Abstract
Objective
To investigate the association of rare predicted loss-of-function (pLoF) variants within previously reported age-related macular degeneration (AMD) risk loci and sub-phenotypes characteristic of intermediate or advanced AMD, including geographic atrophy (GA), choroidal neovascularization (CNV), pseudoreticular drusen, calcified drusen, drusen area in the in the macula, and the Age-Related Eye Disease Study (AREDS) Extended AMD Severity Scale.
Design
Case-control study
Participants
AREDS, AREDS2, and Michigan Genomics Initiative participants.
Methods
Whole genome sequencing data were analyzed for rare pLoF variants (frequency < 0.1% in the study population) in the regions of previously identified 52 independent risk variants known to be associated with AMD. Frequency of the rare pLoF variants in cases with intermediate or advanced AMD were compared with controls. Variants were also assigned to the complement, extracellular matrix (ECM), lipid, cell survival, immune system, metabolism, or unknown/other pathway. Associations of rare pLoF variant pathways with sub-phenotypes within the AMD population were analyzed using logistic and linear regression, and Cox proportional hazard regression models.
Main Outcomes and Measures
Differences in rare pLoF variant pathway burden and association of rare pLoF variant pathways with sub-phenotypes within the AMD population were evaluated.
Results
Rare pLoF variants were found in 298/1689 (17.6%) cases and 237/1518 (15.6%) controls (OR=1.11 [0.91, 1.36], p=0.310). An enrichment of rare pLoF variants in the complement pathway in cases versus controls (OR=2.94 [1.49, 5.79], p=0.002) was observed. Within cases, associations between all rare pLoF variants and CNV (OR=1.34 [1.04, 1.73], p=0.023), calcified drusen (OR=1.33 [1.04, 1.72], p=0.025), higher scores on the AREDS Extended AMD Severity Scale (Standardized Coefficient Beta (B)=0.346 [0.086, 0.605], p=0.009), and progression to advanced disease (HR=1.25 [1.01, 1.55], p=0.042) were observed. At the pathway level, there were associations between the complement pathway and GA (OR=2.17 [1.12, 4.24], p=0.023), the complement pathway and calcified drusen (OR=3.75 [1.79, 7.86], p<0.001), and the ECM pathway and more severe levels in the AREDS Extended AMD Severity Scale (B=0.62 [0.04, 1.20], p=0.035).
Conclusions
Rare pLoF variants are associated with disease progression. Variants in the complement pathway modify the clinical course of AMD and increase risk of developing specific sub-phenotypes.
Introduction
Age-related macular degeneration (AMD) is a multifactorial neurodegenerative disease that is the most common cause of incurable blindness worldwide.1 The population prevalence of AMD rises to 13% in individuals over the age of 85,2 and thus poses an increasing burden to the healthcare system.3 The strongest non-genetic risk factors include advanced age and smoking status.4 Early AMD is present when medium-sized drusen (≥63 μm and <125 μm) are detected in the retina. The presence of large drusen (≥125 μm) indicates intermediate AMD. This may be accompanied by retinal pigment epithelial changes in the retina.5 Drusen are focal extracellular accumulations of debris between the retinal pigment epithelium (RPE) and Bruch’s membrane. The composition of drusen is similar to atherosclerotic plaques, consisting of apolipoproteins, cholesterol, amyloid, and crystallins.6, 7 AMD progresses to two late forms including geographic atrophy (GA), which is characterized by loss of photoreceptors and RPE, resulting in a large area of depigmentation and visible choroidal vessels, and choroidal neovascularization (CNV), which is characterized by aberrant vessel growth and leakage of fluid into the retina, leading to RPE or sensory retinal detachment, subretinal fibrosis, and atrophy.8 The Age-Related Eye Disease Study (AREDS) AMD scale is often used to classify disease and risk of progression.9 Clinical heterogeneity in AMD lends itself to the study of sub-phenotypes that mark progression though various stages of disease.
The first complex disease Genome-Wide Association Study (GWAS) success story was the discovery of an association between the Y402H polymorphism in Complement Factor H (CFH) and AMD,10 followed by additional strong association with many non-coding variants in CFH.11 The other major susceptibility gene is ARMS2.12, 13 Presence of both CFH and ARMS2 homozygous risk alleles confers a 50-fold increased risk of AMD.14 The most recent large GWAS study of 16,144 cases and 17,832 controls identified 52 common and rare variants distributed across 34 loci that are associated with AMD. 15 The genes at these loci belong to a variety of biological pathways, including complement, lipid transport, and extracellular matrix (ECM), many of which have been implicated in AMD pathogenesis.8 The complement pathway is one of the most well-defined pathways involved in AMD and several of the identified genetic variants are related to the complement pathway. Disruptions in the complement and other pathways are predicted to result in clinical variability observed in AMD patients. Notably, CFH risk variants are reported to increase the risk of GA, while ARMS2 variants enhance the risk of CNV.16
Despite the progress in AMD genetics, variants at risk loci explain only about 50% of the disease heritability, and most of the common variants (except at CFH and ARMS2 loci) identified through GWAS exert small effect size on their own. Moreover, for many of the different GWAS loci, we are not yet certain which gene is affected due to the linkage disequilibrium (LD) region surrounding the variant. One way to investigate which gene is involved, is by identifying rare variants (allele frequency < 1%) within candidate genes. They will usually have a stronger predisposition for disease than common variants and may imply a functional role of that gene in AMD pathogenesis. These rare variants may also provide further insight into the role of the affected gene in AMD by showing phenotype correlations. Previous studies have identified rare variants in various complement genes and have shown associations with drusen load and GA.17–23 However, so far, only very few rare variants in genes unrelated to the complement pathway have been implicated in AMD.24 Possibly, this is due to a smaller effect size of the rare variants in other genes as compared to variants in complement genes. A step beyond looking at rare variants, is investigating rare predicted-loss-of-function (pLoF) variants. As the name implies, these variants are usually very rare (allele frequency < 0.1%) and are predicted to substantially affect the gene function. Therefore, they are expected to have even larger effect sizes. Whole genome sequencing (WGS) allows for the identification of novel rare variants that might have larger effect sizes, and evaluation of AMD-associated regions previously identified by GWAS is a rational approach to identify potential causal genes at different loci as it increases our chances of capturing relevant genes.25 Here we report the identification of rare pLoF variants, their association broadly with AMD and more specifically with AMD sub-phenotypes. Because pLoF variants may be very rare and many are present in no more than a single person in the sequenced population, they usually cannot be analyzed individually. Therefore, we collapsed rare variants into 7 biological pathways relevant to disease pathology based on the gene function and examined the increase in risk of developing AMD and its distinct sub-phenotypes. Our studies demonstrate enhanced risk of the development of calcified drusen, GA, and/or CNV with distinct biological pathways and suggest novel opportunities for diagnosis and treatment of AMD.
