Abstract
Acetaminophen (APAP) overdose is the leading cause of acute liver failure. Yet the mechanisms underlying adaptive tolerance toward APAP-induced liver injury are not fully understood. To better understand molecular mechanisms contributing to adaptive tolerance to APAP is an underpinning foundation for APAP-related precision medicine. In the current study, the mRNA and microRNA (miRNA) expression profiles derived from next generation sequencing data for APAP-treated (5 and 10 mM) Hep-aRG cells and controls were analyzed systematically. Putative miRNAs targeting key dysregulated genes involved in APAP hepatotoxicity were selected using in silico prediction algorithms, un-biased gene ontology, and network analyses. Luciferase reporter assays, RNA electrophoresis mobility shift assays, and miRNA pull-down assays were performed to investigate the role of miRNAs affecting the expression of dysregulated genes. Levels of selected miRNAs were measured in serum samples obtained from children with APAP overdose (58.6–559.4 mg/kg) and from healthy controls. As results, 2758 differentially expressed genes and 47 miRNAs were identified. Four of these miRNAs (hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p) suppressed drug metabolizing enzyme (DME) levels involved in APAP-induced liver injury by downregulating HNF1A, HNF4A and NR1I2 expression. Exogenous transfection of these miRNAs into HepaRG cells effectively rescued them from APAP toxicity, as indicated by decreased alanine aminotransferase levels. Importantly, hsa-miR-320a and hsa-miR-877-5p levels were significantly elevated in serum samples obtained from children with APAP overdose compared to health controls. Collectively, these data indicate that hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p suppress DME expression involved in APAP-induced hepatotoxicity and they contribute to an adaptive response in hepatocytes.
Keywords: MicroRNAs, Acetaminophen, Drug metabolizing enzymes, Adaptation, Pharmacogenomics
Introduction
Acetaminophen (APAP) is a widely used over-the-counter analgesic drug and over-dose of APAP can cause APAP-induced hepatotoxicity (Davidson and Eastham 1966). The majority of APAP is metabolized in the liver through glucuronidation and sulfation, specifically through UDP-glucuronosyltransferases and sulfotransferases. A small portion of APAP is activated by cytochrome P450 enzymes to produce the reactive metabolite N-acetyl-p-benzoquinone imine (NAPQI) that binds to proteins and is regarded as a critical step in the development of hepatotoxicity. Glutathione (GSH) conjugation by GSTT1, GSTP1, and GSTM1 efficiently detoxifies NAPQI, but is depleted by exposures to large doses of APAP, resulting in increased covalent binding of NAPQI to proteins (Zhao and Pickering 2011). Despite this existing body of knowledge, few studies have examined molecular mechanisms underlying inter-individual variability and the adaptive tolerance effect in APAP-induced hepatotoxicity (Adjei et al. 2008; Court et al. 2001; Jetten et al. 2016; McKillop et al. 1999; Strubelt et al. 1979).
Recent studies have shown that miRNAs influence the expression of drug metabolizing enzymes (DMEs) (Jin et al. 2016; Mohri et al. 2010; Tsuchiya et al. 2006; Wang et al. 2017; Yu et al. 2010, 2015a, b, c; Zeng et al. 2017), which may thereby influence drug efficacy and safety (Koturbash et al. 2015). miRNAs have been linked to liver injury and represent candidate biomarkers of liver injury because they are sufficiently stable in a wide range of biofluids, including serum (Baraniskin et al. 2012; Chen et al. 2008). Several studies have attempted to address the relationship of miR-NAs and APAP-induced hepatotoxicity (Krauskopf et al. 2015; Vliegenthart et al. 2015; Wang et al. 2009; Ward et al. 2014; Yang et al. 2015). However, the fine regulatory mechanisms of these miRNAs in relation to APAP-induced hepatotoxicity are still unclear.
Adaptive tolerance, or resistance to APAP toxicity that occurs following “pre-exposure” to APAP, is believed to occur in humans and experimental animals exposed to APAP over a long time period (Eakins et al. 2015; O’Brien et al. 2000; Shayiq et al. 1999). The molecular mechanisms underlying adaptive tolerance are not well understood. In the present study, we hypothesized that miRNAs modulate APAP metabolism/toxicity by targeting genes encoding DMEs and nuclear receptors (NRs), which then in turn mediate adaptation/adaptive tolerance towards APAP exposure. To test our hypothesis, we screened candidate miRNAs by high-throughput sequencing and in silico analyses and then validated the interactions between miRNAs and genes encoding DMEs and NRs by a series of functional experiments. Our study delineated miRNA-mediated mechanisms related to adaptive tolerance in APAP-induced hepatotoxicity.
Methods
Study population
Clinical samples were obtained from 28 children and subjects participating in a prospective clinical study approved by the institutional review board of the University of Arkansas for Medical Sciences. The control group (n = 10; APAP dose = 0 mg/kg) consisted of healthy children without use of APAP in the preceding 14 days; APAP therapeutic group (n = 10; APAP dose = 10.0–17.5 mg/kg) consisted of hospitalized children receiving APAP per standard of care; and APAP overdose group (n = 8; APAP dose = 58.6–559.4 mg/kg) consisted of children requiring hospitalization for treatment of APAP overdose. The clinical and laboratory data of the subjects, including age, gender, APAP dose, ALT and APAP protein adducts, were reported in our former study (Yang et al. 2015).
Cell culture, transfection and treatment
Terminally differentiated HepaRG cells were purchased from Life Technologies (Carlsbad, CA), incubated in Williams’ E medium supplemented with Thaw, Plate, and General Purpose Medium Supplement (Life Technologies) for 1 day, and then cultured in Williams’ E medium supplemented with the Maintenance/Metabolism Medium Supplement (Life Technologies) for an additional 7 days until use. Both HepG2 and HEK 293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA), and maintained in Rosewell Park Memorial Institute 1640 (RPMI 1640) or Dulbecco’s Modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), respectively. All cell lines used in this study were at passage numbers less than 10 when the experiments were performed.
APAP was purchased from Sigma-Aldrich (St. Quentin Fallavier, France), and dissolved in DMSO to produce 3.3 mol/L stock solutions, and then added to the cell culture at the final concentration of 5 and 10 mM, respectively. Hep-aRG cells treated with equal amount of 0.3% DMSO were used as solvent control. Cells were harvested to extract total RNA or proteins for further experiments 48 h after APAP treatment. Assays were performed in triplicate.
The siRNAs to silence HNF1A, HNF4A and NR1I2 genes, and the mimics or inhibitors for hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, hsa-miR-877-5p, and negative control were obtained from GE Dharmacon (Lafayette, CO). HepaRG cells were transiently transfected with siRNA or miRNA mimic/inhibitor (all final concentration: 40 nmol/L) using Lipofectamine reagent, and cultured for 48 h to extract total RNA or proteins for further experiments.
RNA extraction from cells, medium and serum
The miRNeasy Mini Kit (Qiagen, Valencia, CA) was used to extract total RNA from HepaRG cells, according to the manufacturer’s protocol. The miRNeasy Serum/Plasma Kit (Qiagen) was used to purify total miRNAs in cell medium, with the C. elegans miR-39 (cel-miR-39) as the Spike-in control, according to the manufacturer’s protocol. Total RNA in serum was extracted using TRIzol (Invitrogen, Carlsbad, CA) methods as described previously (Yang et al. 2012).
