Abstract
Immune control of HIV-1 infection depends heavily on cytotoxic T-lymphocyte responses restricted by diverse HLA class I molecules. Recent work has uncovered specific amino acid residues (AARs) that seem to dictate the extent of immune control in African Americans, which prompted us to test these emerging hypotheses in seroconverters (SCs) from southern and eastern Africa. Based on data from 196 Zambians and 76 Rwandans with fully resolved HLA alleles and pre-therapy HIV-1 viral loads (VL) in the first 3- to 36-month of infection (>2,300 person-visits), four AARs of primary interest (positions 63, 97, 116 and 245 in the mature HLA-B protein) were found to explain 8.1% and 15.8% of variance in set-point VL for these cohorts (P = 0.024 and 7.5 × 10−6, respectively). Two AARs not reported previously (167S in HLA-B and 116F in HLA-C) also showed relatively consistent associations with VL (adjusted P = 0.009 to 0.069), while many population-specific associations were also noted (false discovery rate <0.05). Extensive and often strong linkage disequilibrium among neighboring AAR variants called for more extensive analyses of AAR haplotypes in diverse cohorts before the structural basis of antigen presentation can be fully comprehended.
Keywords: Africa, HIV-1, immunity, HLA-I, antigen presentation, motifs, viral load
Introduction
For more than three decades now, an enduring topic on HIV/AIDS epidemiology has been dealing with the roles of immunogenetic host factors in determining the wide and evolving spectrum of HIV-1-related outcomes, especially viral load (VL) that has a dual impact on pathogenesis and transmission [1–3]. From early candidate gene approach to more recent genome-wide association studies [4, 5] and fine-mapping efforts [6, 7], the most unequivocal findings center on a few variants of human leukocyte antigen class I (HLA-I) genes within the human major histocompatibility complex (chromosome 6p21.3) [3, 5]. A short list of HLA-I alleles (e.g., HLA-B*57:01 -B*57:03 and -B*81:01) and amino acid residues (e.g., positions 63, 97, 116 and 245 in the mature HLA-B protein) seems to offer valuable insights into the specific interplays between HIV-1 antigens and adaptive immune responses (the equivalent of an immunological hide-and-seek or arms race), but HLA-I factors that can readily extrapolate across human populations and HIV-1 subtypes are still elusive, which severely limits their translational potential either as a mere biomarker or as a drugable target.
The challenge to gathering robust and generalizable immunogenetic data is even greater in African populations because of the diverse circulating HIV-1 subtypes [8–10], as well as heterogeneities in local and regional genetic architectures [11–13]. On the other hand, these populations hold an important key to fine-mapping of confirmed genomic regions because variable structure of linkage disequilibrium (LD) can help resolve local haplotype blocks for in-depth interrogation of causal variants, including those that are not fully captured by routine genotyping [6]. Thus, the objectives of this study were to: i) test the portability of recent observations on HLA-I amino acid residues (AARs) in the context of early HIV-1 control, (ii) assess consensus findings for two distinct African populations, and iii) identify new leads that might be important to African populations with relatively recent infection (with contemporary viruses).
Materials and Methods
Study subjects and HLA-I genotyping
HIV-1 seroconverters (SCs) were recruited by the Rwanda-Zambia HIV-1 Research Group for prospective studies [14, 15]. The research procedures were approved by institutional review boards at University of Alabama at Birmingham and Emory University, with further compliance to guidelines set forth by the United States Department of Health and Human Services. DNA samples derived from buffy coats or peripheral blood mononuclear cells were used for high-resolution HLA-I genotyping, as described elsewhere [9, 10, 14, 16, 17]. The individual amino acid residues (AARs) corresponding to each HLA allele were inferred from the IPD-IMGT/HLA Database [18] compiled by the International ImMunoGeneTics Project (http://www.ebi.ac.uk/ipd/imgt/hla/index.html, last accessed in January 2017), as described in the SNP2HLA program [19]. All polymorphic AARs were mapped to their respective positions in the mature protein products, while polymorphic AARs in the leader peptide (24 amino acids) were given negative numbering (i.e., −24 to −1). Rare AARs found in fewer than five SCs in both cohorts were excluded from analyses (for lack of statistical power).
