Abstract
Protein structure refinement remains a challenging yet important problem as it has the potential to bring already accurate template-based models to near-native resolution. Refinement based on molecular dynamics simulations has been a highly promising approach and the performance of MD-based refinement in the Feig group during CASP12 is described here. During CASP12, sampling was extended well into the microsecond scale, an improved force field was applied, and new protocol variations were tested. Progress over previous rounds of CASP was found to be limited which is analyzed in terms of the quality of the initial models and dependency on the amount of sampling and refinement protocol variations. As current MD-based refinement protocols appear to be reaching a plateau, detailed analysis is presented to provide new insight into the major challenges towards more extensive structure refinement, focusing in particular on sampling with and without restraints.
Keywords: protein structure prediction, structure refinement, molecular dynamics simulation, CASP, Markov state models, homology models
INTRODUCTION
Computational protein structure prediction has become a valuable tool in structural biology1. Template-based modeling is now applicable to ever more folds as the number of structures in the Protein Data Bank (PDB) continues to grow rapidly, especially because of advances with cryo-electron microscopy (Cryo-EM)2 and structure determination via X-ray free electron lasers (XFEL)3. As a result, the chance of finding the structure of a close homolog for a given sequence is increasing so that accurate protein structure prediction is routinely possible4. Moreover, modeling based on co-evolutionary information allows high-resolution modeling even in the absence of a homologous structure5,6. This has been enabled by next-generation genome sequencing (NGS), which rapidly increased the number of known sequences for homologous proteins in different organisms as the basis for predicting residue-residue contacts with high accuracy7.
Complementary to the advances outlined above, protein structure refinement methods have been increasingly successful in further improving initial knowledge-based predictions via ab initio techniques8. Evidence of significant protein structure refinement has first emerged around 2010, at CASP99. In CASP10, refinement based on molecular dynamics (MD) led to a significant step forward after introducing the idea of ensemble averaging rather than attempting to select a single snapshot from a generated structural ensemble10–12. The progress with MD-based methods has also been catalyzed by continuous improvements in force fields and ability to sample extensively11. MD-based refinement so far is able to provide moderate and consistent structure improvements13, and recent protocols have been optimized to limit the computational costs by taking advantage of GPU computing14. MD-based refinement has also contributed to significant improvements in local structure quality such as correct stereochemistry and avoidance of clashes, which is almost always possible even if the global structure cannot be improved15. A number of alternate refinement protocols have also been proposed: In CASP11, Della Corte et al. devised a method where homologous structure information was utilized to smooth the potential energy landscape and lower the energy barriers thereby enhancing sampling during MD simulations16. Lee et al. and Park et al. proposed methods that reconstruct unreliable regions such as loops and termini, followed by global relaxation17,18. However, all of the refinement methods available to date, are still limited to relatively localized structural changes and perform best when the initial model is already relatively close to the native state. It remains very challenging to consistently improve models with significant errors19.
In this article, further tests of MD-based protein structure refinement methods during CASP12 are described. Commonly, weak restraints have to be applied for MD-based refinement to succeed12,20, which limits the extent of possible refinement. Since the protein force field has been improved further21 and GPGPU computing has become more widely available, MD simulations were carried out over longer time scales with weaker restraints during CASP12. The hope was to be able to sample more broadly, and, ultimately, refine structures more extensively. Other ideas that were explored during CASP12 involved iterative refinement along the initial direction of refinement. However, while consistent moderate refinement was again possible, we could not significantly expand the extent of refinement indicating that MD-based refinement methods using current protocols may have reached a plateau where simply running longer simulations is not offering further advantages. So, a main focus of this article is to look back and summarize the lessons we have learned until now and discuss where the key barriers are that need to be overcome to move forward.
METHODS
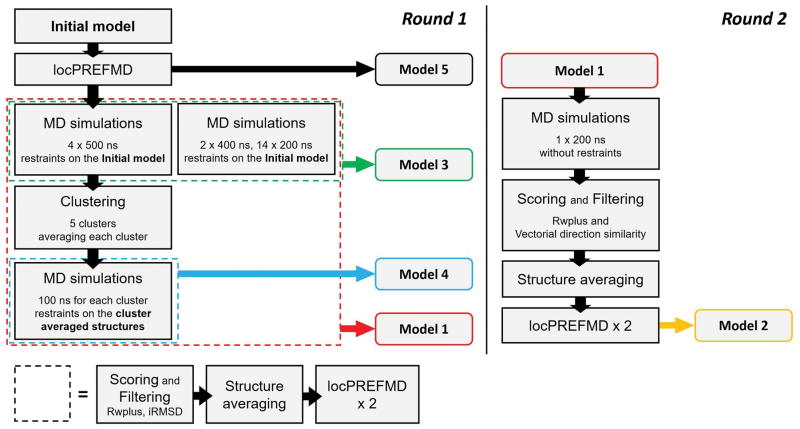
The general approach to protein structure refinement during CASP12 followed our previous protocol that involved long, weakly restrained MD simulations to generate structural ensembles. From the ensembles, a subset of structures was subsequently selected and averaged before finishing with detailed refinement of the local stereochemistry using locPREFMD15. Variants of our previous protocol were tested during CASP12. In the CASP12 protocol, shown as a flow chart in Figure 1, initial models were first subjected to locPREFMD15 to improve the stereochemical quality and obtain better starting structures for the MD simulations. The resulting initial models were submitted as ‘Model 5’ during CASP12 as the most conservative attempt at structure refinement. Afterwards, up to two rounds of MD-based refinement were carried out. Generally, the same scoring and filtering scheme followed by structure averaging was used as described previously and applied in CASP1113 on different ensembles of structures generated from MD trajectories under different conditions (see below). Before submitting final models, locPREFMD was applied twice after ensemble averaging and. depending on which MD ensemble was used, the resulting structures were submitted as Models “1”, “2”, “3”, or “4”, respectively.
Figure 1.
Refinement protocol applied by the FEIG group in CASP12. Each colored box with a dashed line depicts a structural ensemble for which a common protocol of scoring, filtering, averaging, and final local refinement was applied as depicted at the bottom.
