Abstract
Background
We aimed to investigate the effect and mechanism of curcumin (CUR) in Alzheimer's disease (AD).
Methods
Mouse hippocampal neuronal cell line HT-22 was treated with Aβ1–42 and/or CUR, and then cell viability was evaluated by cell counting kit 8, Beclin-l level was detected using western blotting, and the formation of autophagosomes was observed by transmission electron microscopy (TEM). Furthermore, transcriptome sequencing and analysis were performed in cells with Aβ1–42 alone or Aβ1–42 + CUR.
Results
Aβ1–42 treatment significantly inhibited cell viability compared with untreated cells (P < 0.01). After treatment for 48 h, CUR remarkably promoted cell viability compared with cell treated with Aβ1–42 alone (P < 0.01). Compared with cells treated with Aβ1–42 alone, the expression of Beclin-1 was slightly reduced in cells with combined treatment of Aβ1–42 with CUR (P < 0.05). Consistently, TEM results showed that CUR inhibited the formation of autophagosomes in cells treated with Aβ1–42. Furthermore, the protein-protein interaction network showed five key genes, including MYC, Cdh1, Acaca, Egr1, and CCnd1, likely involved in CUR effects.
Conclusions
CUR might have a potential neuroprotective effect by promoting cell viability in AD, which might be associated with cell autophagy. Furthermore, MYC, Cdh1, and Acaca might be involved in the progression of AD.
1. Introduction
Alzheimer's disease (AD), a progressive neurodegenerative disease, is most common type in senile dementia [1]. The morbidity of AD is increasing with the aging population, which endangers physical, psychological, and living quality of old people due to high fatality rate and disability rate [2]. Although a significant progress has been obtained in the pathogenesis of AD, the effective treatments to block the development of AD are unsatisfactory. Therefore, it is urgent to explore the pathogenesis of AD in depth and search for new therapeutic targets and drugs for AD.
Curcumin (CUR) is major polyphenol extracted from the rhizome of curry spice turmeric and is widespread traditional medicine in South and Southeast Asia [3]. Increasing evidences have demonstrated that CUR has the beneficial properties such as antitumor, antioxidant and anti-inflammatory [4–6]. Epidemiological studies have reported that the lowest prevalence rate of AD is found in India, which may be associated with common eating curry spice in India population [7, 8]. Previous study also has revealed that curry consumption is related to better cognitive functions in old people [9]. Several studies have further shown that CUR can significantly improve cognitive functions by reducing oxidative damage and inflammation and then inhibiting amyloid-β-protein (Aβ, especially Aβ-42) aggregation in the experimental AD models [10–12].
It has been well-known that Aβ is a main marker protein in the development of AD [13] and intracellular Aβ-42 aggregation is proved to play a key role in the early stage of AD [14]. Abnormal autophagy can lead to early neuropathic damage in AD [15], which involves the secretion of Aβ [16]. In the early stage of AD, autophagy can eliminate abnormal protein Aβ and has a neuroprotective effect in AD, while continued Aβ aggregation induces dysfunction of lysosomal degradation, which leads to the leakage of lysosomal proteins from autophagic vacuoles and the acidification of cytosol, eventually resulting in neuronal death in the late stage of AD [17]. Wang et al. [18] have reported that CUR induces autophagy by downregulating phosphoinositide 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) signaling pathway and inhibiting the production of Aβ in APP/PS1 double transgenic mice. However, further studies are still necessary to investigate the underlying mechanisms of CUR in AD.
In the present study, mouse hippocampal neuronal cell line HT-22 was treated with Aβ1–42 and/or CUR, and then the cell viability, the expression of autophagy-related protein Beclin-l, and the formation of autophagosomes were detected in HT-22 cells. Furthermore, transcriptome sequencing was performed in cells with Aβ1–42 alone and cells with Aβ1–42 + CUR, respectively, and then the function enrichment and protein-protein interaction (PPT) analysis in differentially expressed genes (DEGs) were conducted, aiming to investigate the underlying mechanisms of CUR in AD.