Gene and miRNA profiling
Purified total RNA from HepaRG cells treated by 0, 5, or 10 mM APAP was divided into two portions. One portion was used for high-throughput RNA sequencing on Illumina HiSeq 1500 systems, according to the manufacturer’s protocol. Raw paired-end sequencing data were mapped to Hg19 reference genome (UCSC) to produce normalized expression data for all transcripts, using TopHat and Cufflinks packages serially (Trapnell et al. 2012). The other portion was used to perform high-throughput miRNA sequencing on Illumina HiSeq 2500. Raw single-end miRNA sequencing data were also mapped to Hg19 reference genome (UCSC) to generate normalized miRNA reads, using mirDeep2 package (Friedlander et al. 2008). The threshold of twofold and tenfold changes (both pFDR < 0.05) was set to generate moderate and severe gene profiles deregulated by 10 mM APAP treatment, respectively. The threshold of twofold change (pFDR < 0.05) was used to obtain miRNA profile induced or repressed by 10 mM APAP treatment.
In silico analyses
The potential miRNA target genes were identified based on the integrated gene and miRNA expression profile, using miRTar.human database (http://mirtar.mbc.nctu.edu.tw/human/). RNAhybrid algorithm (http://bibiserv2.cebitec.unibielefeld.de/rnahybrid) was used to predict the free energy of potential miRNA:mRNA duplexes. Gene ontology and network analyses were carried out by the Ingenuity Pathway Analysis (IPA) software.
Luciferase reporter gene assays
The pGL3-CU vector that expresses Firefly luciferase and described in our previous reports (Yu et al. 2015a, c) was used to create reporter gene plasmids. Firstly, the core 3′-UTRs of CYP3A4, HNF1A, HNF4A and NR1I2 genes that contain the targeting elements for cognate miRNAs were produced by PCR amplification (all primers or oligonucleotides used in this study were purchased from Integrated DNA Technologies (Coraville, IA), and listed in Supplementary Table 7), and subcloned into the linearized nicked pGL3-CU vector to create CYP3A4-CU, HNF1A-CU, HNF4A-CU, and NR1I2-CU constructs, respectively. Site-directed mutagenesis was carried out in the miRNA response elements of the constructs above to generate the corresponding mutant constructs CYP3A4-Mut, HNF1A-Mut, HNF4A-Mut1, HNF4A-Mut2, HNF4A-D-Mut and NR1I2-Mut. All constructs used in this study were sequenced to confirm their authenticity.
HepG2 and HEK 293T cells were seeded into 96-multiwell plates, cultured to 80% confluence, and then transfected with constructed plasmids (100 ng/well), together with miRNA mimic or miRNA negative control (all final concentration: 40 nmol/L), using the Lipofectamine reagent (Life Technologies) per the manufacturer’s instructions. The pRL-SV40 plasmid (Promega, Madison, WI; 1 ng/well) that highly expresses Renilla luciferase was co-transfected, and used to normalize the transfection efficiencies. Dual-Luciferase Reporter 1000 Assay System (Promega) was used to measure the Firefly and Renilla luciferase activities. Three independent experiments were carried out in triplicate.
RNA electrophoretic mobility shift assays (RNA EMSAs)
The hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p oligonucleotides were 5′-modified using IRDye®800 dye, while the cognate mRNA oligonucleotides, i.e. the miRNA response elements in CYP3A4, HNF1A, HNF4A and NR1I2 transcripts, were 2′-O-methyl-modified and 5′-labeled with cy5.5™ dye. Cold miRNA i.e. unlabeled negative control and miRNA oligonucleotides, were used in the competition assays. Cytoplasmic extracts from HepaRG cells were produced using NE-PER Nuclear and Cytoplasmic extraction reagents (ThermoFisher Scientific, Tewksbury, MA) per the manufacturer’s instructions.
The LightShift Chemiluminescent RNA EMSA Kit (ThermoFisher Scientific) was utilized in RNA EMSAs, and the modified protocol was described in our former reports (Yu et al. 2015b, c). Briefly, 200 fmols miRNA and cognate mRNA oligonucleotides were added to basic reaction buffer (1× REMSA binding buffer, 5% glycerol, 200 mM KCl, 100 mM MgCl2) to create miRNA:mRNA complexes, while HepaRG cytoplasmic extracts (2 μg) and non-specific tRNA (1 μg) were subsequently added to form the RNA:protein complexes. Cold oligonucleotides were utilized in competition assays, at 50-fold molar excesses. Antibodies against Ago1, Ago2, Ago3, and Ago4 were purchased from Abcam (Cambridge, MA) and used in supershift assays. All reaction mixtures were adjusted to 20 μL, cultured at room temperature for 20 min, separated by 12% native polyacrylamide gel electrophoresis (PAGE) at 4 °C, and then viewed by Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
miRNA pull-down assays
The miRNA pull-down procedure was modified from the protocol developed by Subramanian et al. (2015). Briefly, 3′-biotin-tagged mismatched miRNA duplexes for hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, hsa-miR-877-5p, and miRNA negative control were synthesized by IDT, and 5′ phosphatized by T4 polynucleotides (New England Biolabs, Ipswich, MA) per the manufacturer’s instructions. HepaRG cells were transiently transfected with the miRNA duplexes (final concentration: 20 nmol/L), cultured for 18 h, and then lysed using the lysis buffer containing 3% IGEPAL ® CA-630 (Sigma-Aldrich) to produce cytoplasmic extracts. The biotin–miRNA–mRNA complex in cytoplasmic extracts was enriched by the Dynabeads MyOne Streptavidin C1 bead (Life technologies), and purified by the miRNeasy Mini Kit.
Real-time PCR validation
QuantiTect Reverse Transcription Kits (Qiagen) and NCode™ microRNA First-Strand cDNA Synthesis Kits (Life Technologies) were used to produce the first-strand cDNA from extracted total RNA and miRNA samples, respectively. Real-time quantitative reverse transcription-PCR (qRT-PCR) was performed using the Quanti-Fast SYBR® Green RT-PCR Kit (Qiagen) with the ABI Prism7900 Sequence Detection System (Applied Biosystems, Foster City, CA).
The comparative Ct method was used to calculate the relative expression of RNAs. The relative mRNA levels for candidate genes were normalized to GAPDH levels, while cellular miRNA levels were normalized to U6 levels. The miRNA levels in serum samples were calculated by comparing to Let-7d levels, as described in our former reports (Yang et al. 2015). The miRNA levels from cell culture medium for HepaRG cells after APAP treatment were adjusted by Spike-in control cel-miR-39, and compared to miRNA levels in the corresponding control samples.
Western blotting
Antibodies against the DMEs and NRs were purchased from Abcam. Total proteins were isolated from HepaRG cells using detergent lysis buffer (RIPA, ThermoFisher Scientific). Quantitative analyses for western blotting were performed using the Odyssey CLx Infrared Imaging System.
ALT assays
As described by Miyakawa et al. (2015), the cell culture medium was collected and the remaining cells were lysed with an equal volume medium containing 1% Triton X-100. To eliminate unbroken cells or cell debris, the medium and cell lysate were centrifuged at 700g for 5 min. The ALT activity was detected by Liquid ALT Reagent (Pointe Scientific, Caton, MI) according to the manufacture’s protocol. Relative ALT in the medium was calculated through a percentage of total ALT activity (medium plus cells lysate). Assays were performed in triplicate.
Lactate dehydrogenase (LDH) assays
Cellular necrosis in HepaRG cells was assessed by measurement of lactate dehydrogenase (LDH) release into media using an ACE Alera Serum Chemistry Analyzer (Alfa Wassermann, Green Brook, NJ) following the manufacturer’s instructions. All experiments were performed in triplicates.
