Summary
The circadian clock drives daily rhythms of many plant physiological responses providing a competitive advantage that improves plant fitness and survival rates [1-5]. While multiple environmental cues are predicted to regulate the plant clock function, most studies focused on understanding the effects of light and temperature [5-8]. Increasing evidence indicates a significant role of plant-pathogen interactions on clock regulation [9, 10], but the underlying mechanisms remain elusive. In Arabidopsis, the clock function largely relies on a transcriptional feedback loop between morning (CCA1 and LHY) and evening (TOC1) expressed transcription factors [6-8]. Here, we focused on these core components to investigate the Arabidopsis clock regulation using a unique biotic stress approach. We found that a single leaf Pseudomonas syringae infection systemically lengthened the period and reduced the amplitude of circadian rhythms in distal uninfected tissues. Remarkably, the low amplitude phenotype observed upon infection was recapitulated by a transient treatment with the defense related phytohormone salicylic acid (SA), which also triggered a significant clock phase delay. Strikingly, despite SA modulated circadian rhythms, we revealed that the master regulator of SA signaling, NPR1 [11, 12], antagonized clock responses triggered by both SA-treatment and P. syringae. In contrast, we uncovered that the NADPH oxidase RBOHD [13] largely mediated the aforementioned clock responses after either SA treatment or the bacterial infection. Altogether, we demonstrated novel and unexpected roles for SA, NPR1 and redox signaling in clock regulation by P. syringae, and revealed a previously unrecognized layer of systemic clock regulation by locally perceived environmental cues.
Keywords: Arabidopsis, Circadian clock, P. syringae, salicylic acid (SA), reactive oxygen species (ROS), CCA1, LHY, TOC1, NPR1, RBOHD
Graphical abstract
Li et al. unravel Arabidopsis clock regulation by a bacterial infection. They report that a localized P. syringae infection affects clock rhythms at the whole plant level, that systemic SA and ROS phenocopy these effects, and that NPR1 (mediates SA responses) and RBOHD (produces apoplastic ROS) antagonize and mediate these responses, respectively.

Results
Localized P. syringae infection triggered a systemic clock amplitude reduction and period lengthening
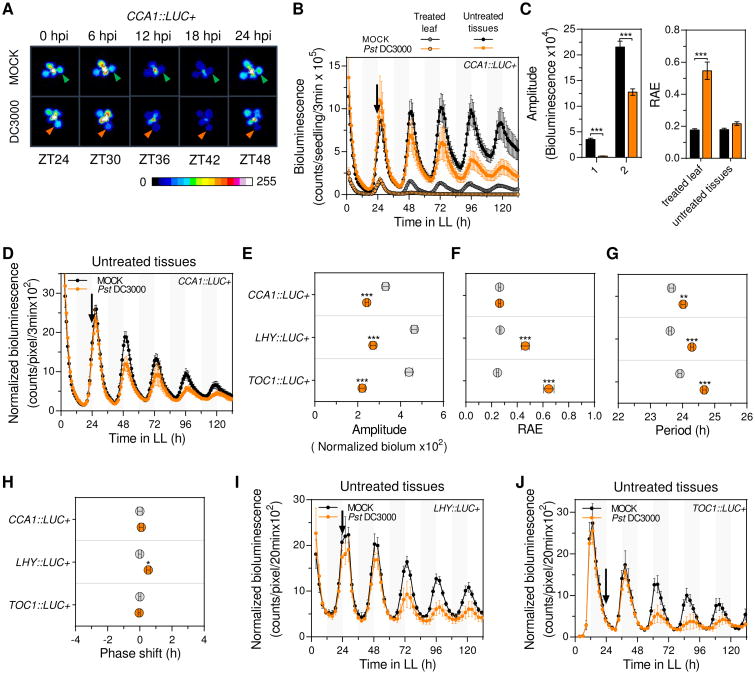
To study plant clock responses to a localized pathogen challenge we used the Arabidopsis-Pseudomonas syringae pathosystem following a leaf-restricted infection. Arabidopsis seedlings carrying a luciferase reporter gene (LUC+) expressed under the control of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) promoter (CCA1∷LUC+) were used to analyze the clock function after a single leaf P. syringae pv. tomato DC3000 (Pst DC3000) or mock treatments (performed at ZT24) (Figure S1A). Analysis of the whole plant luciferase activity after treatment indicated that Pst DC3000 infection results in low amplitude and long period clock rhythms (Figure S1B and data not shown). However, CCA1∷LUC+ activity in the infected leaf rapidly decayed and became arrhythmic, suggesting that the observed phenotypes reflected the clock status in untreated tissues (Figures 1A-1C). We reasoned that the amplitude reduction found after whole plant bioluminescence analysis was likely overestimated due to the inclusion of the infected leaf and the reduction of plant size after single leaf infection (Figure S1C). Thus, we reanalyzed bioluminescence results considering only untreated tissues (of Pst DC3000 and mock treated plants) and normalizing bioluminescence counts to the estimated plant area at each time point (Figure S1D). This analysis indicated that Pst DC3000 infection resulted in systemic clock rhythms of significantly lower amplitude (Figures 1D-1F) and ∼0.5h longer period (Figures 1D and 1G). In addition, the period-normalized phase of CCA1∷LUC+ rhythms was slightly, albeit not significantly, advanced by the infection (Figures 1H and S1E). To confirm these results we performed the same experiment using Arabidopsis reporter lines carrying either the LATE ELONGATED HYPOCOTYL (LHY) or TIMING OF CAB EXPRESSION 1 (TOC1) promoter driving the expression of the LUC+ gene (LHY∷LUC+ or TOC1∷LUC+). Notably, as we observed for the CCA1 promoter activity, both new reporters exhibited a significant decay in the luminescence emitted by the infected leaf as soon as 24h post infection (Figure S1F). When clock rhythms were analyzed in untreated tissues of single leaf Pst DC3000 versus mock treated plants we observed a significant amplitude and robustness reduction, and ∼0.7h longer period upon infection (Figures 1E-1G, 1I and 1J). In addition, LHY (but not TOC1) promoter driven oscillations exhibited a phase advance upon infection (Figures 1H and S1G). To further evaluate the reduced amplitude phenotype, we quantified CCA1, LHY and TOC1 mRNA levels in untreated tissues of single leaf Pst DC3000 and mock treated plants. As shown in Figure S1H, we observed significantly reduced CCA1 and LHY mRNA levels in infected plants supporting the results obtained via bioluminescence assays. Altogether, these findings revealed that a localized Pst DC3000 infection triggered a systemic signal that reduced the amplitude, lengthened the period, and minimally advanced the phase (for morning expressed reporters) of clock rhythms in untreated tissues.
Figure 1. Localized P. syringae infection triggers systemic clock responses.
(A) Representative time course pseudocolored bioluminescence images of soil grown CCA1∷LUC+ plants upon single leaf Pst DC3000 infection or mock treatment (hpi: hours post infection). Triangles (Pst DC3000: orange, mock: green) point to the treated leaf.
(B) Luciferase activity from treated leaf or untreated tissues of soil grown Arabidopsis CCA1∷LUC+ plants upon single leaf Pst DC3000 infection (orange) or mock treatment (black). Treatments were performed at ZT24 (denoted by the black arrow) (see also Figure S1A). Results indicate mean values [± SD, n=6] and are representative of 5 independent experiments.
