 appeared to be increased in the urine supernatant of individuals with AKI78, and the levels of miR-21, miR-200c, and miR-423 were also found to be elevated in individuals with drug-induced AKI79. As for miR-21, an increase in its urinary levels was observed in individuals with AKI7, 9; however, rats with drug-induced AKI showed decreased miR-21 urinary levels9. miR-146a12, 80, miR-1681, miR-30c, and miR-19282 were increased in the urine of patients diagnosed with AKI. In particular, miR-146a and miR-30c were also found to be elevated in the kidneys of patients and mice suffering from AKI12 and in the kidneys of rats with contrast-induced nephropathy23, respectively. In addition, increased urine levels of miR-48983 were reported in rats with gentamicin-induced AKI. Elevated levels of miR-494 were described in mice and individuals developing AKI17, and an increase in miR-494 levels was also detected in the kidney of mice with AKI16, 17, 84. For miR-155, reduced levels in the urine were reported in rats with AKI induced by gentamicin and in patients with AKI9. However, to date, evidence of a truly diagnostic value of the AKI-associated miRNAs found in urinary exosomes is still scarce.
Table 3. miRNAs Showing Altered Levels in the Urine of AKI or CKD.
Urinary miRNA and CKD
In order to identify possible candidate miRNAs to be exploited as biomarkers of CKD, a thorough analysis of the exosomal fractions from human urinary samples was performed. miR-133b, miR-342, miR-30a85, miR-15b, miR-34a, miR-63686, miR-320c, and miR-606887 were found to be increased in urinary exosomes from diabetic patients (Table 3). With regard to lupus nephritis, elevated levels of miR-146a88 and miR-26a62 in urinary exosomes were reported in humans; conversely, the miR-26a level was found to be decreased in the kidneys of mice with lupus nephritis62. An elevated level of miR-146a in the urinary sediment was also observed in a mouse model of lupus nephritis54. Patients with IgA nephropathy showed increased amounts of miR-15 in urinary sediment89 or increased amounts of miR-215 and miR-378i in urinary exosomes90. The detection of miR-15 in urinary sediment also indicates the presence of cells in the urine, which can be of diagnostic significance. However, the miR-15 levels in urinary sediment of diabetic glomerulosclerosis patients were found to be decreased89. Similar to the renal expression of miR-193a51, urinary exosomes containing miR-193a also increased in FSGS compared with in a minimal change group91. Furthermore, miR-1825 and miR-1281 were increased in the urine of patients with CKD92, and the levels of miR-451 and miR-16 in urinary exosomes were also increased in rats with diabetic nephropathy93.
The levels of urinary miR-3201 and miR-1273e94 and of miR-29c in urinary exosomes95 were found to be reduced in patients with lupus nephritis. Altered renal expression of miR-29c has also been reported in mice, humans, and rats67, 68, 71. In agreement with these reports, miR-29c, as well as miR-205, was found to be decreased in the urinary exosomes of individuals developing IgA nephropathy90, and miR-29a, miR-29c, miR-200b, and miR-200c were decreased in the urinary exosomes of CKD patients96. miR-1915 and miR-663 in the urine supernatant from FSGS patients97 and miR-2861, miR-1915, and miR-453294 in urinary exosomes from diabetic nephropathy patients were also found to be decreased. Furthermore, let-7a92 and miR-18198 showed decreased levels in urine supernatant and urinary exosomes of CKD patients, respectively.
