Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating and still untreatable motor neuron disease. Despite the molecular mechanisms underlying ALS pathogenesis that are still far from being understood, several studies have suggested the importance of a genetic contribution in both familial and sporadic forms of the disease. In addition to single-nucleotide polymorphisms (SNPs), which account for only a limited number of ALS cases, a consistent number of common and rare copy number variations (CNVs) have been associated to ALS. Most of the CNV-based association studies use a traditional candidate-gene approach that is inadequate for uncovering the genetic architectures of complex traits like ALS. The emergent paradigm of “systems biology” may offer a new perspective to better interpret the wide spectrum of CNVs in ALS, enabling the characterization of the complex network of gene products underlying ALS pathogenesis. In this review, we will explore the landscape of CNVs in ALS, putting specific emphasis on the functional impact of common CNV regions and genes consistently associated with increased risk of developing disease. In addition, we will discuss the potential contribution of multiple rare CNVs in ALS pathogenesis, focusing our attention on the complex mechanisms by which these proteins might impact, individually or in combination, the genetic susceptibility of ALS. The comprehensive detection and functional characterization of common and rare candidate risk CNVs in ALS susceptibility may bring new pieces into the intricate mosaic of ALS pathogenesis, providing interesting and important implications for a more precise molecular biomarker-assisted diagnosis and more effective and personalized treatments.
Electronic supplementary material
The online version of this article (doi:10.1007/s12035-017-0393-x) contains supplementary material, which is available to authorized users.
Keywords: Amyotrophic lateral sclerosis (ALS), Copy number variations (CNVs), Systems biology, Genomics
Background
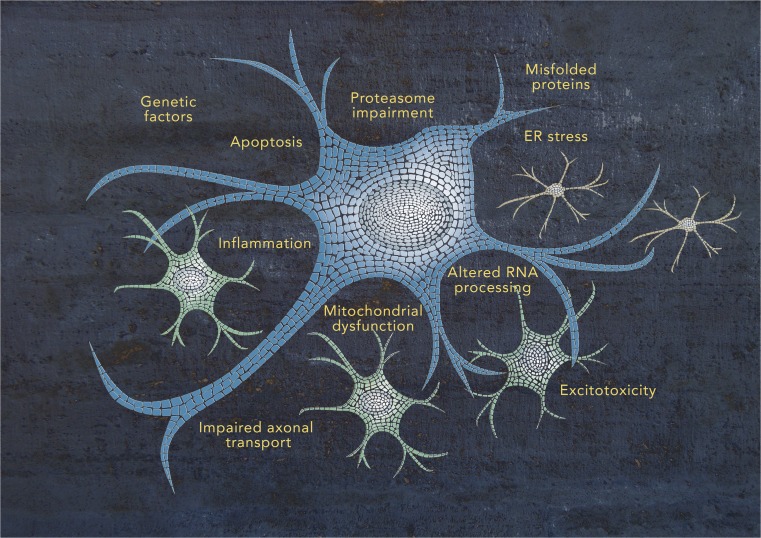
Amyotrophic lateral sclerosis (ALS) is a devastating untreatable neurodegenerative disease characterized by the selective degeneration of motor neurons in the brain and spinal cord, leading to paralysis and death, usually from respiratory failure, within 3–5 years of onset [1]. The disease exists in two forms: familial ALS (FALS) and sporadic ALS (SALS). FALS is a rare monogenic disease that occurs in 5–10% of cases with an autosomal dominant inheritance and for which several causal genes have been identified, including SOD1, ALS2, SETX, SPG11, FUS, VAPB, ANG, TARDBP, FIG4, OPTN, ATXN2, UBQLN2, PGRN, PFN1, DCTN1, and C9ORF72 [2]. SALS comprises the majority (90–98%) of ALS cases and is considered to be a complex multifactorial disorder, involving multiple pathogenic processes, such as oxidative stress, protein aggregation, mitochondrial dysfunction, excitotoxicity, and impaired axonal transport (Fig. 1) [3]. Although there are still missing pieces in the intricate mosaic of SALS pathogenesis, several studies have recognized the important contribution of genetic risk factors, usually associated with incomplete penetrance, and gene-environment interactions for disease susceptibility (Fig. 1).
Fig. 1.
Schematic representation of the complex mosaic of ALS pathogenesis
The remarkable advances in genome technologies over the last years have led to huge progress in the understanding of the genetic mechanisms involved in ALS. The search for pathogenic variants has been initially focused primarily on variations at the single-nucleotide polymorphism (SNP) level. In the last years, several candidate-gene or genome-wide association studies (GWAS) have identified multiple SNPs affecting potentially ALS-associated genes, including VEGFA, ANG, FGGY, DPP6, ITPR2, KIFAP3, and UNC13A [4–7]. However, the contribution of nucleotide sequence abnormalities in SALS pathogenesis remained unclear, with point mutations in the above genes occurring only rarely.
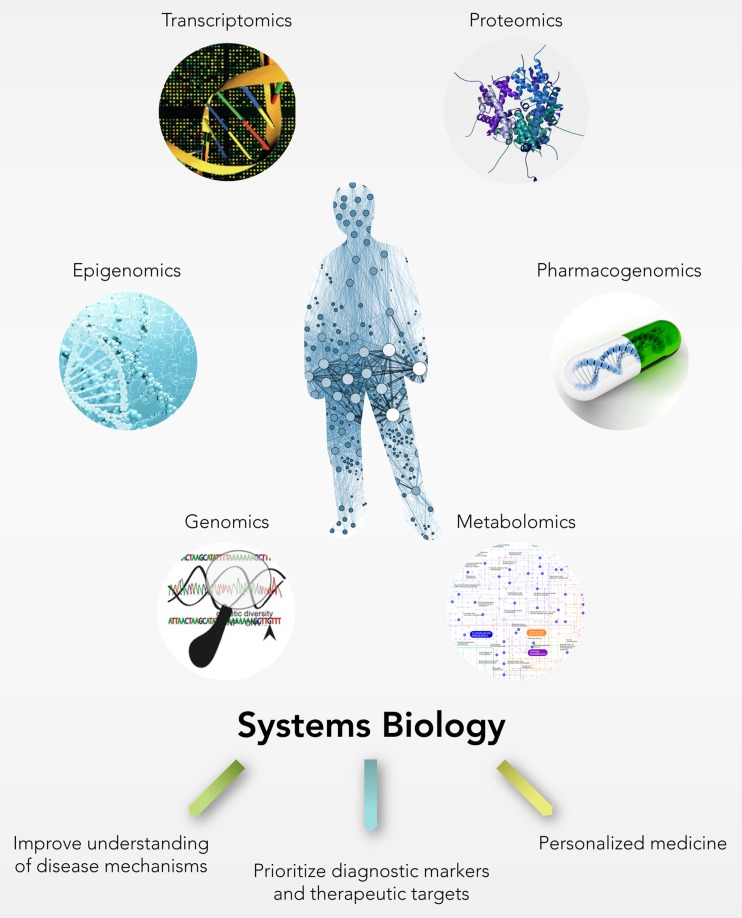
In addition to SNPs, submicroscopic chromosomal changes, also known as copy number variations (CNVs), represent a substantial source of inter-individual genetic variations exerting important phenotypic effects on the expression and function of genes and representing one of the major risk factors for various complex human disorders including ALS [8, 9]. The majority of existing CNV genotyping studies use a traditional single-gene approach that, albeit has provided valuable information regarding the impact of individual common variants, is inadequate for uncovering genetic architectures of complex traits like ALS, often leading to the loss of less-frequently but potentially functionally relevant CNV-driven gene sets. In this context, the holistic “systems biology” approach provides a new perspective in the study of complex disease traits, which goes beyond the conventional one-gene-at-a-time testing scheme, and embraces the entire equilibrium of a biological system undergoing a much more complicated network of molecular components that in association increase the likelihood of developing the disease (Fig. 2) [10].
Fig. 2.
The systems biology approach: from integration of large-scale “omics” data to personalized medicine practice
In this review, we will explore the currently known landscape of CNVs, putting specific emphasis on CNV-driven genes that have been consistently associated with an increased risk of ALS, considering their potential functional impact in the pathophysiology of the disease. In addition, we will examine the potential contribution of multiple rare CNVs in ALS pathogenesis, focusing our attention on the complex mechanisms by which these proteins might impact, both individually or in combination, to the genetic susceptibility of ALS.
Main Text
Copy Number Variations: an Overview
One of the most important scientific discoveries about the human genome study is that, in addition to nucleotide sequence variants such as SNPs, different types of genomic structural variations contribute significantly to genetic heterogeneity [11, 12]. Genomic structural variants include many different types of chromosomal rearrangement encompassing both inversions and balanced translocations as well as genomic imbalances commonly referred to as copy number variations (CNVs).
CNVs are defined as fragments of DNA larger than 1 kb (variants smaller than 1 kb are termed InDels) presenting unbalanced rearrangements in comparison to a reference genome [13–15]. These can be rare (<1%) or common (>5%); de novo or inherited; and include structural gains (duplication or insertional transpositions), losses (e.g., deletions), or complex rearrangements. In particular, deletions can be heterozygous (with only one copy missing), homozygous (with both copies missing), or hemizygous (e.g., X chromosome deletions in males) [16]. In the case of copy number gains, it is possible to distinguish duplications (three copies of a genomic region) from other complex genomic rearrangements, such as homozygous duplications (four copies with two copies on each allele), triplications (four copies with three copies on one allele and one copy on the other allele), and quadruplications (five copies) [17]. Although single-copy deletions and gains of one or two extra copies of DNA are common, the complete genetic loss of both alleles and more extensive amplification of specific DNA regions have been only rarely described.
Methodological Approaches for CNVs Detection and Analysis
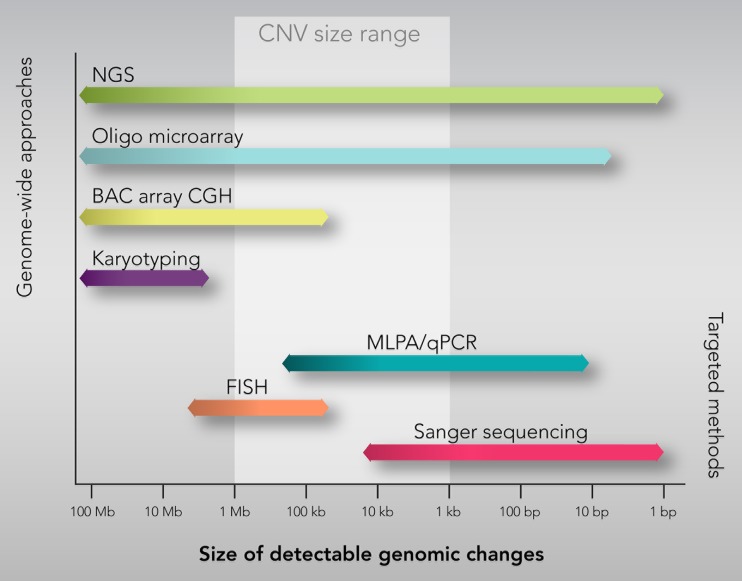
The conventional karyotyping techniques typically detect chromosomal aberrations greater than 5–10 Mb ruling out other forms of submicroscopic CNVs. The recent advent of genome-wide approaches has driven much of the research on CNVs facilitating their identification and characterization at much higher resolution than hitherto. Several methods for detecting CNVs are currently available and can be categorized into genome-wide and targeted detection approaches (Fig. 3) [18].
Fig. 3.
The most widely used methods for genome-wide and targeted CNVs detection and analysis
Genome-wide approaches permit to scan the entire genome for CNVs detection and include microarray-based (array-comparative genomic hybridization (CGH) and SNP array) and next-generation sequencing (NGS)-based analyses (Fig. 3). Originally, array-CGH included only a few hundreds of thousands of large-insert clones (known as bacterial artificial chromosomes (BACs)) with a relatively low spatial resolution (typically >5–10 Mb) and required a relatively large volume of DNA [19]. Subsequently, high-density oligo-synthesized arrays have been developed, offering a much higher resolution with a more accurate definition of CNV boundaries [20]. In addition to array-CGH, high-density SNP genotyping arrays have also gained interest for CNV detection and analysis, mainly due to their dual role for both SNP- and CNV-based association studies [21]. Although microarray technologies currently represent the most powerful available method to identify CNVs, they cannot detect short CNVs. The advent of NGS technologies has allowed to overcome this limitation, offering the possibility to reveal much smaller CNVs (<10 kb) and define CNV breakpoints at the single nucleotide level [22].
Despite genome-wide approaches providing reliable and efficient methodological insights for large-scale association analyses, other CNV detection methods are preferably used for the validation process as well as phenotype/genotype correlation and clinical translation of a relatively small set of known CNVs. These “targeted methods” include fluorescence in situ hybridization (FISH), quantitative polymerase chain reaction (qPCR), and multiplex ligation-dependent probe amplification (MLPA) (Fig. 3) [14]. FISH has been the first molecular method able to detect submicroscopic genomic CNVs, but it is a time-consuming method that requires prior knowledge of the regions of interest, not allowing for genome-wide analyses [23]. qPCR is an efficient method for screening CNV-targeted genomic regions. However, this technique does not allow the simultaneous amplification and quantification of a large number of targets in a single reaction. MLPA is an alternative targeted PCR-based approach that allows simultaneous analysis of multiple targets (up to 40 targets) with one primer pair, reducing the probability of obtaining spurious qPCR results due to different reaction conditions [24].
The Origin of CNVs and their Functional Impact in the Human Genome
Significant progress has been made to date in the field of CNV detection and analysis; however, much remains to be investigated about mechanisms that cause CNVs and their functional consequences. CNVs can arise from a multitude of molecular mechanisms (e.g., non-allelic homologous recombination between repeated elements, non-homologous DNA repair mechanisms, replication errors, and transposable element-mediated mechanisms) and can involve one or multiple genes as well as regulatory regions [25, 26]. Based on their size and the genomic region in which they occur, CNVs can affect gene functions in a variety of ways: by disrupting gene coding sequences (for example by deleting or inserting exonic sequences), producing novel fusion genes and splice variants, as well as by affecting regulatory elements and other functional regions, thus perturbing the assembly of the transcription machinery. These variations at the genomic level may result in alterations at the protein level, leading to eliminated/reduced or increased expression levels or interrupted coding sequences that cause the formation of truncated proteins with altered functions. Moreover, disruption of the regions including transcription regulatory elements and enhancers can also result in long-distance effects (even up to 1 Mb) by altering the expression of genes in neighboring chromosomal regions [27].
Changes in copy number are extremely frequent in the human genome (12–15%) representing one of the most prevalent types of genetic variations [11]. Approximately, more than 66,000 CNVs and 34,000 InDels were identified in various populations and cataloged in the Database of Genomic Variants (DGV) (http://dgv.tcag.ca/dgv/app), a public and comprehensive catalog of human CNVs and genomic structural variations identified in healthy controls [8, 28]. While the majority represents benign polymorphic variants, increasing numbers of CNVs are associated with a higher risk of developing various types of inherited Mendelian and complex disease traits, including neurological disorders [29, 30]. Several recent studies, in fact, have supported the role of CNVs in causing or influencing the susceptibility to many neurological and developmental conditions, such as Charcot–Marie–Tooth neuropathy [31], autism [32], schizophrenia [33], epilepsy [34], Crohn’s disease [35], and neurodegenerative disorders, including Alzheimer’s, Parkinson’s and ALS [36, 37].