The primary set of MD simulations in the first round consisted of four runs at 500 ns, two runs at 400 ns, and 14 runs at 200 ns using weak harmonic restraints applied to Cα atoms with respect to the initial structure (force constant: 0.025 kcal/mol/Å2) for a total of 5.6 μs of sampling. This was about five times the amount of sampling applied during CASP1113. The combination of runs at different lengths was predicated by the availability of GPU resources and the time limit for submitting predictions during CASP. While this ambitious amount of sampling could be reached for many targets within the CASP time frame of a few weeks, some targets were too large and simulation lengths had to be reduced in those cases. Each simulation was carried out in explicit solvent starting with randomly assigned initial momenta. The latest CHARMM force field, CHARMM36m21, was used to take advantage of further improvements in sampling backbone torsions, especially in less-structure segments such as loops. Each protein was solvated in a periodic cubic box with at least a 9 Å solvent buffer to the box edge. The TIP3 water model22 as implemented in CHARMM was used and all systems were neutralized by adding either sodium (Na+) or chloride (Cl−) counter ions, as appropriate. A 10 Å cutoff with switching between 8 Å and 10 Å was applied to the Lennard-Jones potential and the direct part of the electrostatic potential. Particle-mesh Ewald summation was used to evaluate the full electrostatic potential23. Bonds involving hydrogen atoms were kept rigid using holonomic constraints. Each system was prepared by minimization and subsequent heating to 298 K. Langevin dynamics was performed under NVT condition at 298 K with a 2 fs time step and a friction coefficient of 0.01/ps. The ensemble resulting from this set of simulations was filtered and averaged to result in the structure submitted as “Model 3”.
To further enhance the initial conformational ensembles, 200,000 conformations were extracted from the first four 500 ns MD simulations and subsequently clustered into five clusters using the k-center algorithm implemented in MSMBuilder24 using the differences in the pair-wise Cα distance matrix as the distance measure. For each cluster, an averaged structure was generated from which an additional 100 ns of MD simulations were performed in the same manner as explained above except that restraints were applied with respect to the cluster-averaged structure using a reduced force constant of 0.005 kcal/mol/Å2. The ensemble from the five cluster-initiated simulations was filtered and averaged to result in the structure submitted as “Model 4”. The combined set of structures resulting from the initial simulations plus the additional simulations after clustering was used to generate the structure submitted as “Model 1”. In some cases, there was not enough time to run the additional set of simulations after clustering. In this case, “Model 3” was submitted also as “Model 1”.
A second round of refinement began with “Model 1” and involved an additional MD simulation over 200 ns without restraints. From the resulting ensemble, structures were selected that advanced further along the direction of the initial refinement with the idea that those changes would bring the structure closer to the native state. The direction of the initial refinement was determined by comparing “Model 1” with the initial model and the conformations extracted from the second round of MD were accordingly scored using both RWplus25 and the vectorial direction similarity measure Sk calculated for a structure k based on Eqs 1 and 2.
| (1) |
| (2) |
where the dot products of unit vectors of Cα deviation directions between the refined model and a given structure k were averaged over all NCα atoms. The Cα deviation direction was defined as the averaged vector between a given Cα and its two adjacent Cα atoms in a structure k from the initial model after overall least-squares structural superposition according to Eq. 2. virefined was calculated in the same manner.
Only structures generated during round two with Sk > 0.4 and , where r denotes the RWplus score, were selected for structure averaging, to be submitted as “Model 2”. If no such structures were found, i.e. the additional second round sampling did not advance further along the initial direction of refinement, “Model 1” was submitted also as “Model 2”.
The MD production runs were conducted on GPUs using CHARMM26 with OpenMM27 via API integration, except for the second-round single 200 ns simulation that was run using NAMD28. Other MD simulations for equilibration, structure averaging, and locPREFMD were carried out using CHARMM. Structure preparation, selection, and analysis tasks relied on the MMTSB Tool Set29.
RESULTS AND DISCUSSION
Overall performance in CASP12
The overall results during CASP12 with the refinement protocol shown in Figure 1 are given in Table 1. For one target (TR879), the refinement protocol was applied incorrectly and the initial structure disintegrated as a result. Because the results for this target do not reflect the described protocol, the results were excluded from further analysis. Based on all other targets, GDT-HA scores were improved by 1.6 units, and four out of 36 targets could be refined by more than 5 GDT-HA units. Refinement was highly consistent as structures were refined for 30 out of 36 targets (83%). The performance was overall similar to previous years, but the extent of refinement was actually a bit less than in CASP1113 and CASP1011 despite the additional sampling and force field improvements.
Table 1.
Overall results for CASP12 refinement targets
| Target | Improvements for model 1 | ΔGDT-HA for each modela | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| ΔGDT-HA | −ΔRMSD (Å) | ΔGDC-SC | MolProbityb | 1 | 2 | 3 | 4 | 5 | |
| TR862 | 2.96 | 0.06 | −0.85 | 0.51 | 2.96 | 3.50 | 2.96 | 1.61 | −0.81 |
| TR866-D1 | 0.72 | 0.18 | −1.06 | 6.00 | 0.72 | 2.64 | 1.68 | 2.89 | −1.69 |
| TR868-D1 | −1.91 | −0.02 | −2.41 | 0.83 | −1.91 | −2.14 | −2.14 | −4.28 | −0.24 |
| TR869 | 0.00 | −0.12 | −0.10 | 0.80 | 0.00 | −0.49 | 0.00 | −1.92 | −0.24 |
| TR870-D1 | 0.69 | −0.10 | −2.20 | 0.97 | 0.69 | 0.00 | 1.38 | 0.91 | 0.46 |
| TR872 | −1.41 | −0.04 | −0.07 | 0.57 | −1.41 | −0.56 | −1.13 | 3.42 | 0.57 |