2. Materials and Methods
2.1. Cell Culture
HT-22 cells were purchased from JENNIO Bio Technology Co., Ltd. (Guangzhou, China). Cells were cultured in Dulbecco's Modified Eagle Media (DMEM, Gibco Co., Ltd., Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco Co., Ltd.) and 1% penicillin/streptomycin (Gibco Co., Ltd.) in 37°C incubator with 5% CO2.
2.2. Detection of Cell Viability
HT-22 cells (1.0 × 104 cells/well) were seeded into 96-well plates. The second day, the cells were treated with 5 μM Aβ1–42 [oligomer, dissolved in dimethyl sulfoxide (DMSO) and incubated at 37°C for 72 h to induce aggregation, Sigma, Louis, MO, USA] [19], 5 μM Aβ1–42 + 5 μM CUR (dissolved in DMSO, Sigma), 5 μM Aβ1–42 + 10 μM CUR, and 5 μM Aβ1–42 + 15 μM CUR [20], respectively. HT-22 cells without any treatment served as control group. After incubation for 24 h and 48 h, respectively, cells in each well were incubated with 10 μL cell counting kit 8 (CCK8, Dojindo Co., Ltd, Tokyo, Japan) for 2 h. Ultimately, absorbance was read at 450 nm using Synergy H4 microplate reader (BioTek, Winooski, VT, USA).
2.3. Western Blotting
The cells were treated with 5 μM Aβ1–42 or 5 μM Aβ1–42 + 10 μM CUR for 48 h. The cells were collected and treated with RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) on ice for 30 min. After centrifugation at 12,000 rpm for 15 min, supernatant was acquired and protein concentration was measured using the BCA Protein Quantitative Assay (Pierce, Rockford, IL, USA). Protein (30 μg/lane) sample was separated and blotted to polyvinylidene fluoride membranes (Millipore, Belfor, MA, USA), which were blocked in 5% nonfat milk for 1 h. Then, the membranes were incubated with rabbit anti-mouse β-actin polyclonal antibody (1 : 1000, Proteintech, Chicago, IL, USA) or rabbit anti-mouse Beclin-l polyclonal antibody (1 : 500, Abcam, Cambridge, MA, USA) overnight at 4°C, followed by incubation with goat anti-rabbit IgG (H+L)-HRP (1 : 5000, Jackson, West Grove, PA, USA) for 2 h at room temperature. Proteins were expressed using enhanced chemiluminescence (ECL) kit (Millipore) and analyzed by Image J software. The results were quantified from three independent experiments.
2.4. Transmission Electron Microscopy (TEM)
The cells were treated with 5 μM Aβ1–42 or 5 μM Aβ1–42 + 10 μM CUR for 48 h. Then, the formation of autophagosomes in cells was observed using TEM. Briefly, cells were collected and then fixed in 2.5% glutaraldehyde for 2 h at 25°C. After washing with phosphate buffered saline for 3 times, the cells were postfixed in 2% osmium tetroxide for 2 h and then dehydrated in graded alcohols. Subsequently, samples were sectioned and embedded in LX112 plastic. Finally, sections were stained with uranyl acetate and lead citrate, and electron micrographs were obtained by JEM-1230 TEM (JEOL, Japan).
2.5. Transcriptome Sequencing
The cells were treated with 5 μM Aβ1–42 or 5 μM Aβ1–42 + 10 μM CUR for 48 h. Then, the cells were collected and the total RNA was extracted using Trizol (Invitrogen, Gaithersburg, MD, USA). The mRNA-seq library was constructed and then sequenced on the Illumina Genome Analyzer IIx sequencing platform. The raw reads were obtained by the Illumina instrument software and cleaned by removing reads with unknown bases “N” > 5%, adapter sequences, reads with more than 20% Q < 20 bases, and reads with <30 bases. The clean reads were mapped to the mice reference genome based on NCBI by TopHat software. The gene expression values using fragments per kilobase of exon model per million reads were obtained by StringTie tool (V1.2.2) based on mice gene annotation.