Methods
Ethics Statement
The study followed the tenets of the Declaration of Helsinki and complied with the Health Portability and Accountability Act. This study was approved by the local institutional review boards and the local ethics committees at the participating study centers. Written informed consent was obtained from each participant after explanation of the nature and possible consequences of the study.
Population
Age-Related Eye Disease Study (1992–2005)
The Age-Related Eye Disease Study (AREDS), a multicenter, randomized clinical trial of oral supplements of antioxidant vitamins (C, E, beta-carotene) and minerals (zinc and copper) for the treatment of AMD and cataract, was also designed to assess the clinical course, prognosis and risk factors associated with AMD and cataract. Participants (n=4757) ranging between 55 and 80 years of age were described previously.26 The participants were enrolled based on their baseline AMD severity: from no evidence of AMD to advanced AMD in one eye. Participants with WGS and with intermediate AMD or advanced AMD in one eye were included as cases (n=381), while participants with no evidence of AMD served as part of our control group (n=199). Participants were followed every 6 months for at least 7 years. Extensive phenotypic information was gathered, including annual stereoscopic fundus photographs of the macula which were graded by certified and masked graders at a central photograph reading center for AMD severity.
Age-Related Eye Disease Study 2 (2006–2012)
The Age-Related Eye Disease Study 2 (AREDS2) was a multicenter, phase III, randomized, controlled clinical trial enrolling 4203 individuals between 50 and 85 years of age with bilateral large drusen or late AMD in 1 eye as described previously.27 It was designed to evaluate the safety and efficacy of adding supplementation with lutein plus zeaxanthin and omega-3 long-chain polyunsaturated fatty acids to AREDS supplements, as well as changes in the original AREDS supplements (elimination of beta-Carotene and reducing the zinc dose) in reducing the risk of developing advanced AMD. Participants were followed for an average of 5 years. Follow-up study visits were scheduled annually and included standardized stereoscopic fundus photographs that were assessed by masked graders at the same reading center. This population is different from AREDS because it only includes individuals at high risk of progression to late disease. For the purpose of this study, all AREDS2 participants with WGS were included as cases (n=1365).
Michigan Genomics Initiative (MGI)
MGI serves as a repository of DNA and genetic data and was used to derive the remaining control samples for our study (n=1367). The MGI controls were age and sex-matched to the AREDS2 cases, using a 1-to-1 greedy matching algorithm.28 These control participants did not have the extensive ocular phenotyping performed for the AREDS control group, and their rate of AMD development after sampling is expected to be low, equal to the population prevalence of AMD which is 1–3% for people between 50 and 80 years of age.29
Whole-Genome Sequencing
WGS was performed using multiplexed Illumina HiSeq runs. Matched pairs were sequenced together to minimize batch effects. Mapping was performed using BWA-MEM.30 Variants were called using GotCloud snpcall and GotCloud indel, and phased using Beagle 4.31 The chance of false positives was reduced by trimming overlapping sequencing read fragments and support vector machine (SVM) based filtering using GotCloud.32 Contamination was estimated using VerifyBamID, and ancestry was estimated using LASER.33 Samples with high estimated contamination (>3%) and non-European ancestry were removed. An average coverage of 6X was achieved, corresponding to an expected sensitivity for variant calling of approximately 50% for singletons (variants present only once in the sequenced samples). The expected sensitivity for variant calling increases rapidly with allele frequency.34
Rare Variants
PLoF variant annotation categories included stop-gain, start-lost, splice donor, splice acceptor, missense, and frameshift. Variants with an allele frequency < 0.1% in the study population were called from 100 kb windows around variants with R2 ≥ 0.5 to the top 52 independent signals in IAMDGC15 using VEP build 86, GRCh37 coordinates and RefSeq genes. This region contains a total of 253 genes.