Virological outcomes after HIV-1 infection
For SCs identified during frequent (monthly to quarterly) testing [20–22], the estimated dates of HIV-1 infection (EDI) were defined using four criteria that have been detailed before [9, 10, 17]. Viral subtype was defined by partial sequencing of the HIV-1 pol gene [9, 23]. Clinical visits after confirmed HIV-1 infection were scheduled monthly for the first three months after EDI and quarterly for the 3–36 months period (primary infection). In all, 272 SCs (196 Zambians and 76 Rwandans) (Table 1) with known EDI and at least four time points of VL (HIV-1 RNA copies/mL of plasma) in the early chronic phase (3–36 months) of infection were included for this study. Association analyses began with the full dataset (3–36 months of infection), while alternative analyses used the geometric mean VL in each patient as a proxy for the set-point beyond the first 3 months of infection [9, 24].
Table 1.
Characteristics of HIV-1 seroconverters (SCs) from two African countries, with adequate follow-up before antiretroviral therapy.
| Characteristicsa and HLA variants of major interest | Rwanda | Zambia |
|---|---|---|
| Number of SCs with sufficient data | 76 | 196 |
| Sex ratio (M/F) | 1.4 (44/32) | 1.3 (111/85) |
| Age: mean ± SD (yr) | 33.2 ± 9.3 | 33.6 ± 8.0 |
| Age ≥40 yr: no. (%) | 16 (21.1) | 40 (20.4) |
| Estimated dates of infection | ||
| Earliest | May 2005 | June 2005 |
| Latest | Jan. 2011 | Mar. 2011 |
| HIV-1 subtype: no. (%) | ||
| A1 | 62 (81.6) | 0 (0.0) |
| C | 8 (10.5) | 191 (97.4) |
| Others (B, D and recombinants)b | 6 (7.9) | 5 (2.5) |
| HIV-1 viral load (VL) measures: mean (range) | 9 (3–14) | 8 (3–18) |
| Person-visits with eligible VL data (3–36 months) | 669 | 1,636 |
| Duration of infection at VL measurements (range, in moths) | 2.9–32.1 | 2.8–32.8 |
Non-standard abbreviations: SD, standard deviation of the mean.
Including two unknown HIV-1 subtypes in Zambians (viral sequencing failed).
Baseline statistics
The two study cohorts were first summarized for demographics, HIV-1 subtypes, and visits for clinical outcomes (Table 1). Within each cohort, the patterns of linkage disequilibrium (LD) between AAR variants were assessed for each locus, and those with r2 values ≥0.80 were considered as mutually tagging [25]. The LD measures also formed the basis for Bonferroni correction in multi-AAR testing [26], and the number of independent tests was determined using a modified spectral decomposition method [27].
Hypothesis-testing and refinements
Using software from SAS version 9.2 (SAS Institute, Cary, NC) [6, 28], we first tested existing hypotheses regarding four HLA-B AARs (residues 63, 97, 116 and 245). Mixed models evaluated these AARs individually and collectively for their relative impact on repeated measures of VL (after log10-transformation), as reflected by the effect size (mean beta estimate (β) and standard error (SE)) after statistical adjustments for duration of infection (DOI) and demographics (sex and age) [9, 10, 17]. Alternative models assessed the impact of AAR on set-point VL, with a focus on the proportion of variance attributable to each AAR variant. Statistical power for these models was expected to vary by analytical approaches, by effect size and by cohort (marker frequency and distribution of outcome measures) (Figure S1 in Supplemental Materials), but given the well-known strong impact (substantial effect size) of HLA factors on HIV-1 VL [6, 28], our modest sample sizes here had >80% statistical power in detecting associations if: i) the effect size (β) is at least 0.50 log10 and the AAR variant frequency is at least 35% in Rwandans; ii) β is at least 0.50 log10 and the AAR variant frequency is at least 10% in Zambians. Relative consistency between studies (here and elsewhere), as measured by regression β estimates that were in the same direction (positive or negative), was considered as indicative of plausible associations for presentation.
Other association analyses
We also searched for further associations that might be study- or cohort-specific, especially those that were independent of previously reported AARs (residues 63, 97, 116 and 245 for HLA-B) and supported by at least two of the following parameters: (i) statistical significance (P <0.05) accompanied by low false discovery rate (q <0.05), (ii) biologically meaningful effect size |β| >0.30 log10 [29, 30], and (iii) consistency between the two study cohorts. High-dimensional analytical methods, including Bayesian regression models implemented in the BGLR R-package [31], did not reveal additional findings that were consistent between cohorts.