(C) Mean amplitude (left panel) and relative amplitude error (RAE) (right panel) values [± SEM]of CCA1∷LUC+ rhythms in treated leaf or untreated tissues upon single leaf Pst DC3000 infection (orange) or mock treatment (black) for experiments indicated in (B) (n=30).
(D) Luciferase activity (normalized by plant size) from untreated tissues of soil grown Arabidopsis CCA1∷LUC+ plants upon single leaf Pst DC3000 infection (orange) or mock treatment (black). Results indicate mean values [± SD, n=6] and are representative of 5 independent experiments.
(E-H) Mean amplitude (E), relative amplitude error (RAE) (F), period (G) and phase shift values (H) [± SEM] of CCA1∷LUC+ (n=30), LHY∷LUC+ (n=15) and TOC1∷LUC+ (n=15) normalized luciferase activity rhythms upon single leaf Pst DC3000 infection (orange) or mock treatment (gray) for experiments indicated in (D, I and J).
(I-J) Luciferase activity (normalized by plant size) from untreated tissues of soil grown Arabidopsis LHY∷LUC+ and TOC1∷LUC+ plants upon single leaf Pst DC3000 infection (orange) or mock treatment (black). Results indicate mean values [± SD, n=9] and are representative of 2 independent experiments.
Statistical analyses between mock and infected plants were performed using the t test (C, E, F, G and H). Stars indicate the level of significance (*p<0.01, **p<0.001, ***p<0.0001).
Transient SA treatment phenocopied P. syringae triggered amplitude reduction and delayed the phase of clock rhythms
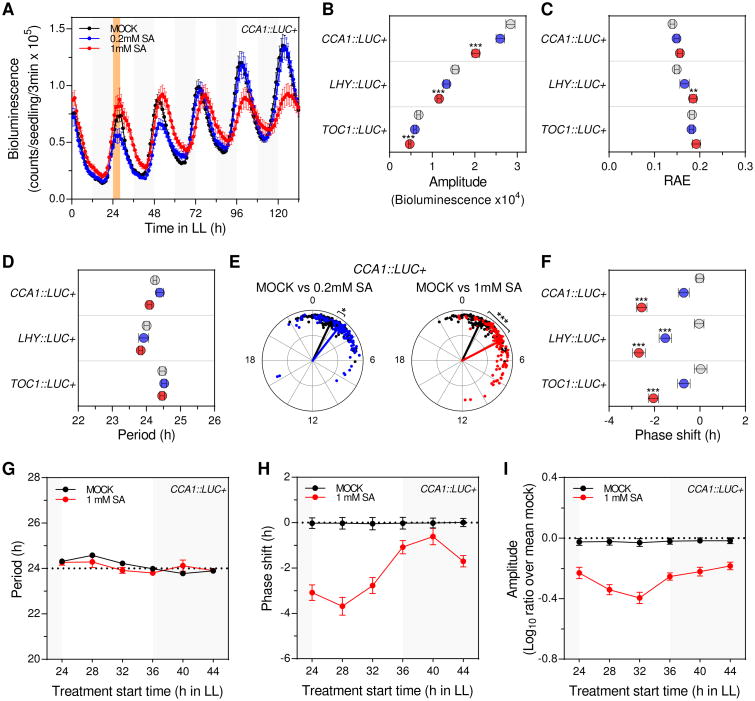
The phytohormone salicylic acid (SA), produced in response to plant infections, regulates many aspects of plant immunity in distal non-infected tissues [12, 14, 15]. While this critical role anticipated that SA could mediate the observed clock phenotypes upon infection, it was previously shown that SA increased (rather than reduced) the amplitude of clock rhythms [10]. Given that this previous finding likely indicated a clock response to long-term plant exposure to SA and that SA is transiently induced after a bacterial pathogen challenge [16], we analyzed Arabidopsis clock responses to a short-term SA treatment (performed at ZT24) using CCA1∷LUC+ seedlings (Figure S2A). Similar to Pst DC3000 infection, a transient SA treatment resulted in low amplitude rhythms (Figures 2A and 2B) although without affecting their robustness (Figure 2C). This amplitude reduction was not influenced by plant size, as plant biomass was not affected by the transient SA treatment (Figure S2B), or by SA effects over luciferase activity, as plants that constitutively express the LUC+ reporter gene displayed same overall bioluminescence levels after SA or mock treatment (Figure S2C). In contrast to the infection context, however, the period of CCA1 promoter driven oscillations was not changed by the SA treatment (Figure 2D) and instead a significant phase delay was observed (Figures 2E and 2F). To evaluate if these clock responses depended on the time at which SA was applied, the experiment outlined above was performed initiating the treatment at different times of the day (ZT24, 28, 32, 36, 40, 44). Interestingly, while the period length remained unaltered in all experiments, we observed a greater phase shift when SA treatment was started close to the subjective dawn (Figures 2G and 2H). In contrast, the amplitude was overall reduced to a similar extent regardless of SA treatment start time (Figure 2I).
Figure 2. Transient SA treatment phenocopies P. syringae-triggered amplitude reduction and delays the phase of clock rhythms.
(A) Luciferase activity from CCA1∷LUC+ seedlings after transient SA treatment. Seedlings were treated with medium alone (mock) or supplemented with SA (0.2mM and 1 mM) at ZT24 for 4h (denoted by the orange shadowed area) (see also Figure S2A). Results indicate mean values [± SEM, n=12] and are representative of 7 independent experiments.
(B-D) Mean amplitude (B), relative amplitude error (RAE) (C) and period (D) values [± SEM] of CCA1∷LUC+ (n=110), LHY∷LUC+ (n=60) and TOC1∷LUC+ (n=60) rhythms after mock (grey), 0.2mM SA (blue) or 1mM SA (red) transient treatments for experiments indicated in (A) and Figure S2D.
(E) Normalized phase and RAE values of CCA1∷LUC+ rhythms after transient SA treatment for experiments indicated in (A) (mock: black, 0.2mM SA: blue and 1mM SA: red) (each dot represents one individual). The angular position of dots and arrows indicates the normalized phase value (0-24h) and the radial position indicates the RAE value (RAE=0 at the outmost radial position and RAE=1 at the center). The arrow points to the mean normalized phase and the arrow length indicates the mean RAE.
(F) Mean phase shift values [± SEM] of CCA1∷LUC+ (n=110), LHY∷LUC+ (n=60) andTOC1∷LUC+ (n=60) rhythms after mock (grey), 0.2mM SA (blue) or 1mM SA (red) transient treatments indicated in (A) and Figure S2D.
(G-I) Period (G), phase shift (H) and normalized amplitude (ratio over mean mock) (I) values of CCA1∷LUC+ rhythms after mock (black) or 1mM SA (red) transient treatments performed at different times of the day. Plants were grown as indicated in (A) and treatments were started at 6 different times during the second day in LL (ZT24, ZT28, ZT32, ZT36, ZT40, ZT44). Results represent mean values [± SEM, n=48] of 4 independent experiments.
