Abstract
RNA molecules can fold into intricate shapes that can provide an additional layer of control of gene expression beyond that of their sequence. In this Review, we discuss the current mechanistic understanding of structures in 5′ untranslated regions (UTRs) of eukaryotic mRNAs and the emerging methodologies used to explore them. These structures may regulate cap-dependent translation initiation through helicase-mediated remodelling of RNA structures and higher-order RNA interactions, as well as cap-independent translation initiation through internal ribosome entry sites (IRESs), mRNA modifications and other specialized translation pathways. We discuss known 5′ UTR RNA structures and how new structure probing technologies coupled with prospective validation, particularly compensatory mutagenesis, are likely to identify classes of structured RNA elements that shape post-transcriptional control of gene expression and the development of multicellular organisms.
In the ancient RNA world, RNA likely served as the main catalytic, self-replicating and information-carrying component pre-dating cellular life1,2. Such intricate activities can be traced to the ability of RNA to fold into complex secondary structures and tertiary structures. In many cases, these RNA structures appear more dynamic than globular protein domains or the double-helical DNA structure. Primarily in bacteria and archaea but also in algae, fungi and plants, prominent examples of RNA structure-directed functions include ribozymes and metabolite-sensing riboswitches3,4. The roles of RNA structures in splicing and in gene regulation by non-coding RNAs may have been a driving force in the evolution of ancient eukaryotes5,6. Most strikingly, the peptidyl transferase centre of ribosomal RNA (rRNA) is a structured remnant of the RNA world that is central to protein synthesis in all living cells.
We focus here on mRNA structures with functions in translation initiation, which is ultimately linked to the ribosome — the macromolecular complex that harbours this peptidyl transferase activity. The great number of protein and RNA components involved in ribosome initiation, scanning, elongation and recycling of mRNAs highlights that translation — especially translation initiation, which is one of the most crucial steps in translation7,8 — is a highly regulated process (BOX 1). Indeed, the ribosome itself, which for a long time was regarded as a constitutive, housekeeping molecular machine, has only recently been appreciated to be functionally heterogeneous with respect to its associated proteins9,10. Translation regulation can also involve structures in the mRNA itself, which are rearranged and unfolded by the ribosome and by RNA remodellers such as RNA helicases. This complex and dynamic interplay of mRNA structures and the translation machinery raises an important question: how much regulatory potential is encoded in mRNA structures or in the structure-mediated sensing and recruitment of interacting factors such as RNA-binding proteins (RBPs) or trans-acting RNAs11–13?
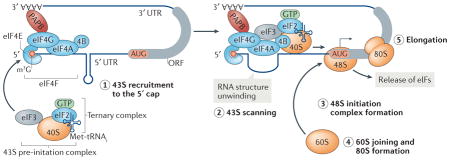
Box 1. Canonical cap-dependent translation initiation.
The translation of most eukaryotic mRNAs is initiated by ribosome recruitment to the 5′ cap, followed by ribosome scanning towards a start codon (see the figure). Canonical cap-mediated initiation mainly occurs by recruitment of the 40S small ribosomal subunit and its associated eukaryotic initiation factors (eIFs) to the 5′ end 7-methylguanosine (m7G) cap structure (reviewed in REFS 7,8,20). The first step of initiation is the assembly of the trimeric eIF4F cap-binding complex at the 5′ cap, wherein cap-binding protein eIF4E interacts with the scaffolding initiation factor eIF4G and the RNA helicase eIF4A. Poly(A) binding protein (PABP) on the 3′ poly(A) tail interacts with eIF4G at the cap, thereby circularizing the mRNA. Through its interaction with eIF3, eIF4G recruits the 43S pre-initiation complex (step 1), which consists of eIF3, the 40S ribosomal subunit, the ternary complex of GTP-bound eIF2 and the initiator tRNA (eIF2–GTP–Met-tRNAi), and eIF1, eIF1A and eIF5 (not shown). Once the 43S complex binds the mRNA near the cap, it travels (‘scans’) along the 5′ untranslated region (UTR) in the 5′ to 3′ direction (step 2) in an ATP-dependent reaction, with partial hydrolysis of the eIF2-bound GTP to GDP in the ternary complex, until it encounters a start codon (AUG). The RNA helicase eIF4A migrates with the 43S complex and unwinds inhibitory RNA secondary structures in the 5′ UTR (a stem–loop structure is shown). Stable binding of the 43S complex at the start codon yields the formation of the 48S initiation complex (step 3) and triggers GTP hydrolysis and release of eIFs. Subsequently, the 60S large ribosomal subunit joins the 40S ribosomal subunit to form the elongation-competent 80S ribosome (step 4), which then proceeds to translation elongation (step 5). Starvation and other stress conditions inhibit the formation of the ternary complex through phosphorylation of eIF2α and block eIF4F assembly by sequestering eIF4E, which is bound by E-binding protein (not shown), thereby suppressing cap-dependent translation initiation. 4B, eIF4B; ORF, open reading frame.
One of the great challenges in answering this question has been to accurately find mRNA structures that modulate translation. Although mRNA primarily transmits genetic information from DNA to protein through its coding sequence (CDS), the 5′ and 3′ untranslated regions (UTRs) are non-coding and do not directly contribute to the protein sequence. Free from the constraints of encoding proteins, UTRs can form considerable Watson–Crick and non-canonical base pairing that can potentially impact every step of translation. Indeed, algorithms for modelling RNA secondary structure14,15 have suggested that UTRs have the potential to engage in intricate RNA base-pairing patterns, which may change in response to protein binding and may impact the recruitment of ribosomes. Although these algorithms have had difficulty in processing long RNA sequences and predicting long-range interactions or complex structures such as pseudoknots and still fail to account for protein interactions, the past decade has witnessed a burst of advances in integrating these methods with experimental RNA structure probing methodologies. Such integration raises the prospect of determining the complete ensembles of folding states of all transcribed RNAs in a given cell type at a given time and of understanding how these structural ensembles may give rise to intricate gene regulation programmes. Expected improvements of these methods would enable precise validation of functional mRNA structures inside cells.
In this Review, we highlight recent research on the roles of functional 5′ UTR structures in eukaryotic mRNAs as modulators of translation initiation. However, it is important to note that regulatory elements elsewhere in the mRNA, especially in the 3′ UTR, can also modulate translation16. It is also important to mention that even within 5′ UTRs, unstructured (linear) regulatory elements are likely to have a crucial impact on translation17–21. Linear elements include upstream open reading frames (uORFs)22–24 and the sequences around the start codon of the main ORF. Such sequence elements have been dissected and discussed elsewhere17,21,25. As a classic example, a strong Kozak sequence26 improves start codon recognition as a feature of highly translated mRNAs. Analysis of the relative strength of AUG codon recognition of all possible translation initiation sites has recently revealed specific responsible sequence motifs around start sites in mammals27.
In this Review, we focus on how 5′ UTR structures in mRNAs may block or recruit ribosomes and other regulatory factors to enable a rapid, dynamic response to diverse cellular conditions to control gene expression. We speculate that such structured elements may be particularly important when newly induced responses through mRNA transcription, processing and nuclear export may be too slow28. Instead, the cell may reversibly change its expression profile by adjusting the stability and translation of pre-existing mRNAs in the cytoplasm. For example, activated macrophages derepress the translation of 90 mRNAs — many of which encode anti-inflammatory regulators — within 1 hour of immune stimulation29. We also highlight how recent technological advances are improving our understanding of RNA structure by chemical probing of RNA conformation in vitro, and especially now inside cells, in vivo. We anticipate that these methods will be crucial for the elucidation of yet unknown structured RNA elements that guide translation control.
5′ UTRs as platforms for RNA structure
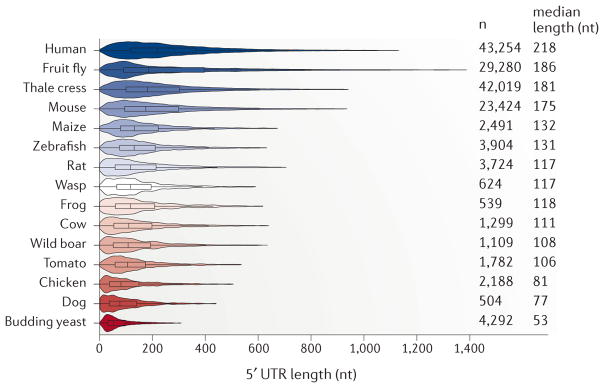
During evolution from invertebrates to humans, genome size greatly increased, and UTRs have especially expanded in length30,31, providing a ‘playground for mRNA evolution’. As UTRs are usually not coated with translating ribosomes, they are presumably more accessible for interactions with regulatory factors. Whereas the length of the 3′ UTR immensely increased during eukaryotic evolution, the 5′ UTR has maintained a median length of approximately 53–218 nucleotides17,19,32,33. The longest known median length of mRNA 5′ UTRs occurs in humans (218 nucleotides)30,31, exceeding those of other mammals and dwarfing that of budding yeast (53 nucleotides) (FIG. 1). However, 5′ UTR lengths vary dramatically among individual genes in higher eukaryotes and can range from a few to thousands of base pairs32,34 (FIG. 1). This large range of 5′ UTR lengths suggests that there may be greater regulation of specific mRNA subsets.
Figure 1. Evolutionary expansion of eukaryotic 5′ UTR lengths.
The length of 5′ untranslated regions (UTRs) has increased in eukaryotes during evolution, with median lengths ranging between 53–218 nucleotides (nt). We compared RefSeq-annotated 5′ UTR lengths of reviewed and validated transcripts (n) between species for which at least 100 5′ UTRs are annotated. For yeast, we used the 5′ UTR lengths as annotated in REFS 34,35. The violin plots depict the distribution of 5′ UTR lengths for 15 species sorted according to decreasing median 5′ UTR length, including human (Homo sapiens), fruit fly (Drosophila melanogaster), thale cress (Arabidopsis thaliana), mouse (Mus musculus), maize (Zea mays), zebrafish (Danio rerio), rat (Rattus norvegicus), wasp (Nasonia vitripennis), western clawed frog (Xenopus tropicalis), cow (Bos taurus), wild boar (Sus scrofa), tomato (Solanum lycopersicum), chicken (Gallus gallus), dog (Canis lupus familiaris) and the budding yeast Saccharomyces cerevisiae. The data range for each species was trimmed to a maximum of the third quartile plus three times the interquartile range (Q3 + 3 × IQR).
High GC content and a highly negative folding free energy (ΔG) level of a 5′ UTR are often used as parameters for predicting 5′ UTR RNA secondary structures. As canonical translation initiation requires that the 43S pre-initiation complex scans the 5′ UTR to reach the start codon (BOX 1), such overall high GC content in the 5′ UTR has been thought to cause inefficient scanning and a lower rate of initiation. Indeed, in classic examples, the prediction of complex secondary structures in GC-rich 5′ UTRs has been correlated with inhibition of translation35, for example, in the mRNA of the metabolic enzyme ornithine decarboxylase36. However, these secondary structure predictions and particularly their functional relevance in cells have not been established. Predictions typically calculate the most stable base pairing of an RNA as the one that has the overall lowest computed ΔG. Although determining the ΔG of an entire RNA structure in an mRNA takes into account the transition of an RNA domain from its fully folded to a completely linear form, scanning is thought to require only local melting of RNA structures rather than linearization of the whole 5′ UTR. Indeed, emerging data indicate that strong local RNA structures and protein interactions may have important roles in impeding ribosome scanning37. Given the limitations of predicting global 5′ UTR mRNA structures in guiding the discovery of RNA structures, the search for individual functional mRNA structures may be more promising. In the next sections, we take a closer look at interesting candidate 5′ UTR RNA structure motifs that regulate mRNA translation and at the methods used to find and confirm them.