miRNAs and Animals
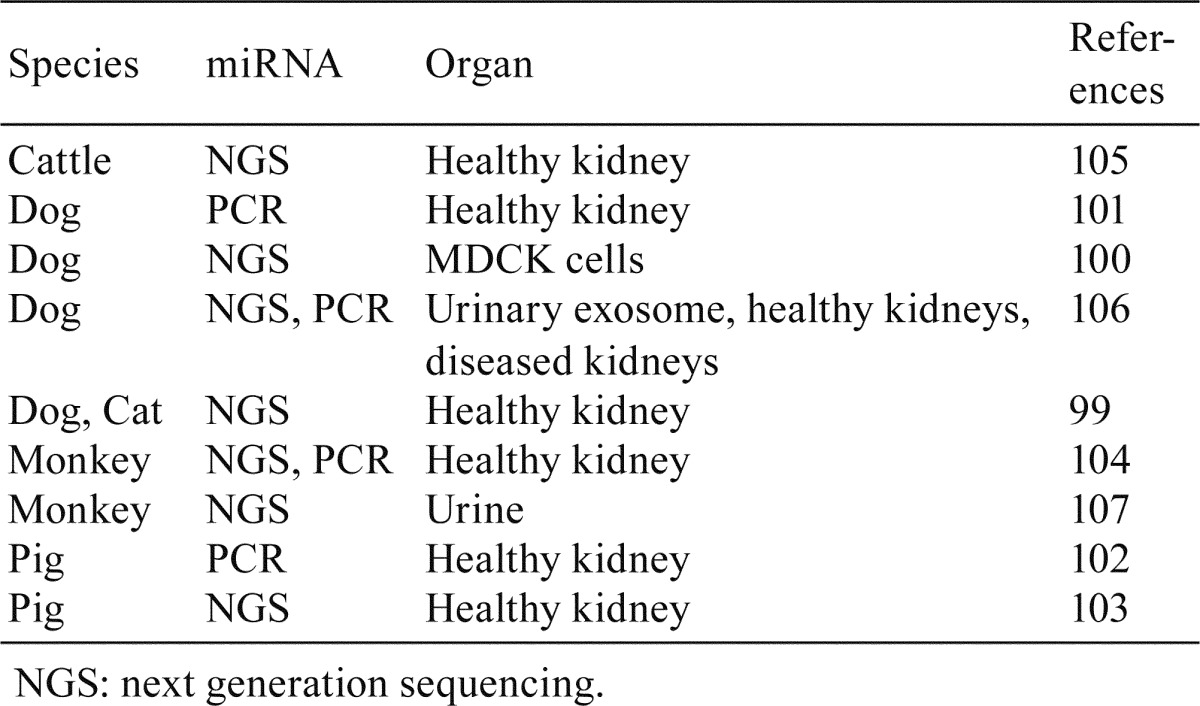
Abundant evidence of altered expression in the kidneys and of abnormal levels in the urine of patients with kidney disease was reported for miR-21 and for the miR-29 family. However, further basic data obtained using rodents as well as other animal species are needed; particularly, the availability of further evidence from dogs, pigs, and monkeys, which are major animal species, would be helpful when extrapolation of experimental results from animals to humans is required. Several studies investigated miRNA expression in the kidneys, including those of dogs99,100,101, pigs102, 103, monkeys104, cats99, and cattle105 (Table 4). In addition, urinary levels of miRNAs were also investigated in dogs106 and monkeys107. As for dog CKD, altered urinary exosome levels of miR-26a and miR-21a were detected106.
Table 4. miRNA Studies Reporting the miRNA Expression in the Kidney or Urine of Various Animal Species.
Conclusion
One kind of miRNA seems to target multiple mRNAs. Therefore, this class of small ncRNAs would strongly affect molecular biological processes by functioning upstream of the central dogma and have a great influence on renal pathogenesis. For the analysis of miRNAs associated with renal pathogenesis, identification of disease type-specific miRNAs and their expressing cells and elucidation of the molecular network controlled by these miRNAs in the cells would be important because the kidneys are structurally and functionally remarkably complex organs. In particular, evidence of miR-21 being associated with both AKI and CKD is strong. Recently, miRNAs have been adopted not only as disease markers but also as therapeutic targets34, 45. We strongly hope that next-generation biomarker diagnosis and/or nucleic acid delivery therapy developed by future miRNA researches will be applied not only for human kidney disease control but also for managing and containing kidney disease in animals.
Acknowledgments
All figures were kindly provided by Dr. Shin-Hyo Lee, Konkuk University School of Medicine.
Footnotes
Disclosure of Potential Conflicts of Interest: This manuscript has not been published or presented elsewhere in part or in entirety and is not under consideration by another journal. Both authors approved the manuscript and agreed to its submission to your esteemed journal. There are no conflicts of interest to declare. This work was partially supported by JSPS KAKENHI Grant Numbers 24688033, 26461231, and 15H05634.