Statistical analyses between mock and SA treated plants were performed using the t test (B, C, D and F) and Watson-Williams test (E). Stars indicate the level of significance (*p<0.01, **p<0.001, ***p<0.0001).
To validate CCA1∷LUC+ results, we next evaluated the clock function upon SA treatment in LHY∷LUC+ and TOC1∷LUC+ reporter lines. Indeed, we observed that a short-term SA treatment caused the same amplitude reduction and phase delay phenotypes in both LHY∷LUC+ and TOC1∷LUC+ rhythms (Figures 2B, 2F, S2D and S2E). To further evaluate these clock phenotypes we quantified CCA1, LHY and TOC1 mRNA levels in SA and mock treated plants. In agreement with bioluminescence experiments, we observed a significant phase delay for all transcripts upon SA treatment (Figures S2F and S2G). Furthermore, although we expected that (due to its limit of sensitivity) real time quantitative PCR would not detect the small amplitude differences found in bioluminescence assays, we did observe a tendency towards a reduced amplitude of mRNA oscillations (for LHY and TOC1 transcripts) (Figures S2F and S2G). Altogether, our results indicated that a pulse of SA significantly affected the phase and amplitude of clock rhythms, indicating a general role for SA in the regulation of Arabidopsis clock function. Furthermore, a SA pulse phenocopied the amplitude reduction that we observed after localized Pst DC3000 inoculation, suggesting that SA was one of the systemic signals that mediated the amplitude phenotype upon infection.
SA effects on the clock function were antagonized by NPR1
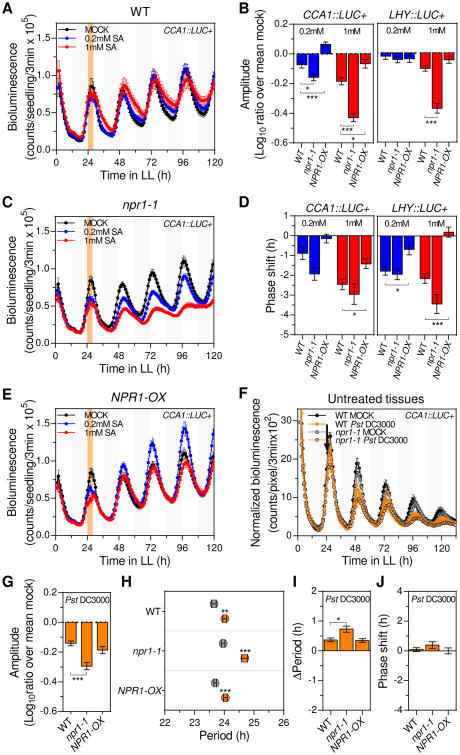
Given that most SA responses are dependent on the transcription cofactor NONEXPRESSER OF PR GENES 1 (NPR1) [12], we next investigated whether clock effects triggered by a transient SA treatment were mediated by NPR1. For that, we introduced the loss-of-function npr1-1 allele into the CCA1∷LUC+ and LHY∷LUC+ reporter backgrounds and treated the resulting lines with SA at ZT24 (as described above, Figure S2A). As expected, we observed that a transient SA treatment caused a significant amplitude reduction in WT plants for both reporter lines (Figures 3A and 3B). Strikingly, in npr1-1 plants the amplitude was reduced to a significantly greater extent in both CCA1∷LUC+ and LHY∷LUC+ reporter backgrounds (Figures 3A-3C and S3A). Consistent with these results, CCA1 and LHY promoter driven oscillations exhibited a decreased robustness when the SA treatment was applied to npr1-1 compared to WT plants (Figure S3B). It should be noted that the amplitude reduction was not influenced by a change in plant size, as same biomass was observed between mock and SA treated npr1-1 plants (Figure S3C). We next determined CCA1, LHY and TOC1 mRNA levels in SA and mock treated npr1-1 plants, and observed that indeed transcript levels for these genes oscillated with a significantly reduced amplitude upon exposure to SA (Figures S3D and S3E). To further validate these findings, we analyzed the clock function after a transient SA treatment in CCA1∷LUC+ and LHY∷LUC+ reporter lines that overexpressed NPR1 (NPR1-OX, Figure S3F). We found that, unlike in WT plants, SA did not reduce the clock amplitude in NPR1-OX plants (Figures 3A, 3B, 3E and S3A).
Figure 3. NPR1 antagonizes clock responses triggered by transient SA treatment or single leaf P. syringae infection.
(A, C and E) CCA1∷LUC+ activity in wild type (WT) (A), npr1-1 (C) and NPR1-OX (E) seedlings after transient SA treatment. Seedlings were treated with medium alone (mock) or supplemented with SA (0.2mM and 1 mM) at ZT24 for 4h (denoted by the orange shadowed area) (see also Figure S2A). Results indicate mean values [± SEM, n=12] and are representative of 4 independent experiments.
(B and D) Mean normalized amplitude (ratio over mean mock) (B) and phase shift (D) values [± SEM] of CCA1∷LUC+ (left panel) (n=72) and LHY∷LUC+ (right panel) (n≥58) rhythms in WT, npr1-1 and NPR1-OX seedlings after 0.2mM (blue) and 1mM (red) transient SA treatment of 4 independent experiments indicated in (A, C and E) (see also Figures S3A, S3H and S3I).
(F) CCA1∷LUC+ activity (normalized by plant size) from untreated tissues of soil grown WT and npr1-1 plants upon a single leaf Pst DC3000 infection (orange) or mock treatment (black). Treatments were performed at ZT24 (denoted by the black arrow) (see also Figure S1A). Results indicate mean values [± SD, n=6] and are representative of 5 independent experiments.
(G, I, J) Mean normalized amplitude (ratio over mean mock) (G), period length change (Δperiod) (I), and phase shift (J) values [± SEM, n=30] of CCA1∷LUC+ rhythms (normalized by plant size) in WT, npr1-1 and NPR1-OX plants after a single leaf Pst DC3000 infection for experiments indicated in (F) (see also Figures S4A and S4C).
(H) Mean period estimates [± SEM, n=30] of CCA1∷LUC+ rhythms (normalized by plant size) in WT, npr1-1 and NPR1-OX plants upon single leaf Pst DC3000 infection (orange) or mock treatment (gray) for experiments indicated in (F).
Statistical analyses compared to WT plants (B, D, G, I and J), or compared mock treated plants (H) were performed using the t test. Stars indicate the level of significance (*p<0.01, **p<0.001, ***p<0.0001).
On the other hand, the period of clock oscillations remained mostly unchanged in all backgrounds and treatment conditions (Figure S3G). Importantly, similar to the aforementioned amplitude phenotypes, analysis of period-normalized phase shifts for CCA1∷LUC+ and LHY∷LUC+ rhythms indicated that SA treatment resulted in greater or milder phase delay in npr1-1 or NPR1-OX backgrounds, respectively, when compared to WT plants (Figures 3D, S3H and S3I). Altogether, these observations revealed that NPR1 loss of function and overexpression respectively enhanced and reduced the impact of SA on amplitude and phase phenotypes, revealing an unexpected role for NPR1, which functioned as an antagonist of transient SA effects on the circadian clock.