5′ UTR structures in ribosome scanning
Translation in eukaryotes typically starts at the 5′ end of the mRNA, which harbours the 5′ cap and a UTR as the entry point for the ribosome (BOX 1; FIG. 2a). Some mRNAs lack a 5′ UTR completely, for example, all mRNA species in mammalian mitochondria are leaderless38, but they are generally rare in higher eukaryotes19. Some human mRNAs with an extremely short 5′ UTR (12 nucleotides on average), known as translation initiator of short 5′ UTR (TISU), undergo scanning-free initiation39. By contrast, some 5′ UTRs are highly structured and can block entry of the ribosome. One of the first and best-studied examples is a small 5′ UTR structural element — the iron responsive element (IRE)40 — which affects the translation of a subset of mRNAs that are important for iron homeostasis41. Briefly, a single conserved IRE stem–loop close to the cap of the mRNAs encoding either the iron storage protein ferritin or iron transporter ferroportin is bound by iron-regulatory protein 1 (IRP1) or IRP2 in low-iron conditions. IRP binding represses translation initiation by preventing the 43S pre-initiation complex from associating with the mRNA42,43 (FIG. 2b). The IRE–IRP ribonucleoprotein (RNP) complex sterically blocks ribosome access to the cap and 5′ UTR. Other stable RNA secondary structures such as cap-proximal hairpins44 might block the assembly of the 43S pre-initiation complex onto the 5′ UTR35,45. The DEAD-box RNA helicase eukaryotic initiation factor 4A (eIF4A), as part of the eIF4F complex that is assembled at the cap, is thought to be crucial for unwinding such structures and therefore preparing a clear path for ribosome scanning46 (BOX 1).
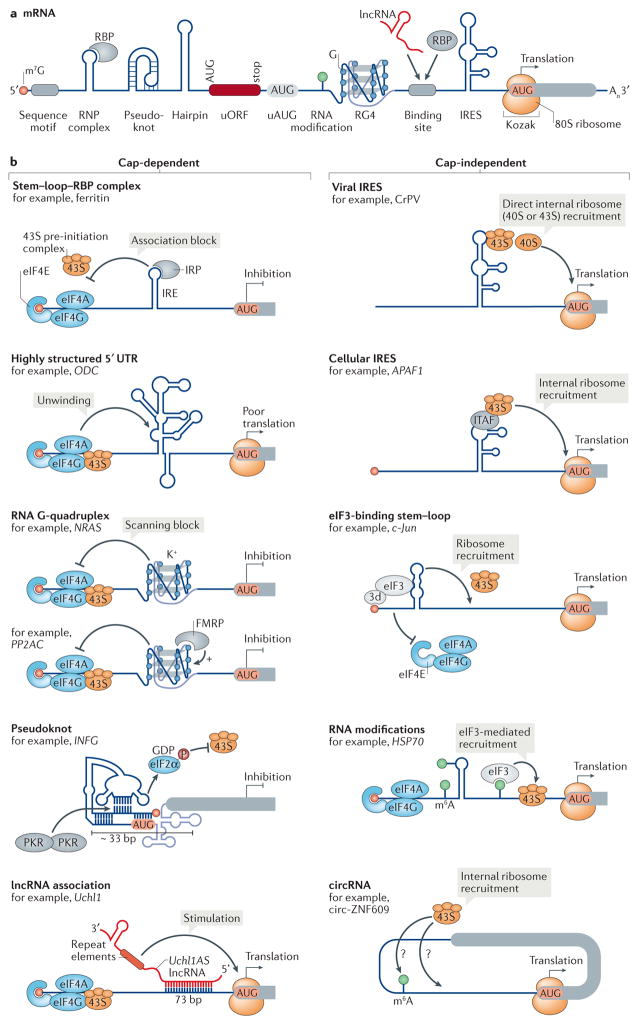
Figure 2. Cis-acting regulatory RNA elements and structures in eukaryotic 5′ UTRs influence mRNA translation.
a | The 7 methylguanosine (m7G) 5′ cap structure (circle) at the 5′ end of the mRNA and the poly(A) tail (An) at the 3′ end stabilize the mRNA and stimulate translation. The 5′ untranslated region (UTR) contains secondary and tertiary structures and other sequence elements. RNA structures such as pseudoknots, hairpins and RNA G-quadruplexes (RG4s), as well as upstream open reading frames (uORFs) and upstream start codons (uAUGs), mainly inhibit translation. Internal ribosomal entry sites (IRESs) mediate translation initiation independently of the cap. RNA modifications, or RNA binding proteins (RBPs) and long non coding RNAs (lncRNAs) that interact with RNA binding sites or form ribonucleoprotein (RNP) complexes, as well as the Kozak sequence around the start codon, can additionally regulate translation initiation. b | Regulatory 5′ UTR RNA structures can influence protein synthesis by promoting or inhibiting either cap-dependent (left) or cap-independent (right) translation. Whereas the structures that regulate cap-dependent initiation — RG4s, stem–loop structures and pseudoknots (but not lncRNAs) — tend to repress initiation, cap-independent regulatory RNA structures, including IRESs, eukaryotic initiation factor 3 (eIF3)-binding stem–loop structures, RNA modifications and circular RNAs (circRNAs), can internally assemble the translation machinery onto the mRNA and generally stimulate translation. 3d, eIF3d; APAF1, apoptotic peptidase activating factor 1; circ-ZNF609, circular-zinc-finger protein 609; CrPV, cricket paralysis virus; FMRP, fragile X mental retardation protein 1; HSP70, heat shock protein 70; IFNG, interferon gamma; IRE, iron responsive element; IRP, iron regulatory protein; ITAF, IRES trans-acting factor; K+, potassium; m6A, N6-methyladenosine; NRAS, NRAS proto oncogene, GTPase; ODC, ornithine decarboxylase; P, phosphorylation; PKR, protein kinase RNA activated; PP2AC, protein phosphatase 2 catalytic subunit alpha (also known asPPP2CA); Uchl1, ubiquitin carboxyl terminal hydrolase L1.
The RNA helicase eIF4A unwinds RNA structures
Recent studies suggest that the function of eIF4A in binding and unwinding RNA can have specific effects on target mRNAs, at least in part through structured RNA elements in 5′ UTRs47–49. Although scanning is assisted by the ATPase-dependent duplex-unwinding activity of eIF4A50, these studies indicate that eIF4A activity is sensitive to both local RNA structures and sequence motifs. Unwinding of local structures may also explain why certain mRNAs that harbour long 5′ UTRs and high GC content are still efficiently translated. For example, the 5′ UTR of the LINE1 mRNA has a high GC content of 60% and is 900 nucleotides in length but is still translated in a cap-dependent manner at a rate similar to that of the very well translated β-actin mRNA51.
Recent unbiased studies based on ribosome profiling have sought to understand the potential specificity of eIF4A in promoting the translation of certain classes of mRNA47–49. Specific small molecule inhibitors of eIF4A are well characterized. For example, silvestrol52,53 and rocaglamide52–54 increase the affinity of eIF4A for RNA. These drugs block the dissociation of eIF4A from RNA and thereby reduce eIF4A recycling. Ribosome profiling of silvestrol-treated human KOPT-K1 leukaemia cells revealed a decrease in the translation efficiency of mRNAs with long 5′ UTRs47, including oncogenes, chromatin modifiers and transcription regulators, which may require tight control of their expression. Interestingly, many of these silvestrol-sensitive transcripts appear to encode a specific structural element in their 5′ UTRs: 12-nucleotide long (CGG)4 motifs that can fold into stable, energetically favourable RNA G-quadruplex (RG4) structures in vitro47 (FIG. 2b), discussed further below. As eIF4A activity is hyperactivated in cancer and silvestrol has been employed in preclinical cancer studies55, RNA structures and the RNA helicase activity of eIF4A could be relevant for cancer research47. Consistent with this possibility, ribosome profiling in silvestrol-treated breast cancer cells revealed that translation initiation was reduced at hundreds of mRNAs, and sensitivity to eIF4A inhibition correlated with the complexity of the inferred structures and the increased length of their 5′ UTRs48. However, only 25% of 5′ UTRs of eIF4A-sensitive mRNAs contain (CGG)4 motifs48, so mRNAs with long 5′ UTRs and structural features other than RG4s may be required for eIF4A sensitivity56. It remains to be determined how many of these 5′ UTR motifs in fact fold into RG4 or other structures. Agreeing in part with the drug-based eIF4A inhibition studies, eIF4A knockdown in MCF7 breast cancer cells followed by polysome profiling and RNA-sequencing analysis revealed eIF4A-sensitive mRNAs that harboured 5′ UTRs with a highly negative ΔG and GC-rich motifs with potential to form RG4s, these measurements also uncovered U-rich and GA-rich sequence motifs57. Moreover, a more recent study using the eIF4A inhibitor rocaglamide A (RocA) found that 5′ UTR structures, including RG4s, contribute little to translation repression49. Rather, by use of toeprinting and RNase I footprinting, RocA was suggested to clamp eIF4A onto polypurine motifs in the 5′ UTRs of target mRNAs. Further increasing this effect of RocA is the possibility that eIF4A ‘trapping’ on certain mRNAs may sequester the helicase from being recycled to other mRNAs that require resolution of 5′ UTR structures, including those with RG4 structures.
In addition to the classic RNA helicase eIF4A, other helicases and initiation factors are also being revealed as key players in translation control. A helicase that may have overlapping activity with eIF4A is the budding yeast helicase Ded1, which appears to be required to scan through long, structured 5′ UTRs58. Other helicases with redundant function in translation, such as the DExH-box protein DHX29, can partially rescue the unwinding of structured 5′ UTRs in the absence of eIF4A activity59. In addition, the eIF4A cofactor eIF4B has been found to stimulate translation of long mRNAs containing structured 5′ UTRs in budding yeast independently of eIF4A, as demonstrated by ribosome profiling, which also correlates with 5′ UTR structure accessibility assessed in vitro60. Finally, following the release of eIF4A upon recognition of the start site, the budding yeast DEAD-box helicase Dhh1 specifically promotes translation of mRNAs that have long and highly structured coding regions61. These examples highlight how diverse RNA helicases and initiation factors can target specific mRNAs with structured regions or motifs, which otherwise may serve as roadblocks to scanning and translation initiation.
