Abstract
In vitro-cultured 3D structures called organoids have become important tools for biological research, but there is little information concerning simple and efficient methods to evaluate organoid morphology. To address this issue, we attempted to establish a simple method by applying conventional histopathology that enables observation of multiple organoids on a single cross section, maintains good morphology, and is applicable to various histopathological stains. By centrifugation in unsolidified agarose solution, we were able to accumulate the organoids onto a single plane. The morphology was well preserved, and the various morphological types and sizes of organoid structures were identified. This method was also applicable for special staining, immunohistochemistry, and immunofluorescence staining. This method makes it possible to utilize the advantages of conventional pathological methods when studying organoids.
Keywords: organoid, pathological method, morphology, single cross section
In vitro culture models play an important role in the field of biological research. Conventional in vitro culture methods incorporate monolayers or suspensions of a uniform type of cell, but with the recent progress of in vitro culture methods, we are now able to culture 3D structures called organoids that consist of multiple cell types and can self-organize to mimic specific organs and tissues1,2,3. Organoids constructed from human cells resemble human organs and tissue to a certain extent, so they have been utilized to model organogenesis, organ function, disease, and drug response, including toxicity3, 4. They are also anticipated to contribute to cell or organ replacement strategies in the clinic3, 4.
Defining characteristics of an organoid are as follows: 1) they consist of multiple organ-specific cell types, 2) they show some organ-specific functionality, and 3) they are grouped together and have a spatial structure that is similar to an organ4. Thus morphological evaluation is thought to be necessary to identify the cell types and structures and furthermore the modifications caused by time course, disease, or drug administration; however there is little information available concerning simple and efficient methods to evaluate organoid morphology. To address this issue, we attempted to establish a method by applying conventional histopathology. For precise morphological evaluation, we deemed it necessary to observe multiple organoids on a single cross section to obtain a comprehensive view of the morphological structures, to maintain good morphology that enables detailed observation, and also to apply various histopathological stains such as special stains, immunohistochemistry, and immunofluorescence for various analyses. In the current report, we evaluated our method from these perspectives.
Organoids can be formed from tissue stem cells of a variety of organs5. Additionally, it has been reported that cancer organoids can be derived from cancer stem cells6. For the current study, we used a human colon cancer stem cell (CSC) line derived from PLR123, which was previously described7, and conducted an organoid culture according to the method of Sato et al.8 Briefly, 2D-cultured PLR123 CSCs were dissociated with accutase (Innovative Cell Technologies, San Diego, CA, USA) and suspended in a culture medium (designated organoid medium) consisted of Advanced DMEM/F12 supplemented with penicillin/streptomycin, 10 mmol/l HEPES, 2 mmol/l Glutamax I, N-2 Supplement, B-27 Supplement (all from Thermo Fisher Scientific Inc., Waltham, MA, USA), and 1 mmol/l N-acetyl cysteine (Sigma-Aldrich, St. Louis, MO, USA)8 at a concentration of 2 × 106 cells/ml. Then Matrigel (Growth Factor Reduced Matrigel Matrix, Phenol Red-free, Corning, NY, USA) was applied to a final concentration of 5 × 103 cells/ml. Next 10 µl of the cell suspension was instilled into each well of a 96 well plate and polymerized at 37°C. Finally, 100 µl of organoid medium was applied, and the plates were incubated at 37°C and 5% CO2 for culture.
For the establishment of a pathological method, we considered a method for observation of multiple organoids on a single cross section. The organoids were distributed diffusely within the Matrigel dome, so in order to concentrate the structures, we examined a method of removing the Matrigel. The Matrigel was mechanically dissociated by pipetting after fixation with 4% paraformaldehyde (PFA, 4% PFA Phosphate Buffer Solution, Nacalai Tesque, Inc., Kyoto, Japan) and examined grossly to determine whether the Matrigel was sufficiently removed.
Next, we attempted to accumulate the organoids onto a single plane by centrifugation in unsolidified agarose solutions. We examined the agarose type, concentration, and material of a mold for the agarose. Then we collected organoids and embedded the samples into paraffin by the AMeX method9, 10. Hematoxylin and eosin-stained (HE) sections were prepared by a routine method and read under a light microscope at each time point to judge whether there was a sufficient number of organoid structures and also to determine the preservation of morphology.