References
- 1.Liu X, Chen X, Yu X, Tao Y, Bode AM, Dong Z, and Cao Y. Regulation of microRNAs by epigenetics and their interplay involved in cancer. J Exp Clin Cancer Res. 32: 96 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, and Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 32: e188 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian Z, Greene AS, Pietrusz JL, Matus IR, and Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 18: 404–411. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oni L, Hawcutt DB, Turner MA, Beresford MW, McWilliam S, Barton C, Park BK, Murray P, Wilm B, Copple I, Floyd R, Peak M, Sharma A, and Antoine DJ. Optimising the use of medicines to reduce acute kidney injury in children and babies. Pharmacol Ther. 174: 55–62. 2017. [DOI] [PubMed] [Google Scholar]
- 5.Palevsky PM, Liu KD, Brophy PD, Chawla LS, Parikh CR, Thakar CV, Tolwani AJ, Waikar SS, and Weisbord SD. KDOQI US commentary on the 2012 KDIGO clinical practice guideline for acute kidney injury. Am J Kidney Dis. 61: 649–672. 2013. [DOI] [PubMed] [Google Scholar]
- 6.Schley G, Köberle C, Manuilova E, Rutz S, Forster C, Weyand M, Formentini I, Kientsch-Engel R, Eckardt KU, and Willam C. Comparison of Plasma and Urine Biomarker Performance in Acute Kidney Injury. PLoS One. 10: e0145042 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du J, Cao X, Zou L, Chen Y, Guo J, Chen Z, Hu S, and Zheng Z. MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PLoS One. 8: e63390 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Güçlü A, Koçak C, Koçak FE, Akçılar R, Dodurga Y, Akçılar A, and Elmas L. MicroRNA-125b as a new potential biomarker on diagnosis of renal ischemia-reperfusion injury. J Surg Res. 207: 241–248. 2017. [DOI] [PubMed] [Google Scholar]
- 9.Saikumar J, Hoffmann D, Kim TM, Gonzalez VR, Zhang Q, Goering PL, Brown RP, Bijol V, Park PJ, Waikar SS, and Vaidya VS. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol Sci. 129: 256–267. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Deng X, Kang Z, Wang Y, Xia T, Ding N, and Yin Y. Elevation of miR-21, through targeting MKK3, may be involved in ischemia pretreatment protection from ischemia-reperfusion induced kidney injury. J Nephrol. 29: 27–36. 2016. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Kriegel AJ, Liu Y, Usa K, Mladinov D, Liu H, Fang Y, Ding X, and Liang M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 82: 1167–1175. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amrouche L, Desbuissons G, Rabant M, Sauvaget V, Nguyen C, Benon A, Barre P, Rabaté C, Lebreton X, Gallazzini M, Legendre C, Terzi F, and Anglicheau D. MicroRNA-146a in Human and Experimental Ischemic AKI: CXCL8-Dependent Mechanism of Action. J Am Soc Nephrol. 28: 479–493. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzen JM, Kaucsar T, Schauerte C, Schmitt R, Rong S, Hübner A, Scherf K, Fiedler J, Martino F, Kumarswamy R, Kölling M, Sörensen I, Hinz H, Heineke J, van Rooij E, Haller H, and Thum T. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J Am Soc Nephrol. 25: 2717–2729. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo MS, Lee CG, Koo JH, and Kim SG. miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell Death Dis. 4: e899 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranganathan P, Jayakumar C, Tang Y, Park KM, Teoh JP, Su H, Li J, Kim IM, and Ramesh G. MicroRNA-150 deletion in mice protects kidney from myocardial infarction-induced acute kidney injury. Am J Physiol Renal Physiol. 309: F551–F558. 2015. [DOI] [PMC free article] [PubMed]
- 16.Wei Q, Liu Y, Liu P, Hao J, Liang M, Mi QS, Chen JK, and Dong Z. MicroRNA-489 Induction by Hypoxia-Inducible Factor-1 Protects against Ischemic Kidney Injury. J Am Soc Nephrol. 27: 2784–2796. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan YF, Chen HH, Lai PF, Cheng CF, Huang YT, Lee YC, Chen TW, and Lin H. MicroRNA-494 reduces ATF3 expression and promotes AKI. J Am Soc Nephrol. 23: 2012–2023. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhatt K, Wei Q, Pabla N, Dong G, Mi QS, Liang M, Mei C, and Dong Z. MicroRNA-687 induced by hypoxia-inducible factor-1 targets phosphatase and tensin homolog in renal ischemia-reperfusion injury. J Am Soc Nephrol. 26: 1588–1596. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatt K, Zhou L, Mi QS, Huang S, She JX, and Dong Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med. 16: 409–416. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CG, Kim JG, Kim HJ, Kwon HK, Cho IJ, Choi DW, Lee WH, Kim WD, Hwang SJ, Choi S, and Kim SG. Discovery of an integrative network of microRNAs and transcriptomics changes for acute kidney injury. Kidney Int. 86: 943–953. 2014. [DOI] [PubMed] [Google Scholar]