Copy Number Variations in ALS
Recent advances in genomic technologies have led to huge progress in understanding the genetic mechanisms underlying ALS, allowing the identification and characterization of novel genetic susceptibility loci, such as CNVs [38]. Various studies have, in fact, highlighted the possible association between common and rare candidate CNV-driven genes or genomic regions and increased risk of developing ALS (Table 1).
Table 1.
Characteristics of the most significant CNVs and overlapped genes from different genome-wide studies showing association with ALS
| Chr | Position start–end (CNV length) | CNV type | CNV detection method | ALS patients | Controls | Overlapped genes¥ | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 41,119,815–41,147,030 | Gain | SNP array (TaqMan qPCR validation) |
8 (no. cases = 406) |
1 (no. controls = 404) |
None | [115] |
| 1 | Not reported (2246 bp) |
Deletion | SNP array | 3 (no. cases = 12) |
0 (no. controls = 24) |
Intron of FMN2 | [138] |
| 1 | 52,994,160–53,423,907 | Gain | BAC array (aCGH validation) |
1 (no. cases = 72) |
0 (no. controls = 700) |
ZYG11B, ECHDC2, SCP2, PODN, SCL1A7, CPT2, C1orf23, MAGOH | [165] |
| 3 | 2,944,819–2,947,844 | Gain | SNP array | 2 (no. cases = 32) |
0 (no. controls = 26 ) |
Intron of CNTN4 | [138, 165] |
| 3 | 89,485,137–89,499,861 | Loss | SNP array (TaqMan qPCR validation) |
2 (no. cases = 117) |
11 (no. controls = 109) |
EPHA3 | [96] |
| 3 | 33,270,957–33,296,620 | Gain | SNP array | 3 (no. cases = 575) |
18 (no. controls = 621) |
FBXL2 | [110, 111] |
| 3 | Not reported (6527 bp) |
Deletion | SNP array | 1 (no. cases = 12) |
0 (no. controls = 24) |
Promoter of CHL1 | [138] |
| 3 | Not reported (53,562 bp) |
Loss | SNP array | 1 (no. cases = 12) |
0 (no. controls = 24) |
52% of DHX30 (+ promoter), 9% of SMARCC1 (+ promoter) | [138] |
| 3 | 60,357,746–60,604,845 | Gain | BAC array (aCGH validation) |
1 (no. cases = 72) |
0 (no. controls = 700) |
FHIT | [165] |
| 4 | 761,587–1,014,752 | Gain | SNP array | 16 (no. cases = 575) |
4 (no. controls = 621) |
CPLX1, GAK, TMEM175, DGKQ, IDUA, SLC26A1, FGFRL1 | [110] |
| 4 | Not reported (144,772 bp) |
Loss | SNP array | 1 (no. cases = 12) |
0 (no. controls = 24) |
3% of RPS3A (+ promoter), 39% of LRBA (+ promoter) | [138] |
| 4 | Not reported (4546 bp) |
Deletion | SNP array | 1 (no. cases = 12) |
0 (no. controls = 24) |
Promoter of UGT8 | [138] |
| 5 | 28,842,013–28,912,873 | Gain | SNP array | 7 (no. cases = 575) |
0 (no. controls = 621) |
None | [110] |
| 5 | 45,850,032–46,384,240 | Gain | SNP array | 264 (no. cases = 575) |
174 (no. controls = 621) |
HCN1 (flanking) | [110] |
| 5 | 70,925,030–70,953,012 | Gain | MLPA assay | 121 (no. cases = 1689) |
68 (no. controls = 1780) |
SMN1–SMN2 | [45–47] |
| Loss | qPCR | 14 (no. cases = 167) |
4 (no. controls = 310) |
||||
| 6 | 109,034,609–109,074,882 | Gain | SNP array | 1 (no. cases = 32) |
0 (no. controls = 26 ) |
Intron of FOXO3 | [99] |
| 6 | 123,569,244–124,360,902 | Gain | BAC array (aCGH validation) |
1 (no. cases = 72) |
0 (no. controls = 700) |
TRDN, TCBA1 | [165] |
| 7 | 153,031,806–154,276,435 | Loss and gain | SNP array (TaqMan qPCR validation) |
10 (9 dup, 1 del) (no. cases = 1875) |
13 (12 dup, 1 del) (no. controls = 8731) |
DPP6 | [84] |
| 7 | 61,663,407–62,155,064 | Gain | SNP array | 177 (no. cases = 575) |
132 (no. controls = 621) |
None | [110] |
| 8 | 47,062,007–47,406,312 | Gain | SNP array | 30 (no. cases = 575) |
8 (no. controls = 621) |
POTEA (flanking) | [110] |
| 8 | 47,062,007–47,711,911 | Gain | SNP array | 3 (no. cases = 575) |
18 (no. controls = 621) |
None | [110] |
| 8 | 43,689,385–43,910,848 | Gain | SNP array | 74 (no. cases = 575) |
52 (no. controls = 621) |
None | [110] |
| 8 | 73,609,541–73,629,084 | Gain | SNP array | 1 (no. cases = 32) |
0 (no. controls = 26 ) |
Region overlaps with 86.50% of KCNB2 | [99] |
| 8 | 144,686,338–144,765,210 | Loss and gain | SNP array | 6 (no. cases = 575) |
0 (no. controls = 621) |
ZC3H3, GSDM, C8orf73, NAPRT1, EEF1D, TIGD5, PYCRL | [110] |
| 10 | 1,050,000–1,090,000 | Gain | aCGH (TaqMan qPCR and high-density customized aCGH validation) |
46 (no. cases = 83) |
10 (no. controls = 100) |
IDI1-IDI2 | [78] |
| 11 | 50,545,00–50,586,426 | Loss | SNP array | 21 (no. cases = 117) |
2 (no. controls = 109) |
None | [96] |
| 11 | 539,119–652,407 | Loss | SNP array | 5 (no. cases = 575) |
0 (no. controls = 621) |
OR4A5, OR4C12 (flanking) LRRC56, C11orf35, RASSF7, PHRF1, IRF7, MUPCDH, SCT, DRD4, DEAF1 | [110] |
| 12 | 36,528,296–36,801,139 | Gain | SNP array | 157 (no. cases = 575) |
8 (no. controls = 621) |
None | [110] |
| 14 | 103,232,016–103,721,150 | Gain | SNP array | 5 (no. cases = 575) |
0 (no. controls = 621) |
KLC1, XRCC3, ZFYVE21, PPP1R13B, C14orf2, TDRD9, ASPG, KIF26A | [110] |
| 14 | Not reported (4384 bp) |
Deletion | SNP array | 5 (no. cases = 12) |
0 (no. controls = 24) |
None | [138] |
| 15 | 20,387,566–44,672,396 | Loss and gain | SNP array (TaqMan qPCR validation) |
12 (4 dup, 8 del) (no. cases = 1875) |
34 (31 dup, 3 del) (no. controls = 8731) |
TUBGCP5, CYFIP1, NIPA2, NIPA1 | [84] |
| 15 | Not reported (5695 bp) |
Deletion | SNP array | 2 (no. cases = 12) |
0 (no. controls = 24) |
None | [138] |
| 15 | 19,818,989–20,084,080 | Deletion | SNP array | 2 (no. cases = 32) |
0 (no. controls = 26 ) |
LOC650137 (+), OR4M2 (+), OR4N4 (+) | [99] |
| 16 | 76,578,045–77,657,555 | Loss | SNP array | 0 (no. cases = 406) |
6 (no. controls = 404) |
CLEC3A, WWOX | [115] |
| 16 | 87,957,353–87,971,263 | Gain | SNP array | 2 (no. cases = 32) |
0 (no. controls = 26) |
Intron of ANKRD11 | [99] |
| 16 | 969,913–1,834,962 | Gain | SNP array | 8 (no. cases = 575) |
15 (no. controls = 621) |
SOX8, SSTR5, C1QTNF8, CACNA1H, TPSG1, TPSB2, TPSAB1, TPSD1, UBE2I, BAIAP3, C16orf42, GNPTG, UNKL, C16orf91, CLCN7, C16orf38, TELO2, IFT140, TMEM204, CRAMP1L, HN1L, MAPK8IP3, NME3, MRPS34, EME2, SPSB3, NUBP2, IGFALS, HAGH, FAHD1, C16orf73 | [110] |
| 17 | Not reported (1159 bp) | Gain | SNP array | 1 (no. cases = 12) |
0 (no. controls = 24) |
AATK, ACTG1, AZI1, BAHCC1, BAIAP2, C17orf55, C17orf56, C17orf70, C17orf89, CHMP6, FSCN2, SLC38A10, TMEM105, 2% of NPLOC4 (+ promoter), 31% of KIAA1303 | [138] |
| 19 | 20,860,930–20,875,787 | Loss | SNP array | 15 (no. cases = 117) |
2 (no. controls = 109) |
None | [96] |
| 19 | 32,615,675-32,935,836 | Gain | SNP array | 165 (no. cases = 575) |
122 (no. controls = 621) |
RDH13 | [110] |
| 19 | Not reported | Loss | SNP array | 2 (no. cases = 12) |
0 (no. controls = 24) |
Intron of ZFP14 | [138] |
| 22 | 23,696,411–24,240,667 | Loss and gain | SNP array | 10 (8 gain 2 loss) (no. cases = 406) |
28 (no. controls = 404) |
CRYBB3, CRYBB2, LOC91353, LRP5L, CRYBB2P1 | [115] |
| 22 | 21,011,312–21,394,287 | Loss | SNP array | 0 (no. cases = 575) |
11 (no. controls = 621) |
ZNF280B, ZNF280A, PRAME, BCR, GGTLC2 | [110, 138] |
| 22 | 29,489,697–29,489,738 | Deletion | SSCP | 5 (no. cases = 530) |
2 (no. controls = 379) |
NEFH | [68] |
| X | 139,400,576–telomere | Deletion | BAC array (aCGH validation) |
1 (no. cases = 72) |
0 (no. controls = 700) |
>100 (No ALS candidate genes) | [165] |
| X | 139,526,743–139,942,807 | Gain | BAC array (aCGH validation) |
1 (no. cases = 72) |
0 (no. controls = 700) |
CDR1 | [165] |
The table shows the most significant CNV loci and relative genes that partially or completely fall within them. Chromosomal positions are referred to the human reference genome assembly corresponding to each individual study: in particular [115], [40], and [81] refer to the NCBI reference sequence build 36, [98] refers to the build 38 and [41] refers to UCSC Genome Browser, May 2004 Freeze. In the case of [71], the authors described deletion coordinates by using the numbering of the published sequence by Lees et al. EMBO J. 1988. Genes that may be reasonable ALS candidates are in bold
Chr Chromosome, SSCP single-strand conformation polymorphism analysis, Loss heterozygous deletion, Deletion homozygous deletion
¥Gene symbols correspond to the NCBI Refseq names
In the following paragraphs, we review the most plausible candidate CNV loci that have been consistently associated with ALS, highlighting their potential functional impact in the pathophysiology of the disease (Tables 1 and 2 and Fig. 4).
Table 2.