RNA G-quadruplex structures
RG4s are stable in vitro, with melting temperatures that are higher than physiological temperature, especially in the presence of potassium ions (K+), which are specifically chelated inside G-quartets. As the cytoplasm contains high concentrations of K+, it has been assumed that RG4s also fold in vivo. The formation of RG4 structures — if validated inside cells — would represent the most stable RNA structure that could block ribosome scanning. Beyond the helicase eIF4A and in contrast to the extensively studied DNA G-quadruplexes62, other physiological roles of RG4s in mRNAs have only fairly recently been explored (reviewed in REFS 63,64) and include roles in mRNA processing and translation regulation (reviewed in REFS 65,66). Most examples of RG4s in 5′ UTRs are linked to translation repression in cis65,67,68 presumably by preventing the 43S pre-initiation complex from binding to mRNA or by slowing down scanning69,70 (FIG. 2b). A scanning block by RG4 structures was first suggested for the human NRAS proto-oncogene mRNA using in vitro translation assays69,71 and for the Zic1 zinc finger protein mRNA in eukaryotic cells72. The stability and position of RG4 structures close to the 5′ end of 5′ UTRs contribute to translation repression in vitro, as tested for the NRAS RG4 (REF. 69), as well as in cells70. However, the inhibitory effect of 5′ UTR RG4s on scanning is still speculative. This effect and the formation of 5′ UTR RG4 structures67 need to be carefully confirmed and studied inside cells. Probing RNA structures inside cells and on a genome-wide scale has only recently been adapted to address the physiological relevance of RG4s in mRNAs. The current data suggest that, inside cells, RG4s in most mRNAs appear mainly unfolded (Supplementary information S1 (box)).
Scanning inhibition is thought to be further increased by recruitment of RG4-stabilizing proteins63,66 such as fragile X mental retardation protein (FMRP)73, which binds to many RG4-harbouring mRNAs74,75 (FIG. 2b). At least in vitro, FMRP appears to bind an RG4 in the CDS of its own mRNA (FMR1)76 and can regulate its own alternative splicing77. However, a clear role for the FMRP–RG4 interaction in translation repression has only been shown for 5′ UTR RG4s in mRNAs other than FMR1 (REFS 76,78). FMRP might inhibit translation initiation or elongation by binding to and stabilizing RG4 structures and recruiting trans-acting factors or by direct binding and stalling of the translating ribosome79,80. Other examples exist of RBPs that promote translation by destabilizing RG4 structures in the CDS81, further highlighting the diverse roles of stable RNA tertiary structures in translation regulation.
Higher-order mRNA structures
RNA secondary structures can form higher-order interactions to assemble tertiary structures or intermolecular RNA complexes. For example, pseudoknots are complex intramolecular RNA structures consisting of at least two intercalated stem–loop structures that form a knot-like three-dimensional shape. A pseudo-knot structure conserved across mammals, along with contiguous helices, has been proposed to reside in the 5′ UTR of human interferon gamma (IFNG) mRNA82,83, on the basis of in vitro ribonuclease mapping, in-line probing and compensatory mutagenesis coupled with structure–function experiments (FIG. 2b). This pseudo-knot signals to another member of the innate immunity pathway, protein kinase R (RNA-activated) (PKR), which is induced by interferon. PKR is typically activated by double-stranded RNAs of >33 bp in length, which do not appear naturally in the cell but are commonly generated by viruses during infection84. Normally, initiating ribosomes unfold the pseudoknot in the IFNG 5′ UTR. The pseudoknot structure refolds as part of a larger, base-paired RNA structure of sufficient length to attract a PKR dimer. The interaction of PKR with the IFNG 5′ UTR is thought to locally activate the kinase, which phosphorylates eIF2α and results in repression of IFNG translation (FIG. 2b). Thus, as part of a feedback loop, this RNA structure adjusts translation of its mRNA to PKR activity levels to prevent excess interferon synthesis82,83.
In addition to pseudoknots, RNAs have the capability to form numerous higher-order interactions, including complexes with trans-acting long non-coding RNAs (lncRNAs) for post-transcriptional control85. In the case of the mouse ubiquitin carboxyl-terminal hydrolase L1 (Uchl1) mRNA, the antisense lncRNA Uchl1AS produced from the same locus undergoes partial base pairing with the Uchl1 mRNA 5′ UTR, and a repeat region of the lncRNA increases ribosome binding and translation by a so far unexplored mechanism86 (FIG. 2b).
To our knowledge, additional examples of 5′ UTR IFNG-like mRNA pseudoknots or Uchl1-like RNA–RNA interactions have not yet been discovered. It is possible, however, that such higher-order interactions, which are difficult to identify, are common in eukaryotic translation initiation. Indeed, pseudoknots constitute a well-known structural motif87 in bacterial riboswitches and ribozymes and have roles in eukaryotic pre-mRNA processing such as in splicing88,89 and adenosine-to-inosine editing90. Furthermore, there are prominent examples of cis-regulatory pseudoknots in the CDS that interact directly with translating ribosomes to induce programmed frameshifting (reviewed in REFS 91–93). Frameshifting pseudoknots can either lead to the production of different polypeptides, as first described in retroviruses94–96, or act as mRNA-destabilizing signals92,97,98 embedded in coding regions that induce no-go decay or nonsense-mediated decay in eukaryotes. Thus, pseudoknots couple translation to trans-acting sensors such as PKR or regulate frameshifting and transcript decay in cis to fine-tune mRNA expression. Finally, as discussed next, higher-order structures, including tertiary structures such as pseudoknots, recur in 5′ UTRs that control translation in a cap-independent rather than a cap-dependent manner.
IRES structures and function
Perhaps the best understood examples of RNA structure and function in translation control are represented by internal ribosome entry sites (IRESs) in viral genomes (Supplementary information S2 (box)). Viruses evolved IRESs to efficiently hijack the host translation machinery for replication and to overcome the cellular block of cap-dependent translation initiation upon viral infection. Many viral mRNAs rely solely on internal initiation of uncapped viral RNA by specific 5′ UTR RNA sequence elements or secondary structures that directly recruit the ribosome for their translation (FIG. 2b). Interestingly, alternative modes of internal but cap-dependent translation initiation are also well established in viruses and are suggested to occur in some eukaryotic mRNAs (Supplementary information S2 (box)).
In contrast to viral RNAs, all cellular mRNAs are capped and thus can undergo cap-dependent translation initiation. However, internal ribosome recruitment by IRES-containing cellular mRNAs is activated or favoured following environmental changes as a means to bypass 5′ cap usage to sustain protein expression when cap-dependent translation is diminished99 (FIG. 2b). Indeed, most proposed cellular IRESs reside in mRNAs that rely on internal initiation for sustained translation in conditions of stress, mitosis or apoptosis, which reduce global cap-dependent translation (reviewed in REFS 100–105). The first cellular IRES, for example, was discovered in the mRNA encoding immunoglobulin heavy chain-binding protein (BiP; also known as GRP78), a stress-induced chaperone106. The IRES sustains BiP translation107 especially during viral infection108.
Cellular IRES activity during stress
Ten to fifteen per cent of mammalian mRNAs are predicted to contain IRESs104, and over 100 proposed IRES-containing mRNAs have been reported109. These mRNAs mostly encode transcription factors, growth factors and transporters105. Nevertheless, after decades of work, few examples have been well characterized. Only in the past 10 years or so have new experimental tools and controls been developed that stringently assess the activity of proposed cellular IRESs110. The importance of RNA structure for IRES activity has only been indicated for some cellular IRESs, and RNA structures have been chemically and enzymatically probed for only a handful of examples (Supplementary information S3 (table)). Overall, cellular IRESs appear to be less structured than viral IRESs111,112, with few structural similarities to each other113, and their mechanisms of action are largely unknown. The activity of cellular IRESs can depend on rather short motifs114 and on the assistance of translation initiation factors or RBPs that serve as IRES trans-acting factors (ITAFs; reviewed in REFS 112,115,116) (FIG. 2b). Although several ITAFs are common to viral and cellular IRESs112,117, only a few ITAFs are well characterized. IRES–ITAF interactions may contribute to stabilizing unstable IRES structures or to inducing a conformational change of the IRES RNA that enables the recruitment and correct positioning of the ribosome. We propose that cellular IRESs can largely be categorized into three groups based on which factors or mRNA elements interact with the IRES structure: assisting ITAFs that remodel IRES structures; uORFs that sequester ribosomes and affect IRES structures; or RG4 structures as part of IRES structures (FIG. 3).
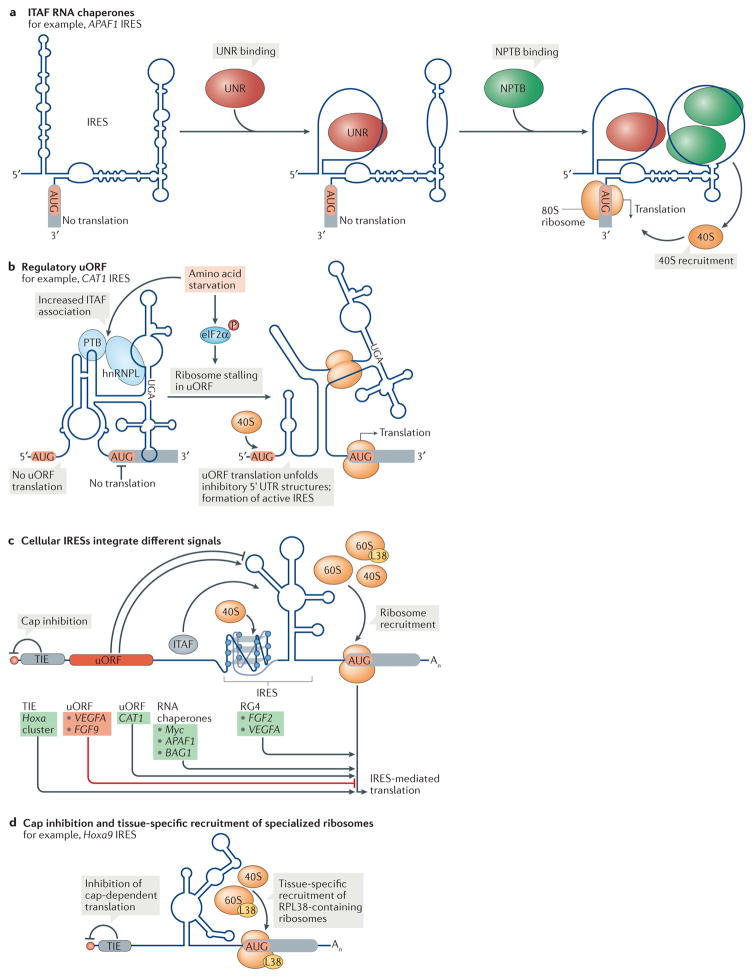
Figure 3. Cellular IRES structures employ different mechanisms for ribosome recruitment.