Pathways
As per definition, the pLoF variants will be present in only a few individuals. Therefore, instead of assessing enrichment on a variant-level, we evaluate effects on a pathway-level based on the gene function. Variants were grouped into 7 pathways: complement, ECM, lipids, cell survival, immune system, metabolism, and unknown/other based on GO35 and REACTOME (Reactome project. “Reactome” http://www.reactome.org/ (March 31, 2017))36, 37 analysis. Genes without annotations or those that did not fit one of the pre-specified pathways were put into the unknown/other category. A rare variant carrier within a pathway was defined as a person who carried at least one rare pLoF variant in a gene assigned to that specific pathway.
Phenotypes
Six phenotypes were assessed in this study: any GA, CNV, pseudoreticular drusen, calcified drusen, and drusen area in the ETDRS grid measured on an ordinal 8-step scale.38 Pseudoreticular drusen were defined as yellowish material with the appearance of soft drusen, arranged in interlacing networks.39 Calcified drusen were graded as chalky-white or shiny drusen, suggestive of calcium deposits.39 The final phenotype assessed was the 12-step AREDS Extended AMD Severity Scale that includes late disease phenotypes for eyes with traits of CNV or central GA, building on the original 9-step AREDS Severity Scale for AMD 9. A score of 9 indicates any GA, a score of 10 denotes the presence of central GA, a score of 11a (recoded here as 11) indicates CNV as diagnosed on color fundus photographs with two of the following characteristics: subsensory retinal detachment, pigment epithelial detachment, subretinal/sub-RPE hemorrhage, hard exudate, or fibrous tissue, and a score of 11b (recoded here as 12) indicates CNV with end-stage disease (disciform scar, photocoagulation scar) or two of the previously mentioned characteristics of end-stage disease. These phenotypes were selected because of our interest in late-stage disease as well as measures of drusen burden. Phenotyping in AREDS and AREDS2 was based on grading of annual stereoscopic fundus photos, performed by masked graders using a standard protocol at a central reading center. Phenotypes were considered to be present if they had ever been reported during the full follow-up time. Drusen area in the ETDRS grid and the AREDS Extended AMD Severity Scale was defined as the highest value recorded during follow-up in the worst affected eye.
Statistical Analysis
Pathway-AMD Study
The burden of rare pLoF variants in each of the seven pathways was compared between cases and controls by counting the number of individuals in the case and control groups that carried at least one rare pLoF variant in that pre-specified pathway. Statistical testing was performed by binary logistic regression. Because these variants are so rare, we have no way of knowing which are risk increasing or protective for AMD. However, since all are loss of function variants, we speculated that on average they would have a negative impact on retinal health, and thus in the burden tests, the assumption was that pLoF variants would be risk factors for AMD. We furthermore identified rare variants unique to cases and present in at least 3 individuals, and compared our study allele frequency to the ExAC database allele frequency40 by chi-square analysis.
Pathway-Phenotype Study
Cox proportional hazards regression with repeated measures for both eyes was used to assess the effect of rare variants on the rate of progression to advanced disease in individuals who started with intermediate disease in at least one eye. The model was adjusted for age, sex, smoking status, rs10490924 (ARMS2), and rs1061170 (CFH), and utilized data from AREDS2 cases. Frequencies of rare variant carriers vs. non-carriers within all cases were compared for each phenotype in the rare variant-phenotype association study. Frequencies of rare variant carriers vs. non-carriers in each of the 7 pathways within cases were compared in the pathway-phenotype association study. Participants with rare variants in other pathways were excluded from the analysis of any given pathway. Binary phenotypes (present or absent) were analyzed by logistic regression. Ordinal phenotypes (drusen area in ETDRS grid and AREDS Extended AMD Severity Scale) were analyzed by linear regression. Top common risk SNPs rs1061170 and rs10490924 were included in the logistic and linear regression models as covariates. Statistical analysis was done using IBM Statistics SPSS 24 and SAS.
Results
Overview of the Analysis
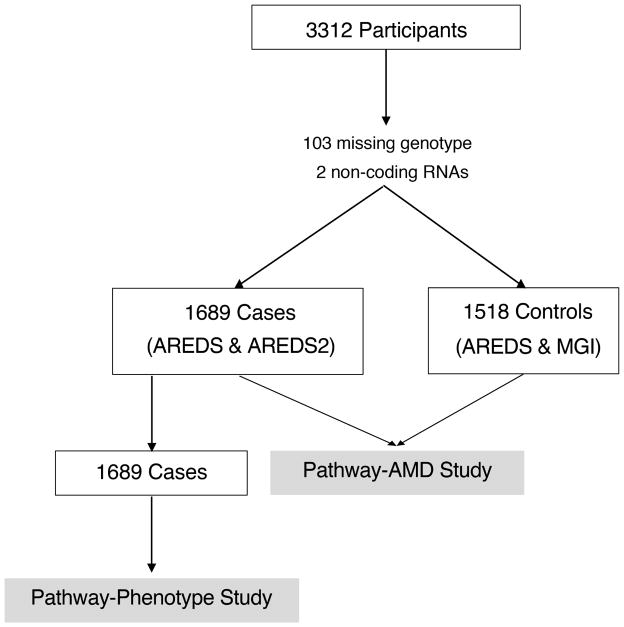
Of the 3312 study participants (1746 AMD cases and 1566 controls (Figure 1)), 55 cases missing genotype information, and two cases with variants within non-coding RNAs (ncRNAs) were excluded from analysis. Forty-eight controls were missing genotype information and were also excluded. In total, 1689 cases and 1518 controls were part of the Pathway-AMD Study. Cases included in the analysis were on average 70.8 (7.1) years of age and 54.4% was female. Controls were slightly older (mean age 72.2 (7.2)) and the proportion of females was 53.2%. We detected a significant difference in rs1061170 (CFH) and rs10490924 (ARMS2) genotype distribution between cases and controls (p<0.001) (Supplementary table 1). Within cases, we observed a difference in ARMS2 genotype (p=0.029) between rare variant carriers and non-carriers, but not in CFH genotype (p=0.449) (Supplementary table 2). To keep analysis consistent, both CFH and ARMS2 were included as covariates in all association studies. We did not find significant difference in mean age, gender, or smoking status between rare variant carriers and non-carriers (p>0.05) (Supplementary table 2).