Results
General characteristics of 272 HIV-1 seroconverters (SCs)
As summarized earlier [9, 10, 17], Rwandan and Zambian SCs (n = 76 and 196, respectively) were similar in terms of sex ratio, age, EDI, frequency of VL measures, and DOI (Table 1). The only notable distinction between the two SC groups was the HIV-1 subtype ― predominantly subtype A1 in Rwanda and subtype C in Zambia, which was also expected [9, 10, 17]. In all, Rwandan and Zambian SCs contributed 669 and 1,636 person-visits, respectively, for analyses of repeated VL measurements.
HLA-I AAR alignment
For the three HLA-I loci with high-resolution molecular genotyping, a total of 312 AAR variants (allele frequency <0.50) could be aligned for at least 90% of subjects (Table S1 in Supplemental Materials). The AAR positions ranged from residues −14 to 334 for HLA-A (113 AAR variants), −22 to 325 for HLA-B (106 AAR variants), and −17 to 339 for HLA-C (93 AAR variants). The distribution of AAR variants clearly differed between Rwanda and Zambia (P = 0.001 by likelihood ratio test), although principal component analysis of these AAR variants was unable to separate the two cohorts into distinct clusters (data not shown but available for J.T.).
LD structure of HLA-I AAR variants
Many AAR variants showed strong pairwise LD (r2>0.80) in individual cohorts or both (Figure S2 in Supplemental Materials). Overall, a stepwise (random) analysis of the 312 AAR variants would imply 88 independent tests for both cohorts, suggesting that a P value <5.7 × 10−4 (0.05/88) would qualify as statistically significant after a study-wide Bonferroni correction. This conservative P value threshold was applied to new findings from each cohort, unless the observed false discovery rates (q values) were <0.05 in both cohorts.
Hypothesis-testing: analyses of HLA-I AAR variants reported earlier for African Americans
In multivariable models that evaluated four HLA-B AARs of primary interest, 63E, 97V, 116F and 245T showed internal consistencies that supported earlier reports for African Americans. Collectively, these variants explained 15.8% and 8.1% of variance in set-point VL for Rwandans (P = 7.5 × 10−6) and Zambians (P = 0.024), respectively (Table 2). The estimated effect size of individual AAR variants on VL did vary widely between cohorts, with just a single strict consensus for 97V (P <0.0001 in Rwanda and P = 0.001 in Zambia), which exclusively tagged HLA-B*57 (a well-known factor in Africans) (Table S2 in Supplemental Materials). Apart from 97V, both 63E and 116F were independent correlates of VL in Rwanda (P = 0.001 and 0.019, respectively), while 245T showed an independent association with VL in Zambia (P = 0.014). In terms of biological significance, the relative effect of 97V, 63E and 116F exceeded the threshold of 0.30 log10 in VL differential (the β estimate) for Rwandans, while 97V and 245T reached the same threshold for Zambians (Table 2).
Table 2.
Partial confirmation of four HLA-B amino acid residue (AAR) variants associated with HIV-1 control.
| Rwandans | Zambians | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Factorsa | Protein | n | β ± SEb | Pb | Cumulativec R2 |
n | β ± SEb | Pb | Cumulativec R2 |
| 97V | HLA-B | 6 | −1.12 ± 0.24 | <0.0001 | 0.133 | 19 | −0.39 ± 0.11 | <0.001 | 0.074 |
| 63E | HLA-B | 61 | −0.59 ± 0.18 | 0.001 | 0.164 | 142 | −0.09 ± 0.07 | 0.225 | 0.073 |
| 116F | HLA-B | 7 | −0.54 ± 0.23 | 0.019 | 0.181 | 30 | −0.07 ± 0.09 | 0.460 | 0.072 |
| 245T | HLA-B | 3 | −0.09 ± 0.32 | 0.771 | 0.158 | 11 | −0.35 ± 0.14 | 0.014 | 0.081 |
As reported earlier for African Americans [6].
For multivariable analyses of repeated HIV-1 viral load measures (after log10-transformation) in the 3- to 36-month intervals after estimated date of infection, the estimates of effect size (β and standard error, SE) and P values are based on omnibus tests; three non-HLA factors, including age (at estimated date of HIV-1 infection), sex, and duration of infection (per quarter), are retained as covariates (potential cofactors).
For analyses of set-point (geometric mean) viral load, the focus is shifted to variance explained by each AAR variant.