P. syringae-triggered amplitude reduction of clock rhythms was antagonized by NPR1
Given the antagonistic effect of NPR1 on the regulation of circadian rhythms by SA, and that both Pst DC3000 infections and SA treatment decreased amplitude of clock rhythms, we next hypothesized that NPR1 may also counteract the amplitude phenotype that we observed after infection. To evaluate this possibility, we performed single leaf Pst DC3000 infections in WT, npr1-1 and NPR1-OX plants carrying the CCA1∷LUC+ reporter. Analysis of the luciferase activity in untreated tissues showed that the amplitude of clock controlled rhythms in uninfected tissues was more significantly reduced by the infection in npr1-1 compared to WT plants, albeit a similar amplitude reduction was observed in NPR1-OX versus WT backgrounds (Figures 3F, 3G and S4A). Consistent with the lower amplitude, a greater decrease in the robustness of clock rhythms was observed after infection in npr1-1 versus WT plants (Figure S4B). Likewise, the period of CCA1∷LUC+ rhythms was lengthened to a significantly greater extent after infection in npr1-1 versus WT and NPR1-OX plants (Figures 3H and 3I). Of note, as described above for WT plants (Fig. 1H), the period-normalized phase of CCA1∷LUC+ rhythms was only minimally changed after infection in both NPR1 genetic backgrounds (Figures 3J and S4C). These results indicated that (as described for a transient SA treatment) NPR1 antagonized the systemic amplitude reduction, while additionally counteracting period lengthening, of clock rhythms observed after single leaf Pst DC3000 infection.
SA effects on the circadian clock were phenocopied by H2O2 treatment and partly mediated by RBOHD
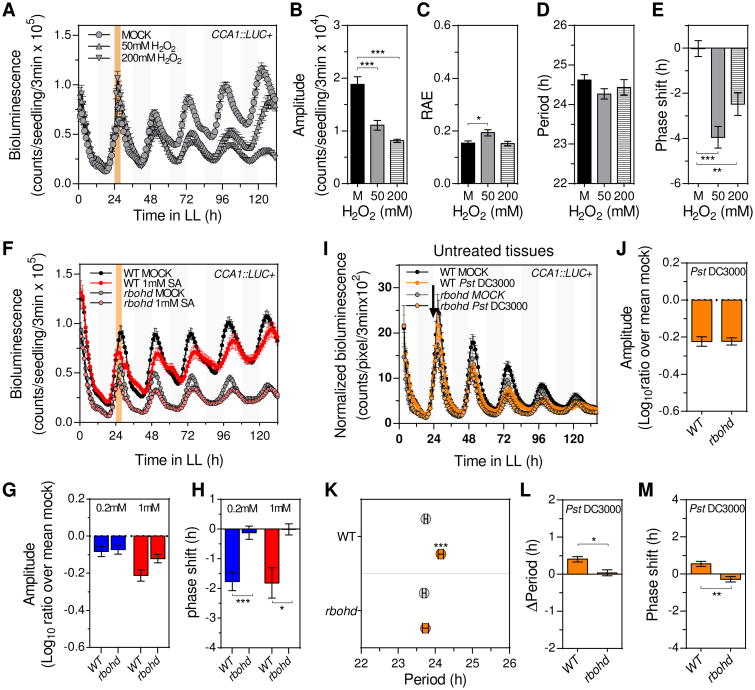
Considering that SA responses are partly mediated by a rapid increase of reactive oxygen species (ROS) [15, 17], we next hypothesized that ROS may mediate the clock phenotypes observed after transient SA treatment. To test this possibility, we first analyzed the effect of a transient H2O2 treatment on CCA1∷LUC+ rhythms (Figure S2A). Remarkably, we found that (as observed for SA, Figures 2 and S2) a transient H2O2 treatment did not affect the robustness and period of circadian rhythms but did cause a profound amplitude reduction and phase delay (Figures 4A-4E). Given that SA could induce apoplastic H2O2 production through the nicotinamide adenine dinucleotide phosphate (NADPH)-oxidase RESPIRATORY BURST OXIDASE HOMOLOGUE D (RBOHD) [13, 18], we next tested the possibility that RBOHD may mediate SA effects on the clock function. For that, we analyzed CCA1∷LUC+ rhythms after a transient SA treatment in rbohd mutant plants (Figure S2A). Strikingly, when bioluminescence was analyzed in mock treated plants (without SA or H2O2 treatment), amplitude reduction and phase advance phenotypes were observed in rbohd mutant compared to WT plants (Figures 4F, S4D and S4E). Upon SA treatment, however, the degree of amplitude reduction was similar in rbohd versus WT plants (Figures 4F, 4G and S4D), while the robustness and period continued being mostly unchanged in both genetic backgrounds (Figures S4F and S4G). In sharp contrast, while SA treatment continued to delay the clock phase in WT plants, we did not observe any SA-induced phase change in the rbohd mutant background (Figures 4H and S4E). Altogether, these results revealed that SA effects on the clock were fully mimicked by the H2O2 treatment, suggesting that ROS likely mediated SA-induced clock regulation. Furthermore, the absence of SA-induced phase, but not amplitude, phenotype in rbohd mutant plants suggested that clock regulation by SA relied on RBOHD-dependent (i.e. apoplastic ROS) and RBOHD-independent ROS sources.
Figure 4. Apoplastic ROS partly mediates clock responses triggered by transient SA treatment or single leaf P. syringae infection.
(A) Luciferase activity of CCA1∷LUC+ seedlings after transient H2O2 treatment. Seedlings were treated with medium alone (mock) (n=24) or supplemented with 50mM (n=12) or 200mM (n=12) H2O2 at ZT24 for 4h (denoted by the orange shadowed area) (see also Figure S2A). Results indicate mean values [± SEM] and are representative of 2 independent experiments.
(B-E) Mean amplitude (B), relative amplitude error (RAE) (C), period (D) and phase shift (E) values [± SEM] of CCA1∷LUC+ rhythms after mock (black bars) (n=49), 50mM H2O2 (gray bars) (n=33) or 200mM H2O2 (striped bars) (n=30) transient treatments for experiments indicated in (A).
(F) CCA1∷LUC+ activity in wild type (WT) (n=24) and rbohd (n=12) seedlings after transient 1mM SA treatment as indicated in Figure S2A. Results indicate mean values [± SEM] and are representative of 4 independent experiments.
(G and H) Mean normalized amplitude (ratio over mean mock) (G) and phase shift (H) values [± SEM] of CCA1∷LUC+ rhythms in WT (n=80) and rbohd (n=64) seedlings after 0.2mM (blue) and 1mM (red) transient SA treatment for experiments indicated in (F) (see also Figures S4D-E).
(I) CCA1∷LUC+ activity (normalized by plant size) from untreated tissues of soil grown WT and rbohd plants upon single leaf Pst DC3000 infection (orange) or mock treatment (black). Treatments were performed at ZT24 (denoted by the black arrow) (see also Figure S1A). Results indicate mean values [± SD, n=6] and are representative of 3 independent experiments.
(J, L, M) Mean normalized amplitude (ratio over mean mock) (J), period length change (Δperiod) (L), and phase shift (M) [± SEM, n=18] for CCA1∷LUC+ rhythms in WT and rbohd after single leaf Pst DC3000 infection for experiments indicated in (I) (see also Figures S4H and S4J).