Statistical analyses
Student’s t test was used to detect the differences between subgroups in luciferase activity, while one-way ANOVA on ranks test was used to detect the differences between subgroups for protein or RNA levels. P < 0.05 was considered statistically significant.
Accession codes
RNA-seq and miRNA-seq data have been deposited at the NCBI SRA (http://www.ncbi.nlm.nih.gov/sra), with the accession number SRP094716.
Results
Differentially expressed mRNAs and miRNAs in HepaRG cells exposed to APAP
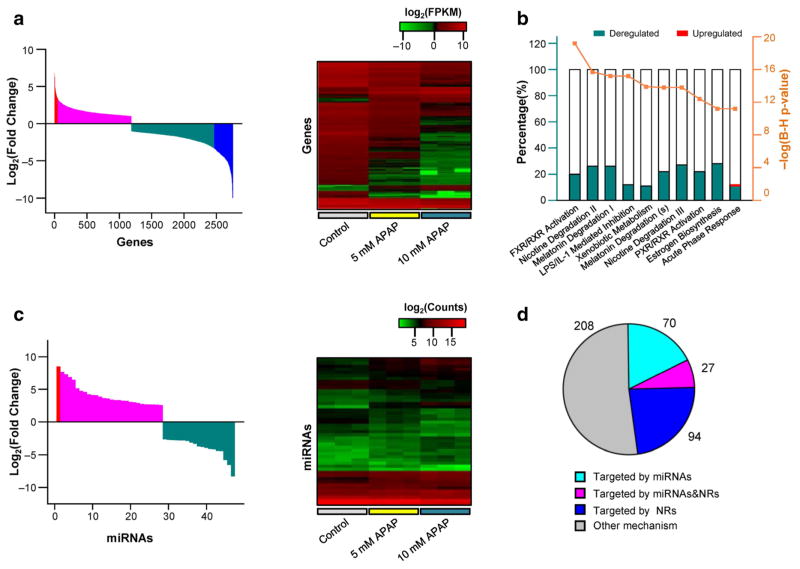
Previous research (McGill et al. 2011) and our preliminary data supported the selection of 5 and 10 mM concentrations for examining APAP toxicity in HepaRG cells (Supplementary Fig. 1). Under our experimental conditions, 2758 differentially expressed genes were observed in HepaRG cells exposed to 10 mM APAP compared with those in cells exposed to vehicle control, using twofold change (pFDR < 0.05) as the threshold (Supplementary Table 1). Gene expression analyses using un-biased gene ontology and network analyses showed that 1194 genes (43.3%, involved in hepatic fibrosis, cell apoptosis, cell migration, inflammation response) were up-regulated, while 1564 genes were down-regulated (56.7%, involved in FXR/RXR activation, LXR/RXR activation, acute phase response) (Supplementary Fig. 2; Fig. 1a, left panel). Furthermore, 339 genes (286 down-regulated and 53 up-regulated, Supplementary Table 2) had a fold change greater than tenfold, and consisted of genes involved in signal transduction, gene expression regulation, and xenobiotic metabolism (Fig. 1b; Supplementary Table 3). In addition, 47 miRNAs had a fold change greater than 2 (pFDR < 0.05; 19 down-regulated and 28 up-regulated) in the treated cells (APAP 10 mM treatment), compared to the untreated cells (Fig. 1c; Supplementary Table 4). When the HepaRG cells were treated with APAP (5 mM), 1311 or 22 deregulated genes were identified using twofold or tenfold change as the threshold, respectively (Supplementary Table 1 and 2); while only 17 miRNAs were deregulated with a fold change greater than twofold (Supplementary Table 5). We then selected the gene and miRNA profiles obtained by 10 mM APAP for further analysis, due to its more significant effects on gene expression or miRNA production compared to those obtained by treatment with 5 mM APAP.
Fig. 1.
Deregulated global gene and miRNA expression in HepaRG cells exposed to APAP. a Distribution of log-fold changes of 2758 genes (fold change > 2, left panel), and heat map of 339 dramatically deregulated genes (fold change > 10, right panel). b Top 10 pathways of 339 deregulated genes listed in IPA analysis. The y-axis (left) indicates the ratio of number of deregulated genes mapping to specific pathway, and the y-axis (right) indicates the P value calculated by Benjamini–Hochberg multiple testing correction. c Distribution of log-fold changes (left panel), and heat map (right panel) of 47 miR-NAs (fold change > 2). d Venn diagram of the overlap of deregulated genes (fold change > 10) regulated by miRNAs and NRs
In silico analysis was subsequently used to further examine the relationship of miRNAs to gene dysregulation. miRNAs were selected for in silico analysis according to two criteria: (1) miRNAs that down-regulate the cognate genes in a opposite direction, and (2) the free energy of the miRNA:mRNA hybridization was ≤ −20 kcal/mol (based on our previous studies (Yu et al. 2015a, c)). As shown in Table 1, 29 miRNAs were predicted to target 97 genes, and detailed prediction results are summarized in Supplementary Table 5. In addition, HNF1A, HNF4A and NR1I2 (also known as PXR), three key nuclear receptors responsible for the regulation of DMEs were down-regulated (93, 98 and 97%, respectively). Ingenuity Pathway Analysis showed that HNF1A, HNF4A and NR1I2 may play important roles in the expression of 121 deregulated genes, including 27 genes predicted to be regulated by the candidate miRNAs (Fig. 1d and Supplementary Table 6).
Table 1.