Evaluation of HLA-I AARs regardless of prior hypotheses
Aside from the four HLA-B AAR variants of primary interest, 89 other AAR variants in Rwandans and three in Zambians reached statistical significance (i.e., P <0.05 and q ≤0.05) when tested in separate cohorts (Figure 1 and Table S1 in Supplemental Materials). The vast majority of these association signals either had conflicting relationships between cohorts (in opposing directions) or reflected strong or even exclusive LD (r2 = 0.80–1.0) between AAR variants. For example, 45M and 46A in HLA-B were in exclusive LD (r2 = 1.0), and their associations with VL (P <5.7 × 10−4 and q <0.01 in both cohorts) was explained by their respective, partial LD with 97V (r2 = 0.527 and 0.699 in Rwanda and Zambia, respectively). Among clusters of AAR variants with identical summary statistics and exclusive LD (Table S1 in Supplemental Materials), some covered a wide range. For instance, one cluster of 12 AAR variants in HLA-C began with −17Q (at residue −17) and ended with 309H. Another cluster in HLA-A included six AAR variants between 44K and 158V (Table S1 in Supplemental Materials).
Figure 1.
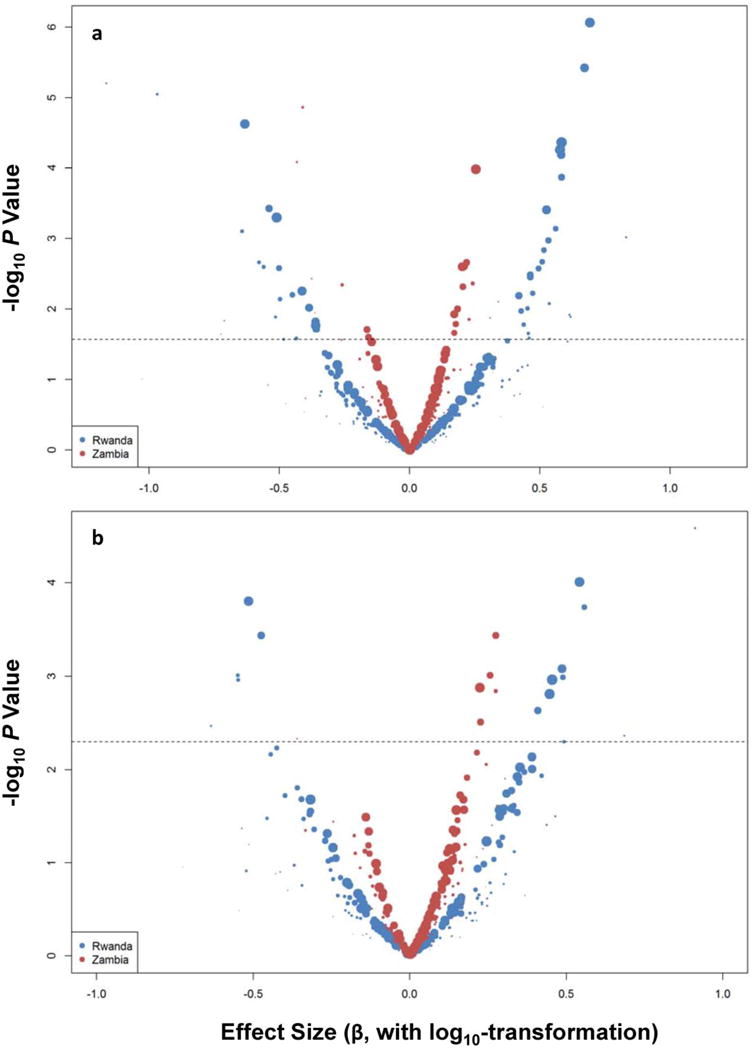
Screening of individual HLA-I amino acid variants for potential associations with repeated measures of HIV-1 viral load (VL). For HIV-1 serconverters from Rwanda and Zambia, the effect size (beta estimate on the X-axis, on a log10 scale) and statistical significance (P value on the Y-axis) are based on two rounds of analyses (with different scales for P values). In round 1 (panel a), demographics (age and sex) and duration of HIV-1 infection at the time of VL measurements are retained as covariates in analyses of 312 individual variants. In round 2 (panel b), analyses are limited to 308 individual variants and further conditioned on four previously reported HLA-B residues (listed in Table 1). Circles (individual signals) above the dotted line have false discovery rates <0.05. The sizes of circles are proportional to population frequencies seen in each color-coded cohort.