| TR874 | 1.35 | −0.06 | 0.33 | 0.50 | 1.35 | 1.35 | 1.57 | 3.60 | 0.22 |
| TR875 | 1.72 | −0.02 | 1.66 | 0.60 | 1.72 | 1.08 | 2.37 | 2.37 | −0.21 |
| TR876-D1 | 0.74 | 0.04 | −0.29 | 0.55 | 0.74 | 0.74 | 0.49 | 0.98 | 0.00 |
| TR877 | 2.11 | 0.02 | −1.63 | 0.53 | 2.11 | 0.17 | 1.94 | −0.35 | 0.35 |
| TR879 | −18.97 | −0.19 | −16.36 | 2.21 | −18.97 | −18.86 | −18.18 | −36.36 | −1.93 |
| TR881 | 2.35 | 0.01 | 5.80 | 0.85 | 2.35 | 2.72 | 2.72 | 0.74 | 0.50 |
| TR882 | 3.80 | 0.09 | −0.52 | 0.60 | 3.80 | 4.12 | 3.80 | 5.39 | 0.64 |
| TR884 | 5.63 | −0.05 | 0.98 | 0.50 | 5.63 | 5.28 | 5.98 | 4.93 | −0.70 |
| TR885-D1 | 3.13 | 0.05 | 3.03 | 0.50 | 3.13 | 1.69 | 3.13 | 1.69 | 0.97 |
| TR887-D9 | 5.43 | 0.12 | 4.88 | 0.50 | 5.43 | 4.19 | 4.50 | 3.26 | 0.47 |
| TR890 | −0.54 | −0.12 | −1.25 | 0.95 | −0.54 | - | 0.26 | −4.13 | −0.40 |
| TR891 | 0.22 | 0.08 | −1.06 | 0.72 | 0.22 | −2.68 | 0.00 | 1.34 | 0.89 |
| TR893 | 0.00 | −0.03 | −11.62 | 1.42 | 0.00 | - | −1.92 | −1.77 | 0.00 |
| TR894 | 1.85 | 0.03 | 6.16 | 0.50 | 1.85 | - | 1.39 | 8.80 | 1.39 |
| TR594 | 1.13 | −0.05 | 1.01 | 0.50 | 1.13 | - | 0.57 | 2.81 | −0.56 |
| TR694 | 1.24 | −0.05 | 1.78 | 0.85 | - | - | 1.24 | - | −0.38 |
| TR895 | 3.96 | 0.01 | 7.14 | 0.69 | 3.96 | - | 5.42 | 4.17 | 0.42 |
| TR896 | −1.45 | −0.06 | 0.35 | 0.90 | −1.45 | −3.19 | −1.45 | −2.03 | 0.29 |
| TR898 | 2.37 | −0.05 | 0.25 | 0.50 | 2.37 | 2.13 | 0.48 | 0.95 | −0.23 |
| TR901 | −1.01 | −0.04 | 0.78 | 0.65 | −1.01 | - | −0.56 | −1.34 | −1.12 |
| TR905 | 0.30 | 0.02 | −2.05 | 1.68 | 0.30 | - | −0.52 | −1.76 | −0.83 |
| TR909 | 1.65 | 0.00 | 1.14 | 1.47 | 1.65 | - | 1.87 | −2.18 | −0.15 |
| TR910 | 1.87 | 0.02 | −1.74 | 0.69 | - | - | 1.87 | - | −0.37 |
| TR912 | 1.21 | 0.07 | 1.39 | 0.85 | 1.21 | - | 2.53 | 5.43 | 0.84 |
| TR913 | 2.29 | 0.01 | 1.56 | 1.11 | 2.29 | - | 2.51 | −11.32 | 0.37 |
| TR917 | 5.43 | 0.08 | 5.88 | 0.50 | 5.43 | - | 5.37 | −9.84 | −0.06 |
| TR920 | 0.79 | 0.04 | 1.21 | 0.50 | - | - | 0.79 | - | −0.35 |
| TR520 | 1.40 | 0.05 | −1.37 | 0.98 | - | - | 1.40 | - | −0.15 |
| TR921 | 3.98 | −0.04 | 1.12 | 0.98 | 3.98 | 4.53 | 3.44 | 5.79 | 0.54 |
| TR922-D1 | 3.23 | 0.00 | −0.61 | 0.71 | 3.23 | 1.21 | 2.83 | 3.23 | 1.61 |
| TR928 | −2.79 | −0.15 | 0.45 | 1.56 | - | - | −2.79 | - | −1.32 |
| TR942 | 2.72 | −0.09 | 3.11 | 0.66 | - | - | 2.72 | - | 0.33 |
| TR944 | 0.79 | 0.01 | 0.90 | 0.85 | - | - | 0.79 | - | 0.19 |
| TR945 | 0.53 | −0.02 | 0.05 | 0.94 | - | - | 0.53 | - | −0.27 |
| TR947 | 2.29 | 0.03 | 0.67 | 0.78 | - | - | 2.29 | - | 0.44 |
| TR948 | 5.38 | −0.07 | 0.94 | 0.71 | 5.38 | - | 5.21 | 0.51 | 2.52 |
|
| |||||||||
| Average (full)c | 1.78 | 0.00 | 0.38 | 0.93 | 1.78 | 1.31 | 1.73 | 1.73 | 0.17 |
| Average (all)d | 1.61 | 0.00 | 0.58 | 0.91 | |||||
Models submitted as “Model 1” are shown in bold characters.
MolProbity scores for refined models;
ΔGDT-HA averages for 20 targets shown in bold for which the full protocol was applied;
refinement protocol was not applied correctly for this target and it was excluded from further analysis. If a method was not applied to a target, the corresponding value is shown as a hyphen (-)
The new protocols tested in CASP12 did not provide significant advantages. Although “Model 1” submissions (that included the snapshots from the enhanced sampling following clustering, see above) seemed to be slightly better than just following the CASP11 protocol, statistical analysis indicates that none of the MD-refinement methods were significantly different from each other (based on p-values > 0.05 from the paired t-test analysis on the common targets). An interesting observation is that the refined models using just the snapshots from the enhanced sampling following clustering (“Model 4”) showed a greater variation with some models much better and others much worse than the “Model 1” structures. Pearson’s correlation coefficients for GDT-HA improvements between “Model 1” and “Model 3” and “Model 4” are 0.96 and 0.68, respectively. This reflects broader sampling as a result of the weaker restraints and the application of restraints with respect to structures generated in the first round of sampling. However, overall, the extent of refinement did not improve as structures moved away from the native as much as they came closer.
The second round of the refinement protocol produced models only for about half of the targets, either because there was not enough time to run additional sampling after the first round was complete or because no structures were generated along the direction of the initial refinement (see Methods). In general, where available, models after the second round were refined to a similar extent as after the first round. Although restraints were not used in the second round sampling, the resulting models did not drift away from the native structure except for two targets (TR891 and TR922-D1, where more than 2 GDT-HA units were lost during the second round). This confirms an earlier conclusion11 that after initial refinement, models are in a deep energy minimum from which escape and additional refinement are very challenging.
Finally, models submitted as “Model 5” for which only the local stereochemistry was refined with locPREFMD did not change much in terms of GDT-HA and RMSD scores. However, although locPREFMD does not target global structure refinement, there were still small improvements in GDT-HA scores suggesting that just the improvement of local structure may also slightly improve the global structure.
Sampling vs. refinement
Under the assumption that the force field is good enough to distinguish the native state as the global free energy minimum, sampling is the limiting factor for being able to traverse the energy landscape until the native state is found. While it remains unclear how much sampling is exactly needed to reach the native state from a nearby initial model on a given landscape, the general expectation is that more sampling (more trajectories as well as longer trajectories) would improve the chances for refinement. In CASP12, we extended the simulation time to more than 5.6 μs per target. At the same time, the restraint force constant was reduced by half and we expected to sample more diverse structures on non-canonical regions (i.e., loops) by introducing the CHARMM36m force field.