Deregulated miRNAs and target genes by acetaminophen exposure
| miRNA name | FCsa | Target genesb |
|---|---|---|
| hsa-miR-10b-5p | 2.306 | ALPL, ANXA13, BDH1, CHST4, ECM2, ETNK2, IYD, MTMR3, SDPR, SLC10A1, VNN1 |
| hsa-miR-122-5p | 0.201 | GPR1, MAFF, NCAM1, RGS16, SESN2, SLC7A11, SLC7A5, SLC9A1, SLCO5A1 |
| hsa-miR-1246 | 5.994 | HMGCS2 |
| hsa-miR-1268a | 0.458 | MAFF, SLC6A9, UNC5B |
| hsa-miR-128-1-5p | 0.469 | CPA4, DCLK1, HFE, ICOSLG, NCAM1, NGFR, SLC1A4, SLC6A17, SLC7A11, SLC7A5, TPRXL, TPRXL, ASNS |
| hsa-miR-1290 | 2.520 | HNMT, MTMR3, PLGLB2 |
| hsa-miR-129-5p | 2.156 | ABCA6, MTTP, PIPOX |
| hsa-miR-130b-5p | 0.335 | MAFF, SESN2, SLC7A11, UPP2 |
| hsa-miR-139-5p | 3.133 | ALDH1A1, CDH1, HNMT, MYLK, SERPIND1, ST6GAL1, TNFSF10 |
| hsa-miR-143-3p | 2.234 | MASP1 |
| hsa-miR-181d-5p | 2.049 | ACSS1, AQP9, IYD, KCNK5, SLC5A9 |
| hsa-miR-185-5p | 0.421 | CPA4, HCN4, HFE, NCAM1, RGS16, SESN2, SLCO5A1, SYP |
| hsa-miR-192-3p | 0.448 | DUSP5, ICOSLG |
| hsa-miR-192-5p | 0.465 | ARL4C, ATF3, NGFR, PSAT1 |
| hsa-miR-194-3p | 0.297 | ARHGEF2, ARL4C, FOSL1, HAP1, ICOSLG, MAFF, NCAM1, NGFR, RFX2, SERPINE1, SLC1A4, SLC6A17, SLC7A5 |
| hsa-miR-199a-3p | 2.103 | PLGLB2, SGK2 |
| hsa-miR-210-5p | 0.316 | HCN4, IGFBP3, NCAM1, NGFR |
| hsa-miR-224-5p | 3.737 | CDH1, CYP3A4, DIO1, IFITM2, KLHDC7A, MTMR3, OLFML3, SDS, SLC47A2 |
| hsa-miR-3065-5p | 3.564 | DDC, ECM2 |
| hsa-miR-30b-3p | 0.320 | ARL4C, CPA4, HAP1, MAFF, NGFR, SLC7A5, SYP, TNFRSF9, UNC5B |
| hsa-miR-3144-3p | 2.933 | SORBS2, UGT2A3 |
| hsa-miR-320a | 2.481 | C6, CYP8B1, DDC, EDN2, FYN, HNF1A, HOXA3, MAP2K6, MTMR3, NQO1, RCN3, RORC, SELL, SLC17A4, SORBS2, TM4SF4 |
| hsa-miR-320b | 6.750 | C6, CYP8B1, DDC, EDN2, FYN, HNF1A, HOXA3, MAP2K6, MTMR3, NQO1, RCN3, RORC, SELL, SLC17A4, TM4SF4 |
| hsa-miR-320c | 7.557 | C6, CYP8B1, DDC, EDN2, FYN, HNF1A, HOXA3, MAP2K6, NQO1, RCN3, RORC, SELL, SLC17A4, TM4SF4 |
| hsa-miR-320d | 10.65 | C6, CYP8B1, EDN2, HOXA3, MAP2K6, NQO1, RCN3, RORC, SELL, TM4SF4 |
| hsa-miR-33b-3p | 0.358 | ARL4C, SLC6A17, UPP2 |
| hsa-miR-449a | 2.586 | CEACAM1, ESPN, FETUB, GBA3, HNF4A, IYD, KRT5, MASP1, NQO1, NRXN2, TM4SF4 |
| hsa-miR-877-5p | 2.120 | AQP9, CYP1A1, G6PC, MASP1, MAT1A, NR1I2, OLFML3 |
| hsa-miR-92b-3p | 0.482 | SLC9A1 |
Fold changes of miRNAs in HepaRG cells treated by 10 mM acetaminophen
Predicted by IPA software
Candidate miRNAs targeting DMEs and NRs involved in APAP metabolism
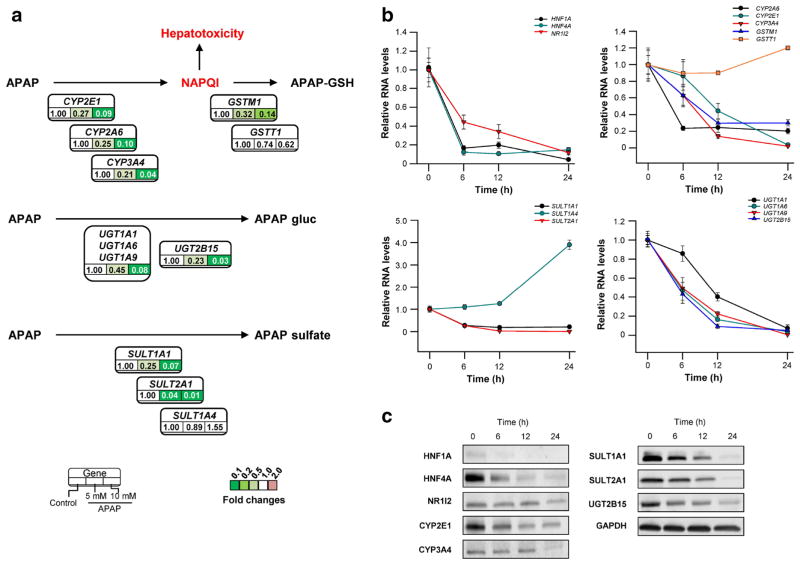
Previous studies suggested that DMEs were the most important proteins accounting for the inter-individual variability of APAP-induced hepatotoxicity and the development of adaptive tolerance (Adjei et al. 2008; Court et al. 2001; Jetten et al. 2016; McKillop et al. 1999; Strubelt et al. 1979). Since un-biased gene expression analyses revealed that genes encoding DMEs and NRs were highly dysregulated after APAP treatment, detailed expression profiles of key DMEs involved in the APAP metabolism were examined. Expression data for genes encoding metabolic activation (CYP2E1, CYP3A4, and CYP2A6), glutathione conjugation (GSTM1, and GSTT1), glucuronidation (UGT1A1, UGT1A6, UGT1A9 and UGT2B15), and sulfation (SULT1A1, SULT1A4 and SULT2A1) were extracted and reanalyzed. As shown in Fig. 1b, the expression of genes encoding DMEs was decreased significantly (> 10 fold change) in HepaRG cells treated with 10 mM APAP, with the exception of GSTT1 and SULT1A4. RNA-seq data were subsequently confirmed by qRT-PCR and/or western blot assays using HepaRG samples obtained at multiple time points after APAP exposure. As shown in Fig. 2b, c, time-dependent reduction of gene expression was observed.
Fig. 2.
The “shut-down” of APAP-metabolism system in HepaRG cells under APAP exposure. a RNA-seq data for genes encoding metabolic activation (CYP2E1, CYP3A4, and CYP2A6), glutathione conjugation (GSTM1, and GSTT1), glucuronidation (UGT1A1, UGT1A6, UGT1A9 and UGT2B15), and sulfation (SULT1A1, SULT1A4 and SULT2A1) were extracted and reanalyzed. The “shut-down” of APAP-metabolism system were also identified using qRT-PCR (b) and western blot (c) in HepaRG samples obtained at multiple time points after APAP exposure
In silico analyses showed that that CYP3A4 was a targeted gene of hsa-miR-224-5p, HNF4A and NR1I2, while CYP2E1, SULT1A1, SULT2A1, UGT1A1 and UGT2B15 were regulated by only NRs. Neither NRs nor miRNAs were predicted to regulate CYP2A6 or GSTM1 genes. Of interest, HNF1A, HNF4A and NR1I2 were identified to be modulated by hsa-miR-320 family, hsa-miR-449a and hsa-miR-877-5p, respectively.
CYP3A4, HNF1A, HNF4A and NR1I2 are targets of miRNAs
Biochemical experiments were conducted to validate the regulatory roles of miRNAs in the expression of CYP3A4, HNF1A, HNF4A and NR1I2. First, reporter gene constructs containing the core 3′-UTRs of CYP3A4, HNF1A, HNF4A and NR1I2, or the corresponding mutated constructs as negative controls were transfected into HepG2 and HEK 293T cells, together with the cognate miRNA mimics or miRNA negative control. As shown in the Supplementary Fig. 3, transfections of hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p efficiently suppressed luciferase activities, reflecting the expression of wild-type 3′-UTRs of CYP3A4, HNF1A, HNF4A or NR1I2 (all P < 0.05), but not the activities corresponding to the expression of mutated 3′UTRs for these genes.