Finally, various stains were examined to determine the potential for the application of a broad range of histopathological methods. For special staining, an Alcian blue-periodic acid-Schiff reaction (AB-PAS) method was tested. Briefly, 1% Alcian blue solution (pH2.5, Muto Pure Chemicals Co., Ltd., Bunkyo, Tokyo, Japan) was applied according to the instructions of the manufacturer. Then the slides were serially treated with 0.5% sodium periodate (Orthoperiodic Acid, Wako Pure Chemical Industries, Ltd., Osaka, Japan) solution, Schiff’s solution (Muto Pure Chemicals Co., Ltd.), and aqueous sodium hydrogen sulfite (Wako Pure Chemical Industries, Ltd.). For immunohistochemistry, a polymer method was tested. Briefly, an anti-human villin antibody (Clone 1D2 C3, Agilent, Santa Clara, CA, USA) and an anti-human Ki-67 antibody (Clone MIB-1, Agilent) were applied as the primary antibodies after antigen retrieval in Target Retrieval Solution (Agilent). A labeled polymer reagent (EnVision+ Single Reagents, HRP. Mouse, Agilent) was applied as the secondary antibody, and the reaction was visualized with a 0.02% 3,3′-diaminobenzidine (Wako Pure Chemical Industries, Osaka, Japan) solution. Isotype-matched control antibodies were applied as negative controls for the primary antibodies. The slides were counterstained with hematoxylin and read under a light microscope. Immunofluorescence was also tested using the Tyramide Signal AmplificationTM method. Briefly, an anti-human LGR5 antibody (Clone 2U2E-2)7 was applied as the primary antibody after antigen retrieval in Target Retrieval Solution (Agilent). Then the sections were incubated with a labeled polymer reagent (EnVision+ Single Reagents, HRP. Mouse, Agilent) as the secondary antibody, and the reaction was visualized by Alexa Fluor 488-labeled tyramide (Thermo Fisher Scientific Inc.). The slides were read under a Nikon A1+ confocal microscope (Nikon Corporation, Tokyo, Japan) according to the method described by Yamazaki et al.11.
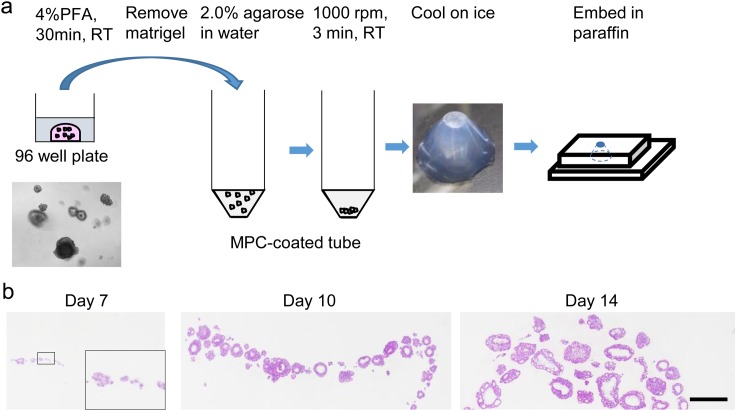
Based on the above examinations, the optimal method for observation of multiple organoids on a single cross section was determined as described in Fig. 1a. Briefly, the organoids were fixed in 4% PFA at room temperature for 30 minutes, dissociated from Matrigel, washed with PBS supplemented with 0.2% bovine serum albumin and 0.1% NaN3, and suspended in agarose. Agarose, Type IX (Sigma-Aldrich, St. Louis, MO, USA) was suited for this method at 2.0%. Then the samples were centrifuged at 1,000 rpm for 3 minutes at room temperature in 50 ml 2-methacryloyloxyethyl phosphorylcholine polymer-coated tubes (Sarstedt AG & Co., Nümbrecht, Germany), and the samples were cooled on ice, removed from the tubes, and embedded into paraffin by the AMeX method. Next to determine whether there was a limit in size for application of this method, HE-stained slides from culture days 7 (small organoids only), 10 (small to mid-sized organoids), and 14 (mainly large organoids) were examined. There were multiple organoids on the same cross section at all time points, which shows that the method can be applied to organoids of various sizes (Fig. 1b).
Fig. 1.
Sample preparation. a: Scheme for sample preparation of organoids. b: Images of slides prepared by the method described in a. A higher magnification is shown for Day 7 in the insert. HE stain. Bar = 500 µm (insert, 120 µm).