Cellular structured internal ribosome entry sites (IRESs) use diverse modes to recruit ribosomes, and their activity is often induced following a change in cellular condition. a | Many IRESs rely on binding by RNA-binding proteins known as IRES trans-acting factors (ITAFs) for ribosome recruitment. Such RNA chaperones remodel the IRES structure and thereby prepare a landing platform for the ribosome. In the IRES of apoptotic peptidase activating factor 1 (APAF1) for example, binding of NRAS upstream gene protein (UNR) to a purine-rich region in a stem–loop opens two stem–loop structures and allows the binding of neural polypyrimidine tract binding protein (NPTB)119, which creates a ribosome- accessible site for translation. b | Cellular IRESs can also use upstream open reading frames (uORFs) to regulate IRES activity. In the 5′ untranslated region (UTR) of the arginine–lysine transporter amino acid transporter, cationic 1 (CAT1) mRNA, translation of a uORF within the IRES is induced upon amino acid stress. This stalls ribosomes in the uORF and causes a structural switch in the IRES to an active conformation, which enables the translation of the main ORF124,125. In addition, the association of the ITAFs PTB and heterogeneous nuclear ribonucleoprotein L (hnRNPL) with the IRES increases and is required for translation during starvation200. c | Cellular IRESs can integrate signals in cis and trans to modulate internal ribosome recruitment. Whereas a translation inhibitory element (TIE) blocks cap-dependent initiation, uORF translation, ITAFs or RNA G-quadruplexes (RG4s) can all increase (green) IRES- mediated translation in a transcript-specific manner; uORF translation can also inhibit (red) IRES activity. d | In a subset of homeobox a (Hoxa) mRNAs in the mouse embryo, an IRES recruits ribosomes in a tissue-specific manner133. Several of these Hoxa IRESs additionally depend on the ribosomal protein RPL38 (L38) for their activity and a TIE at the cap that blocks cap-dependent initiation. BAG1, BCL2 associated athanogene 1; eIF2α, eukaryotic initiation factor 2α; FGF, fibroblast growth factor; P, phosphorylation; VEGFA, vascular endothelial growth factor A.
ITAFs remodel cellular IRES structures
Several cellular IRESs are thought to be activated through a structural change in their RNA motifs following a change in cellular conditions. This often requires RBPs to act as RNA chaperones. Examples include the IRESs in the MYC118, apoptotic peptidase activating factor 1 (APAF1)119 and BCL-2 associated athanogene 1 (BAG1)120,121 mRNAs. The proto-oncogene MYC IRES is activated by genotoxic stress, viral infection or apoptosis. A point mutation found in the MYC IRES structure122 in cell lines derived from individuals with multiple myeloma was predicted to lead to the formation of an additional stem–loop that increased both IRES activity123 and binding of the ITAFs Y-box-binding protein and polypyrimidine tract-binding protein 1 (PTB)118. In the highly structured APAF1 IRES RNA, binding of NRAS upstream gene protein (UNR) to a purine-rich region in a stem–loop opens two stem–loop structures and allows neural PTB (NPTB) binding119, which generates a single-stranded site for 40S recruitment (FIG. 3a). Similarly, poly(rC)-binding protein 1 (PCBP1) opens the structure of the BAG1 IRES, but ribosome recruitment requires the subsequent binding of PTB, for example upon heat stress120,121. Thus, different IRESs require sequential or combinatorial binding by ITAFs to induce the structural changes that serve to recruit ribosome subunits in a cap-independent manner. At present, only a handful of ITAFs have been characterized. In the future the identification of additional RBPs that can either stabilize IRES structures or favour the internal recruitment of ribosomes will be an important area of investigation.
Cap-inhibitory elements favour IRES-dependent translation
A different mechanism to favour IRES-mediated translation over cap-dependent translation is stalling of cap-initiated ribosomes in a short uORF24. Examples include the uORFs upstream of amino acid transporter, cationic 1 (CAT1; also known as SLC7A1)124,125 and fibro-blast growth factor 9 (FGF9)126 IRESs and within the vascular endothelial growth factor A (VEGFA)127 IRES, which are all in mRNAs that encode regulators of differentiation and cell growth. In the CAT1 mRNA, structural remodelling of the 5′ UTR in cis by uORF translation mediates unfolding of inhibitory 5′ UTR structures and a switch to a translationally active state of the IRES structure upon amino acid starvation124,125,128 (FIG. 3b). In contrast to IRES activation by uORF translation, uORFs can also repress IRES activity, as seen in the VEGFA127 and FGF9 (REF. 126) mRNAs. In a VEGFA mRNA isoform, a uORF embedded in the IRES is translated in a cap-independent manner and this uORF suppresses IRES-mediated expression of the main ORF127, possibly by unfolding the IRES structure. In the FGF9 mRNA, translation of a uORF upstream of the IRES suppresses FGF9 protein synthesis in normal conditions, whereas hypoxia-induced inhibition of cap-dependent translation and a switch to IRES-dependent translation increases FGF9 protein levels126.
In addition to ITAF-induced structural remodelling and uORF translation, RG4s have also been implicated in the regulation of IRES-mediated translation (FIG. 3c). Sequences in 5′ UTRs capable of forming RG4s are a functionally important part of the VEGFA IRES129,130 and a structural feature of the FGF2 IRES131, contributing to IRES-mediated translation of both mRNAs. Although the exact functional role of the FGF2 RG4 has not yet been studied, the VEGFA RG4 within the IRES, which in vitro folds independently of the IRES, is required to directly recruit the 40S ribosomal subunit, as shown by in vitro footprinting and structure mapping132, although this remains to be confirmed in vivo.
In summary, structured cellular IRESs in 5′ UTRs employ different strategies to overcome translation silencing in response to environmental change (FIG. 3c).
The physiological importance of cellular IRESs
Although several cellular IRESs have been investigated mainly under cellular stress conditions, IRES elements are emerging as important regulators of normal gene expression programmes that underlie embryonic development. A recent study identified and characterized conserved structured IRESs in the 5′ UTRs of a subset of genes in the homeobox (HOX) gene family133, which encode key regulators of embryonic development and tissue patterning (FIG. 3d). The HOX 5′ UTRs repress their cap-dependent initiation in normal physiological conditions through the cooperation of RNA elements — an IRES and a newly described 5′ proximal translation inhibitory element (TIE)133, which acts as a potent inhibitor of cap-dependent translation (FIG. 3c,d). The TIE acts in a highly modular fashion, as its placement upstream of the well-initiated 5′ UTR of the mRNA encoding β-globin suppresses cap-dependent translation of a reporter mRNA. Thus, translation initiation of the HOX mRNAs is enabled from IRESs in physiological conditions, as TIE motifs enable these mRNAs to bypass the cap-dependent pathway. This TIE–IRES coupling perhaps enables more intricate control of expression in time and space by ribosomes, as suggested by the finding that some HOX IRESs selectively require a specific ribosomal protein, RPL38, for their activity133 (FIG. 3d). For example, RPL38 expression is markedly enriched in specific regions of the embryo, such as developing somites (precursors of vertebral elements), and is required at these locations to control HOX gene expression at the post-transcriptional level through IRES elements within HOX 5′ UTRs. Thereby, certain ribosomal proteins, such as RPL38 (REF. 133) and RPS25 (REF. 134), as well as the pseudouridylation modification of rRNA135, can specifically regulate IRES-dependent translation and highlight the specific contribution of the ribosome itself to gene expression136.
HOX IRESs are examples of structured RNA elements that physiologically regulate gene expression during embryonic development. Another example is the IRES in the human FGF1 mRNA. A domain of the FGF1 IRES is conserved in sequence and structure among six mammals137. The importance of the HOX and FGF1 IRES structures for their function was confirmed by structural and compensatory mutagenesis. Additionally, the Hoxa9 IRES structure was characterized in vitro by selective 2′-hydroxyl acylation analysed by primer extension (SHAPE), mutate-and-map133 and multiplexed •OH cleavage analysis with paired-end sequencing (MOHCA-seq)138 analyses.
The targeted knockout of the Hoxa9 IRES in mice reveals the crucial role for this element in translation of the mRNA in specific regions of the embryo, such as the developing somites and neural tube133. The Fgf2 and Myc IRESs in reporter constructs are also active in the developing mouse embryo, and their abnormal IRES activation has been linked to cellular transformation123,139,140. VEGFC IRES activity is specifically increased in metastatic lymph nodes141, for example, whereas Fgf2 IRES activity may be important for the spatiotemporal regulation of FGF2 expression in neuronal maturation during brain development in mice142 and in hypoxia143. The finding of cellular IRESs that are active in physiological conditions paves the way for systematic approaches144 aimed at discovering functional, tissue-specific IRESs genome-wide (Supplementary information S2 (box)). A central question, especially in the case of cellular IRESs, is whether their activity relies on local RNA structures or sequences and/or on assisting proteins. Analysis of IRES structures by structure probing (Supplementary information S3 (table)) should be complemented with compensatory mutagenesis, ideally for every base pair in proposed structural models, to assess their contribution to function. It is also tempting to speculate that structural changes in each IRES RNP are likely to occur in a tissue-specific or stimuli-dependent manner and therefore indicate potentially important 5′ UTR elements whose RNA structure should be determined in vivo.
Direct eIF3 recruitment by RNA structures
More evidence of translation initiation bypassing the 5′ cap of cellular mRNAs comes from two recent studies focusing on the multisubunit initiation factor eIF3. Outside its central role in the 43S pre-initiation complex — physically connecting eIF4G at the 5′ cap to the 40S ribosomal subunit — eIF3 appears to repress or activate a specific subset of mRNAs by directly binding to stem–loop structures in their 5′ UTRs145 (FIG. 2b). Transcriptome-wide analysis in human cells by photo-activatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR–CLIP) showed that eIF3 directly interacts with 3% of all mRNAs, particularly in the 5′ UTR. These interactions were studied further in two mRNAs encoding the cell proliferation regulators c-Jun (also known as AP1) and B cell translocation gene 1 protein, which are subject to eIF3-mediated translation induction and repression, respectively. Characterization of the Jun 5′ UTR led to the identification of a conserved hairpin that may serve to directly recruit eIF3 for translation activation. Although disruptive mutations in the hairpin abolish eIF3-dependent translation activation, it remains unknown whether the RNA structure is necessary for function, as compensatory mutations are required to demonstrate the relevance of the structure. Interestingly, a putative IRES in the Jun 5′ UTR that directs internal translation in glioblastoma cells146 has previously been mapped to the same region as the suggested eIF3-bound stem–loop. It remains unclear whether the putative stem–loop is part of this IRES or is required for IRES activity. Analysis of additional eIF3 target mRNAs is necessary to determine how widespread this alternative initiation pathway is and to what extent 5′ UTR RNA structures are required for it. In this respect, a specific eIF3 subunit, eIF3d, was surprisingly found to directly bind to the 5′ cap147 (FIG. 2b), a role previously thought to be exclusive to eIF4E. This non-canonical cap interaction may serve to regulate transcript-specific translation of certain eIF3-bound mRNAs such as Jun147. Overall, eIF3-specific translation represents an additional role for eIF3 in selective translation.
eIF3 binds RNA modifications to mediate cap-independent initiation
Recent work discovered that RNA modifications also employ the eIF3 complex for cap-independent ribosome recruitment (FIG. 2b). Reversible RNA modifications have long been overlooked with respect to their impact on mRNA structure and translation (reviewed in REFS 148,149). The most prevalent internal chemical modification of RNA, N6-methyladenosine (m6A)150 (FIG. 4a), was recently shown to stimulate internal ribosome recruitment151 (FIGS 2b,4b). In vitro footprinting and translation assays showed that initiation complexes bind to m6A-containing 5′ UTRs, which stimulates cap-independent translation through the direct interaction of m6A with eIF3 (REF. 151) (FIG. 4b). This type of initiation requires scanning and a free 5′ end but is independent of the 5′ cap itself or cap-binding proteins. This mechanism may indicate that m6A in the 5′ UTR can serve as an alternative to the cap and is selectively bound by eIF3 to stimulate initiation at sites termed ‘m6A-induced ribosome engagement sites’ (REF. 151). This effect of 5′ UTR m6A on translation becomes especially important in stress conditions, such as heat shock152, which selectively increases and redistributes m6A in 5′ UTRs, especially in those of newly transcribed mRNAs (FIG. 4b). During heat stress, the m6A ‘reader’ YTH domain-containing family protein 2 (YTHDF2) localizes to the nucleus and promotes the maintenance of 5′ UTR m6A levels by inhibiting binding of an m6A ‘eraser’, fat mass and obesity-associated protein (FTO)152. This process mediates selective cap-independent translation of stress response transcripts, such as those of the heat shock-induced heat shock protein 70 (HSP70; also known as HSPA) gene family, which are m6A-modified at a single site152. A single m6A in a 5′ UTR is also sufficient to recruit eIF3, which by itself can attract the 43S pre-initiation complex151. As m6A content increases in stress conditions, which also activate IRES-dependent translation of specific mRNAs, it would be interesting to determine to what extent m6A modifications in cellular IRESs are responsible for mediating cap-independent translation.