Genes overlapped with CNV loci with a potential relevance for ALS susceptibility
| Gene | Description | Chromosomal location | GO Processes Associated | Type of CNV |
|---|---|---|---|---|
| SMN1 and SMN2 | Survival motor neurons 1 and 2 | 5q13.2 | mRNA processing, spliceosomal complex assembly, synaptic transmission | Gain and loss |
| NEFH | Neurofilament heavy subunit | 22q12.2 | Cellular response to oxidative stress, neurofilament cytoskeleton organization | Deletion |
| IDI1 and IDI2 | Isopentenyl diphosphate delta isomerase 1 and 2 | 10p15.3 | Cholesterol biosynthesis | Gain |
| DPP6 | Dipeptidyl-peptidase 6 | 7q36.2 | Regulation of neuronal action potential, regulation of potassium ion transport, protein localization to plasma membrane, proteolysis | Gain and loss |
| NIPA1 | Non-imprinted in Prader-Willi/Angelman syndrome 1 | 15q11.2 | Transmembrane transport | Loss |
| EPHA3 | Ephrin type-A receptor 3 | 3p11.2 | Axon guidance, cell adhesion, cell migration, signaling, cytoskeleton organization | Loss |
| AATK | Apoptosis-associated tyrosine kinase | 17q25.3 | Brain development, regulation of axon extension, neuron apoptotic process | Gain |
| BAIAP2 | BAI1-associated protein 2 | 17q25 | G protein coupled receptor signaling pathway, neurotransmitter secretion | Gain |
| CHMP6 | Charged multivesicular body protein 6 | 17q25.3 | Endosomal transport, membrane organization, nucleus organization, protein transport | Gain |
| IGFALS | Insulin-like growth factor binding protein acid labile subunit | 16p13.3 | Cell adhesion, cellular protein metabolic process, signal transduction | Gain |
| CACNA1H | Calcium channel, voltage-dependent, T type, alpha 1H subunit | 16p13.3 | Axon guidance, calcium ion import, membrane depolarization during action potential, muscle contraction, regulation of membrane potential | Gain |
| MAPK8IP3 | Mitogen-activated protein kinase 8 interacting protein 3 | 16p13.3 | JNK cascade, axon guidance, neuron projection development, positive regulation of neuron differentiation, regulation of gene expression, vesicle-mediated transport | Gain |
| BAIAP3 | BAI1-associated protein 3 | 16p13.3 | G-protein coupled receptor signaling pathway, neurotransmitter secretion | Gain |
| UBE2I | Ubiquitin-conjugating enzyme E2I | 16p13.3 | DNA repair, cell cycle, cell division, cellular protein metabolic process, chromosome segregation, nucleotide-excision repair, post-translational protein modification, protein ubiquitination | Gain |
| SPSB3 | splA/ryanodine receptor domain and SOCS box containing 3 | 16p13.3 | Intracellular signal transduction, protein ubiquitination | Gain |
| KCNIP4 | Kv channel interacting protein 4 | 4p15.32 | Potassium ion transport | Deletion |
| KCNB2 | Potassium channel, voltage gated Shab related subfamily B, member 2 | 8q13.2 | Potassium ion transport, synaptic transmission | Gain |
| KCNQ5 | Potassium channel, voltage gated KQT-like subfamily Q, member 5 | 6q14 | Potassium ion transport, synaptic transmission | Loss |
| GSDMD | Gasdermin D | 8q24.3 | Cellular response to extracellular stimulus | Gain & Loss |
| GRIK1 | Glutamate receptor, ionotropic, kainate 1 | 21q22.11 | Central nervous system development, glutamate receptor signaling pathway, ion transport, membrane depolarization, regulation of synaptic transmission, regulation of synaptic plasticity | Loss |
| GRIK2 | Glutamate receptor, ionotropic, kainate 2 | 6q16.3 | Central nervous system development, glutamate receptor signaling pathway, ion transport, membrane depolarization, regulation of synaptic transmission, regulation of synaptic plasticity, regulation of neuron apoptotic process, regulation of JNK cascade | Gain |
| EEF1D | Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | 8q24.3 | Cellular protein metabolic process, gene expression, mRNA transcription, positive regulation of I-kappaB kinase/NF-kappaB signaling, regulation of cell death, signal transduction | Gain & Loss |
| ATXN1 | Ataxin 1 | 6p23 | RNA processing, excitatory postsynaptic potential, regulation of transcription, regulation of glial cell proliferation | Loss |
| ATXN3L | Ataxin 3-like | Xp22.2 | Cellular response to misfolded protein, protein deubiquitination, regulation of transcription | Gain |
| SLC1A7 | Solute carrier family 1 (glutamate transporter), member 7 | 1p32.3 | L-glutamate transmembrane transport, neurotransmitter secretion, synaptic transmission | Gain |
| TRDN | Triadin | 6q22.31 | Cellular calcium ion homeostasis, cytoplasmic microtubule organization, endoplasmic reticulum membrane organization, muscle contraction, transmembrane transport | Gain |
| CPLX1 | Complexin 1 | 4p16.3 | Exocytosis, glutamate secretion, regulation of exocytosis, synaptic transmission, synaptic vesicle exocytosis, transport | Gain |
| ANKRD11 | Ankyrin repeat domain 11 | 16q24.3 | Multicellular organism growth, tissue homeostasis | Gain |
| PPP1R13B | Protein phosphatase 1 regulatory subunit 13B | 14q32.33 | Apoptotic process | Gain |
| FOXO3 | Forkhead box O3 | 6q21 | DNA damage response, apoptotic process, cell differentiation, cellular response to oxidative stress, epidermal growth factor receptor signaling pathway, immune response, regulation of neuron differentiation, transcription, tumor necrosis factor-mediated signaling pathway | Gain |
| HFE | Hemochromatosis | 6p21.3 | Antigen processing and presentation, immune response, ion transport, regulation of proteasomal ubiquitin-dependent protein catabolic process, regulation of receptor activity, positive regulation of gene expression, regulation of protein binding, transport | Gain |
| GGTLC2 | Gamma-glutamyltransferase light chain 2 | 22q11.22 | Glutathione metabolic process | Loss |
| ATG7 | Autophagy related 7 | 3p25.3 | Autophagy, cellular homeostasis, cellular protein modification process, cellular response to hyperoxia, central nervous system neuron axonogenesis, cerebral cortex development, membrane organization, mitochondrion organization, regulation of apoptotic process, protein transport, protein ubiquitination | Loss |
| ANXA5 | Annexin A5 | 4q27 | Calcium ion transmembrane transport, regulation of apoptotic process, signal transduction | Loss |
| GEMIN6 | Gem nuclear organelle-associated protein 6 | 2p22.1 | Gene expression, mRNA processing, mRNA splicing | Loss |
| MTMR7 | Myotubularin-related protein 7 | 8p22 | Dephosphorylation | Loss |
| ACYP2 | Acylphosphatase 2, muscle type | 2p16.2 | Phosphate-containing compound metabolic process | Loss |
| ZFP14 | ZFP14 zinc finger protein | 19q13.12 | Regulation of transcription | Loss |
| FMN2 | Formin 2 | 1q43 | Actin filament assembly, cellular response to DNA damage stimulus, cellular response to hypoxia, intracellular signal transduction, intracellular transport, multicellular organismal development, regulation of apoptotic process, protein transport, vesicle-mediated transport | Deletion |
Loss heterozygous deletion; Deletion homozygous deletion
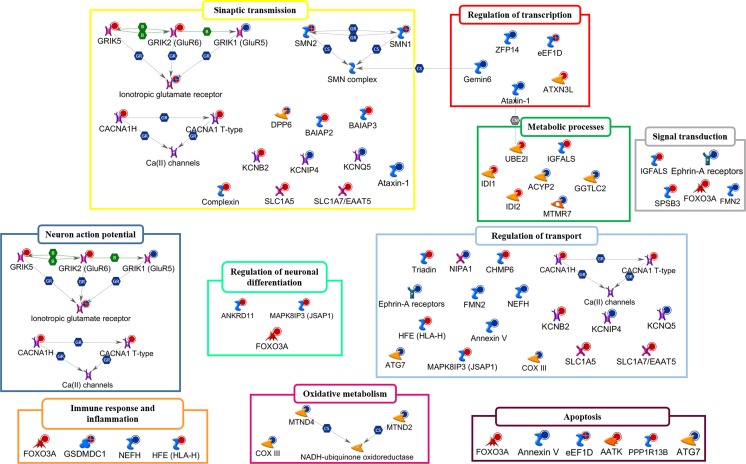
Fig. 4.
A representative illustration showing the functional correlation between ALS-associated CNV-affected genes and their biological processes. Interaction map represents the most promising candidate genes overlapping CNVs that have been consistently associated with ALS, grouped on the basis of the main biological processes associated with them. The map was created using the MetaCore Pathway Map Creator tool (GeneGo). Genes associated with CNV gain regions are labeled with red dots while genes associated with homozygous or heterozygous deleted CNVs are labeled with blue dots. The “checkerboard” color indicates genes displayed both CNV gains and losses. Detailed information about genes depicted in the figure and related biological processes are reported in Table 2. A detailed legend for the network objects is shown in Supplementary Fig. 1
Survival Motor Neuron
Among candidate genes that may modulate susceptibility and disease course in ALS, a great deal of attention has been given to the survival motor neuron (SMN) gene. It maps in a highly duplicated region of chromosome 5 (5q13.3) and is present in two homologous copies: a telomeric copy called SMN1 and a centromeric copy called SMN2 that produces approximated only 20% of full-length transcripts coding for SMN [39]. This is a housekeeping protein that is essential for the efficient assembly and regeneration of spliceosomal small nuclear ribonucleoproteins (snRNP) in various human cell types, including the axons of spinal motor neurons, exerting important effects in protein translation, as well as in the mitochondria and cytoskeleton maturation and synaptic transmission regulation (Table 2 and Fig. 4) [40].
Aberrant CNVs in SMNs result in a defective protein, leading to deficits in snRNP assembly, disruption of normal cellular RNA metabolism, and motor neuron degeneration [41]. Homozygous deletions of SMN1 are responsible for the pathogenesis of spinal muscular atrophy (SMA), an autosomal recessive motor neuron disease characterized by lower motor neuron loss and muscle atrophy, while SMN2 copy numbers seem to modulate the severity of the SMA phenotype and survival [42].
Because of the phenotypic similarities between SMA and ALS, various studies have focused on the possible association between copy number abnormalities in SMN genes and ALS, but controversial results have been obtained (Table 1). Indeed, although several studies have demonstrated that high or low SMN1 copy number together with the absence of SMN2 increased risk of developing ALS [43–48], others have not found any significant association [49–51]. These apparently contradictory data, at least in part, may be due to the use of inadequate size populations and different assays used to assess CNVs in the SMN genes. Similar conflicting results were also observed at the SMN protein level. Indeed, both low and high levels of SMN production seem related to the risk of ALS. Upregulated expression of SMN genes was recently found in the motor cortex of SALS patients, supporting the evidence that overexpression of SMN protein may be toxic for motor neurons [52, 53]. Decreased levels of SMN result, instead, in a reduction of its chaperone-like activity against mutant SOD1-mediated toxicity in vitro, contributing to increase-free radical injury and oxidative stress, two well-established features of ALS [54]. In support of this theory, it has been demonstrated that an increase in the levels of SMN improved motor functions and delayed motor neuron loss in different cellular and animal models of ALS [55, 56].
Further evidence for the contribution of SMN signaling in ALS pathogenesis is based on the observation that SMN and some ALS-associated proteins share common biochemical pathways. This is the case, for example, of FUS and TDP-43, two gene-encoding proteins extensively associated with both sporadic and familial ALS that colocalize with SMN in subnuclear organelles known as gems (gemini of Cajal bodies). ALS-related mutations in these genes seem to contribute to motor neuron dysfunctions also by reducing the localization of SMN to axons with the consequent disruption of the SMN-mediated assembly and integrity of the splicing machinery [57–59].
Considering its potential impact on ALS pathogenesis, several studies have investigated the role of SMN as a pharmacological target in ALS. To this regard, the systemic administration of valproate (VPA), a drug able to increase the SMN levels in the SNC through an epigenetic mechanism, has shown neuroprotective effects both on SMA and ALS patients [60].
In addition to alterations in the activity of SMN complex, aberrant copy numbers in the SMN genes might confer susceptibility for ALS also through SMN-independent mechanisms, acting, for example, as markers of linkage disequilibrium with other ALS susceptibility loci, such as the neuronal apoptosis inhibitory protein (NAIP) gene. A more detailed and accurate molecular genetic investigation of the SMN gene region could reveal additional genetic variants with potential relevance for ALS, providing new insights into the common pathogenic mechanisms underlying ALS and other motor neuron diseases [61].
Neurofilament Heavy Subunit
Neurofilaments (NFs) are the most prominent cytoskeletal components in neurons. They are composed of a globular head, α-helical rod region, and a globular tail and are expressed under three different neuron-specific subunits with different molecular weights: light (NEFL), medium (NEFM), and heavy (NEFH). NFs, together with microtubules and microfilaments, form part of the slow axonal transport and contribute to the formation and maintenance of the neuronal structure (Table 2 and Fig. 4) [62].
Although the role of NFs in ALS is not yet fully clarified, dysregulated expression and point mutations of NFs have been reported in human and animal models of ALS [52, 63, 64]. The principal pathogenic mechanism of these variations seems to be related to defects of the normal NFs assembling and phosphorylation. These alterations, in addition to dysfunction of the ubiquitine-proteasome system, may cause changes in the cross-linking properties of NFs, resulting in the aberrant accumulation of these proteins in motor neurons, which represents an established pathologic hallmark of ALS [65, 66].
Several studies have investigated the role of NF structural chromosomal alterations in ALS (Table 1). Deletions in NEFL, accompanied by a concomitant altered expression of NEFH and NEFM in the cell bodies and axons of motor neurons, have been associated with a significant delay of the disease onset and progression in ALS animal models [67]. In addition, point mutations (C2232T and C2414A) were also found in the short allele of NEFH, suggesting that these may act as markers for deletion mutants [68].
Significant associations were also established between the sporadic form of ALS and deletions in the C-terminal tail of NEFH [68]. This region is composed of a unique functional domain consisting of 43–45 repeat motifs of the amino acids lysine-serine-proline (KSP) [69]. The phosphorylation of these motifs in the C-terminal tail allows NEFH to interact with other cytoskeletal components (e.g., microtubule-associated proteins), regulating the interfilament spacing and thereby the axonal caliber. Deletions of the NEFH KSP domain have been detected in SALS patients as well as in some patients with a FALS pedigree, even if it was not yet determined whether the variant segregates with the disease or not [70, 71]. Interestingly, loss of a KSP motif, or multiples of this, seems to affect a recognition sequence for the neurofilament kinase CDK5, suggesting that deletions at this level may result in aberrant interactions of NEFH with other cytoskeletal elements, leading to axonal and cytoskeleton integrity destruction and motor neuron degeneration.
Isopentenyl Diphosphate Isomerase
Although the exact association between alterations in lipid metabolism and ALS remains unknown, there are several evidence of possible involvement of cholesterol and other lipids in the disease pathogenesis. Decreased cholesterol levels as well as dysfunctions in lipids transport and metabolism have been found in both human and ALS animal models while increasing dietary lipid content appears to be associated with neuroprotective effects [72–75].
Among proteins involved in the lipid metabolism is the isopentenyl diphosphate isomerase (IDI), a cytoplasmic enzyme belonging to the mevalonate pathway that plays an essential role in the biosynthesis of cholesterol and other lipophilic molecules (Table 2 and Fig. 4). IDI has two isoforms in humans, IDI1 and IDI2 that are encoded by two tandemly duplicated genes [76, 77].
Several studies support the role the mevalonate pathway and IDIs in the pathogenesis of sporadic ALS. Downregulated expression of IDI1 was recently found in the motor cortex of a specific subgroup of SALS patients [52]. Moreover, a segmental copy number gain, encompassing the IDI1/IDI2 gene region on 10p15.3 subtelomere, was found in SALS patients. This duplication leads to the disruption of the genetic composition and reduction of the IDI1 expression (Table 1) [78]. The association between IDI1/IDI2 copy number alterations and the sporadic form of ALS may be explained by the instability of this genomic region that contains multiple low-copy repeats in a small region of the genome. These segmental duplications can arise from an unequal crossing-over or end-joining events, suggesting the possibility that de novo CNVs may occur [79]. Further investigation about the function of IDI1/IDI2 in motor neurons may allow to obtain new insights into the pathogenesis of SALS and to identify also novel promising therapeutic targets for this disease.
Dipeptidyl-Peptidase 6
Dipeptidyl aminopeptidase-like protein 6 (DPP6) is a member of the prolyl oligopeptidase family of serine proteases. It is predominantly expressed in the central nervous system which modulates the function and expression of potassium channels as well as excitability at the glutamatergic synapse (Table 2 and Fig. 4).
Although the role of DPP6 in ALS pathogenesis remains unclear, genetic variations in DPP6 have been identified as potential risk factors for different human diseases, including ALS [6, 80–82]. Several genome-wide studies, in fact, have reported an association between an SNP (rs10260404) in DPP6 and ALS susceptibility and increased expression of DPP6 was found in motor cortex of SALS patients [6, 52, 83]. In addition, deletions and duplications affecting DPP6 were also found in some cases of ALS (Table 1) [84]. These copy number aberrations occur mainly at the 5′ end of the gene, a known CNV region flanked by segmental duplications mediating genomic rearrangements [85]. The functional impact of these genomic aberrations seems to involve the generation of altered or truncated transcripts as well as the disruption of regulatory elements in this region that may alter gene expression. Further investigations are necessary to better clarify the function of DPP6 in the nervous system and its potential role in ALS susceptibility.
Non-imprinted in Prader-Willi/Angelman Syndrome
Non-imprinted in Prader–Willi/Angelman syndrome (NIPA1, also known as SPG6) is an inhibitor of bone morphogenic protein signaling, one pathway that plays an important role in the formation, maintenance, and function of synapses as well as in transmembrane transport (Table 2 and Fig. 4). Despite mutations in NIPA1 not being directly linked to ALS pathogenesis, they are known to cause hereditary spastic paraplegia (HSP) type 6, a neurodegenerative disease characterized by the selective degeneration of upper motor neurons [86]. Although ALS and HSP are clinically different, they share important clinical features, including selective motor neuron degeneration. Mutations in some genes causing various forms of HSP, such as SPAST and SPG11, were also associated with ALS phenotype [87]. In particular, SPG11 encodes spatacsin, another inhibitor of bone morphogenic protein signaling, supporting the involvement of this pathway in motor neuron degeneration and pathogenesis of both ALS and HSP.