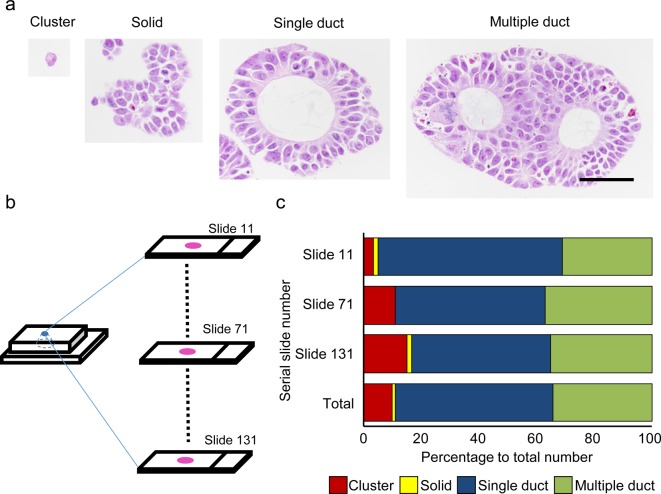
To consider the maintenance of good morphology, we examined organoids at culture day 10. We were able to morphologically identify 4 types of organoid structures: single cells or small clusters of cells consisting of no more than 5 cells (cluster), larger structures consisting of more than 5 cells with unclear polarity and no duct formation (solid), and single (single duct) or multiple (multiple duct) ductal structures (Fig. 2a). From this result, we judged that the morphology was sufficient for detailed morphological observation; however, the mixed population of organoids raised the question of the difference in the level of sedimentation according to the size or shape of the organoid in a single sample. This was thought to be an issue because it would be difficult to determine whether a single cross section would contain all representative structures. To clarify this matter, we examined 3 different cross sections from different levels of a single sample and determined the ratio of each type of structure (Fig. 2b and c). As a result, we found a slight difference in ratio, but we judged that it was largely consistent throughout all 3 levels (Fig. 2c). Thus the method is applicable for samples containing organoids of various shapes and sizes. If an accurate structure count is necessary, examination at multiple levels would be recommended.
Fig. 2.
Morphology of the organoids at day 10. a: The organoids were classified into 4 morphological types. HE stain. Bar = 50 µm. b: Scheme for examining the difference in sedimentation levels. Serial sections were prepared, and slides from the surface (slide 11), middle (slide 71), and deep (slide 131) areas of the block were examined. c: Results of the examination for the difference in sedimentation levels.
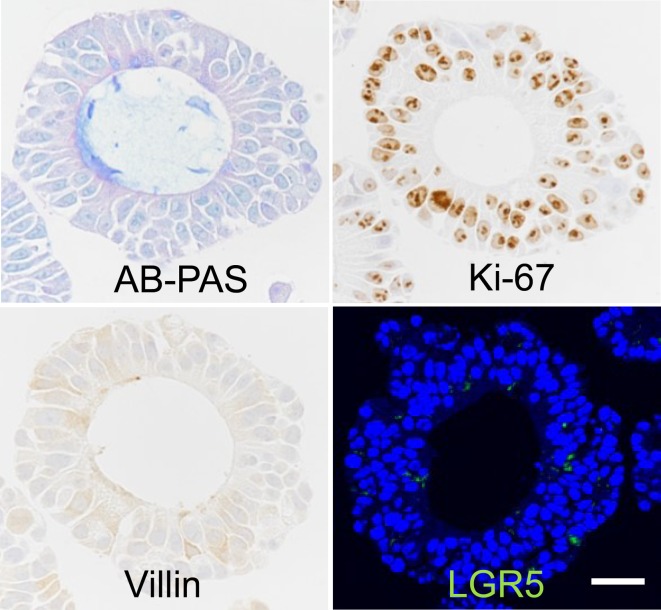
For the histopathological methodology, we evaluated each type of stain in organoids collected at culture day 10. With the AB-PAS method, a substance retained within the ductal lumen that was basophilic with HE stains was stained blue, and a similar basophilic substance seen in the cytoplasm of tumor cells was stained a bluish purple color. Thus we were able to detect mucin production in the organoids (Fig. 3). With immunohistochemistry, villin was found to be expressed in the cytoplasm and apical membrane of some of the tumor cells (Fig. 3). Ki-67 expression was localized in the nucleus of tumor cells, and a majority of the cells were positive (Fig. 3). With immunofluorescence staining of LGR5, there was a slight granular signal in the cytoplasm of some of the tumor cells (Fig. 3). From these results, we considered that there was positive staining that is consistent with the features of each molecule2, 7. Thus we showed the potential of applying various histopathological methods.