In multivariable models that were conditioned on the four HLA-B AAR variants of primary interest (63E, 97V, 116F and 245T), the majority of putative association signals either faded or vanished (Figure 1). Only 167S in HLA-B and 116F in HLA-C were worth noting because of consistency between cohorts (adjusted P = 0.009–0.069 (Table 3). In each case, the evidence for independent association in one cohort gained weak support from the other. Overall, the six HLA-I variants explained 33.8% and 14.8% of variance in set-point VL for Rwandans and Zambians (P <0.001 for both), representing a substantial improvement over the estimates based on the four HLA-B AAR variants alone. Of note, 167S in HLA-B was restricted to B*44 and B*45 alleles, while 116F in HLA-C was present in six allele groups (C*04, C*05, C*07, C*08, C*17 and C*18) (Table S2 in Supplemental Materials), with B*45 and C*04 already known as unfavorable in HIV-1-infected Africans or African Americans [32–34].
Table 3.
Identification of 167S in HLA-B and 116F in HLA-C as putative factors associated with HIV-1 viral load.
| Rwandans | Zambians | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Factors | Protein | n | β ± SEa | Pa | n | β ± SEa | Pa |
| 97V | HLA-B | 6 | −0.99 ± 0.23 | <0.0001 | 19 | −0.40 ± 0.11 | <0.0001 |
| 63E | HLA-B | 61 | −0.51 ± 0.18 | 0.005 | 142 | −0.15 ± 0.08 | 0.073 |
| 116F | HLA-B | 7 | −0.57 ± 0.22 | 0.011 | 30 | −0.09 ± 0.09 | 0.315 |
| 245T | HLA-B | 3 | −0.18 ± 0.31 | 0.555 | 11 | −0.38 ± 0.14 | 0.008 |
| 167Sb | HLA-B | 14 | 0.34 ± 0.18 | 0.059 | 62 | 0.21 ± 0.08 | 0.009 |
| 116Fb | HLA-C | 48 | 0.36 ± 0.14 | 0.012 | 165 | 0.13 ± 0.07 | 0.069 |
Using the same analytical approaches as shown in Table 2.
Not reported elsewhere.
Discussion
Despite a relatively modest sample size, immunogenetic data from two African countries did reveal that HLA-I AARs can be readily associated with early virological outcomes in untreated HIV-1 infection, as measured by their individual or collective impact on VL in the 3–36 months period of infection. Given the known biology of HLA-I molecules in epitope-specific antigen presentation and regulation of natural killer cell functions, these findings are not surprising. On the other hand, our detailed analyses also identified multiple analytical issues that deserve some close attention.
First, in a stepwise selection process, which is commonly applied to high-throughput genomic data and focuses solely on statistical significance (the P value), many potentially informative AARs (Figure 1) can be overlooked as a result of strong or exclusive LD (Table S1 and Figure S1). By nature, DNA sequences encoding many AARs are tightly linked on the same chromosome (the very basis for routine HLA allele assignments) [12, 18, 35]. A joint assessment of statistical significance and the location of AARs relative to the HLA-I protein crystal structure may occasionally pinpoint the underlying biology, but in situations where multiple AAR variants are in strong LD, only cohorts with varying genetic structure (LD patterns) can offer unequivocal conclusions. To this end, a full list of potential associations in each study (e.g., Table S1) should benefit the efforts for fine-mapping.
Second, strict consensus between studies is still elusive. Although a previous study has convincingly shown that four common HLA-B AARs can sufficiently capture the effects of HLA-I alleles on set-point VL in African Americans [6], at least 63E and 116F have not been implicated in cohorts of European ancestry [36]. By emphasizing two estimates of effect size (R2 and β) and internal consistency between both cohorts, we were able to highlight six AAR variants of interest (Table 3), including two (167S in HLA-B and 116F in HLA-C, Table 3) that have not been reported previously. These putative AAR variants could not be readily tracked to specific alleles that are well characterized in the context of HIV-1 control, but they are frequent enough that follow-up studies will not require a prohibitively large cohort for validation. Conversely, verification of various population-specific observations will be much more difficult, as the assembly of new cohorts in similar locations is no longer feasible under the current, global treatment guidelines [37].