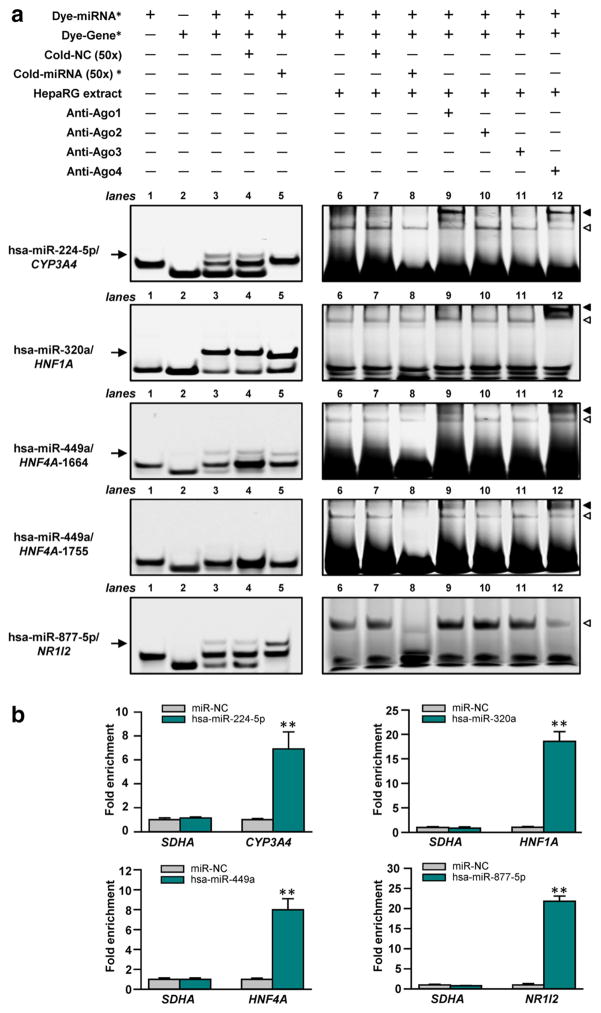
RNA electrophoretic mobility shift assays (RNA EMSA) and miRNA pull-down assays were conducted to test the direct interactions between miRNAs and cognate response elements in the CYP3A4, HNF1A, HNF4A and NR1I2 genes in vitro and in vivo. As shown in Fig. 3a (lane 3), hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p each formed distinct complexes with the response elements in 3′UTRs of CYP3A4, HNF1A, HNF4A and NR1I2 mRNA transcripts. Cytoplasmic extracts from HepaRG cells were able to bind to the miRNA:mRNA complexes created by the hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p oligonucleotides and their cognate mRNA oligonucleotides, to form electrophoretically stable protein–RNA complexes (Fig. 3a, lane 6). In competition assays, excess unlabeled miRNA oligonucleotides attenuated or eliminated both miRNA:mRNA complexes and protein–RNA complexes (Fig. 3a, lanes 5 and 8, respectively), while excess unlabeled nonspecific oligonucleotides failed to show any inhibitory effects (Fig. 3a, lane 4 and 7), suggesting sequence-specific interactions between these miRNAs and their cognate mRNA targets. Moreover, an antibody against Ago4 formed a new supershift band when co-incubated with protein–RNA complexes containing hsa-miR-224-5p, hsa-miR-320a, and hsa-miR-449a oligonucleotides, respectively (Fig. 3a, lane 12). Similarly, an antibody against Ago2 showed the ability to interact with protein–RNA complexes containing hsa-miR-224-5p and hsa-miR-449a oligonucleotides (Fig. 3a, lane 9).
Fig. 3.
miRNAs interacted with the cognate target mRNAs in vitro and in vivo. a RNA EMSAs using 5′-dyed miRNA or mRNA oligonucleotide, cytoplasmic extracts, and antibodies against Ago1-4. Asterisk represents specific gene or miRNA labeled at the left of each panel; arrow represents the miRNA/ mRNA complex; hollow triangle represents miRNA/mRNA/ protein complex; solid triangle represents supershift complex. b The mRNAs of CYP3A4, HNF1A, HNF4A or NR1I2 was enriched by streptavidin pull-down for biotinylated hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p, respectively. **P < 0.001
A modified miRNA pull-down method was used to capture the miRNA:mRNA complex in vivo. As shown in Fig. 3b, after transfections of the 3′-biotinylated oligonucleotides of hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p, the miRNA:mRNA complexes formed with their cognate CYP3A4, HNF1A, HNF4A or NR1I2 mRNA were significantly pulled down by streptavidin-coated beads (6.9-fold enriched for CYP3A4, 18.6-fold enriched for HNF1A, 7.9-fold enriched for HNF4A, and 21.8-fold enriched for NR1I2; all P < 0.001), compared with transfection of miRNA negative controls. Non-target control SDHA mRNAs were pulled down by the streptavidin beads, indicating the specificity of the interaction between miRNA and the target genes.
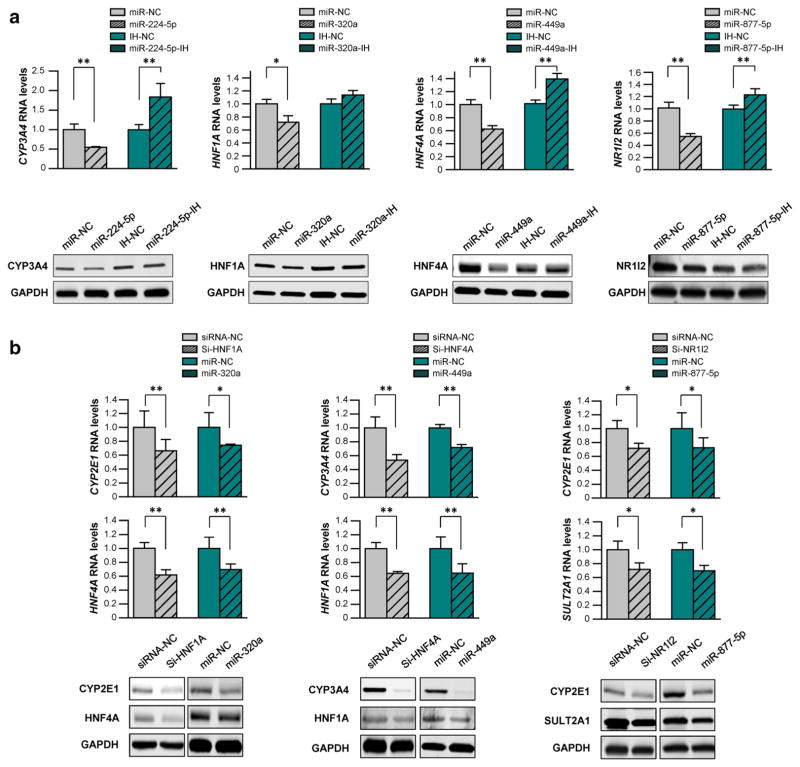
We further transfected hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p mimics into HepaRG cells to test the inhibitory efficacy of miRNAs against the endogenous production of CYP3A4, HNF1A, HNF4A and NR1I2. As shown in Fig. 4a, hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p mimics significantly reduced endogenous mRNA and protein levels of CYP3A4, HNF1A, HNF4A and NR1I2 (45 and 51% for CYP3A4, 28 and 56% for HNF1A, 40 and 66% for HNF4A, and 46 and 25% for NR1I2, all P < 0.05). In addition, the transfection of the specific inhibitors against hsa-miR-224-4p or hsa-miR-449a effectively rescued the suppression of mRNA expression and protein production of CYP3A4 or HNF4A by endogenous miRNAs (1.83-fold expression and 1.43-fold production for CYP3A4, and 1.40-fold expression and 1.53-fold production for HNF4A, all P < 0.05). A marginal rescue effect was observed with the transfection of hsa-miR-320a or hsa-miR-877-5p inhibitors, probably due to competition among hsa-miR-320 family members, the low expression of endogenous hsa-miR-877-5p, or other unknown epigenetic mechanisms. In summary, these data support the postulation that miRNAs target CYP3A4, HNF1A, HNF4A and NR1I2.