(K) Mean period estimates [± SEM, n=18] for CCA1∷LUC+ rhythms in WT and rbohd plants upon single leaf Pst DC3000 infection (orange) or mock treatment (gray) for experiments indicated in (I).
Statistical analyses compared to mock treated plants (B-E and K), or compared to WT plants (G, H, J, L and M) were performed using the t test. Stars indicate the level of significance (*p<0.01, **p<0.001, ***p<0.0001).
P. syringae infection effects on the circadian clock were partly mediated by RBOHD
Given that SA-effects on clock regulation were partly mediated by RBOHD and that pathogen recognition rapidly triggers RBOHD-dependent ROS production [19, 20], we next investigated a putative role for RBOHD on the regulation of systemic clock phenotypes induced by a localized Pst DC3000 infection. For that, we performed single leaf infection experiments in WT and rbohd mutant plants carrying the CCA1∷LUC+ reporter (Figure S1A). As described above for the SA amplitude effect in rbohd mutant plants (Figure 4G), we observed that the amplitude and robustness of CCA1 promoter driven rhythms were equally affected by the infection in both WT and rbohd plants (Figures 4I, 4J, S4H and S4I). Remarkably, we found that both the slight phase advance and the significant period lengthening that we observed in WT plants upon infection were no longer detected in the rbohd mutant background (Figures 4K-4M and S4J). These results supported the notion that while RBOHD-dependent ROS did not mediate the infection-induced amplitude reduction, it did trigger the systemic clock period and phase phenotypes observed after a localized Pst DC3000 infection.
Discussion
The circadian clock is an endogenous timekeeping mechanism that orchestrates daily rhythms in most plant biological processes [1, 4, 7]. This function, critical for sessile plants, requires daily adjustments by multiple environmental cues predicted to signal into the core clock mechanism through a network of heavily integrated circuits [5, 8]. While plants are exposed to a wide range of abiotic and biotic environmental signals, there was a gap in knowledge on how biotic interactions affect plant circadian rhythms and what are the pathways involved in these responses. Our work provided solid evidence indicating that a single leaf Pst DC3000 infection (despite being localized) significantly modulated the clock function at the whole plant level and identified the mechanisms involved, revealing novel roles for SA, NPR1 and redox signaling in the regulation of circadian rhythms (see model in Figure S4K).
We found that the clock function was systemically affected in response to a local Pst DC3000 infection, displaying low amplitude and long period phenotypes (Figures 1D, 1E, 1G, 1I, 1J and S1H). These findings were unexpected as a previous study using P. syringae pv. maculicola ES4326 (PmaDG3) reported a short (rather than long) period phenotype after infection [9]. It should be noted that (in contrast to the local leaf infection and soil grown conditions that we used) in the last-mentioned study whole Arabidopsis seedlings were fully soaked into a bacterial cell suspension and plants were subsequently grown in sucrose supplemented tissue culture medium [9]. Given that a pathogen challenge (i.e. P. syringae inoculation) reduces the photosynthetic capacity in the infected tissues [21, 22] and that sucrose treatment influences the clock function in plants with reduced photosynthesis [23], it is possible that the differences in infection protocols and sucrose availability may have accounted for the disparate results. Furthermore, it has been reported that PmaDG3 and Pst DC3000 infections trigger distinct transcriptional responses in Arabidopsis [24], which could have also contributed to the contrasting observations. Nonetheless, together with the study published by Zhang et al., our work raises the possibility that multiple variables such as the affected tissues, the energy status of the plant and the pathogen strain may influence the outcome of clock regulation by a bacterial infection.
One of the most valuable contributions from our study was to expose an unrecognized regulatory layer within the plant circadian system in which the overall clock function is modulated by environmental cues that locally affect specific plant tissues (Figures 1 and S1). This is an exciting observation since a recent report indicated that the plant circadian system has a hierarchical organization with a master oscillator located at the shoot apical meristem (SAM) and peripheral subordinate clocks in other tissues [25]. Thus, our findings suggest that a systemic signal originated at the infected leaf may regulate the SAM clock and therefore affect the clock function at the whole plant level. Alternatively, it is possible that a systemic signal triggered after the localized infection directly modulates peripheral clocks.
Importantly, our studies support the view that SA, which is produced transiently upon infections [16], was one of the aforementioned systemic signals that regulated the circadian clock in non-infected tissues (Figures 2A, 2B and S2D). It should be noted that previous work by Zhou et al. found that SA increases (rather than decreases) the amplitude of clock rhythms [10]. While this result is in sharp contrast with the sustained amplitude reduction that we observed after both Pst DC3000 infection and transient SA treatment (Figures 1D, 1E, 1I, 1J, S1H, 2A, 2B and S2D), it is possible that these discrepancies reflect the overall dynamic nature of SA responses. For example, it was suggested that spatiotemporal changes in SA concentrations could lead to opposite responses [26] and that SA responses are diminished as plants age [27]. Thus, different hormone concentrations, longer exposure time, and/or slightly older plants used by Zhou et al. may have accounted for the disparate results. Remarkably, our results also showed for the first time that a transient SA treatment impinged a profound phase delay on clock oscillations (Figures 2E, 2F, 2H and S2E-S2G). Notably, such phase delay phenotype was not detected in Pst DC3000 infected plants, which instead exhibited a slight phase advance (Figures 1H, S1E and S1G), raising the possibility that SA-independent pathways neutralized and/or overruled the SA induced phase delay during infection. Likewise, only Pst DC3000, but not SA, lengthened the clock period (Figures 1G and 2D), further suggesting SA-independent pathways that also modulated the clock period upon infection. Given that we observed the same amplitude reduction and phase delay phenotypes even after a longer (12h) transient SA treatment (data not shown), altogether our results suggested that SA-dependent and -independent pathways are integrated to determine the ultimate clock phenotypes in infected plants (depicted in Figure S4K and further discussed below). However, we cannot completely rule out the possibility that (in the context of Pst DC3000 infection) SA might regulate the clock in a manner that was not fully recapitulated by the transient SA treatment used in our study.
Our work also revealed an unexpected antagonistic role for NPR1 in the regulation of circadian rhythms by both SA and P. syringae infection. While NPR1 mediates most SA-induced transcriptional responses [12], we counterintuitively found that NPR1 prevented the amplitude reduction and phase delay triggered by a transient SA treatment (Figures 3A-3E, S3A, S3D and S3E). Importantly, the amplitude reduction induced by Pst DC3000 infection was also enhanced in npr1-1 mutant plants (Figures 3F-G and S4A), supporting the biological significance of the aforementioned antagonistic NPR1 effect on clock regulation by SA (modeled in Figure S4K). These results are in line with previous work suggesting that NPR1 not only mediates SA responses but also provides a safeguard mechanism to counteract SA signaling. For example, it was reported that NPR1 negatively regulates SA synthesis [28] and that NPR1 proteasome-mediated degradation plays dual roles in plant immunity (required for both inactivation and activation of target genes) [29].