NIPA1 polyalanine repeat expansions have been identified as potential risk factors for ALS and appear involved in the modulation of the disease course. Moreover, several CNVs affecting NIPA1 were also found in SALS patients (Table 1) [84, 88]. In particular, these copy number alterations affect a known CNV region of chromosome 15 (15q11.2) flanked by segmental duplications. While duplications in this region do not seem to contribute to ALS susceptibility, deletions overlapping four adjacent genes, including NIPA1, showed a strong statistical association (Table 1) [84]. Recently, CNVs in the NIPA1 region have been also associated with other neurological diseases, including schizophrenia and epilepsy, suggesting that NIPA1 and other genes inside the same CNV-driven region, in association with other risk factors, may concur to cause these different pathologic phenotypes [89].
Ephrin Type-A Receptor 3
Ephrin receptors (Ephs) represent the largest known subfamily of tyrosine kinase receptors. Ephrin receptors and their ligands are involved in a variety of important functions, including axonal outgrowth, cytoskeletal structure development, neuronal connectivity, neuronal apoptosis, synaptic maturation, and plasticity (Table 2 and Fig. 4) [90, 91].
Alterations in the expression or function of ephrins and their receptors induce pathological changes in the motor neuron circuitry, contributing to the initiation and progression of ALS pathogenesis [92]. In accordance with this theory, deregulated expression of ephrins and their receptors was found in the motor cortex and spinal cord of SALS patients [3, 93] and several SNPs affecting Eph/ephrin genes have been associated with ALS susceptibility [94]. It is of interest to note that some of these alterations often result in beneficial effects. Indeed, loss-of-function mutations in EPHA4 or complete knockdown of this gene have shown to rescue and prolong survival in ALS animal models [95]. In addition, a heterozygous deletion of EPHA3 was found significantly higher in controls as compared to ALS patients, supporting a potential protective role of this variant against the risk of developing ALS (Table 1) [96]. All these evidence support the importance of Eph/ephrin signaling in modulating the vulnerability of motor neurons to axonal degeneration, highlighting the necessity to further investigate this protein family as promising therapeutic candidates for ALS.
Mitochondrial DNA Genes
Impaired mitochondrial bioenergetics function plays an important role in several neurodegenerative diseases, including ALS. In particular, dysfunctions in mitochondria result in a reduction in bioenergetics efficiency, increasing the risk of neuronal death when energy demands exceed cellular energy production [97, 98].
One of the genes primarily involved in oxidative phosphorylation encodes cytochrome c oxidase (CO), the terminal component of the mitochondrial respiratory chain that transfers electrons from reduced cytochrome c to molecular oxygen. CO is a multiheteromeric enzyme composed of 13 protein subunits, some of which coded by mitochondrial DNA (mtDNA) genes [99, 100]. Reduced levels of CO histochemical activity as well as mutations and heterozygous deletions in mtDNA genes encoding some CO subunits (such as CO3 and COX7C) have been detected in patients with ALS phenotype, contributing to reduce bioenergetic functions observed in ALS motor neurons (Fig. 4) [101–104].
In addition to genetic variations of mtDNA CO genes, deletions in other mtDNA genes were found in substantia nigra neurons, where it seems to cause cellular respiration deficits, reduced capacity to modulate synaptic activity, and swings in cytosolic calcium levels as well as reduced capacity to survive to excitotoxic stresses [105, 106]. Although it is not yet fully clarified how these mtDNA deletions arise, their origin may involve failures in the mitochondrial biogenesis programs as well as genetic, epigenetic, or post-translational causes (or their combinations) [107, 108]. Similar chromosomal rearrangements have been also observed in ALS phenotype. Indeed, high abundances of deletions involving ND2 and ND4, two mtDNA genes that code for NADH-ubiquinone oxidoreductase, an essential respiratory protein complex, were found in the spinal cord of ALS patients, supporting the concept that some motor neurons in ALS may die because of a deficit in the mitochondrial ATP production and cellular energy metabolism (Fig. 4) [102].
It is interesting to note that while some ALS patients were characterized by an accumulation of mtDNA deletions, other ALS cases showed abundant mtDNA copy numbers and relatively low levels of deletions. This observation is corroborated by our previous work that described a coordinated decrease of several gene-encoding proteins involved in the oxidative phosphorylation pathway only in a specific subgroup of SALS patients, sustaining the existence of a molecular heterogeneity in ALS patients [52]. Based on these results, it is evident that mitochondria-targeted therapies, aimed to provide and/or express intact mtDNA in motor neurons, may represent a helpful strategy for the management of bioenergetics defects in specific subgroups of ALS patients [109].
Multiple Rare CNVs: Assessing their Role through a System Biology Approach
Despite their importance in deciphering the genetic cause underlying ALS, common CNVs account for only a relatively small fraction of the genetic variation, while the contribution of less frequent but potentially functionally important variants is not yet fully elucidated.
Recent advances in high-throughput genomic technologies enabled the identification of a large number of rare and novel ALS-specific CNV loci, which were found in ALS patients but absent or extremely rare in individual controls of each study and/or in >2500 controls present in DGV (Table 1 and Supplementary Table 1) [110, 111]. However, no significant association was found between these rare variants and the risk of developing ALS. This may be mainly due to the effect size of most of these variants that is too small to be detected by using the traditional statistical methods. In addition, currently, the majority of genome-wide CNV studies typically apply a single-gene or single-variant approach, focusing their attention on the identification of a restricted list of potential candidates, previously implicated in the disease, without taking into account the potential joint impact of these rare variants in the etiopathogenesis of ALS [112]. To this regard, the new systems biology perspective may represent a powerful tool to overcome these limitations, enabling to characterize, in a comprehensive or high-throughput manner, the collective effects of these variants that, both individually or in combination, can contribute to increase the likelihood of developing ALS (Fig. 2).
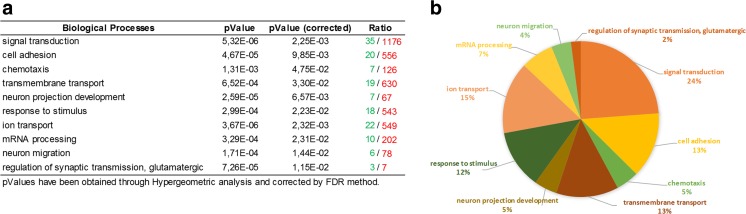
Investigating the potential functional role of rare ALS-related CNVs, it emerges that, albeit these variants are individually rare, the target set of genes and related products may be annotated to one or more common biochemical pathways relevant to ALS pathogenesis, including signal transduction (p value = 5.32 E−06), cell adhesion (p value = 4.67 E−05), ion transport (p value = 3.67 E−06), and messenger RNA (mRNA) processing (p value = 3.29 E−04) (Table 2 and Figs. 4 and 5). Moreover, many of these genes show a functional correlation also with the most known causative ALS genes (e.g., SOD1, ALS2, SETX, FUS, TARDBP), supporting the evidence that multiple common and rare CNVs may exert their pathogenic effect by different multifactorial combinations, jointly contributing to the genetic susceptibility of ALS (Fig. 6). Interestingly, some of the genes affected by rare CNVs encode proteins that are differentially expressed in motor cortex of SALS patients, sustaining the theory that the genomic structural variants frequently affect the transcriptional regulation [52]. Below, we provide a brief description of the most plausible candidate genes affected by rare ALS-related CNVs in the light of the main biological processes in which are involved, in order to investigate their contribution in ALS pathogenesis and their relevance as potential diagnostic biomarkers (Fig. 4, Tables 1 and 2 and Supplementary Table 1).
Fig. 5.
The functional enrichment analysis of the most plausible candidate genes overlapping ALS-specific CNV loci reveals biological processes relevant to ALS pathogenesis. a Representation of the top 10 most significantly enriched (p value <0.05) canonical GO biological processes associated with genes significantly enriched in rare and novel ALS-specific copy number changes (not reported in controls of each of the individual studies and/or in >2500 controls present in DGV). The analysis was performed using the Gene Ontology and KEGG databases and the list is arranged in descending order with the most significant GO biological processes at the top. Detailed information about the entire list of genes affected by ALS-specific CNV loci are reported in Supplementary Table 1. b GO term pie chart of the top 10 enriched (p < 0.05) “Biological processes” for genes overlapping ALS-specific CNV loci. GO terms or biological features of candidate CNV-affected genes and the percentage of genes represented in each category are indicated
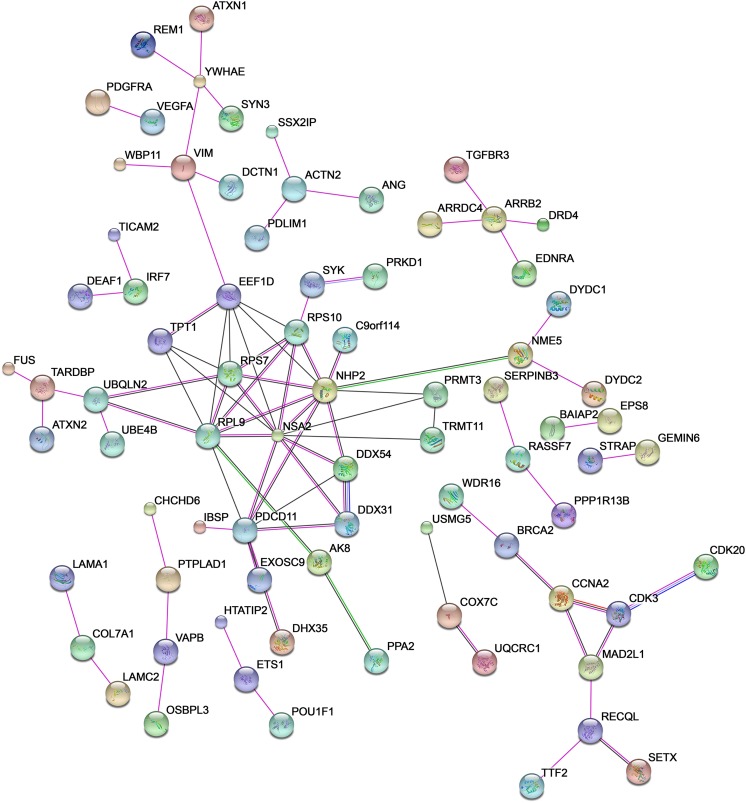
Fig. 6.
Functional network of known and predicted interactions between proteins encoded by genes affected by rare ALS-specific CNV loci and the most known causative ALS genes. The network was produced by the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) v10 (http://string-db.org/) using default settings. Proteins are represented by spheres. Lines linking proteins indicate evidence for interactions: a red line indicates the presence of gene fusion (genes that are sometimes fused into single open reading frames); a green line gene neighborhood (genes that reside within 300 bp on the same strand in the genome); a blue line co-occurrence (gene families whose occurrence patterns across genomes show similarities); a purple line experimental evidence (interaction extracted from protein-protein interaction databases); a yellow line text mining (interaction extracted from scientific literature); a light blue line database (interaction extracted from curated databases); a black line co-expression (proteins whose genes are co-expressed in the same or in other species)
Regulation of Synaptic Transmission and Neuronal Action Potential
Neurons are physically connected to each other to form extensive neural networks; the synaptic transmission is the process by which information rapidly flows in neuronal networks. This is essential for maintaining normal brain function as well as muscle tone and muscle movements coordination [113]. The disruption of the synaptic stability result in a disruption of neuronal circuits that represents almost certainly one of the major underlying causes of many neurologic diseases, including ALS, Parkinson’s and Alzheimer’s disease [114–116].
Multiple GWAS studies have described ALS-specific gains and losses in several genes encoding proteins involved in synaptic transmission and the regulation of neuronal action potential (KCNIP4, KCNB2, KCNQ5, GRIK1, GRIK2, ATXN1, BAIAP2, CPLX1, CACNA1H, SLC1A7) (Tables 1 and 2 and Supplementary Table 1) [110, 117].
Different CNVs in some genes that code for potassium and calcium channels (KCNIP4, KCNB2, KCNQ5, CACNA1H) were found in ALS patients and seem to have a particular interest in the deciphering the genetic cause of ALS (Tables 1 and 2 and Fig. 4). In fact, alterations in the homeostasis and signaling pathways of these ions seem to be responsible for some important toxic pathways underlying ALS, such as oxidative stress, mitochondrial dysfunction, and neuroinflammation [118]. In particular, an autoimmune attack on potassium channels has been identified as a possible risk factor for ALS and alterations in other potassium channel-related genes (e.g., DPP6) were found in several cases of ALS [119]. Moreover, deregulated expression of multiple genes encoding calcium and potassium channels were also found in motor cortex of SALS patients [52].
Changes in copy number and in the expression levels of genes encoding glutamate ionotropic receptors (GRIK1, GRIK2) were identified in several cases of ALS (Table 2 and Fig. 4) [52, 120]. Glutamate receptors mediate fast excitatory synaptic transmission in the central nervous system and their excessive stimulation as well as deregulations in their expression and activity are responsible for excitotoxicity, a phenomenon that has been regarded as one firm mechanism implicated in both acute and chronic neurodegenerative diseases, including ALS [120]. A copy number gain in SLC1A7 (also known as EAAT5), a gene encoding a member of the glutamate transporter family, was found in a case of ALS but not in controls or DGV. Altered expression of this gene was found in the brains of patients with both familial and sporadic forms of ALS, supporting the role of an aberrant glutamate signaling in ALS pathogenesis (Table 2, Fig. 4, and Supplementary Table 1) [121].
Deletions in ATXN1 and a duplication of the related gene ATXN3L were identified in some ALS patients (Supplementary Table 1) [110]. ATXN1 encodes a protein mainly implicated in mRNA processing (Table 2 and Fig. 4) that normally contains a polyglutamine (polyQ) tract with 22–23 repeats; the occurrence of an expanded polyQ repeat has been associated with an increased genetic risk for ALS [122]. In addition to structural aberration, decreased expression of ATXN1 was recently found in a specific subgroup of SALS patients [52]. ATXN1 seems also to be involved in transcriptional repression of angiogenic factors whose deregulations were widely associated with ALS pathogenesis [123]. Another gene involved in the regulation of angiogenic signaling pathway is BAIAP2, a gene encoding a brain-specific angiogenesis inhibitor that is able to modulate synaptic transmission as well as learning and memory (Table 2 and Fig. 4) [124]. Segmental copy number gains in BAIAP2 and other structurally related genes (e.g., BAIAP3), were found in some cases of ALS patients (Table 1) [110, 111]. In addition, increased expression of BAIAP2 and BAIAP3 was found in motor cortex of SALS patients, supporting the theory that an excessive angiogenesis may contribute to ALS pathogenesis.
Copy number gains in CPLX1 have been identified at a significantly higher frequency in ALS patients compared to controls (Table 1) [110]. CPLX1 encodes a protein involved in synaptic vesicle exocytosis whose deregulation has been implicated in the pathogenesis of several neurological and neurodegenerative diseases, including ALS (Table 2 and Fig. 4) [52, 125].
Transcriptional Regulation
Mounting evidence has emerged to support an important role of transcription dysregulation in the initiation and progression of several neurodegenerative diseases, including ALS. Indeed, widespread aberrations in the molecular machinery that regulate gene expression and mRNA splicing have been reported both in sporadic and familial forms of ALS [126, 127]. Although the mechanisms and causal relationships have not yet been fully elucidated, several genome-wide studies have identified ALS-linked mutations and ALS-specific copy number variations in genes involved in the regulation of RNA processing and, thus, in the control of gene expression. In particular, the deletion of an intron of ZFP14 was found in two ALS patients but not in individual controls (Table 1) [111]. The functional group of ZFP14 proteins exerts diverse functions, many of which have been extensively implicated in ALS, such as RNA packaging, transcriptional activation, regulation of apoptosis, as well as protein folding and assembly (Table 2 and Fig. 4).