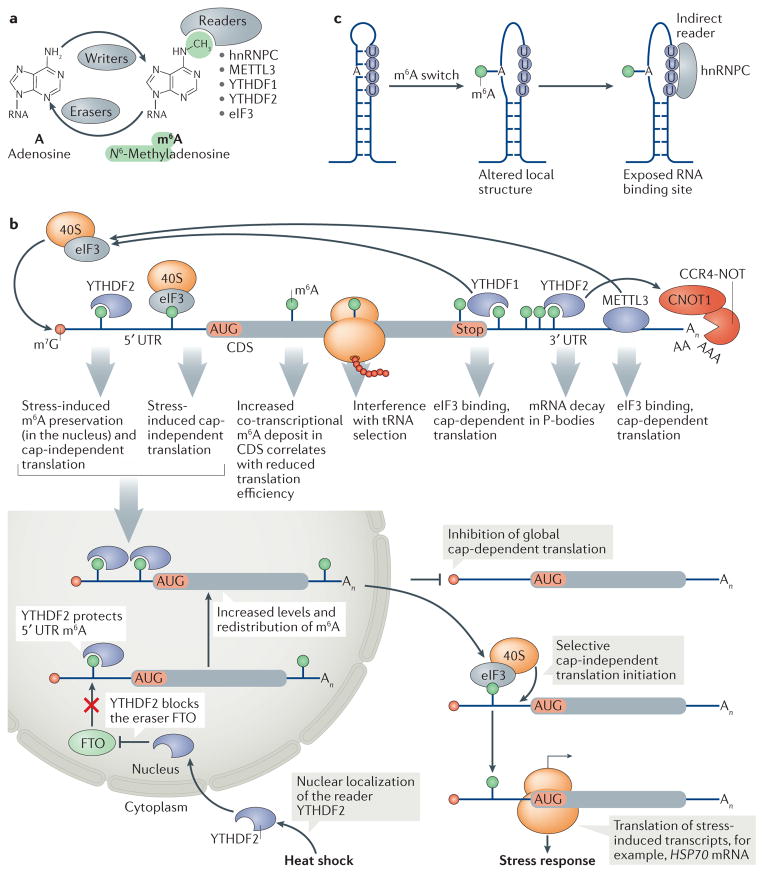
Figure 4. The effects of N6-methyladenosine on mRNA translation and decay.
a | ‘Writer’ proteins establish N6-methyladenosine (m6A) at internal RNA sites, ‘eraser’ proteins remove them, and ‘reader’ proteins directly bind the N6-methyl group of m6A (reviewed in REF. 148). The listed readers affect the translation and stability of m6A-modified mRNAs. b | m6A modifications in mRNAs occur with a preference for the last exon and 3′ untranslated region (UTR)201 and are increased during stress152. According to its position in the mRNA, m6A is bound by readers that can induce cap-dependent translation or internal ribosome recruitment. In the 5′ UTR, eukaryotic initiation factor 3 (eIF3) can directly bind m6A and facilitate internal translation initiation151. Stress-responsive 5′ UTR N6-adenosine methylation, for example during heat shock, is preserved by YTH domain-containing family protein 2 (YTHDF2), which blocks binding of the eraser fat mass and obesity-associated protein (FTO), thereby promoting cap-independent translation initiation of stress response mRNAs152. m6A in the coding sequence (CDS) is linked to tRNA selection163, and at the 3′ UTR it is linked to increased translation owing to YTHDF1 binding to m6A and eIF3 recruitment for cap-dependent translation158. The writer methyltransferase like 3 protein (METTL3) can also directly bind to eIF3 to increase translation of m6A-containing mRNAs independently of its m6A writer activity159. By contrast, YTHDF2 promotes degradation of m6A-modified mRNAs by recruiting the deadenylase complex CCR4–NOT157. Together, increased translation efficiency and activated decay of m6A-modified mRNAs allow dynamic regulation of protein synthesis. c | m6A mRNA modifications are associated with unfolded RNA structures. N6 adenosine methylation in a stem disrupts based paired regions (‘m6A switch’), which allows binding of the ‘indirect reader’ heterogeneous nuclear ribonucleoprotein C (hnRNPC) to exposed U rich motifs in the nucleus153. CNOT1, CCR4–NOT transcription complex subunit 1; HSP70, heat shock protein 70; P body, processing body. Part c is adapted from REF. 153, Macmillan Publishers Limited.
RNA modifications promote translation
RNA m6A modifications may also increase translation efficiency by unfolding RNA structures to recruit proteins153,154 or to aid ribosome scanning154. Using nuclease probing, m6A was recently found to disrupt local RNA structures termed m6A switches (REF. 153) in mRNAs and lncRNAs153–155, thereby rendering previously paired RNA motifs less structured and thus accessible to ‘indirect readers’ (FIG. 4c). The RNA duplex-destabilizing effect of m6A is supported by nuclear magnetic resonance and thermodynamic measurements in vitro156 and by the correlation of in vivo click SHAPE (icSHAPE)-determined local RNA structural changes at or near predicted m6A sites in the transcriptome154. It will be crucial to define to what extent the locally unfolded 5′ UTR sequences around m6A modifications favour translation initiation by the scanning ribosome154. Furthermore, m6A readers integrate many cues from the 5′ UTR152 and 3′ UTR157–159 that have been associated with increased translation efficiency or mRNA decay (FIG. 4b). In addition to m6A, the less abundant N1-methyladenosine (m1A), which occurs mainly in structured regions of mRNA 5′ UTRs160, and hydroxymethylcytosine (hmrC) in coding regions161 have been associated with increased translation initiation and elongation, respectively. However, increased co-transcriptional methylation of A to m6A in mRNA coding regions during slow transcription162 as well as disrupted tRNA selection by m6A in the CDS163 was recently shown to result in decreased translation efficiency and elongation dynamics, respectively, of m6A-modified mRNAs (FIG. 4b).
Recently, circular RNAs (circRNAs) that are formed by the joining of 5′ and 3′ ends through back-splicing, were found to be translated into functional proteins164,165 (FIG. 2b). As they do not have a cap, ribosomes need to be internally recruited onto circRNAs, which may be mediated by m6A modifications before the start codon164,166. Together, these recent data suggest that reversible RNA methylation, directly or through its impact on local RNA structural topology, is a general feature of mRNA function and acts to increase translation initiation.
5′ UTR RNA structure probing in vivo
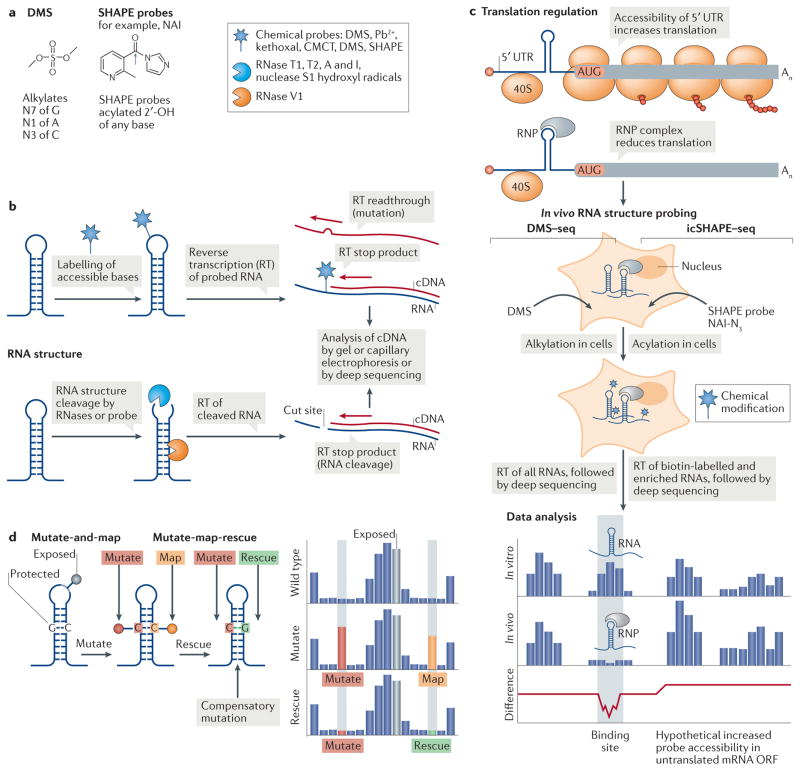
RNA structures are presumably more dynamic in living cells than in typical in vitro experiments, owing to the engagement of trans-acting RNAs, small molecules and proteins. However, most RNA structural data of 5′ UTRs have in fact been obtained in vitro (Supplementary information S1 (box)). Although the cell membrane is an obvious barrier to getting large RNA-modifying molecules into cells, cell-permeable chemicals such as dimethyl sulfate (DMS) and SHAPE reagents have proved successful for RNA structure probing inside living cells (FIG. 5a,b), and they are now being applied to map the folding of the transcriptome in vivo (reviewed in REFS 167–171) (FIG. 5c). Transcriptome-wide comparison of RNA structures in vivo and in vitro by DMS treatment followed by deep sequencing (DMS–seq) has revealed that in cells, mRNAs are generally less structured than they are in vitro172 (Supplementary information S1 (box)). Analysis of living cells by icSHAPE has also shown that RNAs are less folded in vivo, although the extent of unfolding varies between RNA classes154, and RNA structural features in vivo may be used to distinguish between coding and regulatory RNAs. For example, the region just upstream of the start codon appears particularly less structured in cells173. Nevertheless, the structural interpretation of these data remains uncertain, as cellular mRNAs are covered with RBPs, RNA helicases and translating ribosomes that constantly remodel the mRNP and influence the RNA structure172–174. Indeed, the extent of unfolding of the transcriptome is thought to be ATP dependent172, which indicates a major role for RNA helicases in shaping RNA structures.
Figure 5. Global RNA structure probing to assess translation regulation.