Third, for HLA-I factors that has a substantial influence on HIV-1 control, a modest sample size (statistical power) can still offer important insights into generalizable findings, especially after the initial discovery phase is completed (lifting penalty for random testing). HLA-B 97V is one such example (Table 1, P <0.0001 and q <0.05 based on six Rwandans). On the other hand, 97V exclusively tags HLA-B*57 in Rwandans and Zambians here (Table S2), making it impossible to separate the effect of 97V (as part of a functional domain) from HLA-B*57 (as an entire allele). For other HLA-B AAR variants that do cover a wide range of individual alleles (Table S2), one extra issue relates to extended LD in the entire MHC region [35]. Eventually, fine-mapping may further require the analyses of variants well beyond the classic approach to HLA genotyping.
One major limitation in analyzing HLA-I AARs is the current disparity in allelic sequence alignments, as many common alleles are still based on partial sequences, mostly from exons 2–4 [18]. With deep sequencing technologies now readily available [38, 39]}, non-coding sequences and AARs beyond exons 2–4 may provide new dimensions for fine-mapping, assuming that their potential contribution to human health and disease has not been adequately captured by conventional analyses of HLA-I alleles, haplotypes, and supertypes [16, 40, 41], as well as genome-wide (low-density) scans of single nucleotide polymorphisms [5]. Such refinements should strengthen the well-established notion that HLA-I alleles differentially mediate immune control of HIV-1 infection and subsequent immune escape [42–44].
Beyond HIV/AIDS, many studies have consistently revealed compelling evidence that a handful of amino acid residues in diverse HLA alleles, especially those of class II DRβ1 and DQβ1 chains, may play critical roles in mediating autoimmune disorders [45–47]. For example, five amino acids in three HLA proteins (DRβ1, HLA-B, and DQβ1) appear to explain most of the association between human MHC variants and seropositive rheumatoid arthritis [48], and more recent work has uncovered additional evidence that HLA-DRβ1 amino acid variants and their haplotypes may dictate IgG response to viral antigens [49]. If these intriguing observations prove to be true across population boundaries, there would be a strong rationale for manipulating these residues in experimental studies [50, 51]. To this end, HLA-B residue 97 is of particular interest because of its strong and consistent associations with autoimmune and infectious diseases [6, 52].
Supplementary Material
Part 1. Supplemental Tables (Excel file)
Table S1. Analyses of 312 individual HLA-I AAR variants for potential associations with HIV-1 viral load (VL) in two African cohorts (an expanded version of Figure 1).
Table S2. Relationships of HLA-B and HLA-C alleles to several AAR variants of interest (as described in the manuscript).
Part 2. Supplemental Figures (PDF file)
Figure S1. Statistical power in confirming previously reported HLA-I AAR variants associated with HIV-1 viral load (alpha = 0.05).
Figure S1. A graphical view of HLA-I AAR variants that are in strong linkage disequilibrium (r2 >0.80) in either cohort (green = Rwanda only; blue = Zambia only) or both cohorts (red).
Acknowledgments
This work was funded primarily by the United States National Institute of Allergy and Infectious Diseases (NIAID), through two R01 grants (AI071906 to J.T. and AI064060 to P.A.G/E.H./J.T.). The funder has no role in study design, data collection or data analysis. We thank members of the Rwanda-Zambia HIV-1 Research Group for their valuable contributions to cohort assembly and collection of prospective data. We are also grateful to several former associates, especially Travis Porter, Heather A. Prentice and Wei Song, for assistance with genotyping and data management.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare no conflict of interests.