- 21.Liu XJ, Hong Q, Wang Z, Yu YY, Zou X, and Xu LH. MicroRNA-34a suppresses autophagy in tubular epithelial cells in acute kidney injury. Am J Nephrol. 42: 168–175. 2015. [DOI] [PubMed] [Google Scholar]
- 22.Aguado-Fraile E, Ramos E, Sáenz-Morales D, Conde E, Blanco-Sánchez I, Stamatakis K, del Peso L, Cuppen E, Brüne B, and Bermejo ML. miR-127 protects proximal tubule cells against ischemia/reperfusion: identification of kinesin family member 3B as miR-127 target. PLoS One. 7: e44305 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutiérrez-Escolano A, Santacruz-Vázquez E, and Gómez-Pérez F. Dysregulated microRNAs involved in contrast-induced acute kidney injury in rat and human. Ren Fail. 37: 1498–1506. 2015. [DOI] [PubMed] [Google Scholar]
- 24.Inker LA, Astor BC, Fox CH, Isakova T, Lash JP, Peralta CA, Kurella Tamura M, and Feldman HI. KDOQI US commentary on the 2012 KDIGO clinical practice guideline for the evaluation and management of CKD. Am J Kidney Dis. 63: 713–735. 2014. [DOI] [PubMed] [Google Scholar]
- 25.Clinical Practice Guidebook for Diagnosis and Treatment of Chronic Kidney Disease 2012. (in Japanese). [PubMed]
- 26.Ichii O, Konno A, Sasaki N, Endoh D, Hashimoto Y, and Kon Y. Altered balance of inhibitory and active Fc gamma receptors in murine autoimmune glomerulonephritis. Kidney Int. 74: 339–347. 2008. [DOI] [PubMed] [Google Scholar]
- 27.Ito H, Yan X, Nagata N, Aritake K, Katsumata Y, Matsuhashi T, Nakamura M, Hirai H, Urade Y, Asano K, Kubo M, Utsunomiya Y, Hosoya T, Fukuda K, and Sano M. PGD2-CRTH2 pathway promotes tubulointerstitial fibrosis. J Am Soc Nephrol. 23: 1797–1809. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lan HY, Mu W, Tomita N, Huang XR, Li JH, Zhu HJ, Morishita R, and Johnson RJ. Inhibition of renal fibrosis by gene transfer of inducible Smad7 using ultrasound-microbubble system in rat UUO model. J Am Soc Nephrol. 14: 1535–1548. 2003. [DOI] [PubMed] [Google Scholar]
- 29.Kaimori JY, Isaka Y, Hatanaka M, Yamamoto S, Ichimaru N, Fujikawa A, Shibata H, Fujimori A, Miyoshi S, Yokawa T, Kuroda K, Moriyama T, Rakugi H, and Takahara S. Visualization of kidney fibrosis in diabetic nephropathy by long diffusion tensor imaging MRI with spin-echo sequence. Sci Rep. 7: 5731 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Recio C, Lazaro I, Oguiza A, Lopez-Sanz L, Bernal S, Blanco J, Egido J, and Gomez-Guerrero C. Suppressor of cytokine signaling-1 peptidomimetic limits progression of diabetic nephropathy. J Am Soc Nephrol. 28: 575–585. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai JY, Luo J, O’Connor C, Jing X, Nair V, Ju W, Randolph A, Ben-Dov IZ, Matar RN, Briskin D, Zavadil J, Nelson RG, Tuschl T, Brosius FC, 3rd , Kretzler M, and Bitzer M. MicroRNA-21 in glomerular injury. J Am Soc Nephrol. 26: 805–816. 2015. [DOI] [PMC free article] [PubMed]
- 32.Zarjou A, Yang S, Abraham E, Agarwal A, and Liu G. Identification of a microRNA signature in renal fibrosis: role of miR-21. Am J Physiol Renal Physiol. 301: F793–F801. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennino MF, Buob D, Van der Hauwaert C, Gnemmi V, Jomaa Z, Pottier N, Savary G, Drumez E, Noël C, Cauffiez C, and Glowacki F. miR-21-5p renal expression is associated with fibrosis and renal survival in patients with IgA nephropathy. Sci Rep. 6: 27209 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kölling M, Kaucsar T, Schauerte C, Hübner A, Dettling A, Park JK, Busch M, Wulff X, Meier M, Scherf K, Bukosza N, Szénási G, Godó M, Sharma A, Heuser M, Hamar P, Bang C, Haller H, Thum T, and Lorenzen JM. Therapeutic miR-21 silencing ameliorates diabetic kidney disease in mice. Mol Ther. 25: 165–180. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, Li J, Tran PT, Kaimal V, Huang X, Chang AN, Li S, Kalra A, Grafals M, Portilla D, MacKenna DA, Orkin SH, and Duffield JS. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 4: 121ra18 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glowacki F, Savary G, Gnemmi V, Buob D, Van der Hauwaert C, Lo-Guidice JM, Bouyé S, Hazzan M, Pottier N, Perrais M, Aubert S, and Cauffiez C. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS One. 8: e58014 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong X, Chung AC, Chen HY, Meng XM, and Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol. 22: 1668–1681. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loboda A, Sobczak M, Jozkowicz A, and Dulak J. TGF-β1/Smads and miR-21 in Renal Fibrosis and Inflammation. Mediators Inflamm. 2016: 8319283 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R, Chung AC, Dong Y, Yang W, Zhong X, and Lan HY. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-β/Smad3-Azin1 pathway. Kidney Int. 84: 1129–1144. 2013. [DOI] [PubMed] [Google Scholar]
- 40.Kriegel AJ, Liu Y, Cohen B, Usa K, Liu Y, and Liang M. MiR-382 targeting of kallikrein 5 contributes to renal inner medullary interstitial fibrosis. Physiol Genomics. 44: 259–267. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Q, Fan J, Ding X, Peng W, Yu X, Chen Y, and Nie J. TGF-beta-induced MiR-491-5p expression promotes Par-3 degradation in rat proximal tubular epithelial cells. J Biol Chem. 285: 40019–40027. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morizane R, Fujii S, Monkawa T, Hiratsuka K, Yamaguchi S, Homma K, and Itoh H. miR-34c attenuates epithelial-mesenchymal transition and kidney fibrosis with ureteral obstruction. Sci Rep. 4: 4578 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung AC, Huang XR, Meng X, and Lan HY. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol. 21: 1317–1325. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, and Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 104: 3432–3437. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Putta S, Lanting L, Sun G, Lawson G, Kato M, and Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 23: 458–469. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun L, Zhang D, Liu F, Xiang X, Ling G, Xiao L, Liu Y, Zhu X, Zhan M, Yang Y, Kondeti VK, and Kanwar YS. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J Pathol. 225: 364–377. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato M, Arce L, Wang M, Putta S, Lanting L, and Natarajan R. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int. 80: 358–368. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, and Natarajan R. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 11: 881–889. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato M, Wang L, Putta S, Wang M, Yuan H, Sun G, Lanting L, Todorov I, Rossi JJ, and Natarajan R. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-beta-induced collagen expression in kidney cells. J Biol Chem. 285: 34004–34015. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, and Quigg RJ. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 22: 4126–4135. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gebeshuber CA, Kornauth C, Dong L, Sierig R, Seibler J, Reiss M, Tauber S, Bilban M, Wang S, Kain R, Böhmig GA, Moeller MJ, Gröne HJ, Englert C, Martinez J, and Kerjaschki D. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat Med. 19: 481–487. 2013. [DOI] [PubMed] [Google Scholar]
- 52.Lu J, Kwan BC, Lai FM, Tam LS, Li EK, Chow KM, Wang G, Li PK, and Szeto CC. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology (Carlton). 17: 346–351. 2012. [DOI] [PubMed] [Google Scholar]
- 53.Wang G, Kwan BC, Lai FM, Chow KM, Li PK, and Szeto CC. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis Markers. 30: 171–179. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ichii O, Otsuka S, Sasaki N, Namiki Y, Hashimoto Y, and Kon Y. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int. 81: 280–292. 2012. [DOI] [PubMed] [Google Scholar]
- 55.Alipour MR, Khamaneh AM, Yousefzadeh N, Mohammad-nejad D, and Soufi FG. Upregulation of microRNA-146a was not accompanied by downregulation of pro-inflammatory markers in diabetic kidney. Mol Biol Rep. 40: 6477–6483. 2013. [DOI] [PubMed] [Google Scholar]