A large deletion affecting GEMIN6, another gene involved in the control of neuronal gene expression, was found in a single ALS patient (Supplementary Table 1). GEMIN6 is mainly implicated in the regulation of mRNA processing and belongs to the SMN complex, which as previously discussed is a potential risk factor in ALS susceptibility (Table 2 and Fig. 4). In particular, a reduced expression of GEMIN6 has been associated with a reduced activity of the SMN complex, contributing to alter normal snRNP assembly and leading to motor neuron degeneration [128].
A process that has been directly implicated in genetic causes of various motor neuron diseases is the enzymatic delivery of aminoacyl transfer RNAs (tRNAs) to the ribosome. Among genes involved in this process is EEF1D, a gene encoding a subunit of the elongation factor-1 complex. Copy number gains and losses in EEF1D were found in a modest number of ALS patients and increased expression of EEF1D was recently found in SALS patients, confirming the potential role of this factor in ALS pathogenesis (Tables 1 and 2 and Fig. 4) [52, 110].
Immune Response and Inflammation
Immune processes inside the central nervous system contribute to the maintenance of homeostasis and play important roles in resolving inflammation and mediating neuroprotection and repair [129]. Immune responses are finely regulated by multiple checkpoints that are responsible to ensure the protection of neuronal tissue from harmful events.
There is strong evidence that the acute neuroinflammation and the dysregulated immune response are potentially pathogenic factors in a number of neurodegenerative diseases, including ALS [130]. To this regard, aberrant gene expression and copy number variations in genes involved in neuroinflammation and cellular response to injurious stimuli (FOXO3, HFE, GSDMD) have been identified in ALS patients with respect to controls (Table 1 and Supplementary Table 1) [99, 110, 117]. In particular, FOXO3 encodes a transcription factor involved in apoptosis, cellular metabolism, and resistance to oxidative stress, while variations in HFE, a gene encoding a protein involved in antigen presentation and processing, have been reported to significantly increase the risk of SALS in a number of different individuals (Table 2 and Fig. 4) [52, 131, 132]. ALS-specific copy number gains and losses were found also in a genomic region encompassing GSDMD, a gene encoding a member of the gasdermin family that appears to play a role in neuroinflammatory and caspase-induced apoptotic processes (Tables 1 and 2 and Fig. 4).
Signal Transduction
Signal transduction refers to the process through which an extracellular biological molecule, by interating with specific cell-surface receptors, activates a biochemical chain of events inside the cell, eventually eliciting a response. Neuron-specific alterations in various signal transduction pathways represent one of the principal pathological hallmarks of several neurological diseases, including ALS [133, 134]. An elevated number of genes encoding several trophic factors and their receptors as well as proteins involved in the regulation of intracellular signaling cascades were found differentially expressed in SALS motor cortex [52]. In addition, rare copy number aberrations in genes involved in multiple signaling cascades (IGFALS, SPSB3, FOXO3 and FMN2) were also found in some cases of ALS, suggesting that alterations in these genes, either alone or in combination, may concur to generate motor neuron injury in ALS.
IGFALS encodes a serum protein that binds insulin-like growth factors (IGFs), increasing their half-life and their vascular localization. Increased expression of genes encoding IGF receptors and IGFALS was found in motor cortex of SALS patients [52]. In addition, structural gains in the genomic region encompassing IGFALS gene were identified in some ALS cases, supporting the hypothesis that dysregulation of this signaling cascade may be critical for ALS pathogenesis (Tables 1 and 2 and Fig. 4) [110, 135]. It has been also observed that pharmacological inhibition of the IGF signaling pathway exerts neuroprotective effects in different models of neurodegeneration [136]. In addition to IGFALS, another gene involved in the regulation of the intracellular signal transduction maps within the region of gain at chromosome 16 (Tables 1 and 2 and Fig. 4) [110]. This is SPSB3, a gene encoding a protein involved in the ubiquitine-proteasome pathway, the major proteolytic quality control system in cells, whose deregulation has been associated with numerous neurodegenerative diseases, like ALS, leading to abnormal protein aggregation and consequent neuron death [137].
Several GWAS studies have confirmed the presence of a homozygous deletion affecting an intron of FMN2 gene in some cases of ALS but not in controls (Table 1) [80, 110, 138]. FMN2 is expressed in brain and spinal cord and plays an important role in the regulation of cytoskeletal assembly, signal transduction, and protein transport (Table 2 and Fig. 4). Decreased expression of this gene has been related to age-dependent memory loss in mice and SNPs in this gene have been detected in SALS patients [139, 140].
Metabolic Homeostasis
Maintaining a metabolic homoeostasis requires a balance between energy intake and expenditure. Growing evidence highlights that an imbalance in energy metabolism may contribute to the selective death of neurons in ALS [141]. To this regard, copy number abnormalities in some genes involved in metabolic processes (IGFALS, UBE2I, ACYP2, MTMR7) have shown to be potential risk factors for ALS (Table 1 and Supplementary Table 1). In particular, a structural gain on chromosome 16 encompassing multiple genes potentially involved in ALS, including UBE2I, was found in a moderate number of ALS patients (Table 1) [110]. UBE2I encodes an ubiquitin-conjugating enzyme involved in the ubiquitin-proteasome system that is crucial to a vast array of cellular processes, such as cell cycle control, immune responses, and metabolic regulation [142]. Recently, an upregulated expression of UBE2I was identified in astrocytes from ALS animal models and a differentially expression of its gene was also found in motor cortex of SALS patients [52, 143, 144].
Heterozygous deletions of MTMR7 and ACYP2 genes were found in a limited number of ALS patients, but not in controls or DGV (Supplementary Table 1) [84, 96, 145]. MTMR7 is expressed specifically in the brain and is involved in the lipid metabolism and phosphatidylinositol signaling pathway, a mechanism implicated in ALS pathogenesis (Table 2 and Fig. 4) [146]. ACYP2 encodes a protein belonging to enzyme family that acts as a phosphatase and is implicated in metabolic processes and Ca+2 modulation (Table 2 and Fig. 4). Interestingly, deletions of ACYP2 have been associated with a relatively early age of disease onset (32–44) in ALS patients, and its reduced expression has been found in the motor cortex of SALS patients [52].
Another interesting heterozygous deletion encompassing GGTLC2 was found significantly higher in controls as compared to ALS patients, suggesting a potential protective role of this variant against the risk of developing ALS (Table 1). GGTLC2, whose decreased expression was found in SALS patients [52], encodes a protein involved in glutathione metabolism, a process associated with altered energy metabolism in ALS pathogenesis (Table 2 and Fig. 4) [147, 148].
Apoptosis
Apoptosis is a type of programmed cell death that plays an essential role during embryogenesis, cellular homeostasis, immune system maturation, and cellular response to external stimulus, including DNA damage and oxidative stress. There is strong evidence to suggest that apoptosis is responsible for motor neuron degeneration in ALS [149]. Apart from genetic mutations and changes in gene expression, copy number aberrations in genes encoding factors involved in the control of apoptosis were detected only in ALS patients and absent in controls, suggesting a possible contribution of these variants in increased risk of developing ALS (Table 1 and Supplementary Table 1). In particular, heterozygous deletions in regions encompassing ATG7, ANXA5, and PPP1R13B genes and a segmental gain in the AATK gene were observed (Table 1 and Supplementary Table 1) [111, 138, 145]. ATG7 is a crucial factor for the induction of autophagy and ubiquitin-related activities as well as in apoptosis and the maintenance of axonal homeostasis (Table 2 and Fig. 4) [150]. Interestingly, deletions of ATG7 have been reported to induce axonal degeneration in a mouse model, leading to accumulation of polyubiquitinated aggregates and severe neurodegeneration. ANXA5 encodes a protein that plays an essential role in apoptosis and survival as well as neurite growth of in vitro cortical neurons and has been previously indicated as a candidate gene for ALS (Table 2 and Fig. 4) [151, 152]. PPP1R13B encodes an apoptosis-stimulating protein that is able to interact with p53, regulating neuronal differentiation and specifically enhancing p53-induced apoptosis that was identified as an apoptotic mode of motor neuron cell death in the spinal cord of ALS patients (Table 2, Fig. 4, and Supplementary Table 1) [153–155]. AATK encodes a protein that plays an essential role in the induction of mature neuron apoptosis, neuronal genesis and differentiation, as well as axon outgrowth. Copy number gains in AATK are in accordance with the evidence that an increased expression of this gene is induced during apoptosis, supporting a role of this signaling pathway in ALS (Table 2 and Fig. 4) [156].
Neuronal Development and Differentiation
Neuronal development is a process strongly dependent from a balanced and tightly regulated control of several trophic factors capable of regulating important physiological processes, including neuronal survival, migration, and differentiation as well as synapse and dendrite maintenance and axonal outgrowth [157]. Alterations to crucial genes regulating neural development and differentiation can lead to impairments in the cellular homeostasis and have been associated with numerous neurodegenerative diseases, such as Huntington’s and ALS [158, 159]. In addition to changes in genetic and epigenetics factors, through mutation or altered gene expression, copy number variations were associated as potential risk factors for ALS also. In fact, segmental duplications in genes involved in the regulation of neuronal cell death and differentiation (FOXO3, ANKRD11, MAPK8IP3) were found in some cases of ALS (Table 1).
ANKRD11 is a large nuclear protein that regulates transcription, potentially by binding chromatin-modifying enzymes and is involved in apoptotic processes by enhancing the transcriptional activity of p53 proteins (Table 2, Fig. 4 and Supplementary Table 1) [153, 154]. A structural gain in an intron of ANKRD11 was identified only in two ALS patients compared to control subjects [99] (Table 1). In addition to structural aberrations, mutations and altered expression of ANKRD11 have been associated with perturbations in neural development occurring in some neurological conditions, such as autism [160].
MAPK8IP3 encodes a protein that binds and regulates the activity of numerous protein kinase components of the c-Jun N-terminal kinase (JNK) signaling pathway, facilitating JNK activation and thus playing a role in the regulation of many cellular events, including growth control, apoptosis, and axonal transport [161]. Increased levels of MAPK8IP3 seem to be required for neurodegeneration in different models of neurological disorders, including Parkinson’s and Alzheimer’s diseases, suggesting that accumulations of these proteins may represent a response to oxidative stress conditions [162, 163].
Neuronal Transport
The trafficking and transport of vital cellular components is critical for correct neuronal function and axonal maintenance. Increasing evidence has highlighted a correlation between dysfunction of the cellular transport machinery, for example caused by an oxidative damage, and many neurodegenerative conditions, including ALS [164]. Besides point mutations in genes extensively related to ALS pathophysiology (e.g., C9orf72, TDP-43, VAPB, DCTN1), several copy number aberrations encompassing genes involved in the regulation of neuronal transport were found in ALS patients. In addition to some genes that were previously discussed in the above paragraphs (ATG7, NIPA1, CACNA1H, KCNIP4, KCNQ5, KCNB2, SLC1A7, CO3, MAPK8IP3, ANXA5, HFE, FMN2, NEFH, EPHA3), two interesting segmental duplications in TRDN and CHMP6 were found in some cases of ALS and in none of patient controls (Supplementary Table 1) [84, 138, 165]. TRDN encodes an integral membrane protein highly expressed in skeletal muscle with a recognized role in calcium release, and whose low expression has been found in brain and nerve tissue, suggesting its potential role in motor neuron diseases (Table 2, Fig. 4, and Supplementary Table 1) [166]. CHMP6 encodes a member of CHMP subfamilies involved in the formation of endocytic multivesicular bodies that are required for autophagic clearance of protein aggregates (Table 2 and Fig. 4). Deregulation in this process leads to abnormal ubiquitin-positive protein deposits in neurons that represent a hallmark of several neurodegenerative diseases, including ALS. In addition, mutations in another component of CHMP family (CHMP2B) have been widely implicated in frontotemporal dementia and ALS [167], sustaining the role of this protein family in ALS pathogenesis.
Rare ALS-Related CNVs in Intergenic Regions
In addition to CNVs overlapping with genes, several rare non-polymorphic structural variants overlapping with promoters or intergenic regions have been reported as potential ALS candidate CNVs. Although these regions are difficult to decode, the possibility of long-acting gene regulatory zones residing in these regions cannot yet be excluded. Among these, of particular interest are two heterozygous deletions affecting intergenic regions of chromosome 11, in the proximity of the centromere, and chromosome 19 (Table 1) [96]. Despite CNVs overlapping, telomeric and centromeric chromosome regions should be treated with more caution because of the lower probe density coverage that makes them more prone to false CNV calls than other regions of the genome, a significant association was found between CNVs in these regions and an increased risk of developing ALS (Table 1).
Several interesting CNV-driven genomic regions encompassing the promoter region of RPS3A, DHX30, UGT8 and CHL1 were also found in ALS patients (Table 1) [138]. RPS3A encodes a ribosomal protein that plays roles in regulating cell growth, transformation and death. DHX30 encodes an RNA helicase that is thought to be an accessory subunit of the mitochondrial ribosome and whose depletion has been associated with alterations in the levels of mitochondrial mRNAs [168, 169]. UGT8 is a protein belonging to a glycosyltransferase family involved in lipid biosynthesis and metabolism, a process implicated in ALS pathogenesis. CHL1 is a neural cell adhesion molecule that plays a crucial role in axonal guidance and maintenance of neural circuits [170].
Other ALS-related CNVs were found in regions of chromosomes 5, 8, and 11 flanking HCN1, POTEA, OR4A5, and OR4C12, respectively (Table 1) [138]. HCN1 encodes a major component of the hyperpolarization-activated cyclic nucleotide-gated channels that seems to enhance hippocampal-dependent learning and memory and modulate synaptic transmission and plasticity [171]. Duplications in POTEA have been already reported in some cases of early onset Alzheimer’s disease [172]. Olfactory receptor genes (e.g., OR4A5 and OR4C12) have been directly or indirectly involved in neuronal injury and nerve regeneration, and their dysregulation was associated with several neurodegenerative conditions, like Alzheimer’s and Parkinson’s diseases [173, 174].
Conclusions
Despite intensive researches and incredible technological advancements, ALS still remains a fatal incurable neurodegenerative disease in which the majority of cases are diagnosed in advanced stages with limited treatment options and poor prognosis. The most important challenges in the clinical management of ALS are the better understanding of causes and mechanisms underlying motor neuron degeneration as well as the identification of precise diagnostic biomarkers and effective therapeutic strategies.
To achieve a comprehensive understanding of the complex genetic-environmental interactions underlying ALS susceptibility, all forms of genetic variation need to be addressed. In addition to the contribution of SNPs, which account for only a limited number of familial and sporadic ALS cases, evidence suggests that other genomic variants, such as CNVs, appear to have a more dramatic impact on human disease phenotype and represent important clues for the deciphering genetic susceptibility to complex diseases, like ALS. However, the traditional single-gene analysis generally accounts for only a small proportion of the phenotypic variation in ALS while is often inadequate for evaluating the collective effects of multiple rare CNVs on disease risk.