Fig. 4.
miRNAs inhibited endogenous DMEs expression, directly (a) or indirectly (b). The siRNAs specific for HNF1A, HNF4A and NR1I2, or mimics and inhibitors of hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p were transfected into the differentiated HepaRG cells, respectively. Each assay was carried out in triplicate. Data are shown as relative mRNA and protein levels normalized by GAPDH levels. *P < 0.05; **P < 0.001; NC miRNA negative control
miRNAs indirectly reduced the expression of DMEs by targeting NRs
Considering the key regulatory roles of HNF1A, HNF4A and NR1I2 in the expression of DMEs, we hypothesized that hsa-miR-320a, hsa-miR-449a and hsa-miR-877-5p suppressed the production of DMEs through their inhibitory effects on the expression of NRs. Specific siRNAs of HNF1A, HNF4A and NR1I2 were transfected into HepaRG cells (positive control), resulting in efficiently decreased expression of their target genes (Supplementary Fig. 4). As shown in Fig. 4b, the mRNA and protein levels of CYP2E1 and HNF4A, genes regulated by HNF1A, were significantly decreased by HNF1A-specific siRNA and hsa-miR-320a (34 and 26% for mRNA levels, 54 and 45% for protein levels, in the regulation of CYP2E1; 38 and 30% for mRNA levels, 60 and 41% for protein levels, in the regulation of HNF4A; all P < 0.05). Similarly, HNF4A-specific siRNA and hsa-miR-449a indirectly decreased mRNA and protein levels of CYP3A4 and HNF1A (44 and 30% for mRNA levels, 90 and 82% for protein levels in the regulation of CYP3A4; 38 and 38% for mRNA levels, 50 and 48% for protein levels in the regulation of HFN1A; all P < 0.05), while NR1I2-specific siRNA and hsa-miR-877-5p inhibited mRNA expression and protein production of CYP2E1 and SULT2A1 (36 and 35% for mRNA levels, 28 and 30% for protein levels in the regulation of CYP2E1; 28 and 30% for mRNA levels, 39 and 21% for protein levels in the regulation of HFN1A; all P < 0.05). Taken together, the data illustrate that the miRNAs modulated the production of DMEs directly and/ or indirectly.
miRNAs hsa-miR-320a and hsa-miR-877-5p are elevated in the serum of APAP-overdose patients
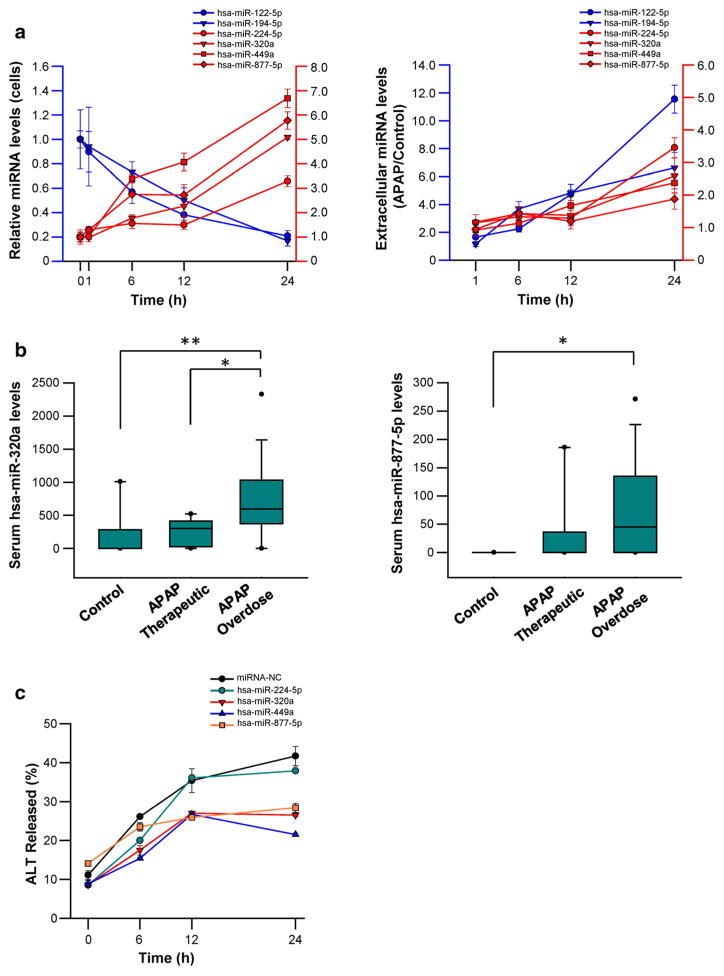
In initial studies, we measured the levels of hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p, together with the levels of hsa-miR-122-5p and hsa-miR-194-5p (two well-known biomarkers for APAP-induced toxicity) in Hep-aRG cells and medium samples harvested at multiple time points after 10 mM APAP exposure. As shown in Fig. 5a, cellular and extracellular levels of hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p significantly increased in response to 10 mM APAP exposure. In contrast, hsa-miR-122-5p and hsa-miR-194-5p levels were reduced in cells but increased in media (Fig. 5a).
Fig. 5.
miRNAs served as serum biomarkers to predict APAP-induced hepatotoxicity. a Time-dependent deregulation of miRNAs in HepaRG cells or medium, respectively. b Increased serum miRNA levels of hsa-miR-320a, or hsa-miR-877-5p in APAP overdose group, compared to the control and/or therapeutic dose group. *P < 0.05; **P < 0.001. c Exogenous transfection of hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p reduced ALT levels induced by APAP
We subsequently quantified serum levels of hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p in human subject samples from controls (no APAP exposure), therapeutic (“low dose” exposure), and APAP overdose. Compared to the control or APAP therapeutic groups (0 [0–1010.10] or 301.25 [0–521.97]; median [range]), serum hsa-miR-320a levels were significantly increased in APAP overdose subjects (9595.75 [0–2330.44], P < 0.05). In addition, levels of hsa-miR-877-5p were also higher in the APAP overdose group than therapeutic groups (45.31 [0–271.15] versus 0 [0–0]), P < 0.05) (Fig. 5b). No expression of hsa-miR-224-5p or hsa-miR-449a was observed in any groups.
Exogenous transfection of hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p rescued hepatic cells from APAP-induced hepatotoxicity
Finally, we transfected hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p mimics into HepaRG cells, and treated the cells with 10 mM APAP. Exogenous transfection of hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p effectively rescued the cells from APAP-induced hepatotoxicity, as indicated by significantly decreased ALT levels (Fig. 5c).