Figure 1. Analysis flow diagram.
Of the 3312 participants with whole genome sequencing, 1689 cases and 1518 controls were included in the Pathway-AMD Study, and all 1689 cases were included in the Pathway-Phenotype Study.
Rare Variant Analysis
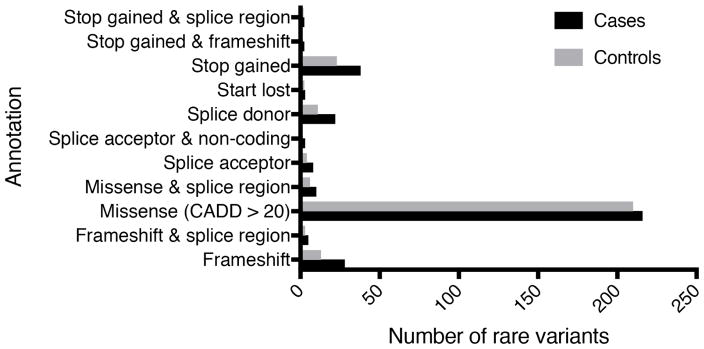
We identified 138 genes containing a total of 337 rare pLoF variants in 308 cases (17.6%) and 273 rare pLoF variants in 244 (15.6%) controls (Supplementary table 3, Supplementary table 4a and 4b). Not counting duplicates, 125 rare variants (35.5%) were unique to cases, 100 rare variants (28.4%) were unique to controls, and 127 rare variants (36.1%) were shared between the groups. Within cases, 27 individuals had two rare variants, one individual had three rare variants, and the remainder had one rare variant. In controls, 23 individuals had two rare variants, three individuals had three rare variants, and the remainder had one rare variant. Of those participants found to have rare pLoF variants in GWAS loci, approximately 90% of them had only 1 variant. The majority of rare variants, approximately 70%, were classified as missense with a Combined Annotation Dependent Depletion (CADD) score > 20 (Figure 2). The next largest category was stop-gain (10.0%), followed by frameshift (6.7%) and splice donor (5.4%). The remaining categories each contain less than 5% of all rare variants. We found two previously reported rare variants,15 including a missense variant in Complement Factor I (CFI) at position 4:110681679 (rs199688124) in two cases, and a missense variant in SLC16A8 at position 22:38478793 (rs113748161) in four cases and two controls. In order to identify potential candidates of interest for follow-up studies, we filtered for rare variants unique to cases and present in at least three individuals (allele frequency ≥ 0.1%) and compared our study frequency to the ExAC database allele frequency (Table 1). One variant showed significant allele enrichment in AMD cases: a splice donor variant in SLC12A3 rs199974259 (p=0.023).
Figure 2. Rare predicted Loss of Function variant types in cases and controls.
The number of rare variants in cases and controls included within each pLoF category
CADD = Combined Annotation Dependent Depletion.
Table 1.
Exome Aggregation Consortium (ExAC) allele frequency comparison of variants detected in at least 3 independent cases and no controls.
| GENE | VARIANT ANNOTATION | CADD | PATHWAY | FREQUENCY IN CASES | FREQUENCY IN EXAC | P |
|---|---|---|---|---|---|---|
|
SLC12A3 rs199974259 |
splice donor | 16.11 | Unknown/other | 4/3378 (0.12%) | 19/66486 (0.03%) | 0.023 |
|
|
||||||
|
HGS rs145607073 |
missense & splice region | 24.8 | Unknown/other | 3/3378 (0.09%) | 84/61604 (0.14%) | 0.630 |
|
|
||||||
|
CFI rs146444258 |
missense | 21.3 | Complement | 3/3378 (0.09%) | 33/66698 (0.05%) | 0.250 |
CADD = Combined Annotation Dependent Depletion
Pathway-AMD Study
Of the 1689 cases included in the Pathway-AMD Study, 298 (17.6%) were rare variant carriers, compared to 237 out of 1518 (15.6%) controls (OR=1.11 [0.91, 1.36], p=0.310) (Table 2). We observed an enrichment for pLoF variants in the complement pathway in cases versus controls (OR=2.94 [1.49–5.79], p=0.002). We did not identify significant association between AMD and pLoF rare variants in any other pathways.
Table 2.