The systems biology paradigm represents an innovative way for analyzing the complex underlying biological processes, providing new instruments to fill interstices of the intricate mosaic of ALS pathogenesis and generate a more definite molecular picture of this disease.
Here, we provided some insights into the associations between common and rare CNVs and ALS pathogenesis, focusing on the characterization of the potential CNVs’ impact on different signaling pathways whose deregulation seems to contribute to motor neuron degeneration in ALS. Overall, the use of this integrative genomics approach promises to enlarge our knowledge about genetic and molecular risk factors for ALS and, even in the light of the molecular heterogeneity emerged from our previous work [52], offers an important stepping-stone for the understanding, diagnosis and treatment of complex and multifactorial diseases, like ALS.
We believe that future comprehensive analyses of structural genomic variations together with other high-resolution genotyping data, could help to provide a better definition of the molecular signatures of ALS, laying the basis for a more accurate and precise molecular biomarker-assisted diagnosis and the ultimate development of more effective and personalized therapeutic strategies (Fig. 2).
Cu/Zn superoxide-dismutase gene (SOD1); alsin (ALS2); senataxin (SETX); spastic paraplegia type 11 (SPG11); FUS RNA binding protein (FUS); vesicle-associated protein B (VAPB); angiogenin (ANG); TAR DNA binding protein (TARDBP); FIG4 phosphoinositide 5-phosphatas (FIG4); optineurin (OPTN); ataxin 2 (ATXN2); ubiquilin 2 (UBQLN2); granulin (PGRN); profilin 1 (PFN1); dynactin 1 (DCTN1); chromosome 9 open reading frame 72 (C9ORF72); vascular endothelial growth factor A (VEGFA); FGGY carbohydrate kinase domain containing (FGGY); dipeptidyl-peptidase like 6 (DPP6); inositol 1,4,5-trisphosphate receptor type 2 (ITPR2); kinesin associated protein 3 (KIFAP3); unc-13 homolog A (UNC13A); apoptosis-associated tyrosine kinase (AATK); brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2); charged multivesicular body protein 6 (CHMP6); insulin-like growth factor binding protein acid labile subunit (IGFALS); calcium channel, voltage-dependent, T type, alpha 1H subunit (CACNA1H); mitogen-activated protein kinase 8 interacting protein 3 (MAPK8IP3); BAI1-associated protein 3 (BAIAP3); ubiquitin-conjugating enzyme E2I (UBE2I); splA/ryanodine receptor domain and SOCS box containing 3 (SPSB3); Kv channel interacting protein 4 (KCNIP4); potassium channel, voltage gated Shab related subfamily B; member 2 (KCNB2); potassium channel, voltage gated KQT-like subfamily Q, member 5 (KCNQ5); gasdermin D (GSDMD); glutamate receptor, ionotropic, kainate 1 (GRIK1); glutamate receptor, ionotropic, kainate 2 (GRIK2); eukaryotic translation elongation factor 1 delta (EEF1D); ataxin 1 (ATXN1); ataxin 3-like protein (ATXN3L); solute carrier family 1 member 7 (SLC1A7); triadin (TRDN); complexin-1 (CPLX1); ankyrin repeat domain 11 (ANKRD11); protein phosphatase 1 regulatory subunit 13B (PPP1R13B); forkhead box O3 (FOXO3); hemochromatosis (HFE); gamma-glutamyltransferase light chain 2 (GGTLC2); autophagy related 7 (ATG7); annexin A5 (ANXA5); gem-associated protein 6 (GEMIN6); myotubularin related protein 7 (MTMR7); acylphosphatase 2 (ACYP2); ZFP14 Zinc Finger Protein (ZFP14); formin 2 (FMN2); ribosomal protein S3A (RPS3A); DEAH-box helicase 30 (DHX30); UDP glycosyltransferase 8 (UGT8); cell adhesion molecule L1 like (CHL1); hyperpolarization-activated cyclic nucleotide-gated channel 1 (HCN1); POTE ankyrin domain family member A (POTEA); olfactory receptor family 4 subfamily A member 5 (OR4A5); olfactory receptor family 4 subfamily C member 12 (OR4C12); NADH dehydrogenase subunit 2 (ND2); cytochrome oxidase subunit-3 (CO3); NADH dehydrogenase subunit 4 (ND4).
Electronic supplementary material
(GIF 178 kb)
.
.
(PDF 560 kb)
.
Acknowledgements
This work was supported by the Italian Ministry of Education, Universities, and Research through grant CTN01_00177_817708. The authors gratefully acknowledge Cristina Calì, Alfia Corsino, Maria Patrizia D’Angelo, and Francesco Marino for their administrative and technical assistance.
Authors’ Contributions
Each author contributed parts of the overall text. GM and SC integrated the parts into the final version. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12035-017-0393-x) contains supplementary material, which is available to authorized users.
References
- 1.Statland JM, Barohn RJ, McVey AL, Katz JS, Dimachkie MM. Patterns of weakness, classification of motor neuron disease, and clinical diagnosis of sporadic amyotrophic lateral sclerosis. Neurol Clin. 2015;33(4):735–748. doi: 10.1016/j.ncl.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iguchi Y, Katsuno M, Ikenaka K, Ishigaki S, Sobue G. Amyotrophic lateral sclerosis: an update on recent genetic insights. J Neurol. 2013;260(11):2917–2927. doi: 10.1007/s00415-013-7112-y. [DOI] [PubMed] [Google Scholar]
- 3.Figlewicz DA, Orrell RW. The genetics of motor neuron diseases. Amyotrophic lateral sclerosis and other motor neuron disorders: official publication of the World Federation of Neurology, Research Group on Motor Neuron Diseases. 2003;4(4):225–231. doi: 10.1080/14660820310011287. [DOI] [PubMed] [Google Scholar]
- 4.van Es MA, Van Vught PW, Blauw HM, Franke L, Saris CG, Andersen PM, Van Den Bosch L, de Jong SW, et al. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. The Lancet Neurology. 2007;6(10):869–877. doi: 10.1016/S1474-4422(07)70222-3. [DOI] [PubMed] [Google Scholar]
- 5.Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, Joshipura K, Halperin RF, Stamper C, et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med. 2007;357(8):775–788. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- 6.van Es MA, van Vught PW, Blauw HM, Franke L, Saris CG, Van den Bosch L, de Jong SW, de Jong V, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40(1):29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- 7.van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught PW, Birve A, Lemmens R, Schelhaas HJ, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41(10):1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 8.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39(7 Suppl):S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 10.Auffray C, Chen Z, Hood L. Systems medicine: the future of medical genomics and healthcare. Genome medicine. 2009;1(1):2. doi: 10.1186/gm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurles ME, Dermitzakis ET, Tyler-Smith C. The functional impact of structural variation in humans. Trends in genetics : TIG. 2008;24(5):238–245. doi: 10.1016/j.tig.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 13.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 14.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 15.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16(3):172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 16.Hwang MY, Moon S, Heo L, Kim YJ, Oh JH, Kim YJ, Kim YK, Lee J, et al. Combinatorial approach to estimate copy number genotype using whole-exome sequencing data. Genomics. 2015;105(3):145–149. doi: 10.1016/j.ygeno.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43(11):1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aten E, White SJ, Kalf ME, Vossen RH, Thygesen HH, Ruivenkamp CA, Kriek M, Breuning MH, et al. Methods to detect CNVs in the human genome. Cytogenetic and genome research. 2008;123(1-4):313–321. doi: 10.1159/000184723. [DOI] [PubMed] [Google Scholar]
- 19.Kallioniemi OP, Kallioniemi A, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization: a rapid new method for detecting and mapping DNA amplification in tumors. Semin Cancer Biol. 1993;4(1):41–46. [PubMed] [Google Scholar]
- 20.Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, Pursley AN, Kang SH, Yang Y, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31(12):1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CF, Naj AC, Wang LS (2013) Analyzing copy number variation using SNP array data: protocols for calling CNV and association tests. Current protocols in human genetics/editorial board, Jonathan L Haines [et al.] 79:Unit 1 27. doi:10.1002/0471142905.hg0127s79 [DOI] [PMC free article] [PubMed]
- 22.Abel HJ, Duncavage EJ. Detection of structural DNA variation from next generation sequencing data: a review of informatic approaches. Cancer genetics. 2013;206(12):432–440. doi: 10.1016/j.cancergen.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao Y, Liu X, Harvard C, Nolin SL, Brown WT, Koochek M, Holden JJ, Lewis ME, et al. Large-scale copy number variants (CNVs): distribution in normal subjects and FISH/real-time qPCR analysis. BMC Genomics. 2007;8:167. doi: 10.1186/1471-2164-8-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sellner LN, Taylor GR. MLPA and MAPH: new techniques for detection of gene deletions. Hum Mutat. 2004;23(5):413–419. doi: 10.1002/humu.20035. [DOI] [PubMed] [Google Scholar]
- 25.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464(7289):704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Current opinion in genetics & development. 2012;22(3):211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Przybytkowski E, Ferrario C, Basik M. The use of ultra-dense array CGH analysis for the discovery of micro-copy number alterations and gene fusions in the cancer genome. BMC Med Genet. 2011;4:16. doi: 10.1186/1755-8794-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52(1):103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 30.Buchanan JA, Scherer SW. Contemplating effects of genomic structural variation. Genetics in medicine: official journal of the American College of Medical Genetics. 2008;10(9):639–647. doi: 10.1097/GIM.0b013e318183f848. [DOI] [PubMed] [Google Scholar]
- 31.Pehlivan D, Beck CR, Okamoto Y, Harel T, Akdemir ZH, Jhangiani SN, Withers MA, Goksungur MT, et al. The role of combined SNV and CNV burden in patients with distal symmetric polyneuropathy. Genetics in medicine : official journal of the American College of Medical Genetics. 2016;18(5):443–451. doi: 10.1038/gim.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merikangas AK, Segurado R, Heron EA, Anney RJ, Paterson AD, Cook EH, Pinto D, Scherer SW, et al. The phenotypic manifestations of rare genic CNVs in autism spectrum disorder. Mol Psychiatry. 2015;20(11):1366–1372. doi: 10.1038/mp.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gulsuner S, McClellan JM. Copy number variation in schizophrenia. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2015;40(1):252–254. doi: 10.1038/npp.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mefford HC. Copy number matters in epilepsy. Epilepsy currents/American Epilepsy Society. 2015;15(4):180–182. doi: 10.5698/1535-7511-15.4.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fransen K, Mitrovic M, van Diemen CC, Weersma RK. The quest for genetic risk factors for Crohn’s disease in the post-GWAS era. Genome medicine. 2011;3(2):13. doi: 10.1186/gm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szigeti K, Lal D, Li Y, Doody RS, Wilhelmsen K, Yan L, Liu S, Ma C, et al. Genome-wide scan for copy number variation association with age at onset of Alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2013;33(2):517–523. doi: 10.3233/JAD-2012-121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toft M, Ross OA. Copy number variation in Parkinson’s disease. Genome medicine. 2010;2(9):62. doi: 10.1186/gm183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schymick JC, Talbot K, Traynor BJ (2007) Genetics of sporadic amyotrophic lateral sclerosis. Human molecular genetics 16 Spec No. 2:R233-242. doi:10.1093/hmg/ddm215 [DOI] [PubMed]
- 39.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 40.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298(5599):1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 41.Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J Child Neurol. 2007;22(8):990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 42.Butchbach ME. Copy number variations in the survival motor neuron genes: implications for spinal muscular atrophy and other neurodegenerative diseases. Front Mol Biosci. 2016;3:7. doi: 10.3389/fmolb.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corcia P, Camu W, Praline J, Gordon PH, Vourch P, Andres C. The importance of the SMN genes in the genetics of sporadic ALS. Amyotrophic lateral sclerosis: official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases. 2009;10(5-6):436–440. doi: 10.3109/17482960902759162. [DOI] [PubMed] [Google Scholar]
- 44.Veldink JH, van den Berg LH, Cobben JM, Stulp RP, De Jong JM, Vogels OJ, Baas F, Wokke JH, et al. Homozygous deletion of the survival motor neuron 2 gene is a prognostic factor in sporadic ALS. Neurology. 2001;56(6):749–752. doi: 10.1212/WNL.56.6.749. [DOI] [PubMed] [Google Scholar]
- 45.Corcia P, Mayeux-Portas V, Khoris J, de Toffol B, Autret A, Muh JP, Camu W, Andres C, et al. Abnormal SMN1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann Neurol. 2002;51(2):243–246. doi: 10.1002/ana.10104. [DOI] [PubMed] [Google Scholar]
- 46.Veldink JH, Kalmijn S, Van der Hout AH, Lemmink HH, Groeneveld GJ, Lummen C, Scheffer H, Wokke JH, et al. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology. 2005;65(6):820–825. doi: 10.1212/01.wnl.0000174472.03292.dd. [DOI] [PubMed] [Google Scholar]
- 47.Corcia P, Camu W, Halimi JM, Vourc’h P, Antar C, Vedrine S, Giraudeau B, de Toffol B, et al. SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology. 2006;67(7):1147–1150. doi: 10.1212/01.wnl.0000233830.85206.1e. [DOI] [PubMed] [Google Scholar]
- 48.Wang XB, Cui NH, Gao JJ, Qiu XP, Zheng F. SMN1 duplications contribute to sporadic amyotrophic lateral sclerosis susceptibility: evidence from a meta-analysis. J Neurol Sci. 2014;340(1-2):63–68. doi: 10.1016/j.jns.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 49.Crawford TO. Skolasky RL., Jr The relationship of SMN to amyotrophic lateral sclerosis. Ann Neurol. 2002;52(6):857–858. doi: 10.1002/ana.10378. [DOI] [PubMed] [Google Scholar]
- 50.Gamez J, Barcelo MJ, Munoz X, Carmona F, Cusco I, Baiget M, Cervera C, Tizzano EF. Survival and respiratory decline are not related to homozygous SMN2 deletions in ALS patients. Neurology. 2002;59(9):1456–1460. doi: 10.1212/01.WNL.0000032496.64510.4E. [DOI] [PubMed] [Google Scholar]