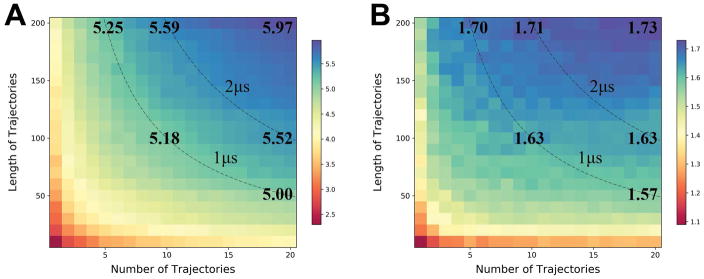
As already mentioned above, the more extended sampling did not result in better refinement compared to previous rounds of CASP, but we performed post-analysis based on simulation subsets to determine how the simulation length affected the results just within CASP12. Figure 2 illustrates how GDT-HA improvements depend on the amount of sampling. This analysis is based on averages over 24 targets for which we have generated at least 20 trajectories over 200 ns. Generally, increased sampling resulted in higher GDT scores, consistent with expectations, and using the full 4 μs resulted in the maximum improvements. However, the GDT-HA scores for the top 5% percentile of the distribution was almost converged after 1–2 μs of sampling, and additional sampling did not make much of a difference.
Figure 2.
Average improvements in GDT-HA score as a function of simulation time. Improvements in GDT-HA for the best (A) and top 5% percentile (B) of the sampled structure are shown as heat maps. Blue colors indicate the largest improvements in GDT-HA while red colors indicate the least improvement. Contours indicate the same total simulation times of 1 μs and 2 μs. Values in bold are given for: 5 × 200ns, 10 × 100ns, 20 × 50ns (on the 1 μs line), 10 × 200ns, 20 × 100ns (on the 2 μs line), and 20 × 100ns (upper right corner).
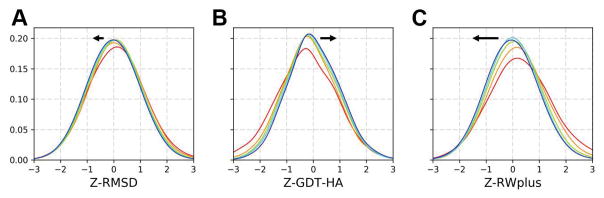
The next question we investigated was to what degree RMSD and GDT-HA metrics as well as the RWplus score improved with increasing simulation time. As shown in Figure 3, average RMSD values did not change much but the distribution of GDT-HA scores shifted to larger values and RWplus scores moved to lower values up until 50–100 ns, after which there was little change. The significant shift in the RWplus score towards lower energies implies that the interaction within proteins and internal packing were getting better as the simulations progressed. Therefore, it appears that extending simulations beyond 200 ns would not provide much benefits, at least in the presence of positional restraints.
Figure 3.
Z-score distributions for Cα RMSD (A), GDT-HA scores (B), and RWplus scores (C) averaged over 24 targets using 20 trajectories over 200 ns as a function of simulation time (red: 0–10 ns; orange: 10–20 ns; green: 20–50 ns; cyan: 50–100 ns; blue: 100–200 ns). Arrows indicate the direction of change towards longer simulation times.
The optimal balance between the number and length of simulation with the same amount of sampling is another issue. Many short simulations may explore different regions of conformational space based on different initial momenta and are better suited to typical parallel computing resources than a few long simulations. However, it is more difficult to cross significant kinetic barriers with short simulations so that an optimal balance may be a compromise of a moderate number of moderate-length simulations. Figure 2 confirms this as 5 × 200 ns (1 μs in total) or 10 × 200 ns (2 μs in total) provide slightly better results than 20 × 50 ns or 20 × 100 ns. As there is little improvement in GDT-HA and RWplus scores during 100–200 ns (see Figure 3), the optimal sampling strategy seems to be to run as many as possible 200 ns simulations within the CASP12 refinement protocol.
Scoring, filtering, and structure averaging
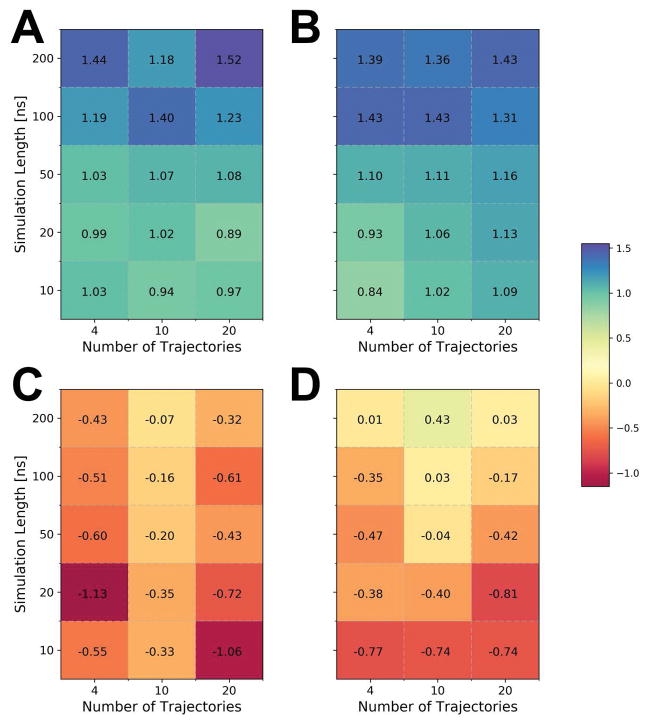
The general approach towards scoring, filtering, and structure averaging was established first during CASP10. In CASP11, DFIRE30 was replaced with RWplus25 and the filtering criteria were slightly adjusted13. In CASP12 the same protocol was applied as in CASP11. Briefly, the filtering step involves the selection of a subset of structures that have both low RWplus scores and are closer to the initial model. In this section, we reassessed whether this scoring, filtering, and averaging protocol is still optimal for the CASP12 test set. We compared refined models with and without filtering and extracted models at the center of a given ensemble based on the lowest Cα RMSD sum to all other structures instead of averaging. The resulting GDT-HA improvements are summarized in Figure 4. It can be seen, that averaging still provides a significant benefit, either with or without filtering, but, surprisingly, the filtering step did not offer a clear advantage anymore. With averaging, the results were similar whether the ensemble was filtered or not. Moreover, when averaging was not applied, the use of the filter actually resulted in worse performance. This suggests that it may be necessary to reassess when and how to filter MD-generated ensembles as sampling has increased, force fields have improved, and the generation of the initial models may have changed (see below) from when we first optimized our refinement protocol.