- 56.Lee HW, Khan SQ, Khaliqdina S, Altintas MM, Grahammer F, Zhao JL, Koh KH, Tardi NJ, Faridi MH, Geraghty T, Cimbaluk DJ, Susztak K, Moita LF, Baltimore D, Tharaux PL, Huber TB, Kretzler M, Bitzer M, Reiser J, and Gupta V. Absence of miR-146a in podocytes increases risk of diabetic glomerulopathy via up-regulation of ErbB4 and Notch-1. J Biol Chem. 292: 732–747. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H, Hasni SA, Perez P, Tandon M, Jang SI, Zheng C, Kopp JB, Austin H, 3rd , Balow JE, Alevizos I, and Illei GG. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. J Am Soc Nephrol. 24: 1073–1087. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang B, Koh P, Winbanks C, Coughlan MT, McClelland A, Watson A, Jandeleit-Dahm K, Burns WC, Thomas MC, Cooper ME, and Kantharidis P. miR-200a Prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes. 60: 280–287. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Y, Tong J, He F, Yu X, Fan L, Hu J, Tan J, and Chen Z. miR-141 regulates TGF-β1-induced epithelial-mesenchymal transition through repression of HIPK2 expression in renal tubular epithelial cells. Int J Mol Med. 35: 311–318. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang H, Liu W, Shi Y, Harley JB, Shen N, and Qian Y. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat Med. 18: 1077–1086. 2012. [DOI] [PubMed] [Google Scholar]
- 61.Zhao B, Li H, Liu J, Han P, Zhang C, Bai H, Yuan X, Wang X, Li L, Ma H, Jin X, and Chu Y. MicroRNA-23b targets ras GTPase-activating protein SH3 domain-binding protein 2 to alleviate fibrosis and albuminuria in diabetic nephropathy. J Am Soc Nephrol. 27: 2597–2608. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ichii O, Otsuka-Kanazawa S, Horino T, Kimura J, Nakamura T, Matsumoto M, Toi M, and Kon Y. Decreased miR-26a expression correlates with the progression of podocyte injury in autoimmune glomerulonephritis. PLoS One. 9: e110383 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koga K, Yokoi H, Mori K, Kasahara M, Kuwabara T, Imamaki H, Ishii A, Mori KP, Kato Y, Ohno S, Toda N, Saleem MA, Sugawara A, Nakao K, Yanagita M, and Mukoyama M. MicroRNA-26a inhibits TGF-β-induced extracellular matrix protein expression in podocytes by targeting CTGF and is downregulated in diabetic nephropathy. Diabetologia. 58: 2169–2180. 2015. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Z, Guan M, Jia Y, Wang D, Pang R, Lv F, Xiao Z, Wang L, Zhang H, and Xue Y. The coordinated roles of miR-26a and miR-30c in regulating TGFβ1-induced epithelial-to-mesenchymal transition in diabetic nephropathy. Sci Rep. 6: 37492 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiong M, Jiang L, Zhou Y, Qiu W, Fang L, Tan R, Wen P, and Yang J. The miR-200 family regulates TGF-β1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol. 302: F369–F379. 2012. [DOI] [PubMed] [Google Scholar]
- 66.Oba S, Kumano S, Suzuki E, Nishimatsu H, Takahashi M, Takamori H, Kasuya M, Ogawa Y, Sato K, Kimura K, Homma Y, Hirata Y, and Fujita T. miR-200b precursor can ameliorate renal tubulointerstitial fibrosis. PLoS One. 5: e13614 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qin W, Chung AC, Huang XR, Meng XM, Hui DS, Yu CM, Sung JJ, and Lan HY. TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol. 22: 1462–1474. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, Koh P, Thomas M, Jandeleit-Dahm K, Gregorevic P, Cooper ME, and Kantharidis P. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 23: 252–265. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin CL, Lee PH, Hsu YC, Lei CC, Ko JY, Chuang PC, Huang YT, Wang SY, Wu SL, Chen YS, Chiang WC, Reiser J, and Wang FS. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J Am Soc Nephrol. 25: 1698–1709. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu YC, Chang PJ, Ho C, Huang YT, Shih YH, Wang CJ, and Lin CL. Protective effects of miR-29a on diabetic glomerular dysfunction by modulation of DKK1/Wnt/β-catenin signaling. Sci Rep. 6: 30575 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang Y, Yu X, Liu Y, Kriegel AJ, Heng Y, Xu X, Liang M, and Ding X. miR-29c is downregulated in renal interstitial fibrosis in humans and rats and restored by HIF-α activation. Am J Physiol Renal Physiol. 304: F1274–F1282. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, and Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 56: 1733–1741. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnstone RM, Adam M, Hammond JR, Orr L, and Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 262: 9412–9420. 1987. [PubMed] [Google Scholar]