References
- 1.Mellors JW, Kingsley LA, Rinaldo CR, Jr, Todd JA, Hoo BS, Kokka RP, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 2.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 3.Prentice HA, Tang J. HIV-1 dynamics: A reappraisal of host and viral factors, as well as methodological issues. Viruses. 2012;4:2080. doi: 10.3390/v4102080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartha I, Carlson JM, Brumme CJ, McLaren PJ, Brumme ZL, John M, et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. Elife. 2013;2:e01123. doi: 10.7554/eLife.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLaren PJ, Ripke S, Pelak K, Weintrob AC, Patsopoulos NA, Jia X, et al. Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum Mol Genet. 2012;21:4334. doi: 10.1093/hmg/dds226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prentice HA, Pajewski NM, He D, Zhang K, Brown EE, Kilembe W, et al. Host genetics and immune control of HIV-1 infection: fine mapping for the extended human MHC region in an African cohort. Genes Immun. 2014;15:275. doi: 10.1038/gene.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Ni R, Song W, Shao W, Shrestha S, Ahmad S, et al. Clear and independent associations of several HLA-DRB1 alleles with differential antibody responses to hepatitis B vaccination in youth. Hum Genet. 2009;126:685. doi: 10.1007/s00439-009-0720-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prentice HA, Price MA, Porter TR, Cormier E, Mugavero MJ, Kamali A, et al. Dynamics of viremia in primary HIV-1 infection in Africans: Insights from analyses of host and viral correlates. Virology. 2014;449:254. doi: 10.1016/j.virol.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prentice HA, Lu H, Price MA, Kamali A, Karita E, Lakhi S, et al. Dynamics and correlates of CD8 T-cell counts in Africans with primary human immunodeficiency virus type 1 infection. J Virol. 2016;90:10423. doi: 10.1128/JVI.01467-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 12.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13:175. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 14.Merino AM, Song W, He D, Mulenga J, Allen S, Hunter E, et al. HLA-B signal peptide polymorphism influences the rate of HIV-1 acquisition but not viral load. J Infect Dis. 2012;205:1797. doi: 10.1093/infdis/jis275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J, Li X, Price MA, Sanders EJ, Anzala O, Karita E, et al. CD4:CD8 lymphocyte ratio as a quantitative measure of immunologic health in HIV-1 infection: findings from an African cohort with prospective data. Front Microbiol. 2015;6:670. doi: 10.3389/fmicb.2015.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang J, Malhotra R, Song W, Brill I, Hu L, Farmer PK, et al. Human leukocyte antigens and HIV type 1 viral load in early and chronic infection: Predominance of evolving relationships. PLoS ONE. 2010;5:e9629. doi: 10.1371/journal.pone.0009629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Price MA, He D, Kamali A, Karita E, Lakhi S, et al. Host genetics and viral load in primary HIV-1 infection: clear evidence for gene by sex interactions. Hum Genet. 2014;133:1187. doi: 10.1007/s00439-014-1465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karita E, Price M, Hunter E, Chomba E, Allen S, Fei L, et al. Investigating the utility of the HIV-1 BED capture enzyme immunoassay using cross-sectional and longitudinal seroconverter specimens from Africa. AIDS. 2007;21:403. doi: 10.1097/QAD.0b013e32801481b7. [DOI] [PubMed] [Google Scholar]
- 21.Prentice HA, Porter TR, Price MA, Cormier E, He D, Farmer PK, et al. HLA-B*57 versus HLA-B*81 in HIV-1 infection: slow and steady wins the race? J Virol. 2013;87:4043. doi: 10.1128/JVI.03302-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amornkul PN, Karita E, Kamali A, Rida WN, Sanders EJ, Lakhi S, et al. Disease progression by infecting HIV-1 subtype in a seroconverter cohort in sub-Saharan Africa. AIDS. 2013;27:2775. doi: 10.1097/QAD.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price MA, Wallis CL, Lakhi S, Karita E, Kamali A, Anzala O, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses. 2011;27:5. doi: 10.1089/aid.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S, et al. Prospective study of acute HIV-1 infection in adults in east Africa and Thailand. N Engl J Med. 2016;374:2120. doi: 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95:221. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 28.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 29.Saag MS, Holodniy M, Kuritzkes DR, O’Brien WA, Coombs R, Poscher ME, et al. HIV viral load markers in clinical practice. Nat Med. 1996;2:625. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 30.Modjarrad K, Chamot E, Vermund SH. Impact of small reductions in plasma HIV RNA levels on the risk of heterosexual transmission and disease progression. AIDS. 2008;22:2179. doi: 10.1097/QAD.0b013e328312c756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez P, de los Campos G. Genome-wide regression and prediction with the BGLR statistical package. Genetics. 2014;198:483. doi: 10.1534/genetics.114.164442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koehler RN, Walsh AM, Saathoff E, Tovanabutra S, Arroyo MA, Currier JR, et al. Class I HLA-A*7401 is associated with protection from HIV-1 acquisition and disease progression in Mbeya, Tanzania. J Infect Dis. 2010;202:1562. doi: 10.1086/656913. [DOI] [PubMed] [Google Scholar]