- 51.Corcia P, Ingre C, Blasco H, Press R, Praline J, Antar C, Veyrat-Durebex C, et al. Homozygous SMN2 deletion is a protective factor in the Swedish ALS population. European Journal of Human Genetics: EJHG. 2012;20(5):588–591. doi: 10.1038/ejhg.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aronica E, Baas F, Iyer A, ten Asbroek AL, Morello G, Cavallaro S. Molecular classification of amyotrophic lateral sclerosis by unsupervised clustering of gene expression in motor cortex. Neurobiol Dis. 2015;74:359–376. doi: 10.1016/j.nbd.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Blauw HM, Barnes CP, van Vught PW, van Rheenen W, Verheul M, Cuppen E, Veldink JH, van den Berg LH. SMN1 gene duplications are associated with sporadic ALS. Neurology. 2012;78(11):776–780. doi: 10.1212/WNL.0b013e318249f697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou T, Ilangovan R, Yu F, Xu Z, Zhou J. SMN protects cells against mutant SOD1 toxicity by increasing chaperone activity. Biochem Biophys Res Commun. 2007;364(4):850–855. doi: 10.1016/j.bbrc.2007.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner BJ, Parkinson NJ, Davies KE, Talbot K. Survival motor neuron deficiency enhances progression in an amyotrophic lateral sclerosis mouse model. Neurobiol Dis. 2009;34(3):511–517. doi: 10.1016/j.nbd.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Turner BJ, Alfazema N, Sheean RK, Sleigh JN, Davies KE, Horne MK, Talbot K. Overexpression of survival motor neuron improves neuromuscular function and motor neuron survival in mutant SOD1 mice. Neurobiol Aging. 2014;35(4):906–915. doi: 10.1016/j.neurobiolaging.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerbino V, Carri MT, Cozzolino M, Achsel T. Mislocalised FUS mutants stall spliceosomal snRNPs in the cytoplasm. Neurobiol Dis. 2013;55:120–128. doi: 10.1016/j.nbd.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 58.Yu Y, Chi B, Xia W, Gangopadhyay J, Yamazaki T, Winkelbauer-Hurt ME, Yin S, Eliasse Y, et al. U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish. Nucleic Acids Res. 2015;43(6):3208–3218. doi: 10.1093/nar/gkv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groen EJ, Fumoto K, Blokhuis AM, Engelen-Lee J, Zhou Y, van den Heuvel DM, Koppers M, van Diggelen F, et al. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum Mol Genet. 2013;22(18):3690–3704. doi: 10.1093/hmg/ddt222. [DOI] [PubMed] [Google Scholar]
- 60.Tremolizzo L, Sala G, Conti E, Rodriguez-Menendez V, Fogli A, Michelucci A, Simi P, Penco S, et al. Valproate treatment in an ALS patient carrying a c.194G > A spastin mutation and SMN2 homozygous deletion. Case reports in neurological medicine. 2014;2014:216094. doi: 10.1155/2014/216094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kunst CB, Messer L, Gordon J, Haines J, Patterson D. Genetic mapping of a mouse modifier gene that can prevent ALS onset. Genomics. 2000;70(2):181–189. doi: 10.1006/geno.2000.6379. [DOI] [PubMed] [Google Scholar]
- 62.Petzold A, Keir G, Warren J, Fox N, Rossor MN. A systematic review and meta-analysis of CSF neurofilament protein levels as biomarkers in dementia. Neurodegener Dis. 2007;4(2-3):185–194. doi: 10.1159/000101843. [DOI] [PubMed] [Google Scholar]
- 63.Garcia ML, Singleton AB, Hernandez D, Ward CM, Evey C, Sapp PA, Hardy J, Brown RH, Jr, et al. Mutations in neurofilament genes are not a significant primary cause of non-SOD1-mediated amyotrophic lateral sclerosis. Neurobiol Dis. 2006;21(1):102–109. doi: 10.1016/j.nbd.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 64.Tu PH, Gurney ME, Julien JP, Lee VM, Trojanowski JQ. Oxidative stress, mutant SOD1, and neurofilament pathology in transgenic mouse models of human motor neuron disease. Laboratory investigation; a journal of technical methods and pathology. 1997;76(4):441–456. [PubMed] [Google Scholar]
- 65.Delisle MB, Carpenter S. Neurofibrillary axonal swellings and amyotrophic lateral sclerosis. J Neurol Sci. 1984;63(2):241–250. doi: 10.1016/0022-510X(84)90199-0. [DOI] [PubMed] [Google Scholar]
- 66.Leigh PN, Dodson A, Swash M, Brion JP, Anderton BH. Cytoskeletal abnormalities in motor neuron disease. An immunocytochemical study. Brain : a journal of neurology. 1989;112(Pt 2):521–535. doi: 10.1093/brain/112.2.521. [DOI] [PubMed] [Google Scholar]
- 67.Williamson TL, Bruijn LI, Zhu Q, Anderson KL, Anderson SD, Julien JP, Cleveland DW. Absence of neurofilaments reduces the selective vulnerability of motor neurons and slows disease caused by a familial amyotrophic lateral sclerosis-linked superoxide dismutase 1 mutant. Proc Natl Acad Sci U S A. 1998;95(16):9631–9636. doi: 10.1073/pnas.95.16.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, Shaw CE, Powell JF, et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet. 1999;8(2):157–164. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- 69.Lees JF, Shneidman PS, Skuntz SF, Carden MJ, Lazzarini RA. The structure and organization of the human heavy neurofilament subunit (NF-H) and the gene encoding it. EMBO J. 1988;7(7):1947–1955. doi: 10.1002/j.1460-2075.1988.tb03032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Figlewicz DA, Krizus A, Martinoli MG, Meininger V, Dib M, Rouleau GA, Julien JP. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet. 1994;3(10):1757–1761. doi: 10.1093/hmg/3.10.1757. [DOI] [PubMed] [Google Scholar]
- 71.Rooke K, Figlewicz DA, Han FY, Rouleau GA. Analysis of the KSP repeat of the neurofilament heavy subunit in familiar amyotrophic lateral sclerosis. Neurology. 1996;46(3):789–790. doi: 10.1212/WNL.46.3.789. [DOI] [PubMed] [Google Scholar]
- 72.Huang R, Guo X, Chen X, Zheng Z, Wei Q, Cao B, Zeng Y, Shang H. The serum lipid profiles of amyotrophic lateral sclerosis patients: a study from south-west China and a meta-analysis. Amyotrophic lateral sclerosis & frontotemporal degeneration. 2015;16(5-6):359–365. doi: 10.3109/21678421.2015.1047454. [DOI] [PubMed] [Google Scholar]
- 73.Kim SM, Kim H, Kim JE, Park KS, Sung JJ, Kim SH, Lee KW. Amyotrophic lateral sclerosis is associated with hypolipidemia at the presymptomatic stage in mice. PLoS One. 2011;6(3):e17985. doi: 10.1371/journal.pone.0017985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhai J, Strom AL, Kilty R, Venkatakrishnan P, White J, Everson WV, Smart EJ, Zhu H. Proteomic characterization of lipid raft proteins in amyotrophic lateral sclerosis mouse spinal cord. FEBS J. 2009;276(12):3308–3323. doi: 10.1111/j.1742-4658.2009.07057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dupuis L, Corcia P, Fergani A, Gonzalez De Aguilar JL, Bonnefont-Rousselot D, Bittar R, Seilhean D, Hauw JJ, et al. Dyslipidemia is a protective factor in amyotrophic lateral sclerosis. Neurology. 2008;70(13):1004–1009. doi: 10.1212/01.wnl.0000285080.70324.27. [DOI] [PubMed] [Google Scholar]
- 76.Nakamura K, Mori F, Tanji K, Miki Y, Yamada M, Kakita A, Takahashi H, Utsumi J, et al. Isopentenyl diphosphate isomerase, a cholesterol synthesizing enzyme, is localized in Lewy bodies. Neuropathology: official journal of the Japanese Society of Neuropathology. 2015;35(5):432–440. doi: 10.1111/neup.12204. [DOI] [PubMed] [Google Scholar]
- 77.Ramos-Valdivia AC, van der Heijden R, Verpoorte R. Isopentenyl diphosphate isomerase: a core enzyme in isoprenoid biosynthesis. A review of its biochemistry and function. Nat Prod Rep. 1997;14(6):591–603. doi: 10.1039/np9971400591. [DOI] [PubMed] [Google Scholar]
- 78.Kato T, Emi M, Sato H, Arawaka S, Wada M, Kawanami T, Katagiri T, Tsuburaya K, et al. Segmental copy-number gain within the region of isopentenyl diphosphate isomerase genes in sporadic amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2010;402(2):438–442. doi: 10.1016/j.bbrc.2010.10.056. [DOI] [PubMed] [Google Scholar]
- 79.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77(1):78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cronin S, Berger S, Ding J, Schymick JC, Washecka N, Hernandez DG, Greenway MJ, Bradley DG, et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet. 2008;17(5):768–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- 81.Garber K. Genetics. The elusive ALS genes. Science. 2008;319(5859):20. doi: 10.1126/science.319.5859.20. [DOI] [PubMed] [Google Scholar]
- 82.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Del Bo R, Ghezzi S, Corti S, Santoro D, Prelle A, Mancuso M, Siciliano G, Briani C, et al. DPP6 gene variability confers increased risk of developing sporadic amyotrophic lateral sclerosis in Italian patients. J Neurol Neurosurg Psychiatry. 2008;79(9):1085. doi: 10.1136/jnnp.2008.149146. [DOI] [PubMed] [Google Scholar]
- 84.Blauw HM, Al-Chalabi A, Andersen PM, van Vught PW, Diekstra FP, van Es MA, Saris CG, Groen EJ, et al. A large genome scan for rare CNVs in amyotrophic lateral sclerosis. Hum Mol Genet. 2010;19(20):4091–4099. doi: 10.1093/hmg/ddq323. [DOI] [PubMed] [Google Scholar]
- 85.Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79(2):275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am J Hum Genet. 2003;73(4):967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen S, Sayana P, Zhang X, Le W. Genetics of amyotrophic lateral sclerosis: an update. Mol Neurodegener. 2013;8:28. doi: 10.1186/1750-1326-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blauw HM, van Rheenen W, Koppers M, Van Damme P, Waibel S, Lemmens R, van Vught PW, Meyer T, et al. NIPA1 polyalanine repeat expansions are associated with amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21(11):2497–2502. doi: 10.1093/hmg/dds064. [DOI] [PubMed] [Google Scholar]
- 89.Cox DM, Butler MG. The 15q11.2 BP1-BP2 microdeletion syndrome: a review. Int J Mol Sci. 2015;16(2):4068–4082. doi: 10.3390/ijms16024068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huot J. Ephrin signaling in axon guidance. Prog Neuro-Psychopharmacol Biol Psychiatry. 2004;28(5):813–818. doi: 10.1016/j.pnpbp.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 91.Klein R. Eph/ephrin signaling in morphogenesis, neural development and plasticity. Curr Opin Cell Biol. 2004;16(5):580–589. doi: 10.1016/j.ceb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 92.Schmidt ER, Pasterkamp RJ, van den Berg LH. Axon guidance proteins: novel therapeutic targets for ALS? Prog Neurobiol. 2009;88(4):286–301. doi: 10.1016/j.pneurobio.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 93.Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57(2):236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 94.Lesnick TG, Sorenson EJ, Ahlskog JE, Henley JR, Shehadeh L, Papapetropoulos S, Maraganore DM. Beyond Parkinson disease: amyotrophic lateral sclerosis and the axon guidance pathway. PLoS One. 2008;3(1):e1449. doi: 10.1371/journal.pone.0001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Hoecke A, Schoonaert L, Lemmens R, Timmers M, Staats KA, Laird AS, Peeters E, Philips T, et al. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat Med. 2012;18(9):1418–1422. doi: 10.1038/nm.2901. [DOI] [PubMed] [Google Scholar]
- 96.Uyan O, Omur O, Agim ZS, Ozoguz A, Li H, Parman Y, Deymeer F, Oflazer P, et al. Genome-wide copy number variation in sporadic amyotrophic lateral sclerosis in the Turkish population: deletion of EPHA3 is a possible protective factor. PLoS One. 2013;8(8):e72381. doi: 10.1371/journal.pone.0072381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee J, Boo JH, Ryu H. The failure of mitochondria leads to neurodegeneration: do mitochondria need a jump start? Adv Drug Deliv Rev. 2009;61(14):1316–1323. doi: 10.1016/j.addr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moreira PI, Zhu X, Wang X, Lee HG, Nunomura A, Petersen RB, Perry G, Smith MA. Mitochondria: a therapeutic target in neurodegeneration. Biochim Biophys Acta. 2010;1802(1):212–220. doi: 10.1016/j.bbadis.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Borthwick GM, Johnson MA, Ince PG, Shaw PJ, Turnbull DM. Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Ann Neurol. 1999;46(5):787–790. doi: 10.1002/1531-8249(199911)46:5<787::AID-ANA17>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 100.Capaldi RA. Structure and assembly of cytochrome c oxidase. Arch Biochem Biophys. 1990;280(2):252–262. doi: 10.1016/0003-9861(90)90327-U. [DOI] [PubMed] [Google Scholar]
- 101.Comi GP, Bordoni A, Salani S, Franceschina L, Sciacco M, Prelle A, Fortunato F, Zeviani M, et al. Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann Neurol. 1998;43(1):110–116. doi: 10.1002/ana.410430119. [DOI] [PubMed] [Google Scholar]
- 102.Keeney PM, Bennett JP., Jr ALS spinal neurons show varied and reduced mtDNA gene copy numbers and increased mtDNA gene deletions. Mol Neurodegener. 2010;5:21. doi: 10.1186/1750-1326-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rahman S, Taanman JW, Cooper JM, Nelson I, Hargreaves I, Meunier B, Hanna MG, Garcia JJ, et al. A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am J Hum Genet. 1999;65(4):1030–1039. doi: 10.1086/302590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang H, Brosel S, Acin-Perez R, Slavkovich V, Nishino I, Khan R, Goldberg IJ, Graziano J, et al. Analysis of mouse models of cytochrome c oxidase deficiency owing to mutations in Sco2. Hum Mol Genet. 2010;19(1):170–180. doi: 10.1093/hmg/ddp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38(5):515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 106.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 107.Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, et al. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40(3):275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 108.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18(6):1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keeney PM, Quigley CK, Dunham LD, Papageorge CM, Iyer S, Thomas RR, Schwarz KM, Trimmer PA, et al. Mitochondrial gene therapy augments mitochondrial physiology in a Parkinson’s disease cell model. Hum Gene Ther. 2009;20(8):897–907. doi: 10.1089/hum.2009.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wain LV, Pedroso I, Landers JE, Breen G, Shaw CE, Leigh PN, Brown RH, Tobin MD, et al. The role of copy number variation in susceptibility to amyotrophic lateral sclerosis: genome-wide association study and comparison with published loci. PLoS One. 2009;4(12):e8175. doi: 10.1371/journal.pone.0008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pamphlett R, Morahan JM, Luquin N, Yu B. Looking for differences in copy number between blood and brain in sporadic amyotrophic lateral sclerosis. Muscle Nerve. 2011;44(4):492–498. doi: 10.1002/mus.22095. [DOI] [PubMed] [Google Scholar]
- 112.Zhu M, Zhao S. Candidate gene identification approach: progress and challenges. Int J Biol Sci. 2007;3(7):420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Knight RA, Verkhratsky A. Neurodegenerative diseases: failures in brain connectivity? Cell Death Differ. 2010;17(7):1069–1070. doi: 10.1038/cdd.2010.23. [DOI] [PubMed] [Google Scholar]
- 114.Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298(5594):789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 115.Calabresi P, Picconi B, Parnetti L, Di Filippo M. A convergent model for cognitive dysfunctions in Parkinson’s disease: the critical dopamine-acetylcholine synaptic balance. The Lancet Neurology. 2006;5(11):974–983. doi: 10.1016/S1474-4422(06)70600-7. [DOI] [PubMed] [Google Scholar]