The distribution of rare predicted loss of function (pLoF) variants in each pathway in cases and controls.
| PAHWAY | % CASES WITH RARE VARIANTS | % CONTROLS WITH RARE VARIANTS | P | OR [95% CI] |
|---|---|---|---|---|
| RARE VARIANT (ALL) | 297/1689 (17.6%) | 237/1518 (15.6%) | 0.310 | 1.11 [.91, 1.36] |
| COMPLEMENT | 36/1689 (2.1%) | 14/1518 (0.9%) | 0.002 | 2.94 [1.49, 5.79] |
| ECM | 52/1689 (3.1%) | 51/1518 (3.4%) | 0.610 | 0.90 [.58, 1.37] |
| LIPIDS | 49/1689 (2.9%) | 38/1518 (2.5%) | 0.777 | 1.07 [.67, 1.71] |
| IMMUNE SYSTEM | 14/1689 (0.8%) | 6/1518 (0.4%) | 0.265 | 1.78 [.65, 4.89] |
| METABOLISM | 13/1689 (0.8%) | 9/1518 (0.6%) | 0.838 | 1.10 [.43, 2.86] |
| CELL SURVIVAL | 28/1689 (1.7%) | 18/1518 (1.2%) | 0.250 | 1.46 [.77, 2.76] |
| UNKNOWN/OTHER | 126/1689 (7.5%) | 115/1518 (7.6%) | 0.555 | 0.92 [.69, 1.22] |
From logistic regression models adjusted for rs1061170 (CFH) and rs10490924 (ARMS2).
ECM = extracellular matrix, OR = odds ratio, CI = confidence interval.
Pathway-Phenotype Study
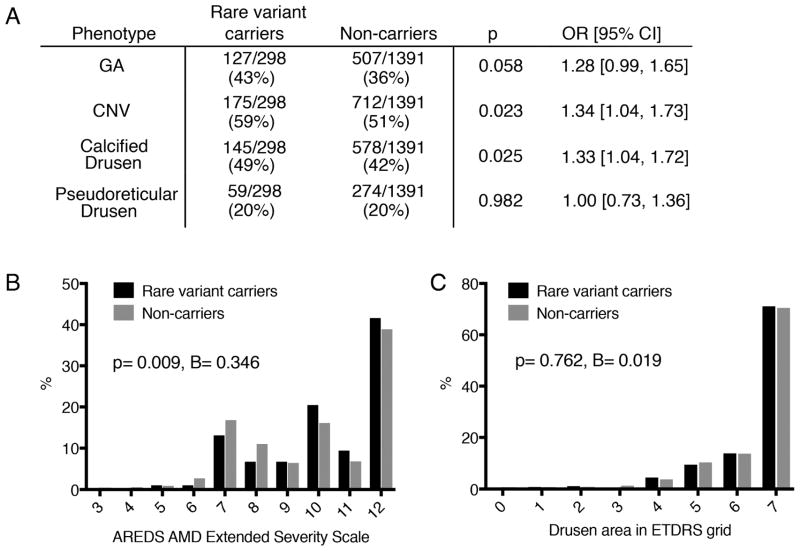
We compared the prevalence of AMD sub-phenotypes in rare variant carriers and non-carriers within cases, not stratified per pathway (Figure 3). We observed associations between rare variants and sub-phenotypes, including a trend towards increased risk of GA (OR=1.28 [0.99, 1.65]; p=0.058), a significantly increased risk of CNV (OR=1.34 [1.04, 1.73]; p=0.023), and significantly more calcified drusen in rare variant carriers (OR=1.33 [1.04, 1.72], p=0.025) (Figure 3A). We also observed that rare variants were associated with higher scores on the AREDS Extended AMD Severity Scale (B=0.346 [0.086, 0.605], p=0.009) (Figure 3B). In this population, owing to the enrollment criteria for AREDS2, the majority of individuals (63%) had advanced disease as indicated by score of 9 or higher. In AREDS2, rare variant carriers had an increased risk of progressing to late AMD with a HR of 1.25 [1.01, 1.55] (p=0.042).
Figure 3. Rare Variant-Phenotype Study.
A. Distribution of binary phenotypes (graded as present or absent) in rare variant carriers and non-carriers, analyzed by binary logistic regression. B. Distribution of AREDS AMD Extended Severity Scale scores in rare variant carriers and non-carriers, analyzed by linear regression. C. Distribution of drusen area in the ETDRS grid in rare variant carriers and non-carriers, analyzed by linear regression.
GA = geographic atrophy, CNV = choroidal neovascularization.
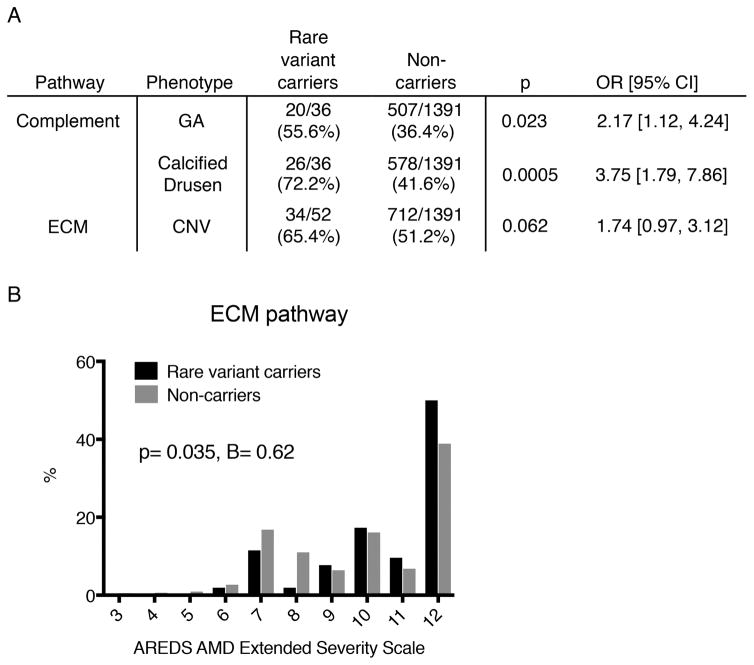
Finally, we looked for associations between the seven pathways and six AMD sub-phenotypes (Supplementary table 5). We observed significant associations between the complement pathway and GA (OR=2.17 [1.12, 4.24]; p=0.023), the complement pathway and calcified drusen (OR=3.75 [1.79, 7.86]; p=0.0005), and the ECM pathway and higher scores on the AREDS Extended Severity Scale (B=0.62 [0.04, 1.20], p=0.035), which seemed to be mostly driven by an association between the ECM pathway and CNV (OR=1.74 [0.97, 3.12]; p=0.062) (Figure 4). The unknown/other pathway also showed several significant associations: GA (p=0.014), calcified drusen (p=0.046) and the AREDS Extended Severity Scale (p=0.004) (Supplementary table 5). After Bonferroni correction for multiple testing (p<0.05/42 = 0.0012), the association of calcified drusen with rare variants in the complement pathway remained significant.