2.6. Identification and Analysis of DEGs
DEGs between cells with Aβ1–42 samples and cells with Aβ1–42 + CUR samples were obtained using the Linear Model for Microarray package in R [21]. The cutoff criteria for DEGs were set up as follows: |log2 fold change| value > 2 and the P value < 0.05. For functional analysis for DEGs, gene ontology terms (GO; http://www.geneontology.org) in biological process (BP) were performed based on the Database for Annotation, Visualization and Integrated Discovery [22]. In addition, PPI network for DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes online database [23] and visualized using the Cytoscape [24] software.
2.7. Statistical Analysis
Statistical analysis was performed by SPSS 19.0 statistical analysis software (SPSS Inc., Chicago, IL, USA). Data were expressed as the mean ± SEM and analyzed by t-test. A value of P < 0.05 was considered significant and P < 0.01 was considered highly significant.
3. Results
3.1. Effect of CUR on Cell Viability in Aβ1–42 Treated HT-22 Cells
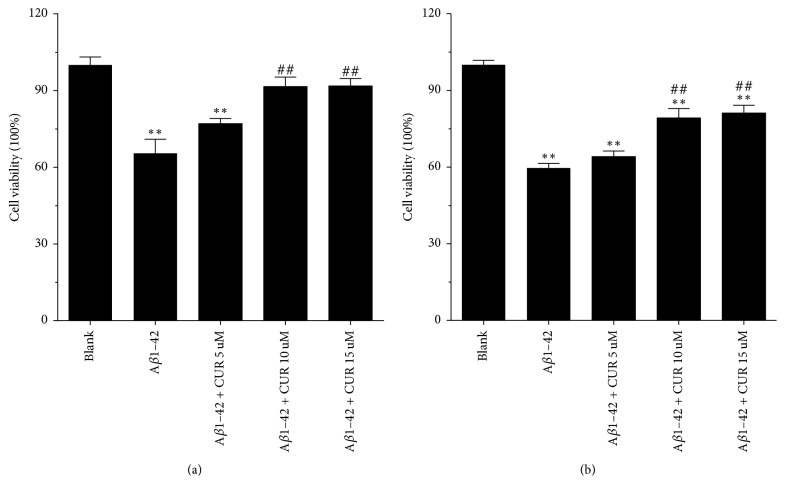
CCK8 assay results showed that compared with untreated cells, cell viability was significantly inhibited in cells treated with Aβ1–42 alone (P < 0.01), while cell viability was remarkably increased after treatment with 10 μM (P < 0.01) or 15 μM (P < 0.01) but not 5 μM CUR for 24 h (Figure 1(a)). Similarly, after treatment for 48 h, CUR (10, 15 μM) significantly promoted cell viability in comparison with cell treated with Aβ1–42 alone (P < 0.01, Figure 1(b)). Based on CCK8 assay, the combined treatment with 5 μM Aβ1–42 + 10 μM CUR for 48 h was used in subsequent experiment.
Figure 1.
Curcumin (CUR) promoted cell viability in Aβ1–42 induced cells. Cell viability in untreated cells (blank), Aβ1–42 induced cells, and CUR + Aβ1–42 treated cells at 24 h (a) and 48 h (b) using CCK-8 assay. The experiment was repeated for three times. ∗∗P < 0.01 versus Blank group; ##P < 0.01 versus Aβ1–42 group.