Discussion
Through the integration of high-throughput mRNA and miRNA sequencing data, we found that hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p were able to suppress the production of key DMEs involved in APAP metabolism, either directly or indirectly. We also observed elevations of these miRNAs in serum samples from APAP overdose patients. Further, we demonstrated that introduction of exogenous hsa-miR-320a, hsa-miR-449a, or hsa-miR-877-5p rescued hepatic cells from APAP-induced hepatotoxicity. Compared to previous studies reporting the gene or miRNA expression profiles associated with APAP-induced hepatotoxicity (Jetten et al. 2015; Morishita et al. 2006; Wang et al. 2009), the present study comprehensively and systematically examined the whole-genome expression of miRNAs and protein-coding genes affected by APAP-induced hepatotoxicity, validated the regulatory network between genes and miRNAs, and thus is relevant to understanding the role of miRNAs in regulating CYP450 expression in development of adaptive tolerance in APAP-induced hepatotoxicity.
Studies have suggested that repeated exposures to APAP in animal models or human subjects resulted in an adaptive tolerance effect against APAP-induced hepatotoxicity (adaptation) (Eakins et al. 2015; O’Brien et al. 2000; Shayiq et al. 1999); however, the mechanisms by which “adaptation” develops are unclear. Eakins et al. observed that specific enzymes involved in APAP metabolism, such as CYP2E1, UGT1A1 and SULT2A1, were significantly depleted in the adaptation to APAP, and speculated that the “shut-down” of APAP-metabolism system should represent a key facet of adaptation (Eakins et al. 2015). Thorgeirsson et al. reported that APAP treatment decreased the production of P450s during initial exposure to APAP (Thorgeirsson et al. 1976), similar results were reported in the mouse model and in studies using human liver microsomes (Snawder et al. 1994; Zhang et al. 2004). In contrast, sub-cytotoxic of APAP increased the expression of CYP2E1, CYP3A4 and other P450s (Kim et al. 2007), via unclear mechanisms.
In this study we observed this “shut-down” phenomenon; with the most striking changes in gene expression being those associated with metabolism. To validate the effects of APAP on UDP-glucuronosyltransferases and sulfotransferases, we extracted the expression data of the key genes, including genes encoding UDP-glucuronosyltransferases and sulfotransferases, in HepaRG cells from the GSE74000 data set (gene expression data affected by sub-cytotoxic concentration of APAP) (Rodrigues et al. 2016), and observed similar “shut-down” response (data not shown).
Our study delineated molecular mechanisms for the hypothesis of Eakins et al., and indicated that the miRNAs and NRs contributed to the suppression of APAP-metabolism system. The emergent “shut-down” of P450s system by miRNAs and NRs is biologically reasonable, because it can directly abolish the production of NAPQI to protect cells against APAP-induced hepatotoxicity, while the dramatic reduction of UDP-glucuronosyltransferases and sulfotransferases could be attributed to the “side effect” of the reduction of DMEs, largely due to the uniform modulation of DMEs by miRNAs and NRs.
Our results showed that hsa-miR-224-5p targeted CYP3A4, while hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p indirectly modulated the expression of downstream P450s, glucuronosyltransferases and sulfotransferases by targeting HNF1A, HNF4A and NR1I2. Though previous studies reported that several miRNAs are involved in the expression of DMEs and NRs, such as miR-27b for CYP3A4 (Pan et al. 2009), miR-378 for CYP2E1 (Mohri et al. 2010), miR-24 and miR-34a for HNF4A (Wang and Burke 2013), and miR-148a for NR1I2 (Smutny et al. 2013), no deregulation of these miRNAs was observed in HepaRG cells exposed to APAP under our experimental conditions. Despite differences in cell types, algorithms for miRNA-targeting prediction, and experimental strategies, these results indicated the complexity of regulatory networks between miRNAs and their cognate targets by which miRNAs play different roles in versatile cellular processes including drug responses.
Levels of hsa-miR-320a and hsa-miR-877-5p were significantly elevated in the serum samples of APAP overdosed patients, compared to serum samples from “low dose” exposures to APAP and controls. In addition, the exogenous transfection of hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p effectively rescued HepaRG cells from APAP-induced toxicity. Previous studies (Vliegenthart et al. 2015; Wang et al. 2009; Ward et al. 2014; Yang et al. 2015) showed that multiple miRNAs, including miR-122, let-7d, and miR-29a, were increased in the serum of humans or mice overdosed with APAP, and speculated that these miR-NAs were transported into serum by some specific mechanism (Wang et al. 2009). We found that hsa-miR-122-5p and hsa-miR-194-5p were down-regulated in HepaRG cells exposed to 10 mM APAP, and yet were increased in cell medium and patients’ serum samples, which supports the speculation by Wang et al. that miR-122 might be eliminated from cells via exportation, thus interfering with its suppressive role in hepatotoxicity. Collectively, these data suggest that extracellular elevations hsa-miR-320a and hsa-miR-877-5p in cell media or serum samples represent a cellular response to APAP injury and that miRNAs are released from cells during necrosis or leakage of cellular contents. Exogenous transfection of hsa-miR-320a, hsa-miR-449a or hsa-miR-877-5p was able to rescue HepaRG cells from APAP-induced hepatotoxicity, further indicating the protective role of these miRNAs.
In conclusion, by RNA and miRNA sequencing and functional characterization, we identified multiple miRNAs that regulate DMEs involved in APAP metabolism, either directly or indirectly, through the modulation of HNF1A, HNF4A or NR1I2. More importantly, our findings suggest that hsa-miR-224-5p, hsa-miR-320a, hsa-miR-449a, and hsa-miR-877-5p play a self-protective or adaptive role in APAP-induced liver injury.
Supplementary Material
Acknowledgments
This work was supported by the US FDA, protocol # E0731311. B. N., W. T., R. D. B. and L. J. proposed and organized the study; D. Y., P. G. and B. N. designed the study; D. Y., S. C., Y. J. and J. S. conducted the total RNA extraction; L. W., W. X., H. H., L. S., S. B., S. M., H. F. and T. S. analyzed the gene and miRNA expression data; D. Y., Z. R., Y. W., Y. C., and L. Z. conducted the Reporter assays and RNA EMSA assays; D. Y., V. T., I. P., S. M. and S. B. conducted miRNA pull-down experiment; B. K., J. S. and L. S. provided the technical support; X. Y., K. D., Y. G, L. J. and L. S analyzed the miRNA levels in serum samples. D. Y. and B. N. wrote the manuscript; W. H. T, W. X., L. G., Y. G., W. M., L. J., P. G., S. B., S. M., N. M., I. P. and W. T. contributed to data interpretation and revised the manuscript. All authors reviewed the manuscript.
Footnotes
Disclaimer: The views presented in this paper are those of the authors and do not necessarily represent those of the U.S. Food and Drug Administration.
Electronic supplementary material: The online version of this article (doi:10.1007/s00204-017-2090-y) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest: The authors declare no competing financial interests.