- 34.Matthews PC, Adland E, Listgarten J, Leslie A, Mkhwanazi N, Carlson JM, et al. HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703. J Immunol. 2011;186:5675. doi: 10.4049/jimmunol.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorak MT, Shao W, Machulla HK, Lobashevsky ES, Tang J, Park MH, et al. Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun. 2006;7:450. doi: 10.1038/sj.gene.6364315. [DOI] [PubMed] [Google Scholar]
- 36.McLaren PJ, Coulonges C, Bartha I, Lenz TL, Deutsch AJ, Bashirova A, et al. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc Natl Acad Sci USA. 2015;112:14658. doi: 10.1073/pnas.1514867112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ngongo PB, Priddy F, Park H, Becker J, Bender B, Fast P, et al. Developing standards of care for HIV prevention research in developing countries – a case study of 10 research centers in Eastern and Southern Africa. AIDS Care. 2012;24:1277. doi: 10.1080/09540121.2012.656572. [DOI] [PubMed] [Google Scholar]
- 38.Mack SJ, Milius RP, Gifford BD, Sauter J, Hofmann J, Osoegawa K, et al. Minimum information for reporting next generation sequence genotyping (MIRING): Guidelines for reporting HLA and KIR genotyping via next generation sequencing. Hum Immunol. 2015;76:954. doi: 10.1016/j.humimm.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schofl G, Lang K, Quenzel P, Bohme I, Sauter J, Hofmann JA, et al. 2.7 million samples genotyped for HLA by next generation sequencing: lessons learned. BMC Genomics. 2017;18:161. doi: 10.1186/s12864-017-3575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazaryan A, Lobashevsky E, Mulenga J, Karita E, Allen S, Tang J, et al. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J Virol. 2006;80:6056. doi: 10.1128/JVI.02119-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, Allen S, et al. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol. 2008;181:2626. doi: 10.4049/jimmunol.181.4.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 43.Tang J, Kaslow RA. The impact of host genetics on HIV infection and disease progression in the era of highly active antiretroviral therapy. AIDS. 2003;17:S51. doi: 10.1097/00002030-200317004-00006. [DOI] [PubMed] [Google Scholar]
- 44.Goulder PJ, Walker BD. HIV and HLA class I: an evolving relationship. Immunity. 2012;37:426. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat Rev Genet. 2016;17:160. doi: 10.1038/nrg.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terao C, Yoshifuji H, Yamano Y, Kojima H, Yurugi K, Miura Y, et al. Genotyping of relapsing polychondritis identified novel susceptibility HLA alleles and distinct genetic characteristics from other rheumatic diseases. Rheumatology (Oxford) 2016;55:1686. doi: 10.1093/rheumatology/kew233. [DOI] [PubMed] [Google Scholar]
- 47.Van Drongelen V, Holoshitz J. A reciprocal HLA-disease association in rheumatoid arthritis and pemphigus vulgaris. Front Biosci (Landmark Ed) 2017;22:909. doi: 10.2741/4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammer C, Begemann M, McLaren PJ, Bartha I, Michel A, Klose B, et al. Amino acid variation in HLA class II proteins is a major determinant of humoral response to common viruses. Am J Hum Genet. 2015;97:738. doi: 10.1016/j.ajhg.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Shi H, Yuan J, Bowness P. Position 97 of HLA-B, a residue implicated in pathogenesis of ankylosing spondylitis, plays a key role in cell surface free heavy chain expression. Ann Rheum Dis. 2017;76:593. doi: 10.1136/annrheumdis-2016-209512. [DOI] [PubMed] [Google Scholar]
- 51.Kaur G, Gras S, Mobbs JI, Vivian JP, Cortes A, Barber T, et al. Structural and regulatory diversity shape HLA-C protein expression levels. Nat Commun. 2017;8:15924. doi: 10.1038/ncomms15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortes A, Pulit SL, Leo PJ, Pointon JJ, Robinson PC, Weisman MH, et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat Commun. 2015;6:7146. doi: 10.1038/ncomms8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Part 1. Supplemental Tables (Excel file)
Table S1. Analyses of 312 individual HLA-I AAR variants for potential associations with HIV-1 viral load (VL) in two African cohorts (an expanded version of Figure 1).
Table S2. Relationships of HLA-B and HLA-C alleles to several AAR variants of interest (as described in the manuscript).
Part 2. Supplemental Figures (PDF file)
Figure S1. Statistical power in confirming previously reported HLA-I AAR variants associated with HIV-1 viral load (alpha = 0.05).
Figure S1. A graphical view of HLA-I AAR variants that are in strong linkage disequilibrium (r2 >0.80) in either cohort (green = Rwanda only; blue = Zambia only) or both cohorts (red).