Figure 4.
Average improvements in GDT-HA scores (see color scale) based on different protocols for generating refined models from MD ensembles as a function of the length and number of MD trajectories. Structures obtained by averaging (A, B) are compared with structures obtained as ensemble centers (C, D), and with (A, C) and without (B, D) ensemble filtering (see text).
Refinement as a function of the initial model
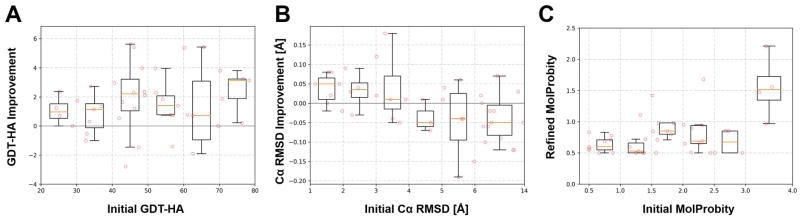
Our finding that refinement was less successful in CASP12 than in previous rounds of CASP despite increased sampling prompted us to examine again13 how the degree of refinement depended on the initial model quality. As shown in Figure 5, we found little correlation of GDT-HA improvements with initial GDT-HA scores, although the highest average improvement was seen for models with initial GDT-HA scores above 70. Improvements in Cα RMSD were more strongly dependent on the initial Cα RMSD. There was a clear trend of structures with lower RMSD values, below 4 Å in Cα RMSD, being improved in terms of RMSD, while structures with larger initial RMSD values became worse in terms of RMSD after refinement. Finally, MolProbity scores were consistently reduced to less than 1.0 in most cases except when the initial MolProbity score was worse than 3.0, although even in those cases, the MolProbity scores were reduced significantly (to about 1.5). This analysis suggests that the reduced amount of refinement seen in CASP12 vs. previous rounds of CASP was probably not because initial models had higher or lower GDT or RMSD values.
Figure 5.
Refinement performance as a function of on initial model quality. GDT-HA improvements are compared to initial GDT-HA scores (A), RMSD improvements are compared to initial RMSD values (B), and MolProbity scores after refinement are shown as a function of initial MolProbity scores (C). Boxplots summarize the results with orange lines inside the boxes indicating median values, while the top and bottom of the boxes represent first and third quartiles. The minimum and maximum values within 1.5 interquartile from the first and third quartiles are indicated as whiskers. Individual data points are overlaid as red circles.
The next question we posed was whether the origin of the initial model played a role. In CASP12, the initial models came from various protein structure prediction servers, but models built by the Lee and Baker groups each contributed 12 initial structures for the 42 refinement targets. Table 2 summarizes the refinement performance as a function of where the initial models came from. Most notably, models originating from the LEE server could only be improved by modest amounts in terms of GDT-HA while more substantial GDT-HA improvements were possible for most other methods. Furthermore, the accuracy of sidechains, measured with the GDC-SC score, actually deteriorated for the LEE models while other models were again improved to different degrees.
Table 2.
Performance dependency on initial model prediction groups
| Server | Na | Initial model quality | Average improvement for model 1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| GDT-HA | RMSD (Å) | GDC-SC | MolProbity | ΔGDT-HA | −ΔRMSD (Å) | ΔGDC-SC | MolProbityb | ||
| Lee | 12 | 50.30 | 5.01 | 29.75 | 1.80 | 0.64 | −0.01 | −1.12 | 0.91 |
| Baker | 12 | 58.58 | 4.13 | 38.69 | 1.01 | 1.96 | 0.04 | 0.60 | 1.12 |
| Bates_BMM | 4 | 41.35 | 5.00 | 18.56 | 2.62 | 2.04 | −0.02 | 4.02 | 0.64 |
| QUARK | 3 | 39.86 | 6.48 | 19.20 | 2.28 | 3.77 | −0.03 | 1.08 | 0.56 |
| Pcons | 3 | 42.72 | 7.49 | 23.29 | 0.85 | 2.13 | 0.02 | 1.69 | 0.52 |
| Otherc | 7 | 43.74 | 7.72 | 25.26 | 2.49 | 1.29 | −0.06 | 0.79 | 1.04 |
Number of targets;
MolProbity scores for refined models;
Less than three targets each came from RaptorX, YASARA, Seok-server, myprotein-me, HHPred1, and BhageerathH-Plus. TR879 (from FFAS-3D) was excluded from the analysis.
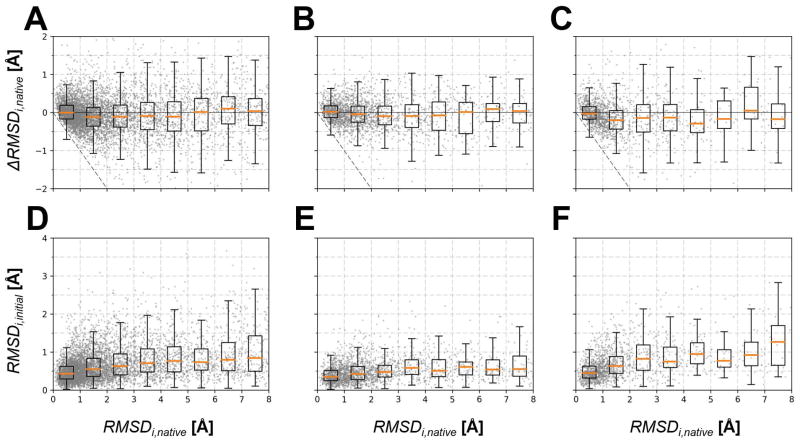
We performed more detailed analysis of the change in per-residue Cα RMSD from the native as well as the per-residue Cα RMSD with respect to the initial model. As shown in Figure 6, most of the improvements were made when the initial Cα RMSD was in the range of 1–5 Å. This suggests that residues with moderate errors are more likely to move toward the native structure, while refinement of substantially deviated residues is more challenging. Residues that were already close to the native (less than 1 Å) did not improve much as there is little room for refinement but these residues also moved less with respect to the initial structure. This means that residues that were essentially correct in the initial model tended to remain at that position during refinement. This would be expected for residues already in the deep global native minimum but residues close to the experimental structure are also more likely part of the highly packed protein core where there is less room for displacement even if the force field energy was not optimal.
Figure 6.