- 116.Maselli RA, Wollman RL, Leung C, Distad B, Palombi S, Richman DP, Salazar-Grueso EF, Roos RP. Neuromuscular transmission in amyotrophic lateral sclerosis. Muscle Nerve. 1993;16(11):1193–1203. doi: 10.1002/mus.880161109. [DOI] [PubMed] [Google Scholar]
- 117.Cronin S, Blauw HM, Veldink JH, van Es MA, Ophoff RA, Bradley DG, van den Berg LH, Hardiman O. Analysis of genome-wide copy number variation in Irish and Dutch ALS populations. Hum Mol Genet. 2008;17(21):3392–3398. doi: 10.1093/hmg/ddn233. [DOI] [PubMed] [Google Scholar]
- 118.Jaiswal MK. Calcium, mitochondria, and the pathogenesis of ALS: the good, the bad, and the ugly. Front Cell Neurosci. 2013;7:199. doi: 10.3389/fncel.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nwosu VK, Royer JA, Stickler DE. Voltage gated potassium channel antibodies in amyotrophic lateral sclerosis. Amyotrophic lateral sclerosis : official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases. 2010;11(4):392–394. doi: 10.3109/17482960903452283. [DOI] [PubMed] [Google Scholar]
- 120.Foran E, Trotti D. Glutamate transporters and the excitotoxic path to motor neuron degeneration in amyotrophic lateral sclerosis. Antioxid Redox Signal. 2009;11(7):1587–1602. doi: 10.1089/ars.2009.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Archiv : European journal of physiology. 2004;447(5):469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- 122.Conforti FL, Spataro R, Sproviero W, Mazzei R, Cavalcanti F, Condino F, Simone IL, Logroscino G, et al. Ataxin-1 and ataxin-2 intermediate-length PolyQ expansions in amyotrophic lateral sclerosis. Neurology. 2012;79(24):2315–2320. doi: 10.1212/WNL.0b013e318278b618. [DOI] [PubMed] [Google Scholar]
- 123.Cvetanovic M, Patel JM, Marti HH, Kini AR, Opal P. Vascular endothelial growth factor ameliorates the ataxic phenotype in a mouse model of spinocerebellar ataxia type 1. Nat Med. 2011;17(11):1445–1447. doi: 10.1038/nm.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Luksys G, Ackermann S, Coynel D, Fastenrath M, Gschwind L, Heck A, Rasch B, Spalek K, et al. BAIAP2 is related to emotional modulation of human memory strength. PLoS One. 2014;9(1):e83707. doi: 10.1371/journal.pone.0083707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Milanese M, Bonifacino T, Fedele E, Rebosio C, Cattaneo L, Benfenati F, Usai C, Bonanno G. Exocytosis regulates trafficking of GABA and glycine heterotransporters in spinal cord glutamatergic synapses: a mechanism for the excessive heterotransporter-induced release of glutamate in experimental amyotrophic lateral sclerosis. Neurobiol Dis. 2015;74:314–324. doi: 10.1016/j.nbd.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 126.Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20(16):2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- 127.Lazo-Gomez R, Ramirez-Jarquin UN, Tovar YRLB, Tapia R. Histone deacetylases and their role in motor neuron degeneration. Front Cell Neurosci. 2013;7:243. doi: 10.3389/fncel.2013.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Feng W, Gubitz AK, Wan L, Battle DJ, Dostie J, Golembe TJ, Dreyfuss G. Gemins modulate the expression and activity of the SMN complex. Hum Mol Genet. 2005;14(12):1605–1611. doi: 10.1093/hmg/ddi168. [DOI] [PubMed] [Google Scholar]
- 129.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140(6):918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McCombe PA, Henderson RD. The role of immune and inflammatory mechanisms in ALS. Curr Mol Med. 2011;11(3):246–254. doi: 10.2174/156652411795243450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goodall EF, Greenway MJ, van Marion I, Carroll CB, Hardiman O, Morrison KE. Association of the H63D polymorphism in the hemochromatosis gene with sporadic ALS. Neurology. 2005;65(6):934–937. doi: 10.1212/01.wnl.0000176032.94434.d4. [DOI] [PubMed] [Google Scholar]
- 132.Praline J, Blasco H, Vourc’h P, Rat V, Gendrot C, Camu W, Andres CR, French ALSSG. Study of the HFE gene common polymorphisms in French patients with sporadic amyotrophic lateral sclerosis. J Neurol Sci. 2012;317(1-2):58–61. doi: 10.1016/j.jns.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 133.Carri MT, Ferri A, Casciati A, Celsi F, Ciriolo MR, Rotilio G. Copper-dependent oxidative stress, alteration of signal transduction and neurodegeneration in amyotrophic lateral sclerosis. Funct Neurol. 2001;16(4 Suppl):181–188. [PubMed] [Google Scholar]
- 134.Gerschutz A, Heinsen H, Grunblatt E, Wagner AK, Bartl J, Meissner C, Fallgatter AJ, Al-Sarraj S, et al. Neuron-specific alterations in signal transduction pathways associated with Alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2014;40(1):135–142. doi: 10.3233/JAD-131280. [DOI] [PubMed] [Google Scholar]
- 135.Chung YH, Joo KM, Shin CM, Lee YJ, Shin DH, Lee KH, Cha CI. Immunohistochemical study on the distribution of insulin-like growth factor I (IGF-I) receptor in the central nervous system of SOD1(G93A) mutant transgenic mice. Brain Res. 2003;994(2):253–259. doi: 10.1016/j.brainres.2003.09.047. [DOI] [PubMed] [Google Scholar]
- 136.Renna M, Bento CF, Fleming A, Menzies FM, Siddiqi FH, Ravikumar B, Puri C, Garcia-Arencibia M, et al. IGF-1 receptor antagonism inhibits autophagy. Hum Mol Genet. 2013;22(22):4528–4544. doi: 10.1093/hmg/ddt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang Q, Figueiredo-Pereira ME. Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis: an international journal on programmed cell death. 2010;15(11):1292–1311. doi: 10.1007/s10495-010-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pamphlett R, Morahan JM, Yu B. Using case-parent trios to look for rare de novo genetic variants in adult-onset neurodegenerative diseases. J Neurosci Methods. 2011;197(2):297–301. doi: 10.1016/j.jneumeth.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 139.Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M, et al. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. The Lancet Neurology. 2007;6(4):322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- 140.Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 141.Ngo ST, Steyn FJ. The interplay between metabolic homeostasis and neurodegeneration: insights into the neurometabolic nature of amyotrophic lateral sclerosis. Cell regeneration. 2015;4(1):5. doi: 10.1186/s13619-015-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guo W, Shang F, Liu Q, Urim L, Zhang M, Taylor A. Ubiquitin-proteasome pathway function is required for lens cell proliferation and differentiation. Invest Ophthalmol Vis Sci. 2006;47(6):2569–2575. doi: 10.1167/iovs.05-0261. [DOI] [PubMed] [Google Scholar]
- 143.Serpente M, Fenoglio C, Cioffi SM, Bonsi R, Arighi A, Fumagalli GG, Ghezzi L, Scarpini E, et al. Profiling of ubiquitination pathway genes in peripheral cells from patients with frontotemporal dementia due to C9ORF72 and GRN mutations. Int J Mol Sci. 2015;16(1):1385–1394. doi: 10.3390/ijms16011385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Oliveira GP, Alves CJ, Chadi G. Early gene expression changes in spinal cord from SOD1(G93A) amyotrophic lateral sclerosis animal model. Front Cell Neurosci. 2013;7:216. doi: 10.3389/fncel.2013.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Blauw HM, Veldink JH, van Es MA, van Vught PW, Saris CG, van der Zwaag B, Franke L, Burbach JP, et al. Copy-number variation in sporadic amyotrophic lateral sclerosis: a genome-wide screen. The Lancet Neurology. 2008;7(4):319–326. doi: 10.1016/S1474-4422(08)70048-6. [DOI] [PubMed] [Google Scholar]
- 146.Mochizuki Y, Majerus PW. Characterization of myotubularin-related protein 7 and its binding partner, myotubularin-related protein 9. Proc Natl Acad Sci U S A. 2003;100(17):9768–9773. doi: 10.1073/pnas.1333958100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.D’Alessandro G, Calcagno E, Tartari S, Rizzardini M, Invernizzi RW, Cantoni L. Glutamate and glutathione interplay in a motor neuronal model of amyotrophic lateral sclerosis reveals altered energy metabolism. Neurobiol Dis. 2011;43(2):346–355. doi: 10.1016/j.nbd.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 148.Weiduschat N, Mao X, Hupf J, Armstrong N, Kang G, Lange DJ, Mitsumoto H, Shungu DC. Motor cortex glutathione deficit in ALS measured in vivo with the J-editing technique. Neurosci Lett. 2014;570:102–107. doi: 10.1016/j.neulet.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 149.Sathasivam S, Ince PG, Shaw PJ. Apoptosis in amyotrophic lateral sclerosis: a review of the evidence. Neuropathol Appl Neurobiol. 2001;27(4):257–274. doi: 10.1046/j.0305-1846.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 150.Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104(36):14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Han S, Zhang KH, Lu PH, Xu XM. Effects of annexins II and V on survival of neurons and astrocytes in vitro. Acta Pharmacol Sin. 2004;25(5):602–610. [PubMed] [Google Scholar]
- 152.Kasperaviciute D, Weale ME, Shianna KV, Banks GT, Simpson CL, Hansen VK, Turner MR, Shaw CE, et al. Large-scale pathways-based association study in amyotrophic lateral sclerosis. Brain: a journal of neurology. 2007;130(Pt 9):2292–2301. doi: 10.1093/brain/awm055. [DOI] [PubMed] [Google Scholar]
- 153.Liu ZJ, Lu X, Zhong S. ASPP—apoptotic specific regulator of p53. Biochim Biophys Acta. 2005;1756(1):77–80. doi: 10.1016/j.bbcan.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 154.Willemsen MH, Fernandez BA, Bacino CA, Gerkes E, de Brouwer AP, Pfundt R, Sikkema-Raddatz B, Scherer SW, et al. Identification of ANKRD11 and ZNF778 as candidate genes for autism and variable cognitive impairment in the novel 16q24.3 microdeletion syndrome. European journal of human genetics : EJHG. 2010;18(4):429–435. doi: 10.1038/ejhg.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ranganathan S, Bowser R. p53 and cell cycle proteins participate in spinal motor neuron cell death in ALS. The open pathology journal. 2010;4:11–22. doi: 10.2174/1874375701004010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tomomura M, Fernandez-Gonzales A, Yano R, Yuzaki M. Characterization of the apoptosis-associated tyrosine kinase (AATYK) expressed in the CNS. Oncogene. 2001;20(9):1022–1032. doi: 10.1038/sj.onc.1204210. [DOI] [PubMed] [Google Scholar]
- 157.Tovar YRLB, Ramirez-Jarquin UN, Lazo-Gomez R, Tapia R. Trophic factors as modulators of motor neuron physiology and survival: implications for ALS therapy. Front Cell Neurosci. 2014;8:61. doi: 10.3389/fncel.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Achour M, Le Gras S, Keime C, Parmentier F, Lejeune FX, Boutillier AL, Neri C, Davidson I, et al. Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington’s disease mice. Hum Mol Genet. 2015;24(12):3481–3496. doi: 10.1093/hmg/ddv099. [DOI] [PubMed] [Google Scholar]
- 159.Mehler MF, Gokhan S. Mechanisms underlying neural cell death in neurodegenerative diseases: alterations of a developmentally-mediated cellular rheostat. Trends Neurosci. 2000;23(12):599–605. doi: 10.1016/S0166-2236(00)01705-7. [DOI] [PubMed] [Google Scholar]
- 160.Gallagher D, Voronova A, Zander MA, Cancino GI, Bramall A, Krause MP, Abad C, Tekin M, et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev Cell. 2015;32(1):31–42. doi: 10.1016/j.devcel.2014.11.031. [DOI] [PubMed] [Google Scholar]
- 161.Kelkar N, Gupta S, Dickens M, Davis RJ. Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol Cell Biol. 2000;20(3):1030–1043. doi: 10.1128/MCB.20.3.1030-1043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hallock P, Thomas MA. Integrating the Alzheimer’s disease proteome and transcriptome: a comprehensive network model of a complex disease. Omics: a journal of integrative biology. 2012;16(1-2):37–49. doi: 10.1089/omi.2011.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ghosh AS, Wang B, Pozniak CD, Chen M, Watts RJ, Lewcock JW. DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J Cell Biol. 2011;194(5):751–764. doi: 10.1083/jcb.201103153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Soo KY, Farg M, Atkin JD. Molecular motor proteins and amyotrophic lateral sclerosis. Int J Mol Sci. 2011;12(12):9057–9082. doi: 10.3390/ijms12129057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shoichet SA, Waibel S, Endruhn S, Sperfeld AD, Vorwerk B, Muller I, Erdogan F, Ludolph AC, et al. Identification of candidate genes for sporadic amyotrophic lateral sclerosis by array comparative genomic hybridization. Amyotrophic lateral sclerosis: official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases. 2009;10(3):162–169. doi: 10.1080/17482960802535001. [DOI] [PubMed] [Google Scholar]
- 166.Marty I. Triadin: a multi-protein family for which purpose? Cellular and molecular life sciences : CMLS. 2004;61(15):1850–1853. doi: 10.1007/s00018-004-4196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cox LE, Ferraiuolo L, Goodall EF, Heath PR, Higginbottom A, Mortiboys H, Hollinger HC, Hartley JA, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS) PLoS One. 2010;5(3):e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Naora H. Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol. 1999;77(3):197–205. doi: 10.1046/j.1440-1711.1999.00816.x. [DOI] [PubMed] [Google Scholar]
- 169.Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 170.Zhou L, Barao S, Laga M, Bockstael K, Borgers M, Gijsen H, Annaert W, Moechars D, et al. The neural cell adhesion molecules L1 and CHL1 are cleaved by BACE1 protease in vivo. J Biol Chem. 2012;287(31):25927–25940. doi: 10.1074/jbc.M112.377465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nolan MF, Malleret G, Dudman JT, Buhl DL, Santoro B, Gibbs E, Vronskaya S, Buzsaki G, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119(5):719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 172.Wiseman FK, Al-Janabi T, Hardy J, Karmiloff-Smith A, Nizetic D, Tybulewicz VL, Fisher EM, Strydom A. A genetic cause of Alzheimer disease: mechanistic insights from down syndrome. Nat Rev Neurosci. 2015;16(9):564–574. doi: 10.1038/nrn3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ansoleaga B, Garcia-Esparcia P, Llorens F, Moreno J, Aso E, Ferrer I. Dysregulation of brain olfactory and taste receptors in AD, PSP and CJD, and AD-related model. Neuroscience. 2013;248:369–382. doi: 10.1016/j.neuroscience.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 174.Garcia-Esparcia P, Schluter A, Carmona M, Moreno J, Ansoleaga B, Torrejon-Escribano B, Gustincich S, Pujol A, et al. Functional genomics reveals dysregulation of cortical olfactory receptors in Parkinson disease: novel putative chemoreceptors in the human brain. J Neuropathol Exp Neurol. 2013;72(6):524–539. doi: 10.1097/NEN.0b013e318294fd76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(GIF 178 kb)
(PDF 560 kb)