Global RNA structure probing inside cells can assess the transcriptome structure in the presence of proteins. a | Chemical schematics of the RNA structure probes dimethyl sulfate (DMS) and the selective 2′-hydroxyl acylation analysed by primer extension (SHAPE) reagent 2 methylnicotinic acid imidazolide (NAI) and their reactivity. The grey arrow indicates the site of 2′-OH attack of the RNA by the probe. Different probes induce single-strand-specific chemical labelling (star) or cleavage by enzymes or probes (blue Pac Man shape, single strand specific; orange Pac Man, double strand specific). b | RNA probing either labels RNA through a covalent reaction of a chemical probe with accessible nucleotides (top) or cleaves the RNA backbone with RNases (bottom). A pool of modified or cleaved RNAs is transcribed into cDNA by reverse transcription (RT), and modified or cleaved sites are identified by their effect on RT. c | A 5′ untranslated region (UTR) ribonucleoprotein (RNP) complex that inhibits ribosome scanning reduces the accessibility of the mRNA for the probe in cells. In in vivo RNA structure probing, shown here for DMS treatment followed by deep sequencing (DMS–seq) and in vivo click SHAPE followed by deep sequencing (icSHAPE–seq), RNA structures areprobedincells bychemicalmodification. Data analysis of in vitro-probed and in vivo-probed RNA can indicate the presence of RNA structures as protein binding sites, owing to masked probe accessibility or to remodelling of the structure by protein interaction. Where a 5′ UTR RNP complex inhibits the translation of an open reading frame (ORF), the accessibility of the probe to poorly translated coding sequences might increase in the absence of ribosomes. d | In multidimensional mutate and map chemical probing, a mutation (red) that eliminates base pairing exposes the mutated nucleotide and its partner nucleotide (orange) to chemical modification (pins). In mutate-map-rescue probing, a mutation is rescued from modification by a compensatory mutation of the partner nucleotide (green). The reactivity profile reflects changes in probe accessibility upon mutation (red), which allows mapping (orange) of the base-paired nucleotide, while rescue (green) confirms base pairing. Nucleotides in loops are exposed and accessible to the probe. Pb2+, lead; CMCT, 1 cyclohexyl (2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate.
New psoralen-based technologies that map sites of cis and trans RNA–RNA interactions (Supplementary information S1 (box)) may also provide new insights into the folding status of the cellular transcriptome and of specific RNA duplexes. As these methods have only recently been established, data remain sparse for RNAs of relatively low abundance, including most mRNAs. As biases in protocols are reduced, sequencing depths increase, data normalization is standardized, and specific mRNAs are targeted for in-depth analysis175–177, it may become possible to investigate the roles of RNA–RNA interactions and RNA structures in translation initiation.
A substantial improvement of SHAPE175 and DMS-based178 RNA structure probing has been achieved by reading out nucleotides affected by chemical probes as mismatches introduced at the adduct site during reverse transcription (FIG. 5b) and is being adapted for application in cells. For example, DMS mutational profiling with sequencing (DMS–MaPseq), an in vivo adaptation of DMS–seq, was recently used for global or targeted RNA structure probing of low-abundance RNAs in Drosophila melanogaster ovaries, as well as in yeast and mammalian cells177. This approach includes compensatory mutations of stem structures modelled in the 5′ UTR of FMR1 autosomal homologue 2 (FXR2) mRNA, which showed rescue of activity, although the effect was weak177.
What are the next steps in RNA structure probing?
The current generation of high-throughput methods for probing RNA accessibility and base pairing inside cells represent substantial advances over prior techniques. However, these methods still appear to be too limited to test the repertoire of proposed mRNA regulatory structures or to establish new ones (Supplementary information S1 (box)). There are two major limitations to these in vivo methods, both inherited from their ‘parent’ in vitro methodologies. First, structure modelling from these data remains mostly tied to chemistries and modelling methods whose accuracies are poor even in vitro179,180. Any such inaccuracies are further exacerbated by the numerous ‘unknowns’ of RNA interactions in cells, from RBP interactions to mRNA domains potentially forming a heterogeneous ensemble of structures. Second, although chemical probing methods coupled with computation can provide base-pair-level structural models, they have generally not been tested in prospective experiments — not even in vitro. This absence of standard validation methods is a major obstacle for our understanding of how mRNA structures impact mRNA translation.
Fortunately, solutions for both problems have recently been found for in vitro RNA structure modelling and could be brought to bear in the next generation of in vivo chemical probing methods. New in vitro methods infer effects of mutations at each RNA nucleotide from changes in chemical accessibility elsewhere in the RNA (mutate-and-map)181,182 (FIG. 5d). These methods have enabled highly accurate structure determination in structure prediction competitions180,183. Numerous strategies are now available to generate RNA libraries with mutations inside cells176,184,185, which raises the prospect of carrying out the same mutate-and-map analysis in vivo, high-throughput versions of methods to correlate hydroxyl radical damage at nearby nucleotides138 to study tertiary structure are now available and were recently applied to study chromatin structure in vivo186. These methods promise a considerable boost in the accuracy of RNA structure modelling from in vivo data, even of molecules with complex secondary and tertiary structures183.
The gold standard for validating RNA structures remains rescue by compensatory mutagenesis, although this approach has typically required quantitative functional readouts that need to be individually developed for each RNA molecule (see, for example, REF. 177). Recently, compensatory mutagenesis has been analysed by using quite general chemical probing readouts (FIG. 5d), even allowing correction of erroneous structures from conventional SHAPE or DMS-based structure mapping182. Correlation of in vivo RNA structures of operon units in bacterial polycistronic mRNAs with their translation efficiency has been supported by in vivo DMS–seq of compensatory mutants in one mRNA187. These early results foreshadow that the detection of secondary and tertiary RNA structures as well as high-throughput compensatory mutagenesis, if adapted in vivo and to eukaryotes, may soon enrich our understanding of structured RNA elements that control eukaryotic translation.
Conclusions
An accumulating body of knowledge highlights the diverse repertoire of mechanisms through which translation initiation can be controlled by 5′ UTR structures in eukaryotic mRNAs. Nevertheless, for decades, research has focused on linear RNA sequence motifs bound by RBPs that control post-transcriptional processes12,188,189. For many of these motifs, it is not clear whether RNA structures improve or hinder access of proteins by exposing or burying them, respectively, in local structure37. Thus, an unstructured element embedded in an RNA structure may actually be the driving force of a structured motif. In the case of IRESs, this notion could provide an explanation for the discovery of short linear poly(U) motifs that harbour IRES activity144, for the finding that the ITAF PTB recognizes and binds to a polypyrimidine-rich motif, (CCU)n, in structured cellular IRESs114, or for the detection of an unstructured region that is important for the activity of the turnip crinkle virus IRES190, despite the fact that viral IRESs are thought to largely be structured. Thus, local mRNA structures as well as unstructured motifs therein can regulate translation.
How can we better find and confirm functional mRNA structures, both in vitro and in vivo? Many compact and rapidly evolving prokaryotic genomes have allowed the discovery of novel RNA motifs by means of genome alignments, conservation and sequence covariation analyses. In eukaryotes, evolutionary covariation analysis has been difficult and may be fundamentally impossible if 5′ UTR sequences are highly divergent191, even as some mRNA structures are suggested to be abundant and conserved enough192 to define distinct families193,194. As new global probing tools can now assess nearly all RNA structures in cells, they may enable the discovery of novel RNA structures in complex genomes and analysis of their contribution to gene regulation167. These tools may also permit revisiting the analysis of decades-old examples of highly translation-ally regulated mRNAs to assess potential underlying 5′ UTR structures such as the long, GC-rich 5′ UTR of cyclin D1 mRNA195. Its translation is thought to depend on eIF4E-assisted structure unwinding by the helicase eIF4A, but the responsible RNA structures are not known.
Just as important as finding regulatory RNA structures based on structure probing or prediction is their experimental validation. We encourage the field to more routinely apply compensatory mutagenesis to evaluate the physiological relevance and functional contribution of candidate mRNA structures. As experimental methods continue to improve, a major goal for global structure probing is their application across diverse cell types and tissues in the context of development, cell differentiation, stress or disease. This approach is especially important as intricate modes of gene regulation such as translation may be highly cell-type specific. Emerging technologies that are highly complementary to in vivo structure probing include advances in cryo-electron microscopy196, which may soon reveal the intricate interactions of the ribosome and structured mRNA elements197, including cellular IRES elements. Moreover, cryo-electron tomography in situ198 may soon provide three-dimensional snapshots of translating ribosomes and their template mRNAs in native cells. These methods may also profit from machine learning algorithms and conventional biophysical modelling to more accurately predict and model structures in mRNA 5′ UTRs199. We speculate that such advances will enable an mRNA-centric view of translation, in which one can visualize how the ribosome and RBPs change the accessibility of RNA structured domains to control translation in time and space. It has long been appreciated that the ribosome is a dynamic, multi-state machine that has originated from the ancient RNA world. Upcoming advances raise the promise of understanding whether mRNAs and their structures provide, on top of the ribosome, a new layer of gene regulation that allows the control of translation to meet the needs of modern eukaryotic biology.
Supplementary Material
Acknowledgments
The authors apologize to those researchers whose work was not cited owing to space limitations and thank G. W. Byeon in the Barna laboratory for data analysis and figure preparation of eukaryotic 5′ UTR lengths, the Barna laboratory members for helpful comments on the manuscript, and H.-G. Wendel, J. Kieft, P. Sarnow and M. Hentze for thoughtful suggestions. RNA research in the authors’ laboratories is supported by the New York Stem Cell Foundation (M.B.), Mallinckrodt Foundation Award (M.B.), Pew Scholars Award (M.B.) and NIH grants R01HD086634 (M.B.), R21HD086730 (M.B.), R01 GM102519 (R.D.) and R35 GM122579 (R.D.). M.B. is a New York Stem Cell Foundation Robertson Investigator. K.L. is supported by an EMBO Long-Term Fellowship (ALTF 539–2015) and is the Layton Family Fellow of the Damon Runyon Cancer Research Foundation (DRG-2237-15).
Glossary
- Secondary structures
Patterns of Watson–Crick base pairs (G–C, A–U and G–U) that define the double helices of an RNA.
- Tertiary structures
Interactions that orient RNA double helices into specific three-dimensional arrangements, often involving non-Watson–Crick base pairs.
- Ribozymes
RNA molecules that catalyse specific biochemical reactions.
- Riboswitches
Non-coding mRNA structures that sense the environmental status of a cell by directly binding to small molecule ligands, such as a metabolite or an ion. This interaction changes the RNA conformation and controls gene expression through alternative splicing, transcription or translation.
- Peptidyl transferase centre
The site in the large ribosomal subunit that catalyses peptide bond formation and peptide release; it is composed entirely of RNA.
- Pseudoknots
RNA tertiary structures formed by base pairing of a single-stranded loop of a hairpin with a complementary sequence outside that hairpin.
- Upstream open reading frames (uORFs)
Small ORFs located in the 5′ UTR of some mRNAs. Translation of uORFs can regulate the translation of the downstream ORF.
- Kozak sequence
A favourable sequence (GCCRCCAUGG in mammals, where R is a purine) surrounding the translation start codon (AUG); also called the Kozak consensus or Kozak context.
- GC content
Percentage of guanine and cytosine nucleotides in an RNA molecule. G–C base pairs are more stable than A–U base pairs and can form more stable RNA structures.
- Free energy (ΔG)
The stability of an RNA secondary structure, estimated as the sum of the free energies assigned to all loops and base pair stacks of a folded RNA, based on a computational folding algorithm.