Figure 4. Pathway-Phenotype Study.
A. Pathways with significantly different distributions of binary phenotypes (graded as present or absent) in rare variant carriers and non-carriers, analyzed by binary logistic regression. B. Distribution of extracellular matrix (ECM) pathway rare variant carriers and non-carriers for the AREDS AMD Extended Severity Scale, analyzed by linear regression.
GA = geographic atrophy, CNV = choroidal neovascularization.
Discussion
We used a pathway-based method to study variants that may be too rare in the population to study individually. Approaching a rare variant analysis from a pathway perspective is a reasonable method to generate more mechanistic hypotheses and achieve sufficient statistical power, as it allows for the grouping of a large number of unique variants into fewer, biologically relevant pathways. Characterizing these pathways and understanding their association to disease incidence and progression is important as we consider rational therapeutic targets, particularly in complex diseases where the culprit is not a single gene. Many variants may not play a causal role in initiating disease, but rather may modify complex disease phenotypes. We show enrichment for rare pLoF variants in the complement pathway in cases, however, we did not observe a significant difference in rare variant frequency between cases and controls overall, or in any other pathway.
There are some limitations that make it difficult to interpret this mostly negative result. The MGI is not a pure control group, but rather a sample of the population. The rate of late AMD is expected to be 1–3% and the rate of large drusen is expected to be around 8%.29 Including missense variants also may have increased noise, but filtering based on CADD score was one way to attenuate this and allowed for the inclusion of additional pathogenic variants in our analysis to increase the power. For a rare variant analysis, this was still a relatively small study, and approximately half of the rare variants were assigned to the unknown/other category in an effort to maintain strictly defined pathways.
The phenotypes assessed in this study were graded by masked graders on annual fundus photographs over the course of the entire follow-up (5 years in AREDS2 and 10 years in AREDS), resulting in overall high-quality phenotype data. Pseudoreticular drusen, however, are best graded on near-infrared imaging. Fundus photography is less sensitive for this phenotype; however, the specificity of detection is 100%.41 Therefore, it should be noted that we may have missed some cases of pseudoreticular drusen, but the ones that we did identify are most likely correctly classified. Furthermore, some phenotypes may be correlated to each other, such as calcified drusen as a precursor of GA, and the phenotype analyses may not all be independent of each other. The relation between rare variants in the complement pathway and progression to GA may therefore be mediated by a process involving calcified drusen formation.
Rare variants in the complement pathway were found in 2.1% of people with AMD compared to 0.9% of the control population and increased the risk of AMD 2.9-fold (p=0.002). Complement dysregulation is a well-studied mechanism of AMD pathogenesis, and variants in a number of complement genes have been associated with increased disease risk. The first risk variant was discovered in the CFH gene in 2005, and it is estimated that individuals carrying the common Y402H variant harbor a 4-fold increase in disease risk.10 Today, rare risk variants in CFH,17 Complement Factor I (CFI),18 C3, and C919–21 have been discovered. There have also been protective alleles reported, including a multiallelic copy-number variant (CNV) in C4A.42 Most of these rare variants in the complement pathway are reported to have large effect sizes, some conferring up to a 20-fold increase in AMD risk.43 In our study, the increased risk was modest compared to previous reports. Important to note is that not all pLoF variants reported in this study may be associated with AMD. Most variants were present in one or two individuals, precluding any robust statistical evaluation of their individual effect. Possibly, the aggregation of pLoF variants with unknown effects within pathways, has diluted some of the results. Hence, it is difficult to estimate the true implications of carrying a rare pLoF variant for any individual. Yet, in a small, but not unsubstantial, subpopulation of individuals with AMD, very rare pLoF variants in the complement pathway may contribute to disease. This needs to be taken into consideration, especially as treatment shifts more and more towards mechanism based approaches such as complement inhibition.
Only three rare pLoF variants were present in at least 3 cases and not in controls, so an assessment of their enrichment in AMD compared to the general population could be performed. We identified a variant in one gene that was significantly enriched in cases: SLC12A3, solute carrier family 12 member 3, a renal sodium-chloride cotransporter, which is found in the CETP locus. Mutations in this gene cause Gitelman’s syndrome, or inherited hypokalemic alkalosis.44 This gene has not been functionally studied in AMD, and received a gene priority score of 2 in the previous GWAS report.15 SLC12A3 received points for having ≥1 variant in 95% credible sets (statistical evidence) and for being a drug target (pathway evidence).