Fig. 3.
Application of various histopathological methods. Bar = 25 µm.
Recently, there has been a large amount of progress in microscopic technologies that involves devices such as high-efficiency confocal microscopy, light sheet fluorescence microscopy, and multi-photon microscopy, and this has contributed to the development of 3D imaging of biological organisms12, 13. Information from 3D imaging obtained by these novel methods can be valuable when analyzing organoids, because they are complex structures and precise observation of cells and molecules in their non-disrupted context is required13, 14.
These methods are suited for evaluating specific cells and molecules, but conventional pathological methods exert their power when attempting to obtain a comprehensive view of the total morphological picture. Another advantage of well-established conventional methods is that they are consistent and efficient because they are based on historical accumulation of knowledge. Our current method enables the utilization of these advantages of conventional pathological methods when studying organoids, and we anticipate that by combining this method with 3D imaging, the accuracy and efficiency of morphological analysis will improve.
Acknowledgments
We would like to thank Ms. Kumiko Nakajima and Ms. Akiko Hasebe of Forerunner Pharma Research Co., Ltd. in assisting the organoid cultures, Ms. Yayoi Takai of Chugai Research Institute for Medical Science, Inc. for assisting in histopathological procedures, and Dr. Sachiko Shioda and Ms. Kaori Nishihara of Chugai Pharmaceutical Co., Ltd. for assisting in the analysis of structure counts.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare that they have no competing interests.
References
- 1.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, and Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 3: 519–532. 2008. [DOI] [PubMed] [Google Scholar]
- 2.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459: 262–265. 2009. [DOI] [PubMed] [Google Scholar]
- 3.Willyard C. The boom in mini stomachs, brains, breasts, kidneys and more. Nature. 523: 520–522. 2015. [DOI] [PubMed] [Google Scholar]
- 4.Lancaster MA, and Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 345: 1247125 2014. [DOI] [PubMed] [Google Scholar]
- 5.Drost J, and Clevers H. Translational applications of adult stem cell-derived organoids. Development. 144: 968–975. 2017. [DOI] [PubMed] [Google Scholar]
- 6.Ohta Y, and Sato T. Intestinal tumor in a dish. Front Med (Lausanne). 1: 14 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi S, Yamada-Okabe H, Suzuki M, Natori O, Kato A, Matsubara K, Jau Chen Y, Yamazaki M, Funahashi S, Yoshida K, Hashimoto E, Watanabe Y, Mutoh H, Ashihara M, Kato C, Watanabe T, Yoshikubo T, Tamaoki N, Ochiya T, Kuroda M, Levine AJ, and Yamazaki T. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells. 30: 2631–2644. 2012. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, and Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141: 1762–1772. 2011. [DOI] [PubMed] [Google Scholar]
- 9.Sato Y, Mukai K, Watanabe S, Goto M, and Shimosato Y. The AMeX method. A simplified technique of tissue processing and paraffin embedding with improved preservation of antigens for immunostaining. Am J Pathol. 125: 431–435. 1986. [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki M, Katsuyama K, Adachi K, Ogawa Y, Yorozu K, Fujii E, Misawa Y, and Sugimoto T. Combination of fixation using PLP fixative and embedding in paraffin by the AMeX method is useful for histochemical studies in assessment of immunotoxicity. J Toxicol Sci. 27: 165–172. 2002. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki M, Kato A, Zaitsu Y, Watanabe T, Iimori M, Funahashi S, Kitao H, Saeki H, Oki E, and Suzuki M. Intensive immunofluorescence staining methods for low expression protein: detection of intestinal stem cell marker LGR5. Acta Histochem Cytochem. 48: 159–164. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carneiro K, de Brito JM, and Rossi MI. Development by three-dimensional approaches and four-dimensional imaging: to the knowledge frontier and beyond. Birth Defects Res C Embryo Today. 105: 1–8. 2015. [DOI] [PubMed] [Google Scholar]
- 13.Susaki EA, and Ueda HR. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem Biol. 23: 137–157. 2016. [DOI] [PubMed] [Google Scholar]
- 14.Kaushik G, Leijten J, and Khademhosseini A. Concise Review: Organ Engineering: Design, Technology, and Integration. Stem Cells. 35: 51–60. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]