- 5′ cap
A modified guanine nucleotide, m7GpppN (where m7G is 7-methylguanosine, p is a phosphate group and N is any base), located at the 5′ end of eukaryotic mRNAs.
- Ribonucleoprotein (RNP)
A complex of proteins in association with an RNA (RNP) or mRNA (mRNP).
- Ribosome profiling
Sequencing of the RNA fragments protected by ribosomes, providing a quantitative signature of the translated mRNAs at a given time.
- RNA G-quadruplex (RG4)
Extremely stable RNA structure formed in G-rich regions by the stacking of at least two G-tetrads, each of them forming a square-shaped structure by non-Watson–Crick interactions between two or more layers of paired G-quartets.
- Toeprinting
Nucleotide-resolution assay that uses primer extension inhibition induced by the complex of a protein, or the ribosome, bound to an mRNA, to report its position on the mRNA.
- In-line probing
Nucleotide-resolution structure probing method that uses the natural instability of RNA: the 2′ hydroxyl of each nucleotide can attack its phosphodiester backbone at a rate dependent on local structure.
- No-go decay
An mRNA and translation quality-control mechanism that recognizes and degrades mRNAs following prolonged ribosome stalling during translation.
- Nonsense-mediated decay
An RNA surveillance mechanism that recognizes and degrades mRNAs containing premature termination codons to prevent their translation.
- Selective 2′-hydroxyl acylation analysed by primer extension (SHAPE)
SHAPE and in vivo click SHAPE (icSHAPE) use cell-permeable reagents to acylate the 2′-OH of accessible, single-stranded RNA at all four nucleotide bases.
- Mutate-and-map
Two-dimensional expansion of chemical probing that infers RNA base pairing by mutating each nucleotide and detecting increased chemical accessibility at other nucleotides.
- m6A switches
mRNA sequences that adopt a different secondary structure depending on N6-adenosine methylation.
- Dimethyl sulfate (DMS)
A cell-permeable reagent that methylates adenine and cytosine nucleotides that are not protected by base pairing. Modification sites stall or induce a mutation during primer extension by reverse transcriptase, and sequencing the resulting complementary DNA indicates the folding state of a nucleotide in an RNA.
- Psoralen
A natural plant product that intercalates into DNA and RNA and reversibly crosslinks interacting RNA duplex molecules at UpA motifs upon irradiation with ultraviolet light.
- Sequence covariation
Nucleotide substitutions that differ between two or more homologous genes but retain the potential for RNA base pairing in each sequence.
Footnotes
Author contributions
All authors substantially contributed to researching data for the article, discussion of the content, writing and editing and/or reviewing the manuscript before submission.
Competing interests statement
The authors declare no competing interests.
See online article: S1 (box) | S2 (box) | S3 (table)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4:1–15. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman JC, Hud NV, Williams LD. The ribosome challenge to the RNA world. J Mol Evol. 2015;80:143–161. doi: 10.1007/s00239-015-9669-9. [DOI] [PubMed] [Google Scholar]
- 3.Nahvi A, et al. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 4.Wachter A. Gene regulation by structured mRNA elements. Trends Genet. 2014;30:172–181. doi: 10.1016/j.tig.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 6.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simsek D, et al. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017;169:1051–1057. doi: 10.1016/j.cell.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Z, et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell. 2017;67:71–83. doi: 10.1016/j.molcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 12.Pichon X, et al. RNA binding protein/RNA element interactions and the control of translation. Curr Protein Pept Sci. 2012;13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning KS, Cooper TA. The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol. 2016;18:102–114. doi: 10.1038/nrm.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenz R, et al. ViennaRNA package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr C. Regulation by 3′-untranslated regions. Annu Rev Genet. 2017;51:171–194. doi: 10.1146/annurev-genet-120116-024704. [DOI] [PubMed] [Google Scholar]
- 17.Pesole G, et al. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 18.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 19.Lynch M, Scofield DG, Hong X. The evolution of transcription-initiation sites. Mol Biol Evol. 2005;22:1137–1146. doi: 10.1093/molbev/msi100. [DOI] [PubMed] [Google Scholar]
- 20.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 21.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira JP, Overton KW, Wang CL. Tuning gene expression with synthetic upstream open reading frames. Proc Natl Acad Sci USA. 2013;110:11284–11289. doi: 10.1073/pnas.1305590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somers J, Pöyry T, Willis AE. A perspective on mammalian upstream open reading frame function. Int J Biochem Cell Biol. 2013;45:1690–1700. doi: 10.1016/j.biocel.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araujo PR, et al. Before it gets started: regulating translation at the 5′ UTR. Comp Funct Genomics. 2012;2012:1–8. doi: 10.1155/2012/475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 27.Noderer WL, et al. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol. 2014;10:1–14. doi: 10.15252/msb.20145136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathews MB, Sonenberg N, Hershey JWB. Origins and principles of translational control. Cold Spring Harbor Monogr Ser. 2007;48:1–40. doi: 10.1101/cshperspect.a032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schott J, et al. Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLOS Genet. 2014;10:e1004368. doi: 10.1371/journal.pgen.1004368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernández G, Altmann M, Lasko P. Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem Sci. 2010;35:63–73. doi: 10.1016/j.tibs.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez G, Osnaya VG, Garcia A, Velasco MX. In: Evolution of the Protein Synthesis Machinery and its Regulation. Hernández G, Jagus R, editors. Springer Int. Pub; 2016. pp. 81–107. [Google Scholar]
- 32.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:4.1–4.10. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. This is one of the first studies to show a role for a 5′ UTR RNA secondary structure in influencing mRNA translation. [DOI] [PubMed] [Google Scholar]
- 36.Manzella JM, Blackshear PJ. Regulation of rat ornithine decarboxylase mRNA translation by its 5′-untranslated region. J Biol Chem. 1990;265:11817–11822. [PubMed] [Google Scholar]
- 37.Taliaferro JM, et al. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol Cell. 2016;64:294–306. doi: 10.1016/j.molcel.2016.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montoya J, Ojala D, Attardi G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 39.Haimov O, Sinvani H, Dikstein R. Cap-dependent, scanning-free translation initiation mechanisms. Biochim Biophys Acta. 2015;1849:1313–1318. doi: 10.1016/j.bbagrm.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Hentze MW, et al. Identification of the iron responsive element for the translational regulation of human ferritin mRNA. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 41.Muckenthaler MU, Rivella S, Hentze MW, Galy B. A red carpet for iron metabolism. Cell. 2017;3:1–18. doi: 10.1016/j.cell.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray NK, Hentze MW. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 44.Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsyan A, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 47.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. Ribosome profiling in cancer cells following eIF4A perturbation identifies stable RG4 structures in the 5′ UTR in eIF4A-sensitive target mRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubio CA, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:1–19. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Özes AR, Feoktistova K, Avanzino BC, Fraser CS. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J Mol Biol. 2011;412:674–687. doi: 10.1016/j.jmb.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dmitriev SE, et al. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol Cell Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bordeleau ME, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cencic R, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadlish H, et al. Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem Biol. 2013;8:1519–1527. doi: 10.1021/cb400158t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu J, Pelletier J. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim Biophys Acta. 2014;1849:781–791. doi: 10.1016/j.bbagrm.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Gandin V, et al. NanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016;26:636–648. doi: 10.1101/gr.197566.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Modelska A, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6:e1603. doi: 10.1038/cddis.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015;25:1196–1205. doi: 10.1101/gr.191601.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pisareva VP, Pisarev AV, Komar AA, Hellen CUT, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DexH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sen ND, Zhou F, Harris MS, Ingolia NT, Hinnebusch AG. eIF4B stimulates translation of long mRNAs with structured 5′ UTRs and low closed-loop potential but weak dependence on eIF4G. Proc Natl Acad Sci USA. 2016;113:10464–10472. doi: 10.1073/pnas.1612398113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jungfleisch J, et al. A novel translational control mechanism involving RNA structures within coding sequences. Genome Res. 2017;27:95–106. doi: 10.1101/gr.209015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol. 2017;5:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 63.Cammas A, Millevoi S. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2016;45:1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fay MM, Lyons SM, Ivanov P. RNA G-quadruplexes in biology: principles and molecular mechanisms. J Mol Biol. 2017;429:2127–2147. doi: 10.1016/j.jmb.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song J, Perreault JP, Topisirovic I, Richard S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation (Austin) 2016;4:e1244031. doi: 10.1080/21690731.2016.1244031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beaudoin JD, Perreault JP. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010;38:7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolduc F, Garant JM, Allard F, Perreault JP. Irregular G-quadruplexes found in the untranslated regions of human mRNAs influence translation. J Biol Chem. 2016;291:21751–21760. doi: 10.1074/jbc.M116.744839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumari S, Bugaut A, Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halder K, Wieland M, Hartig JS. Predictable suppression of gene expression by 5′-UTR-based RNA quadruplexes. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arora A, et al. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Melko M, Bardoni B. The role of G-quadruplex in RNA metabolism: involvement of FMRP and FMR2P. Biochimie. 2010;92:919–926. doi: 10.1016/j.biochi.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 74.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 75.Anderson BR, et al. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016;44:6649–6659. doi: 10.1093/nar/gkw593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Didiot MC, et al. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castets M, et al. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum Mol Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- 79.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benhalevy D, et al. The human CCHC-type zinc finger nucleic acid-binding protein binds G-rich elements in target mRNA coding sequences and promotes translation. Cell Rep. 2017;18:2979–2990. doi: 10.1016/j.celrep.2017.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-γ mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 83.Cohen-Chalamish S, et al. Dynamic refolding of IFN-γ mRNA enables it to function as PKR activator and translation template. Nat Chem Biol. 2009;5:896–903. doi: 10.1038/nchembio.234. The studies in Refs 82 and 83 show how short helical segments assemble into a pseudoknot structure in a 5′ UTR, which is bound by PKR and activates it, lead to inhibition of translation initiation. [DOI] [PubMed] [Google Scholar]
- 84.Nallagatla SR, Toroney R, Bevilacqua PC. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carrieri C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 87.Brierley I, Gilbert RJ, Pennell S. RNA pseudoknots and the regulation of protein synthesis. Biochem Soc Trans. 2008;36:684–689. doi: 10.1042/BST0360684. [DOI] [PubMed] [Google Scholar]
- 88.Osman F, Jarrous N, Ben-Asouli Y, Kaempfer R. A cis-acting element in the 3′-untranslated region of human TNF-α mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 1999;13:3280–3293. doi: 10.1101/gad.13.24.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Namer LS, et al. An ancient pseudoknot in TNF-α pre-mRNA activates PKR, inducing eIF2α phosphorylation that potently enhances splicing. Cell Rep. 2017;20:188–200. doi: 10.1016/j.celrep.2017.06.035. [DOI] [PubMed] [Google Scholar]
- 90.Reenan RA. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434:409–413. doi: 10.1038/nature03364. [DOI] [PubMed] [Google Scholar]
- 91.Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Advani VM, Dinman JD. Reprogramming the genetic code: the emerging role of ribosomal frameshifting in regulating cellular gene expression. Bioessays. 2016;38:21–26. doi: 10.1002/bies.201500131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baril M, Dulude D, Steinberg SV, Brakier-Gingras L. The frameshift stimulatory signal of human immunodeficiency virus type 1 group O is a pseudoknot. J Mol Biol. 2003;331:571–583. doi: 10.1016/S0022-2836(03)00784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Namy O, Moran SJ, Stuart DI, Gilbert RJC, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Belew AT, Advani VM, Dinman JD. Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 2011;39:2799–2808. doi: 10.1093/nar/gkq1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belew AT, et al. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014;512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 100.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 101.Jackson RJ. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol. 2013;5:a011569. doi: 10.1101/cshperspect.a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pestova TV, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 104.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 105.Lacerda R, Menezes J, Romão L. More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol Life Sci. 2016;74:1659–1680. doi: 10.1007/s00018-016-2428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. This study presents the first description of IRES activity in the 5′ UTR of a cellular mRNA. [DOI] [PubMed] [Google Scholar]