Per-residue change in Cα RMSD from the native, ΔRMSDi,native, (A, B, C) and Cα RMSD from the initial model, RMSDi,initial, (D, E, F) vs. initial Cα RMSD from the native for a given residue i. The analysis is compared for all targets (A, D) and for targets from the LEE (B, E) and BAKER (C, F) groups. Boxplots are drawn in the same manner as in Figure 5. Individual data points are shown as gray dots. Dashed lines indicate perfect refinement which makes Cα deviation to zero.
Comparing this analysis for models from the LEE and BAKER servers, there are noticeable differences especially in the amount of displacement from the initial model. With LEE models, there was much less displacement from the initial model even for residues with high initial RMSD values. This suggests that the LEE models were already in a deep energetic minimum, presumably as a result of refinement using a similar protocol and energy function as what we have applied here. Therefore, our refinement of the LEE models was apparently more akin to a second round refinement. This would explain the much more moderate improvements for the LEE models compared to the refinement of other models and the overall lower success with refinement during CASP12 since the LEE models made up a significant fraction of refinement targets.
Limitations towards further progress
Moderate structure refinement via MD-based sampling is now consistently possible and has apparently even become integrated in standard automated modeling platforms as evidenced by the LEE server models during CASP12. However, we seem to have reached a plateau where further progress is difficult to achieve. An obvious issue is the use of restraints that limits how far structures can deviate from a given initial model and, thereby, how much initial models can be refined towards the native state. The application of restraints stems from early lessons during the application of MD to the structure refinement problem that have taught us that entirely unrestrained simulations from homology models are more likely to move away from the native state than towards it12,20,31 although it is not entirely clear why exactly. The use of restraints during refinement simulations has been the key to achieving consistency in structure refinement, but it is clear that with simulations restraint to the initial model it will not be possible to reach the native state for any but those initial models that are already very close to the native state. Thus, understanding why exactly unrestrained sampling from initial homology models generally fails to reach the native state is the key issue in our opinion for advancing structure refinement to the next level.
In order to gain further insights, we examined one of the CASP12 targets (TR872) in detail. Since the initial model for this target was relatively far from the native state, there was significant room for improvement (initial Cα RMSD and GDT-HA scores were 5.59 Å and 56.8, respectively). During CASP12, we could not improve this model using our MD-based protocol (ΔCα RMSD and ΔGDT-HA score are +0.04 Å and −1.4, respectively) and we therefore chose this target to understand the limitations in our protocol better. This target had errors at both termini and at a β-turn. There was also relatively poor packing of sidechains between β-sheets. We complemented the simulations generated during CASP12 with extensive unrestrained simulations that were started either from the native structure or the initial model given during CASP12. The additional MD simulations involved initially a set of 40 runs over 100 ns each. These simulations were carried out in the same manner as the MD simulations during CASP12 (except for the lack of restraints). The generated conformations were then clustered similarly as during CASP12 (see Methods). From each cluster another round of ten additional simulations over 100 ns were started. This resulted in a total of 22 μs of additional sampling (8 μs started from the native, 14 μs started from the initial model). The resulting conformations, as well as the structures generated during CASP12 with restrained sampling, were then combined and projected onto the two principal components from time-structure independent component analysis32 with a lag time 1 ns and using pair-wise Cα distances as the distance metric.
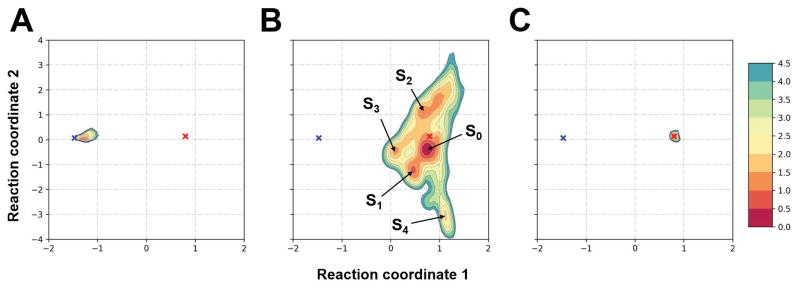
Figure 7 illustrates the conformational sampling for TR872 during our refinement simulations in CASP12 compared with the additional unrestrained simulations started from the native structure and initial model. When started from the native structure, the simulations explore only a relatively narrow conformational space, despite the extensive sampling and a protocol that encourages broad exploration of conformational space. The only significant dynamics is found in the flexible C-terminus and a loop connecting an internal hairpin (see also Fig. 8A). The average structure obtained from the MD simulations remains close to the native state with a GDT-HA score of 79.0 and a Cα RMSD value of 2.24 Å for the average structure. This indicates that the native state coincides with a deep free energy minimum in the MD simulations as expected from the protein folding funnel hypothesis33. While we cannot say for sure based on the still limited sampling whether the native state is indeed at the global free energy minimum, the native state is at least a prominent local minimum with the force field used here from which escape is unlikely if it can be reached during refinement.
Figure 7.
Conformational sampling of TR872 during refinement projected onto the two principal coordinates obtained from time-structure independent component analysis (tICA). Contour plots are drawn for sampling probabilities (as –log P based on probabilities P and shifted to zero at the minimum value). The coordinates for the native and the initial model are indicated by using a blue and red ‘X’, respectively. Sampling distributions for different sets of MD trajectories are shown in different panels: (A) started from the native structure without restraints; (B) started from the initial model without restraints; and (C) started from the initial model with restraints according to our CASP protocol. Representative states (S0–4) are indicated in panel B.
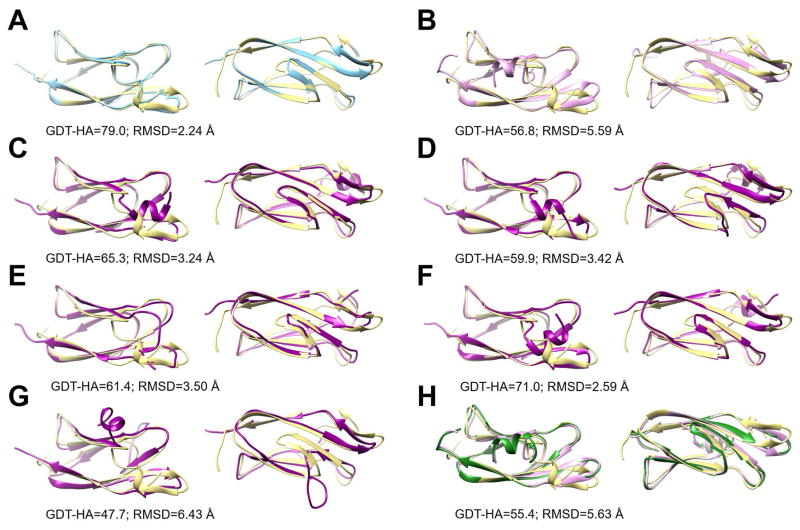
Figure 8.