- 74.Loyer X, Vion AC, Tedgui A, and Boulanger CM. Microvesicles as cell-cell messengers in cardiovascular diseases. Circ Res. 114: 345–353. 2014. [DOI] [PubMed] [Google Scholar]
- 75.Kumar D, Gupta D, Shankar S, and Srivastava RK. Biomolecular characterization of exosomes released from cancer stem cells: Possible implications for biomarker and treatment of cancer. Oncotarget. 6: 3280–3291. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, and Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 9: 654–659. 2007. [DOI] [PubMed] [Google Scholar]
- 77.Alvarez ML, Khosroheidari M, Kanchi Ravi R, and DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 82: 1024–1032. 2012. [DOI] [PubMed] [Google Scholar]
- 78.Ramachandran K, Saikumar J, Bijol V, Koyner JL, Qian J, Betensky RA, Waikar SS, and Vaidya VS. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin Chem. 59: 1742–1752. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pavkovic M, Robinson-Cohen C, Chua AS, Nicoara O, Cárdenas-González M, Bijol V, Ramachandran K, Hampson L, Pirmohamed M, Antoine DJ, Frendl G, Himmelfarb J, Waikar SS, and Vaidya VS. Detection of drug-induced acute kidney injury in humans using urinary KIM-1, miR-21, -200c, and -423. Toxicol Sci. 152: 205–213. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aguado-Fraile E, Ramos E, Conde E, Rodríguez M, Martín-Gómez L, Lietor A, Candela Á, Ponte B, Liaño F, and García-Bermejo ML. A pilot study identifying a set of microRNAs as precise diagnostic biomarkers of acute kidney injury. PLoS One. 10: e0127175 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mousavi MZ, Chen HY, Lee KL, Lin H, Chen HH, Lin YF, Wong CS, Li HF, Wei PK, and Cheng JY. Urinary micro-RNA biomarker detection using capped gold nanoslit SPR in a microfluidic chip. Analyst (Lond). 140: 4097–4104. 2015. [DOI] [PubMed] [Google Scholar]
- 82.Zou YF, Wen D, Zhao Q, Shen PY, Shi H, Zhao Q, Chen YX, and Zhang W. Urinary MicroRNA-30c-5p and MicroRNA-192-5p as potential biomarkers of ischemia-reperfusion-induced kidney injury. Exp Biol Med (Maywood). 242: 657–667. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou X, Qu Z, Zhu C, Lin Z, Huo Y, Wang X, Wang J, and Li B. Identification of urinary microRNA biomarkers for detection of gentamicin-induced acute kidney injury in rats. Regul Toxicol Pharmacol. 78: 78–84. 2016. [DOI] [PubMed] [Google Scholar]
- 84.Yuan J, Benway CJ, Bagley J, and Iacomini J. MicroRNA-494 promotes cyclosporine-induced nephrotoxicity and epithelial to mesenchymal transition by inhibiting PTEN. Am J Transplant. 15: 1682–1691. 2015. [DOI] [PubMed] [Google Scholar]
- 85.Eissa S, Matboli M, and Bekhet MM. Clinical verification of a novel urinary microRNA panal: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed Pharmacother. 83: 92–99. 2016. [DOI] [PubMed] [Google Scholar]
- 86.Eissa S, Matboli M, Aboushahba R, Bekhet MM, and Soliman Y. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J Diabetes Complications. 30: 1585–1592. 2016. [DOI] [PubMed] [Google Scholar]
- 87.Delić D, Eisele C, Schmid R, Baum P, Wiech F, Gerl M, Zimdahl H, Pullen SS, and Urquhart R. Urinary exosomal miRNA signature in type ii diabetic nephropathy patients. PLoS One. 11: e0150154 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perez-Hernandez J, Forner MJ, Pinto C, Chaves FJ, Cortes R, and Redon J. Increased urinary exosomal microRNAs in patients with systemic lupus erythematosus. PLoS One. 10: e0138618 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szeto CC, Ching-Ha KB, Ka-Bik L, Mac-Moune LF, Cheung-Lung CP, Gang W, Kai-Ming C, and Kam-Tao LP. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Dis Markers. 33: 137–144. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Min QH, Chen XM, Zou YQ, Zhang J, Li J, Wang Y, Li SQ, Gao QF, Sun F, Liu J, Xu YM, Lin J, Huang LF, Huang B, and Wang XZ. Differential expression of urinary exosomal microRNAs in IgA nephropathy. J Clin Lab Anal. 2017: e22226 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang Z, Zhang Y, Zhou J, and Zhang Y. Urinary exosomal miR-193a can be a potential biomarker for the diagnosis of primary focal segmental glomerulosclerosis in children. BioMed Res Int. 2017: 7298160 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muralidharan J, Ramezani A, Hubal M, Knoblach S, Shrivastav S, Karandish S, Scott R, Maxwell N, Ozturk S, Beddhu S, Kopp JB, and Raj DS. Extracellular microRNA signature in chronic kidney disease. Am J Physiol Renal Physiol. 312: F982–F991. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mohan A, Singh RS, Kumari M, Garg D, Upadhyay A, Ecelbarger CM, Tripathy S, and Tiwari S. Urinary exosomal microRNA-451-5p is a potential early biomarker of diabetic nephropathy in rats. PLoS One. 11: e0154055 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cardenas-Gonzalez M, Srivastava A, Pavkovic M, Bijol V, Rennke HG, Stillman IE, Zhang X, Parikh S, Rovin BH, Afkarian M, de Boer IH, Himmelfarb J, Waikar SS, and Vaidya VS. Identification, confirmation, and replication of novel urinary microRNA biomarkers in lupus nephritis and diabetic nephropathy. Clin Chem. 63: 1515–1526. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Solé C, Cortés-Hernández J, Felip ML, Vidal M, and Ordi-Ros J. miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol Dial Transplant. 30: 1488–1496. 2015. [DOI] [PubMed] [Google Scholar]
- 96.Lv LL, Cao YH, Ni HF, Xu M, Liu D, Liu H, Chen PS, and Liu BC. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am J Physiol Renal Physiol. 305: F1220–F1227. 2013. [DOI] [PubMed] [Google Scholar]
- 97.Ramezani A, Devaney JM, Cohen S, Wing MR, Scott R, Knoblach S, Singhal R, Howard L, Kopp JB, and Raj DS. Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: a pilot study. Eur J Clin Invest. 45: 394–404. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khurana R, Ranches G, Schafferer S, Lukasser M, Rudnicki M, Mayer G, and Hüttenhofer A. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA. 23: 142–152. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ichii O, Otsuka S, Ohta H, Yabuki A, Horino T, and Kon Y. MicroRNA expression profiling of cat and dog kidneys. Res Vet Sci. 96: 299–303. 2014. [DOI] [PubMed] [Google Scholar]
- 100.Shukla P, Vogl C, Wallner B, Rigler D, Müller M, and Macho-Maschler S. High-throughput mRNA and miRNA profiling of epithelial-mesenchymal transition in MDCK cells. BMC Genomics. 16: 944 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boggs RM, Moody JA, Long CR, Tsai KL, and Murphy KE. Identification, amplification and characterization of miR-17-92 from canine tissue. Gene. 404: 25–30. 2007. [DOI] [PubMed] [Google Scholar]
- 102.Timoneda O, Balcells I, Córdoba S, Castelló A, and Sánchez A. Determination of reference microRNAs for relative quantification in porcine tissues. PLoS One. 7: e44413 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Timoneda O, Balcells I, Núñez JI, Egea R, Vera G, Castelló A, Tomàs A, and Sánchez A. miRNA expression profile analysis in kidney of different porcine breeds. PLoS One. 8: e55402 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Veeranagouda Y, Rival P, Prades C, Mariet C, Léonard JF, Gautier JC, Zhou X, Wang J, Li B, Ozoux ML, and Boitier E. Identification of microRNAs in Macaca fascicularis (cynomolgus monkey) by homology search and experimental validation by small RNA-Seq and RT-qPCR using kidney cortex tissues. PLoS One. 10: e0142708 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jin W, Grant JR, Stothard P, Moore SS, and Guan LL. Characterization of bovine miRNAs by sequencing and bioinformatics analysis. BMC Mol Biol. 10: 90 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ichii O, Ohta H, Horino T, Nakamura T, Hosotani M, Mizoguchi T, Morishita K, Nakamura K, Hoshino Y, Takagi S, Sasaki N, Takiguchi M, Sato R, Oyamada K, and Kon Y. Urinary exosome-derived microRNAs reflecting the changes of renal function and histopathology in dogs. Sci Rep. 7: 40340 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Veeranagouda Y, Léonard JF, Gautier JC, and Boitier E. Next-Generation Sequencing to Investigate Urinary microRNAs from Macaca fascicularis (Cynomolgus Monkey). Methods Mol Biol. 1641: 349–378. 2017. [DOI] [PubMed] [Google Scholar]