References
- Adjei AA, Gaedigk A, Simon SD, Weinshilboum RM, Leeder JS. Interindividual variability in acetaminophen sulfation by human fetal liver: implications for pharmacogenetic investigations of drug-induced birth defects. Birth Defects Res A Clin Mol Teratol. 2008;82(3):155–165. doi: 10.1002/bdra.20535. [DOI] [PubMed] [Google Scholar]
- Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012;14(1):29–33. doi: 10.1093/neuonc/nor169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Court MH, Duan SX, von Moltke LL, et al. Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J Pharmacol Exp Ther. 2001;299(3):998–1006. [PubMed] [Google Scholar]
- Davidson DG, Eastham WN. Acute liver necrosis following overdose of paracetamol. Br Med J. 1966;2(5512):497–499. doi: 10.1136/bmj.2.5512.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakins R, Walsh J, Randle L, et al. Adaptation to acetaminophen exposure elicits major changes in expression and distribution of the hepatic proteome. Sci Rep. 2015;5:16423. doi: 10.1038/srep16423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, et al. Discovering micro-RNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Jetten MJ, Ruiz-Aracama A, Coonen ML, et al. Interindividual variation in gene expression responses and metabolite formation in acetaminophen-exposed primary human hepatocytes. Arch Toxicol. 2015 doi: 10.1007/s00204-015-1545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten MJ, Ruiz-Aracama A, Coonen ML, et al. Interindividual variation in gene expression responses and metabolite formation in acetaminophen-exposed primary human hepatocytes. Arch Toxicol. 2016;90(5):1103–1115. doi: 10.1007/s00204-015-1545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Yu D, Tolleson WH, et al. MicroRNA hsa-miR-25-3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochem Pharmacol. 2016;113:88–96. doi: 10.1016/j.bcp.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SN, Seo JY, Jung DW, Lee MY, Jung YS, Kim YC. Induction of hepatic CYP2E1 by a subtoxic dose of acetaminophen in rats: increase in dichloromethane metabolism and carboxy-hemoglobin elevation. Drug Metab Dispos. 2007;35(10):1754–1758. doi: 10.1124/dmd.107.015545. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Tolleson WH, Guo L, et al. microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark Med. 2015;9(11):1153–1176. doi: 10.2217/bmm.15.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf J, Caiment F, Claessen SM, et al. Application of high-throughput sequencing to circulating microRNAs reveals novel biomarkers for drug-induced liver injury. Toxicol Sci. 2015;143(2):268–276. doi: 10.1093/toxsci/kfu232. [DOI] [PubMed] [Google Scholar]
- McGill MR, Yan HM, Ramachandran A, Murray GJ, Rollins DE, Jaeschke H. HepaRG cells: a human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology. 2011;53(3):974–982. doi: 10.1002/hep.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKillop IH, Schmidt CM, Cahill PA, Sitzmann JV. Altered Gq/G11 guanine nucleotide regulatory protein expression in a rat model of hepatocellular carcinoma: role in mitogenesis. Hepatology. 1999;29(2):371–378. doi: 10.1002/hep.510290201. [DOI] [PubMed] [Google Scholar]
- Miyakawa K, Albee R, Letzig LG, et al. A cytochrome P450-independent mechanism of acetaminophen-induced injury in cultured mouse hepatocytes. J Pharmacol Exp Ther. 2015;354(2):230–237. doi: 10.1124/jpet.115.223537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79(7):1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Morishita K, Mizukawa Y, Kasahara T, et al. Gene expression profile in liver of differing ages of rats after single oral administration of acetaminophen. J Toxicol Sci. 2006;31(5):491–507. doi: 10.2131/jts.31.491. [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Slaughter MR, Swain A, et al. Repeated acetaminophen dosing in rats: adaptation of hepatic antioxidant system. Hum Exp Toxicol. 2000;19(5):277–283. doi: 10.1191/096032700678815918. [DOI] [PubMed] [Google Scholar]
- Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37(10):2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RM, Heymans A, De Boe V, et al. Toxicogenomics-based prediction of acetaminophen-induced liver injury using human hepatic cell systems. Toxicol Lett. 2016;240(1):50–59. doi: 10.1016/j.toxlet.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Shayiq RM, Roberts DW, Rothstein K, et al. Repeat exposure to incremental doses of acetaminophen provides protection against acetaminophen-induced lethality in mice: an explanation for high acetaminophen dosage in humans without hepatic injury. Hepatology. 1999;29(2):451–463. doi: 10.1002/hep.510290241. [DOI] [PubMed] [Google Scholar]
- Smutny T, Mani S, Pavek P. Post-translational and post-transcriptional modifications of pregnane X receptor (PXR) in regulation of the cytochrome P450 superfamily. Curr Drug Metab. 2013;14(10):1059–1069. doi: 10.2174/1389200214666131211153307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snawder JE, Roe AL, Benson RW, Roberts DW. Loss of CYP2E1 and CYP1A2 activity as a function of acetaminophen dose: relation to toxicity. Biochem Biophys Res Commun. 1994;203(1):532–539. doi: 10.1006/bbrc.1994.2215. [DOI] [PubMed] [Google Scholar]
- Strubelt O, Siegers CP, Volpel M, Younes M. Studies on the mechanism of paracetamol-induced protection against paracetamol hepatotoxicity. Toxicology. 1979;12(2):121–133. doi: 10.1016/0300-483x(79)90038-6. [DOI] [PubMed] [Google Scholar]
- Subramanian M, Li XL, Hara T, Lal A. A biochemical approach to identify direct microRNA targets. Methods Mol Biol. 2015;1206:29–37. doi: 10.1007/978-1-4939-1369-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson SS, Sasame HA, Mitchell JR, Jollow DJ, Potter WZ. Biochemical changes after hepatic injury from toxic doses of acetaminophen or furosemide. Pharmacology. 1976;14(3):205–217. doi: 10.1159/000136597. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. Micro-RNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66(18):9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- Vliegenthart AD, Shaffer JM, Clarke JI, et al. Comprehensive microRNA profiling in acetaminophen toxicity identifies novel circulating biomarkers for human liver and kidney injury. Sci Rep. 2015;5:15501. doi: 10.1038/srep15501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burke PA. The role of microRNAs in hepatocyte nuclear factor-4alpha expression and transactivation. Biochem Biophys Acta. 2013;1829(5):436–442. doi: 10.1016/j.bbagrm.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu D, Tolleson WH, et al. A systematic evaluation of microRNAs in regulating human hepatic CYP2E1. Biochem Pharmacol. 2017 doi: 10.1016/j.bcp.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J, Kanchagar C, Veksler-Lublinsky I, et al. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc Natl Acad Sci USA. 2014;111(33):12169–12174. doi: 10.1073/pnas.1412608111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Greenhaw J, Shi Q, et al. Identification of urinary micro-RNA profiles in rats that may diagnose hepatotoxicity. Toxicol Sci. 2012;125(2):335–344. doi: 10.1093/toxsci/kfr321. [DOI] [PubMed] [Google Scholar]
- Yang X, Salminen WF, Shi Q, et al. Potential of extracellular microRNAs as biomarkers of acetaminophen toxicity in children. Toxicol Appl Pharmacol. 2015;284(2):180–187. doi: 10.1016/j.taap.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Dhakal IB, Beggs M, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118(2):391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Marrone A, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015a;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Tolleson W, et al. MicroRNA hsa-miR-29a-3p modulates CYP2C19 production in human liver cells. Biochem Pharmacol. 2015b doi: 10.1061/j.bcp.2015.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Tolleson WH, Knox B, et al. Modulation of ALDH5A1 and SLC22A7 by microRNA hsa-miR-29a-3p in human liver cells. Biochem Pharmacol. 2015c doi: 10.1016/j.bcp.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Chen Y, Wang Y, et al. MicroRNA hsa-miR-370-3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol. 2017 doi: 10.1016/j.bcp.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QX, Melnikov Z, Feierman DE. Characterization of the acetaminophen-induced degradation of cytochrome P450-3A4 and the proteolytic pathway. Basic Clin Pharmacol Toxicol. 2004;94(4):191–200. doi: 10.1111/j.1742-7843.2004.pto940406.x. [DOI] [PubMed] [Google Scholar]
- Zhao L, Pickering G. Paracetamol metabolism and related genetic differences. Drug Metab Rev. 2011;43(1):41–52. doi: 10.3109/03602532.2010.527984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.