Structures for target TR872 during refinement and with additional MD-based sampling. The native structure is shown in yellow in all panels and compared with the MD-simulated average structure of the native state (blue; A), the initial model given during CASP12 for refinement (pink; B), structures sampled via MD from the initial model and designated as states S0–S4 (cf. Figure 7, purple, C–G), and the model submitted during CASP12 (green, H). Structures are shown in two views and for each panel, GDT-HA and Cα RMSD measures relative to the native structure are given.
Our refinement simulations from CASP12 that were started from the CASP-provided initial model but involved weak positional restraints also did not significantly explore conformational space (see Fig. 6C). It is readily apparent from Fig. 6 that the differences between the initial model and the native state are much too large to be overcome in the presence of the restraints. In contrast, the unbiased simulations started from the initial model generated broad sampling where a number of distinct states were visited (see Fig. 7B and Fig. 8C–G). The state S0, closest to the initial model, was most favorable and is already significantly refined with respect to the native state. S0 has a GDT-HA score of 65.3 that is about 8.5 units better than the initial model. The state S0 is easily reachable from the initial model without restraints since the initial model and S0 are close in terms of the tICA principal components. However, as S0 is more than 5 Å Cα RMSD away from our model 1 submitted during CASP12, the restraints in our CASP12 protocol clearly prevented this state from being reached as it lies outside the sampling radius in the restrained simulations (see Figs. 7B and C). Other states in the unrestrained simulations, S1–S4, were only slightly less favorable than S0 and separated by kinetic barriers. One of the states, S3, is even closer to the native state than S0 (see Fig. 8F) with a GDT-HA score of 71.0 and a Cα RMSD value of 2.59 Å. This state closely resembles the native state except for a persistent incorrect N-terminal helix that would need to dissolve for the structure to completely match the native state (see Fig. 8F). On the other hand, the simulations also visit a state (S4) that is located significantly further away from the native state than the initial model. In S4, the N-terminal is opened up significantly (see Fig. 8G). S4 is also about 5 Å Cα RMSD away from our model 1 and, therefore, relaxing the restraints enough to be able to reach S0 would also allow S4 to be visited.
According to Fig. 7B, a transition from S0 to S3, would appear as the most direct path to the native state, but a transition via S2 or S1 is kinetically more likely. Both of these states are slightly unfolded relative to S0 and, so, in this example, further refinement from the initially relaxed state S0 would require partial unfolding (via S1 or S2) and refolding (to S3). The final transition to melt the N-terminal helix (which we did not observe) probably also requires an additional partial unfolding and repacking process.
The initial relaxation to S0 is essentially what we can reach with our current refinement protocol as long as the restraints are weak enough to allow large enough structural changes from the initial model. For the example chosen here, our restraints were too strong, but weaker restraints should make it possible to reach S0. However, further refinement would not simply proceed downhill on a folding funnel along a direct path. Instead, multiple cycles of partial unfolding and refolding may be required that are hindered severely by restraints as larger conformational changes are required but also involve the danger of unfolding towards states that are further away from the native state if restraints are not applied. Once states further away from the native state are sampled (such as S4), a recovery towards the native state becomes increasingly difficult due to the explosion of configurational space.
We believe that the example presented here typifies the general problem of structure refinement. We appear to be well-equipped to achieve modest refinement from an initial model towards locally relaxed structures by using restrained MD simulations aided by conformational averaging. However, further refinement likely requires cycles of partial unfolding and refolding that are essentially impossible in the presence of restraints. As such transitions require the crossing of kinetic barriers to states with similar relative energies, the key challenge going forward seems to be how to favor productive cycles that lead towards the native state while, at the same time, preventing transitions to states that lead away from the native state, but without a priori knowledge of where the native state is located. It does appear, though, that once the native state is reached it can be recognized based on the force field providing a deep free energy minimum. It remains to be seen in future studies how general these findings are and, ultimately, how to transform such insight into successful refinement protocols that can achieve more significant refinement than what is currently possible.
Further challenges
While the protein structure refinement exercise within CASP focuses on the very specific task of improving a given initial model with respect to the experimental structure, the larger goal is the generation of meaningful structural models of proteins in their biologically most relevant form. This includes the modeling of complete structures in their most prevalent oligomeric states, which involves additional challenges. Reliable templates used to generate initial models may not cover the entire sequence while information about the oligomeric state and especially oligomer interfaces are often lacking. Unfortunately, neither the ab initio modeling of missing fragments nor the application of protein-protein docking techniques to determine likely oligomer configurations are trivial. However, carrying out MD-based structure refinement would be expected to benefit from having complete structures in their oligomeric state as that would represent the most realistic physical environment for a given protein structure. It remains to be assessed in detail, though, what the effect of missing fragments and the neglect of oligomeric states have on refinement success with MD-based techniques.
CONCLUSIONS
The refinement of protein structures via MD simulations remains a highly successful approach. Our protocol that combines extensive sampling via MD followed by filtering, scoring and structure averaging remains highly successful in providing moderate but consistent refinement. However, the extent of refinement during CASP12 with our MD-based protocol has seen a decline based on CASP12 targets while we expected improved performance with increased sampling, further improved force fields, and new refinement protocols that were tested during CASP12. To some extent this appears to be a result of MD-based refinement becoming a standard component of modeling pipelines, so that initial models available in the refinement category at CASP are more difficult to refine further. But more generally, we seem to have reached a plateau with the current refinement protocols. A major issue is the use of restraints during sampling which is necessary on one hand to achieve consistency but severely limits the degree to which structures can be refined. A detailed analysis of one of the CASP12 targets suggests that much weaker restraints may be needed to achieve further refinement, and, in particular, to allow partial unfolding and refolding to reach the native state. The key issue remains, however, that very weak, or no restraints, also allow states to be visited that lead away from the native state while it appears that the energy function does not provide strong guidance towards the native state until the native state is actually reached. While extensive sampling is now possible and force fields have become highly realistic, we see the remaining challenge in effectively guiding sampling during refinement to eventually reach the native state rather than drift away towards unfolded states.
Acknowledgments
Funding was provided by the National Institute of Health Grant R01 GM084953 and computing time at NSF XSEDE facilities (TG-MCB090003) was used to carry out this research.