In the pathway-phenotype study, we observed an association between rare pLoF variants and late AMD, and showed that disease progresses at a faster rate in genetically susceptible individuals. Rare variants in the unknown/other pathway also showed an association with late AMD, and are likely the main drivers behind this observation. These rare variants could not be assigned to known AMD-pathways based on their reported gene function. This could indicate that these genes have different functions in the retina or that there are pathways involved in the modification of AMD progression that we are not yet aware of.
We observed several significant associations between pathways and AMD sub-phenotypes, although it should be noted only the association between the complement pathway and calcified drusen remained significant after Bonferroni correction. Rare variants in the complement pathway resulted in a 2.2-fold increased risk of GA, which is consistent with previous results. It has been reported that rare variants in CFI, C9, and C3 are associated with earlier age of disease onset, and GA as opposed to CNV.19, 21, 45 We also report an association between complement and calcified drusen, which has been recently linked to rare variants in CFH.22 Rare pLoF variants in the complement pathway resulted in a nearly 4-fold increased risk of this phenotype. Little has been published about calcified drusen except that they may be precursors for progression to GA as observed by AREDS investigators.46, 47 Further studies are necessary to understand the significance of this observation. We found that rare variants in the ECM pathway resulted in an increased risk of late AMD, and CNV in particular. This finding is also consistent with previous observations of variants involved in the ECM pathway. In the previous GWAS, a variant in Matrix Metalloproteinase 9 (MMP9) was associated with CNV, but not GA.15 In addition, there has been a report of variants in Matrix Metalloproteinase 20 (MMP20) and ARMS2/HTRA1 affecting the growth and ultimate size of neovascular lesions in CNV.48 Rare pLoF variants in various pathways seem to contribute to disease progressions and imply that variation in biological pathways drives the development of heterogeneous clinical phenotypes.
This study is the largest WGS study in AMD to date and yet, sample size could still be considered small. We have attempted to overcome power issues by selecting for potential high impact pLoF variants and grouping variants based on biological pathway. Our results suggest a role of rare pLoF variants in the pathogenesis of AMD and phenotypic variability. Future studies with even larger populations will be necessary in order to determine if the associations we observed here can be replicated in different groups. Ultimately, the goal of such studies is to create more accurate prediction models for disease progression utilizing pathway-level information.
Supplementary Material
Acknowledgments
Financial Support: This study was supported by the Intramural Research Program of the National Eye Institute (EY000474 and EY000546 to A.S., and AREDS contract NOI-EY-02127 to E.Y.C.). The funding organization had no role in the design or conduct of this research.
Footnotes
Conflict of Interest: No conflicting relationship exists for the following: Alexandra Pietraszkiewicz, Freekje van Asten, Alan Kwong, Rinki Ratnapriya, and Emily Y. Chew. For Anand Swaroop: he receives royalties from U. of Michigan and the National Eye Institute for patents related to nephronophthesis and AMD markers:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Congdon N, O’Colmain B, Klaver CC, et al. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004;122(4):477–85. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 2.Smith W, Assink J, Klein R, et al. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology. 2001;108(4):697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 3.Day S, Acquah K, Lee PP, et al. Medicare costs for neovascular age-related macular degeneration, 1994–2007. Am J Ophthalmol. 2011;152(6):1014–20. doi: 10.1016/j.ajo.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratnapriya R, Chew EY. Age-related macular degeneration-clinical review and genetics update. Clin Genet. 2013;84(2):160–6. doi: 10.1111/cge.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14(7):835–46. [PubMed] [Google Scholar]
- 7.Malek G, Li CM, Guidry C, et al. Apolipoprotein B in cholesterol-containing drusen and basal deposits of human eyes with age-related maculopathy. Am J Pathol. 2003;162(2):413–25. doi: 10.1016/S0002-9440(10)63836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritsche LG, Fariss RN, Stambolian D, et al. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–71. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis MD, Gangnon RE, Lee LY, et al. The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS Report No. 17. Arch Ophthalmol. 2005;123(11):1484–98. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Atmaca-Sonmez P, Othman M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38(9):1049–54. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227–36. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 13.Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104(41):16227–32. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaumberg DA, Hankinson SE, Guo Q, et al. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007;125(1):55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 15.Fritsche LG, Igl W, Bailey JN, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–43. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobrin L, Reynolds R, Yu Y, et al. ARMS2/HTRA1 locus can confer differential susceptibility to the advanced subtypes of age-related macular degeneration. Am J Ophthalmol. 2011;151(2):345–52. e3. doi: 10.1016/j.ajo.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri S, Iartchouk O, Chin K, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43(12):1232–6. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Ven JP, Nilsson SC, Tan PL, et al. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat Genet. 2013;45(7):813–7. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 19.Seddon JM, Yu Y, Miller EC, et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013 doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helgason H, Sulem P, Duvvari MR, et al. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat Genet. 2013;45(11):1371–4. doi: 10.1038/ng.2740. [DOI] [PubMed] [Google Scholar]