Finally, our study uncovered a previously unrecognized role for RBOHD (and thus apoplastic ROS) in clock regulation both before and after infection. We first found that rbohd mutant plants exhibited a reduced amplitude and advanced phase in basal conditions (Figures 4F, S4D and S4E), revealing a previously unrecognized and pivotal role for apoplastic ROS in maintaining the overall function of the Arabidopsis clock function under steady state conditions. In addition, consistently with enhanced ROS signaling upon SA treatment or pathogen recognition [15, 17], we observed that clock responses to SA were phenocopied by a transient H2O2 treatment (Figures 4A-E). These results were consistent with a previous study indicating that spraying seedlings with H2O2 or ROS-inducing-chemicals (likely resulting in a prolonged ROS treatment) induces phase delayed clock rhythms [30]. Importantly, despite multiple ROS sources [15, 17], our results indicated that SA-induced phase delay was mediated by apoplastic ROS as it was reverted when apoplastic ROS production was compromised (rbohd mutant plants) (Figures 4H and S4E). In contrast, we found that the amplitude phenotype likely involved other ROS sources since SA and Pst DC3000 equally reduced the clock amplitude in wild-type and rbohd mutant plants (Figures 4G and 4J). Finally, although we could not observe any period phenotype after (transient) H2O2 treatment (Figure 4D), the aforementioned study by Lai et al. showed that (likely sustained) ROS treatments did lengthened the clock period [30] and we observed that the long period phenotype detected after Pst DC3000 infection depended on RBOHD (Figures 4K-4L). Thus, together with the result by Lai et al., our work supported a model in which persistently elevated aplopastic ROS levels lengthened the clock period after infection, which is consistent with the continued ROS accumulation [31, 32] and enhanced RBOHD activity in infected plants [19, 20]. Given that the period phenotype was not observed after SA treatment (Figure 2D), these results suggested that the putative SA-independent pathways that regulated the clock function after infection were, at least partly, mediated by apoplastic ROS. Consistently, our results suggested that SA-independent apoplastic ROS also promoted the slight phase advance observed after infection (Figure 4M). Thus, we propose that RBOHD-dependent ROS production is a key signal that largely mediated the aforementioned SA-dependent and -independent pathways that fine-tuned the overall plant clock after a leaf-restricted infection (Figure S4K). This is in line with previous studies indicating that RBOHD propagates a systemic long distance ROS wave triggered by a localized stress [33, 34]. It remains unclear, however, how apoplastic ROS (or RBOHD) delayed the clock phase downstream SA (i.e. upon sole SA treatment) while having the opposite effect in the infection context (i.e. a net advancing phase effect after infection). Possible explanations include different levels, duration and/or distribution of SA-independent versus SA-dependent apoplastic ROS, in addition to contextual regulation induced by specific signals in infected plants.
In conclusion, our work used a unique approach to investigate the Arabidopsis clock, characterizing for the first time how a virulent pathogen infection modulated circadian rhythms in soil grown plants. Remarkably, we revealed a new layer of regulation within the plant circadian system in which the overall clock function is modulated by locally perceived environmental cues. Period and phase changes predict that upon infection plant endogenous rhythms would be desynchronized from external environmental cycles, anticipating a suboptimal photosynthetic capacity [2]. It is possible that this mechanism may have evolved to prevent ROS hyper accumulation and its negative consequences after infection. In addition, amplitude reduction indicates a weakened clock function upon infection, which may be necessary to allow continuous immunity to contain the infectious bacteria and prevent re-infection. Importantly, we identified critical signaling pathways (SA and ROS) and key components of these pathways (NPR1 and RBOHD) that regulated clock responses after infection. Most saliently, we revealed an unexpected antagonistic role for NPR1 in SA-triggered clock responses, and a novel role for apoplastic ROS as a regulator of circadian rhythms, not only upon biotic stress but also in basal conditions. Given that ROS signaling is found at the crossroad of most plant biotic or abiotic stress signaling pathways [34, 35] our findings may also illuminate how plant circadian rhythms are adjusted by multiple stress responses. Thus, our study disentangles the highly complex regulation of circadian rhythms after infection and paves the way for future studies aiming at potentially tweaking the clock to enhance plant performance in general, and specifically after pathogen encounter.
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jose L. Pruneda-Paz (jprunedapaz@ucsd.edu).
Experimental Model and Subject Details
Plant materials
Arabidopsis thaliana (Arabidopsis) seedlings used in this work were from the Columbia ecotype (Col-0). CCA1∷LUC+ [36], LHY∷LUC+ [37],TOC1∷LUC+ [38] reporter lines, and npr1-1 [39] and rbohd (SALK_070610) [18] loss-of function lines were previously described.
To generate NPR1 overexpression lines, NPR1 protein coding sequence was PCR amplified (primer sequences are indicated in Table S1) and cloned into the pENTR/D-TOPO vector (Life technologies). The resulting pENTR/D-NPR1 vector was used to transfer NPR1 coding sequence into the pMDC32 binary vector [40] using LR Clonase II (Life Technologies). Finally, pMDC32-NPR1 was transferred into the Arabidopsis CCA1∷LUC+ or LHY∷LUC+ backgrounds by Agrobacterium-mediated transformation [41]. For that, Agrobacterium GV3101 cells carrying the pMDC32-NPR1 plasmid were grown overnight in liquid Luria-Bertani medium supplemented with kanamycin (50mg/L) and gentamycin (30mg/L). Cells were harvested by centrifugation (3220 × g for 10min at room temperature) and resuspended in 5% sucrose solution containing 0.02% Silwet L-77 (Lehle seeds). Developing Arabidopsis inflorescences for the aforementioned reporter lines were dipped into the agrobacterium cell suspension for 30sec, and dipped plants were wrapped with a plastic film and incubated horizontally in a growth chamber for 16-24 h. Finally, the plastic film was removed, and plants were returned to the normal growth position and incubated in a growth chamber until seed collection (∼1.5 months). To generate 35S∷LUC+ lines, the multiple cloning site (MCS) from pBluescript KS(-) was PCR amplified (primer sequences are indicated in Table S1) and cloned into pENTR/D-TOPO. The resulting pENTR/D-MCS vector was used to transfer MCS sequence into the pMDCLUC+ vector [42] using LR Clonase II. Finally, pMDC-MCSLUC+ was transferred into the Arabidopsis Col-0 background by Agrobacterium-mediated transformation [41], as described above.
The rbohd(CCA1∷LUC+), npr1-1(CCA1∷LUC+) and npr1-1(LHY∷LUC+) lines were generated by genetic cross and mutations were confirmed by PCR.
Plant growth conditions
For Pseudomonas syringae infection assays, stratified sterile seeds were grown in autoclaved soil (Sunshine professional mix, Sungro) under 12h light (∼100 μmol.m-2.s-1) / 12h dark cycles (LD) for 14 days at 22°C. At the beginning of day 1 5, plants were transferred to constant light (60 μmol.m-2.s-1, 22°C) (LL) for bioluminescence imaging or RNA tim e course tissue collection. Single leaf infections were performed at the beginning of the second day in LL (ZT24).