References
- 1.Zhang Y. Protein Structure Prediction: When Is It Useful? Curr Opin Struct Biol. 2009;19:145–155. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhlbrandt W. Cryo-EM Enters a New Era. Elife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirata K, Shinzawa-Itoh K, Yano N, Takemura S, Kato K, Hatanaka M, Muramoto K, Kawahara T, Tsukihara T, Yamashita E, Tono K, Ueno G, Hikima T, Murakami H, Inubushi Y, Yabashi M, Ishikawa T, Yamamoto M, Ogura T, Sugimoto H, Shen J-R, Yoshikawa S, Ago H. Determination of Damage-Free Crystal Structure of an X-Ray-Sensitive Protein Using an XFEL. Nat Meth. 2014;11:734–736. doi: 10.1038/nmeth.2962. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Skolnick J. The Protein Structure Prediction Problem Could be Solved Using the Current PDB Library. Proc Natl Acad Sci USA. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas J, Ramakrishnan N, Bailey-Kellogg C. Graphical Models of Residue Coupling in Protein Families. IEEE Trans Comput Biol Bioinf. 2008;5:183–197. doi: 10.1109/TCBB.2007.70225. [DOI] [PubMed] [Google Scholar]
- 6.Nugent T, Jones DT. Accurate de novo Structure Prediction of Large Transmembrane Protein Domains using Fragment-Assembly and Correlated Mutation Analysis. Proc Natl Acad Sci USA. 2012;109:E1540–E1547. doi: 10.1073/pnas.1120036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ovchinnikov S, Park H, Varghese N, Huang P-S, Pavlopoulos GA, Kim DE, Kamisetty H, Kyrpides NC, Baker D. Protein Structure Determination Using Metagenome Sequence Data. Science. 2017;355:294–298. doi: 10.1126/science.aah4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feig M. Computational Structure Refinement: Almost There, Yet Still so Far to Go. Wiley Interdiscip Rev: Comput Mol Sci. 2017;7:e1307. doi: 10.1002/wcms.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacCallum JL, Perez A, Schnieders MJ, Hua L, Jacobson MP, Dill KA. Assessment of protein structure refinement in CASP9. Proteins. 2011;79:74–90. doi: 10.1002/prot.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nugent T, Cozzetto D, Jones DT. Evaluation of predictions in the CASP10 model refinement category. Proteins. 2014;82:98–111. doi: 10.1002/prot.24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirjalili V, Noyes K, Feig M. Physics-Based Protein Structure Refinement through Multiple Molecular Dynamics Trajectories and Structure Averaging. Proteins. 2014;82:196–207. doi: 10.1002/prot.24336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirjalili V, Feig M. Protein Structure Refinement through Structure Selection and Averaging from Molecular Dynamics Ensembles. J Chem Theory Comput. 2013;9:1294–1303. doi: 10.1021/ct300962x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feig M, Mirjalili V. Protein Structure Refinement via Molecular-Dynamics Simulations: What Works and What Does Not? Proteins. 2016;84(Suppl 1):282–292. doi: 10.1002/prot.24871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heo L, Feig M. PREFMD: A Web Server for Protein Structure Refinement via Molecular Dynamics Simulations. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx726. under review. [DOI] [PMC free article] [PubMed]
- 15.Feig M. Local Protein Structure Refinement via Molecular Dynamics Simulation with locPREFMD. J Chem Inf Model. 2016;56:1304–1312. doi: 10.1021/acs.jcim.6b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Della Corte D, Wildberg A, Schröder GF. Protein Structure Refinement with Adaptively Restrained Homologous Replicas. Proteins. 2016;84(Suppl 1):302–313. doi: 10.1002/prot.24939. [DOI] [PubMed] [Google Scholar]
- 17.Park H, Seok C. Refinement of Unreliable Local Regions in Template-Based Protein Models. Proteins. 2012;80:1974–1986. doi: 10.1002/prot.24086. [DOI] [PubMed] [Google Scholar]
- 18.Lee GR, Heo L, Seok C. Effective Protein Model Structure Refinement by Loop Modeling and Overall Relaxation. Proteins. 2016;84(Suppl 1):293–301. doi: 10.1002/prot.24858. [DOI] [PubMed] [Google Scholar]
- 19.Modi V, Dunbrack RLJ. Assessment of Refinement of Template-Based Models in CASP11. Proteins. 2016;84(Suppl 1):260–281. doi: 10.1002/prot.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Brooks CL., III Can Molecular Dynamics Simulations Provide High-Resolution Refinement of Protein Structure? Proteins. 2007;67:922–930. doi: 10.1002/prot.21345. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmüller H, MacKerell AD., Jr CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 23.Darden TA, York D, Pedersen LG. Particle-Mesh Ewald: An N log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 24.Beauchamp KA, Bowman GR, Lane TJ, Maibaum L, Haque IS, Pande VS. MSMBuilder2: Modeling Conformational Dynamics at the Picosecond to Millisecond Scale. J Chem Theory Comput. 2011;7:3412–3419. doi: 10.1021/ct200463m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Zhang Y. A Novel Side-Chain Orientation Dependent Potential Derived from Random-Walk Reference State for Protein Fold Selection and Structure Prediction. Plos One. 2010;5:e15386. doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The Biomolecular Simulation Program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eastman P, Friedrichs MS, Chodera JD, Radmer RJ, Bruns CM, Ku JP, Beauchamp KA, Lane TJ, Wang LP, Shukla D, Tye T, Houston M, Stich T, Klein C, Shirts MR, Pande VS. OpenMM 4: A Reusable, Extensible, Hardware Independent Library for High Performance Molecular Simulation. J Chem Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feig M, Karanicolas J, Brooks CL., III MMTSB Tool Set: Enhanced Sampling and Multiscale Modeling Methods for Applications in Structural Biology. J Mol Graph Modell. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Zhou HY, Zhou YQ. Distance-Scaled, Finite Ideal-Gas Reference State Improves Structure-Derived Potentials of Mean Force for Structure Selection and Stability Prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raval A, Piana S, Eastwood MP, Dror RO, Shaw DE. Refinement of Protein Structure Homology Models Via Long, All-Atom Molecular Dynamics Simulations. Proteins. 2012;80:2071–2079. doi: 10.1002/prot.24098. [DOI] [PubMed] [Google Scholar]
- 32.Naritomi Y, Fuchigami S. Slow dynamics in protein fluctuations revealed by time-structure based independent component analysis: the case of domain motions. J Chem Phys. 2011;134:065101. doi: 10.1063/1.3554380. [DOI] [PubMed] [Google Scholar]
- 33.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci U S A. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]