- 107.Thoma C, Bergamini G, Galy B, Hundsdoerfer P, Hentze MW. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol Cell. 2004;15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 108.Buchkovich NJ, Yu Y, Pierciey FJ, Alwine JC. Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. J Virol. 2010;84:11479–11486. doi: 10.1128/JVI.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mokrejs M, et al. IRESite — a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 2010;38:D131–D136. doi: 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baranick BT, et al. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc Natl Acad Sci USA. 2008;105:4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 112.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baird SD, Lewis SM, Turcotte M, Holcik M. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007;35:4664–4677. doi: 10.1093/nar/gkm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mitchell SA, et al. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.King HA, Cobbold LC, Willis AE. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 116.Faye MD, Holcik M. The role of IRES trans-acting factors in carcinogenesis. Biochim Biophys Acta. 2014;1849:887–897. doi: 10.1016/j.bbagrm.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 117.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]
- 118.Cobbold LC, et al. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene. 2010;29:2884–2891. doi: 10.1038/onc.2010.31. [DOI] [PubMed] [Google Scholar]
- 119.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 120.Pickering BM, Mitchell SA, Evans JR, Willis AE. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulates its activity in vitro and in vivo. Nucleic Acids Res. 2003;31:639–646. doi: 10.1093/nar/gkg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pickering BM, Mitchell SA, Spriggs KA, Willis AE, Stoneley M. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol Cell Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Le Quesne JP, Stoneley M, Fraser GA, Willis AE. Derivation of a structural model for the c-myc IRES. J Mol Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 123.Chappell SA, et al. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 124.Yaman I, et al. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 125.Fernandez J, et al. Ribosome stalling regulates ires-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol Cell. 2005;17:405–416. doi: 10.1016/j.molcel.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 126.Chen TM, et al. Overexpression of FGF9 in colon cancer cells is mediated by hypoxia-induced translational activation. Nucleic Acids Res. 2014;42:2932–2944. doi: 10.1093/nar/gkt1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bastide A, et al. An upstream open reading frame within an IRES controls expression of a specific VEGF-A isoform. Nucleic Acids Res. 2008;36:2434–2445. doi: 10.1093/nar/gkn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fernandez J, et al. Regulation of internal ribosome entry site-mediated translation by eukaryotic initiation factor-2a phosphorylation and translation of a small upstream open reading frame. J Biol Chem. 2002;277:2050–2058. doi: 10.1074/jbc.M109199200. [DOI] [PubMed] [Google Scholar]
- 129.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 130.Cammas A, et al. Stabilization of the G-quadruplex at the VEGF IRES represses cap-independent translation. RNA Biol. 2015;12:320–329. doi: 10.1080/15476286.2015.1017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bonnal S, et al. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bhattacharyya D, Diamond P, Basu S. An independently folding RNA G-quadruplex domain directly recruits the 40S ribosomal subunit. Biochemistry. 2015;54:1879–1885. doi: 10.1021/acs.biochem.5b00091. [DOI] [PubMed] [Google Scholar]
- 133.Xue S, et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. The first targeted knockout of a cellular IRES in mice reveals a key role for IRES-driven translation of Hox mRNAs in vivo and identifies a new cis-regulatory element, the TIE, which blocks cap-dependent translation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jack K, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44:660–666. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martineau Y, et al. Internal ribosome entry site structural motifs conserved among mammalian fibroblast growth factor 1 alternatively spliced mRNAs. Mol Cell Biol. 2004;24:7622–7635. doi: 10.1128/MCB.24.17.7622-7635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cheng CY, et al. Consistent global structures of complex RNA states through multidimensional chemical mapping. eLife. 2015;4:1–38. doi: 10.7554/eLife.07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Créancier L, Morello D, Mercier P, Prats AC. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Créancier L, Mercier P, Prats A, Morello D, Cre L. c-Myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol Cell Biol. 2001;21:1833–1840. doi: 10.1128/MCB.21.5.1833-1840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Morfoisse F, et al. Hypoxia induces VEGF-C expression in metastatic tumor cells via a HIF-1a-independent translation-mediated mechanism. Cell Rep. 2014;6:155–167. doi: 10.1016/j.celrep.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 142.Audigier S, et al. Potent activation of FGF-2 IRES-dependent mechanism of translation during brain development. RNA. 2008;14:1852–1864. doi: 10.1261/rna.790608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Conte C, et al. FGF2 translationally induced by hypoxia is involved in negative and positive feedback loops with HIF-1α. PLOS ONE. 2008;3:e3078. doi: 10.1371/journal.pone.0003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Weingarten-Gabbay S, et al. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351:1–24. doi: 10.1126/science.aad4939. This study reports the first genome-wide approach to identify novel IRES-like elements in vivo and finds short poly(U) sequences and small structured elements that harbour IRES activity. [DOI] [PubMed] [Google Scholar]
- 145.Lee ASY, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. This study identifies a novel role for eIF3 in interacting with RNA stem–loop structures in the 5′ UTR of mRNAs to directly recruit the ribosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blau L, et al. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc Natl Acad Sci USA. 2012;109:E2875–E2884. doi: 10.1073/pnas.1203659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee AS, Kranzusch PJ, Doudna JA, Cate JH. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016;536:96–99. doi: 10.1038/nature18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2016;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Harcourt EM, Kietrys AM, Kool ET. Chemical and structural effects of base modifications in messenger RNA. Nature. 2017;541:339–346. doi: 10.1038/nature21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 151.Meyer KD, et al. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. Refs 151 and 152 demonstrate that m6A promotes cap-independent mRNA translation initiation, which is mediated by direct eIF3 recruitment. They also show that m6A levels in 5′ UTRs are increased during stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. This study reports that the incorporation of m6A into a hairpin modulates the local secondary structure of RNA (‘m6A switch’), thereby facilitating the binding of the indirect m6A reader hnRNPC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. This paper presents the first global in vivo RNA structure probing method, icSHAPE, and connects m6A modifications with destabilization of local RNA structure across the transcriptome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schwartz S, et al. High-Resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Roost C, et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang X, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2015;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dominissini D, et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Delatte B, et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 162.Slobodin B, et al. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell. 2017;169:326–337. doi: 10.1016/j.cell.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Choi J, et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol. 2016;23:110–115. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Legnini I, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pamudurti NR, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhou C, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat Rev Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 168.Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci. 2015;40:221–232. doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 169.Kubota M, Tran C, Spitale RC. Progress and challenges for chemical probing of RNA structure inside living cells. Nat Chem Biol. 2015;11:933–941. doi: 10.1038/nchembio.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bevilacqua PC, Ritchey LE, Su Z, Assmann SM. Genome-wide analysis of RNA secondary structure. Annu Rev Genet. 2016;50:235–266. doi: 10.1146/annurev-genet-120215-035034. [DOI] [PubMed] [Google Scholar]
- 171.Incarnato D, Oliviero S. The RNA epistructurome: uncovering RNA function by studying structure and post-transcriptional modifications. Trends Biotechnol. 2017;35:318–333. doi: 10.1016/j.tibtech.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 172.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. Refs 172 and 173 introduce the first methods for in vivo DMS-based chemical modification of accessible RNA nucleotides to globally probe RNA secondary structure. [DOI] [PubMed] [Google Scholar]
- 174.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. Probing xist RNA structure in cells using targeted structure-seq. PLoS Genet. 2015;11:e1005668. doi: 10.1371/journal.pgen.1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zubradt M, et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods. 2016;14:75–82. doi: 10.1038/nmeth.4057. At the current vanguard of in vivo chemical mapping techniques, DMS–MaPseq leverages several methodological advances to pilot applications to probe low-abundance mRNAs and pre-mRNAs and to derive structural models that inform compensatory mutagenesis experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Homan PJ, et al. Single-molecule correlated chemical probing of RNA. Proc Natl Acad Sci USA. 2014;111:13858–13863. doi: 10.1073/pnas.1407306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sergiev PV, Dontsova OA, Bogdanov AA. Chemical methods for the structural study of the ribosome: judgment day. Mol Biol. 2001;35:472–495. [PubMed] [Google Scholar]
- 180.Miao Z, et al. RNA-puzzles round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA. 2017;23:655–672. doi: 10.1261/rna.060368.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kladwang W, VanLang CC, Cordero P, Das R. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat Chem. 2011;3:954–962. doi: 10.1038/nchem.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tian S, Cordero P, Kladwang W, Das R. High-throughput mutate-map-rescue evaluates SHAPE-directed RNA structure and uncovers excited states. RNA. 2014;20:1815–1826. doi: 10.1261/rna.044321.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tian S, Das R. RNA structure through multidimensional chemical mapping. Q Rev Biophys. 2016;49:1–30. doi: 10.1017/S0033583516000020. [DOI] [PubMed] [Google Scholar]
- 184.Smola MJ, Calabrese JM, Weeks KM. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry. 2015;54:6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cheng C, Kladwang W, Yesselman J, Das R. RNA structure inference through chemical mapping after accidental or intentional mutations. Proc Natl Acad Sci USA. 2017;114:9876–9881. doi: 10.1073/pnas.1619897114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Risca VI, Denny SK, Straight AF, Greenleaf WJ. Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature. 2016;541:237–241. doi: 10.1038/nature20781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Burkhardt DH, et al. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife. 2017;6:e22037. doi: 10.7554/eLife.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dreyfuss G, Kim VN, Kataoka N, Medical HH. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 189.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.May J, Johnson P, Saleem H, Simon AE. A sequence-independent, unstructured IRES is responsible for internal expression of the coat protein of Turnip Crinkle Virus. J Virol. 2017;91:e02421–e02416. doi: 10.1128/JVI.02421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Clements J, Eddy SR, Rivas E. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods. 2017;14:45–48. doi: 10.1038/nmeth.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pedersen JS, et al. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:251–262. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Parker BJ, et al. New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Res. 2011;21:1929–1943. doi: 10.1101/gr.112516.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Goodarzi H, et al. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 2012;485:264–268. doi: 10.1038/nature11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hinnebusch AG. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci. 2017;1348:1–23. doi: 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 197.Martin F, et al. Ribosomal 18S rRNA base pairs with mRNA during eukaryotic translation initiation. Nat Commun. 2016;7:1–7. doi: 10.1038/ncomms12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mahamid J, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 199.Eddy SR. Computational analysis of conserved RNA secondary structure in transcriptomes and genomes. Annu Rev Biophys. 2014;43:433–456. doi: 10.1146/annurev-biophys-051013-022950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Majumder M, et al. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol. 2009;29:2899–2912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ke S, et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015;29:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.