For transient SA or H2O2 treatments, Arabidopsis seeds were placed on 60mm plates (12 seeds/plate) containing 1× Murashige & Skoog basal salts (MS) medium (Caisson Labs) supplemented with 3% sucrose overlaid with a nylon mesh (50 micron square opening, white) (Small parts) and stratified for 2-3 days at 4°C. Plates were incubated for 10 days under 12 hour light (100 μmol.m-2.s-1)/12 hour dark cycles (LD) at 22°C. At the beginning of the 11th day, plants were transferred to constant light (60 μmol.m-2.s-1, 22°C) (LL) for bioluminescence imaging or RNA time course tissue collection. Transient SA or H2O2 treatments were performed during the second day in LL.
To determine NPR1 expression level in NPR1 overexpression lines, seedlings were grown in petri dishes containing 1× MS - 3% sucrose medium for 10 days under LD cycles at 22°C.
Pseudomonas syringae culture conditions
Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) [43] liquid cultures (King's B medium: 2% Proteose peptone No.3, 1% Glycerol, 8.6mM K2HPO4 and 6mM MgSO4) were grown in the dark at 28°C (shaking at 175rpm) until OD600 between 0.6 and 0.7 was reached (several dilutions were started to assure that a suitable culture was available at the time of treatment).
Accession numbers
Gene models in this article can be found in The Arabidopsis Information Resource (TAIR) (www.arabidopsis.org) with the following accession numbers: CCA1, AT2G46830; LHY, AT1G01060; TOC1, AT5G61380; RBOHD, AT5G47910; NPR1, AT1G64280; IPP2, AT3G02780; PP2A, AT1G13320.
Method Details
Single leaf Pseudomonas syringae infection
To prepare Pst DC3000 cell suspension inoculum, bacteria from a liquid culture (OD600 between 0.6 and 0.7) were harvested by centrifugation at 3220 × g for 2min, resuspended in sterile water (LabChem), and harvested by centrifugation at 3220 × g for 3min. The bacterial pellet was resuspended in water (LabChem) adjusting OD600 to 0.2 (∼1×10ˆ8 cfu), and Silwet L77 (Lehle seeds) was added to a final concentration of 0.025%. About half of a single leaf was dipped into this Pst DC3000 cell suspension or a mock solution (0.025% Silwet L77) for 1min. After treatment, excess inoculum was blot-dried from the leaf surface using a sterile filter paper strip and plants were returned to LL for bioluminescence imaging (Fig. S1A) or for RNA time course tissue collection.
Transient SA and H2O2 treatment
Nylon meshes with seedlings were transferred from growth plates (1× MS - 3% sucrose) to treatment plates (1× MS - 3% sucrose containing 0.2mM SA, 1mM SA, 50mM H2O2, or 200mM H2O2), or mock plates (MS without SA or H2O2). Treatment and mock plates were incubated in LL for 4 hours. After incubation, nylon meshes with seedlings were briefly blotted on a sterile filter paper, and then placed back to the original MS plate and incubated in LL for bioluminescence imaging (Figure S2A) or for RNA time course tissue collection.
Bioluminescence detection
One day before the imaging period started, plants were sprayed with 5mM of D-lucifernin potassium salt (in 0.01% triton X-100 solution). For soil grown plants, 5mM D-lucifernin potassium salt (in water solution) was also added to the soil at the same time (3ml per plant). Bioluminescence was quantified every 1h (for CCA1∷LUC+ soil or MS grown seedlings), every 2h (for LHY∷LUC+ and TOC1∷LUC+ MS grown seedlings) or every 2.5h (for LHY∷LUC+ and TOC1∷LUC+ soil grown seedlings) using a Pixis 1024 CCD camera (Princeton Instruments).
Bioluminescence data analysis
Bioluminescence images were processed using the MetaMorph image analysis software (Molecular Devices) to determine bioluminescence counts (for plate and soil grown plants) and number of bioluminescent pixels (for soil grown plants) per plant or for a specific tissue section. To estimate plant size across an entire time course experiment, a third-order polynomial curve was regressed from the experimental pixel count data using GraphPad Prism version 6 (GraphPad Software, www.graphpad.com) (curve fitting was used to minimize pixel count bias due to plant movement and unequal signal bleeding due to rhythmic bioluminescence levels). Bioluminescence counts (for plate grown seedlings) and plant size normalized bioluminescence values (for soil grown plants or tissue sections) for each experiment were analyzed by Fast Fourier Transform-Non Linear Least Squares (FFT-NLLS) [44] using the interface provided by the Biological Rhythms Analysis Software System (BRASS) [45]. Amplitude changes within each experiment were calculated as the ratio between the amplitude value obtained for each individual (both for mock and treated plants) and the mean amplitude obtained for mock treated plants (amplitude change = individual amplitude / mean amplitude mock) (log10 transformed ratios were used for statistical analysis). Normalized phase values were calculated as [24*(t-24)/p], where p is the period of the corresponding individual plant calculated by BRASS and t is the fitted acrophase time closest to the second subjective morning (ZT24) extrapolated using BRASS. Normalized phase mean values were calculated using an R circular statistics package [46]. Phase shifts were calculated by subtracting the normalized phase of each individual from the mean normalized phase of mock treated plants (phase shift = mean phase mock – individual phase) (phase advance or delay were indicated by positive or negative values respectively). Normalized phase versus relative amplitude error circular plots were generated using an R “polar.plot” function [47].
Biomass measurement
Tissue collection was performed at the end of single leaf P. syringae infection experiments (single plant aerial tissues) (Figure S1A) or transient SA treatment experiments (pooled whole seedlings from each treatment plate) (Figure S2A). Fresh weight was determined immediately after tissue collection and dry weight was determined after 10 days incubation at 37°C.
mRNA transcript quantification
Pooled tissue samples were collected from MS-grown plants upon mock or SA treatment (12 seedlings per pool), and soil-grown plants upon single leaf mock or P. syringae treatment (5 plants without the treated leaf per pool) and snap freezed in liquid nitrogen. Total RNA from these samples was isolated using the RNeasy plant mini kit (Qiagen) and on-column DNAse (Roche) treatment. For cDNA synthesis, 1μg of total RNA was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad). Transcript levels in each sample were determined by real time quantitative PCR (qPCR) (Bio-Rad CFX96 Real-time PCR detection system) using the Maxima Sybr green qPCR mix (Life Technologies), and the following PCR conditions: 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, 60°C for 25 seconds, and 72°C for 25 seconds. qPCR primer sequences are indicated in Table S1. Gene expression levels were normalized to a reference gene (IPP2 or PP2A) using the comparative Ct method [48] and then to the mean expression of mock samples within each biological replicate. Amplitude values and acrophase times for clock gene expression in mRNA time courses were determined from fitted sine waves of average expression traces by the following formula: Y=Amplitude*sin((2*pi*(t-Acrophase)/period)+pi/2)+baseline (where t is the time in LL and Y is the corresponding gene expression level) using GraphPad Prism version 6.
Quantification and Statistical Analysis
Statistical analyses of circular data (i.e. phase values) were performed using an R circular statistics package [46], and the Watson-Williams test in a matlab circular statistics tool box [49]. All other statistical analyses were performed using GraphPad Prism version 6. Details of statistical tests applied are indicated in figure legends including statistical methods, number of biological replicates, number of individuals, mean and error bar details, and statistical significances.
Supplementary Material
Highlights.