- 21.Zhan X, Larson DE, Wang C, et al. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat Genet. 2013;45(11):1375–9. doi: 10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kersten E, Geerlings MJ, den Hollander AI, et al. Phenotype Characteristics of Patients With Age-Related Macular Degeneration Carrying a Rare Variant in the Complement Factor H Gene. JAMA Ophthalmol. 2017 doi: 10.1001/jamaophthalmol.2017.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrara D, Seddon JM. Phenotypic Characterization of Complement Factor H R1210C Rare Genetic Variant in Age-Related Macular Degeneration. JAMA Ophthalmol. 2015;133(7):785–91. doi: 10.1001/jamaophthalmol.2015.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratnapriya R, Zhan X, Fariss RN, et al. Rare and common variants in extracellular matrix gene Fibrillin 2 (FBN2) are associated with macular degeneration. Hum Mol Genet. 2014;23(21):5827–37. doi: 10.1093/hmg/ddu276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.den Hollander AI. Omics in Ophthalmology: Advances in Genomics and Precision Medicine for Leber Congenital Amaurosis and Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci. 2016;57(3):1378–87. doi: 10.1167/iovs.15-18167. [DOI] [PubMed] [Google Scholar]
- 26.Age-Related Eye Disease Study Research G. The Age-Related Eye Disease Study (AREDS): design implications. AREDS report no. 1. Control Clin Trials. 1999;20(6):573–600. doi: 10.1016/s0197-2456(99)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Group AR, Chew EY, Clemons T, et al. The Age-Related Eye Disease Study 2 (AREDS2): study design and baseline characteristics (AREDS2 report number 1) Ophthalmology. 2012;119(11):2282–9. doi: 10.1016/j.ophtha.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons LS. Reducing bias in a propensity score matched-pair sample using greedy matching techniques. SAS Global Users Group. 2001;26:214, e226. [Google Scholar]
- 29.Klein R, Chou CF, Klein BE, et al. Prevalence of age-related macular degeneration in the US population. Arch Ophthalmol. 2011;129(1):75–80. doi: 10.1001/archophthalmol.2010.318. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jun G, Wing MK, Abecasis GR, Kang HM. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25(6):918–25. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Zhan X, Bragg-Gresham J, et al. Ancestry estimation and control of population stratification for sequence-based association studies. Nat Genet. 2014;46(4):409–15. doi: 10.1038/ng.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rashkin S, Jun G, Chen S, et al. Optimal sequencing strategies for identifying disease-associated singletons. PLoS Genet. 2017;13(6):e1006811. doi: 10.1371/journal.pgen.1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabregat A, Sidiropoulos K, Garapati P, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–7. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milacic M, Haw R, Rothfels K, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 2012;4(4):1180–211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danis RP, Domalpally A, Chew EY, et al. Methods and reproducibility of grading optimized digital color fundus photographs in the Age-Related Eye Disease Study 2 (AREDS2 Report Number 2) Invest Ophthalmol Vis Sci. 2013;54(7):4548–54. doi: 10.1167/iovs.13-11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Age-Related Eye Disease Study Research G. The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study Report Number 6. Am J Ophthalmol. 2001;132(5):668–81. doi: 10.1016/s0002-9394(01)01218-1. [DOI] [PubMed] [Google Scholar]
- 40.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z, Ayton LN, Luu CD, et al. Reticular Pseudodrusen in Intermediate Age-Related Macular Degeneration: Prevalence, Detection, Clinical, Environmental, and Genetic Associations. Invest Ophthalmol Vis Sci. 2016;57(3):1310–6. doi: 10.1167/iovs.15-18682. [DOI] [PubMed] [Google Scholar]
- 42.Grassmann F, Cantsilieris S, Schulz-Kuhnt AS, et al. Multiallelic copy number variation in the complement component 4A (C4A) gene is associated with late-stage age-related macular degeneration (AMD) J Neuroinflammation. 2016;13(1):81. doi: 10.1186/s12974-016-0548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geerlings MJ, de Jong EK, den Hollander AI. The complement system in age-related macular degeneration: A review of rare genetic variants and implications for personalized treatment. Mol Immunol. 2017;84:65–76. doi: 10.1016/j.molimm.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon DB, Nelson-Williams C, Bia MJ, et al. Gitelman’s variant of Bartter’s syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet. 1996;12(1):24–30. doi: 10.1038/ng0196-24. [DOI] [PubMed] [Google Scholar]
- 45.Saksens NT, Geerlings MJ, Bakker B, et al. Rare Genetic Variants Associated With Development of Age-Related Macular Degeneration. JAMA Ophthalmol. 2016;134(3):287–93. doi: 10.1001/jamaophthalmol.2015.5592. [DOI] [PubMed] [Google Scholar]
- 46.Cukras C, Agron E, Klein ML, et al. Natural history of drusenoid pigment epithelial detachment in age-related macular degeneration: Age-Related Eye Disease Study Report No. 28. Ophthalmology. 2010;117(3):489–99. doi: 10.1016/j.ophtha.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein ML, Ferris FL, 3rd, Armstrong J, et al. Retinal precursors and the development of geographic atrophy in age-related macular degeneration. Ophthalmology. 2008;115(6):1026–31. doi: 10.1016/j.ophtha.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 48.Akagi-Kurashige Y, Yamashiro K, Gotoh N, et al. MMP20 and ARMS2/HTRA1 Are Associated with Neovascular Lesion Size in Age-Related Macular Degeneration. Ophthalmology. 2015;122(11):2295–302. e2. doi: 10.1016/j.ophtha.2015.07.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.