A single leaf P. syringae infection affects Arabidopsis clock rhythms systemically.
A pulse of SA phenocopies low amplitude clock rhythms triggered by P. syringae.
NPR1 antagonizes clock responses triggered by P. syringae and SA.
RBOHD partly mediates P. syringae and SA effects on the Arabidopsis clock function.
Acknowledgments
We thank Dr. Steve Briggs for providing Pst DC3000 strain and sharing infection protocols, and Dr. Frank Harmon for sharing the R script to generate polar plots.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health by the National Institutes of Health [R01GM056006 to J.L.P. (co-investigator)], the Hellman Foundation [to J.L.P.] and the UCSD Academic Senate [to J.L.P.].
Footnotes
Author Contributions: Conceptualization, J.L.P.; Methodology, Z.L. and J.L.P.; Validation, Z.L. and J.L.P.; Formal Analysis, Z.L.; Investigation, Z.L., K.B. and F.U.; Writing-Original Draft, Z.L. and J.L.P.; Writing-Review & Editing, Z.L. and J.L.P.; Funding Acquisition, J.L.P.; Resources, Z.L. and J.L.P.; Supervision, J.L.P.
Conflict of Interest: The authors declare no competing conflicts of interest.
Supplemental Information: Supplemental information includes four figures and one table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bendix C, Marshall CM, Harmon FG. Circadian Clock Genes Universally Control Key Agricultural Traits. Molecular plant. 2015;8:1135–1152. doi: 10.1016/j.molp.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 3.Green RM, Tingay S, Wang ZY, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant physiology. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenham K, McClung CR. Integrating circadian dynamics with physiological processes in plants. Nature reviews Genetics. 2015;16:598–610. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez SE, Kay SA. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harbor perspectives in biology. 2016;8 doi: 10.1101/cshperspect.a027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu PY, Harmer SL. Wheels within wheels: the plant circadian system. Trends in plant science. 2014;19:240–249. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nohales MA, Kay SA. Molecular mechanisms at the core of the plant circadian oscillator. Nature structural & molecular biology. 2016;23:1061–1069. doi: 10.1038/nsmb.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends in plant science. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Xie Q, Anderson RG, Ng G, Seitz NC, Peterson T, McClung CR, McDowell JM, Kong D, Kwak JM, et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS pathogens. 2013;9:e1003370. doi: 10.1371/journal.ppat.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, Wang W, Karapetyan S, Mwimba M, Marques J, Buchler NE, Dong X. Redox rhythm reinforces the circadian clock to gate immune response. Nature. 2015;523:472–476. doi: 10.1038/nature14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X. NPR1, all things considered. Current opinion in plant biology. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS pathogens. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalachova T, Iakovenko O, Kretinin S, Kravets V. Involvement of phospholipase D and NADPH-oxidase in salicylic acid signaling cascade. Plant physiology and biochemistry : PPB. 2013;66:127–133. doi: 10.1016/j.plaphy.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annual review of plant biology. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 15.Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a multifaceted hormone to combat disease. Annual review of phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 16.Ederli L, Madeo L, Calderini O, Gehring C, Moretti C, Buonaurio R, Paolocci F, Pasqualini S. The Arabidopsis thaliana cysteine-rich receptor-like kinase CRK20 modulates host responses to Pseudomonas syringae pv. tomato DC3000 infection. Journal of plant physiology. 2011;168:1784–1794. doi: 10.1016/j.jplph.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 17.Herrera-Vasquez A, Salinas P, Holuigue L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Frontiers in plant science. 2015;6:171. doi: 10.3389/fpls.2015.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogany M, von Rad U, Grun S, Dongo A, Pintye A, Simoneau P, Bahnweg G, Kiss L, Barna B, Durner J. Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant physiology. 2009;151:1459–1475. doi: 10.1104/pp.109.141994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant & cell physiology. 2015;56:1472–1480. doi: 10.1093/pcp/pcv063. [DOI] [PubMed] [Google Scholar]
- 20.Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Current opinion in plant biology. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Bonfig KB, Schreiber U, Gabler A, Roitsch T, Berger S. Infection with virulent and avirulent P. syringae strains differentially affects photosynthesis and sink metabolism in Arabidopsis leaves. Planta. 2006;225:1–12. doi: 10.1007/s00425-006-0303-3. [DOI] [PubMed] [Google Scholar]
- 22.de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bolter B, Delfino L, et al. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nature plants. 2015;1:15074. doi: 10.1038/nplants.2015.74. [DOI] [PubMed] [Google Scholar]
- 23.Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502:689–692. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Mitra RM, Hasselmann KD, Sato M, Lenarz-Wyatt L, Cohen JD, Katagiri F, Glazebrook J. The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Molecular plant-microbe interactions : MPMI. 2008;21:1408–1420. doi: 10.1094/MPMI-21-11-1408. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi N, Hirata Y, Aihara K, Mas P. A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell. 2015;163:148–159. doi: 10.1016/j.cell.2015.08.062. [DOI] [PubMed] [Google Scholar]
- 26.Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carella P, Wilson DC, Cameron RK. Some things get better with age: differences in salicylic acid accumulation and defense signaling in young and mature Arabidopsis. Frontiers in plant science. 2014;5:775. doi: 10.3389/fpls.2014.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou YR, Bor M, Yan J, Preuss AS, Jander G. Arabidopsis NATA1 Acetylates Putrescine and Decreases Defense-Related Hydrogen Peroxide Accumulation. Plant physiology. 2016;171:1443–1455. doi: 10.1104/pp.16.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravichandran S, Stone SL, Benkel B, Prithiviraj B. Purple Acid Phosphatase5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC plant biology. 2013;13:107. doi: 10.1186/1471-2229-13-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Science signaling. 2009;2:ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- 34.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. ROS signaling: the new wave? Trends in plant science. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Sewelam N, Kazan K, Schenk PM. Global Plant Stress Signaling: Reactive Oxygen Species at the Cross-Road. Frontiers in plant science. 2016;7:187. doi: 10.3389/fpls.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. The Plant cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 39.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. The Plant cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature protocols. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 42.Farre EM, Kay SA. PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. The Plant journal : for cell and molecular biology. 2007;52:548–560. doi: 10.1111/j.1365-313X.2007.03258.x. [DOI] [PubMed] [Google Scholar]
- 43.Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. The Plant cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. Journal of biological rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 45.Southern MM, Brown PE, Hall A. Luciferases as reporter genes. Methods in molecular biology. 2006;323:293–305. doi: 10.1385/1-59745-003-0:293. [DOI] [PubMed] [Google Scholar]
- 46.Agostinelli C, Lund U. R package ‘circular’: Circular Statistics (version 0.4-93) 2017 https://r-forge.r-project.org/projects/circular/
- 47.Marshall CM, Tartaglio V, Duarte M, Harmon FG. The Arabidopsis sickle Mutant Exhibits Altered Circadian Clock Responses to Cool Temperatures and Temperature-Dependent Alternative Splicing. The Plant cell. 2016;28:2560–2575. doi: 10.1105/tpc.16.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 49.Berens P. CircStat: A MATLAB Toolbox for Circular Statistics. J Stat Softw. 2009;31:1–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.