3.2. Effect of CUR on Cell Autophagy in Aβ1–42 Treated HT-22 Cells
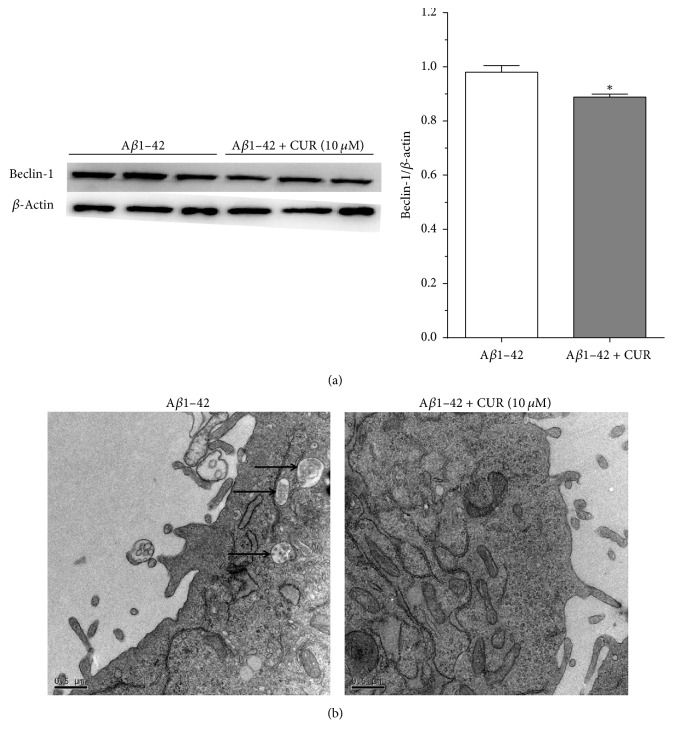
Western blotting results found that autophagy-related protein Beclin-1 was slightly downregulated in cells with combined treatment of Aβ1–42 and CUR compared with cells treated with Aβ1–42 alone (P < 0.05, Figure 2(a)). In addition, TEM results showed that autophagosome could be observed in cells treated with Aβ1–42 alone, while no autophagosome appeared in cells with combined treatment of Aβ1–42 and CUR (Figure 2(b)), which was consistent with the results of downregulated Beclin-1.
Figure 2.
Curcumin (CUR) inhibited cell autophagy in Aβ1–42 induced cells after treatment for 48 h. (a) The protein expression of Beclin-1 in Aβ1–42 induced cells and CUR + Aβ1–42 treated cells using western blotting; (b) cell autophagosome in Aβ1–42 induced cells and CUR + Aβ1–42 treated cells using transmission electron microscopy. Bar = 0.5 μm; ∗P < 0.05 versus Aβ1–42 group.
3.3. Function Enrichment Analysis of DEGs
Totally, 882 DEGs between cells with Aβ1–42 alone and cells with Aβ1–42 + CUR were obtained, including 324 upregulated DEGs and 558 downregulated DEGs. GOBP enrichment analysis showed that upregulated DEGs were significantly related to negative regulation of molecular function, epidermis development, metal ion transport, and keratinocyte differentiation, and downregulated DGEs were mainly correlative to intracellular organelle lumen, membrane-enclosed lumen, organelle lumen, nuclear lumen, and nucleolus. The top 10 GOBP terms with upregulated and downregulated DGEs are shown in Table 1.
Table 1.
The enriched pathways of DEGs between cells with Aβ1–42 alone and cells with Aβ1–42 + CUR.
| DEGs | Terms | Name | Counts | Gene | P value |
|---|---|---|---|---|---|
| Upregulated DEGs | GO:0044092 | Negative regulation of molecular function | 7 | ATP7A, MYC… |
0.005451488 |
| GO:0008544 | Epidermis development | 6 | ATP7A, GRPC5D… |
0.018159092 | |
| GO:0030001 | Metal ion transport | 12 | ATP7A, MCOLN1… |
0.019076392 | |
| GO:0030216 | Keratinocyte differentiation | 4 | GPRC5D, EVPL… |
0.020403199 | |
| GO:0052548 | Regulation of endopeptidase activity | 4 | CDH1, MYC… |
0.022720082 | |
| GO:0043281 | Regulation of caspase activity | 4 | CDH1, MYC… |
0.022720082 | |
| GO:0007398 | Ectoderm development | 6 | ATP7A, GPRC5D… |
0.023056172 | |
| GO:0006812 | Cation transport | 13 | ATP7A, MCOLN1… |
0.023211511 | |
| GO:0009913 | Epidermal cell differentiation | 4 | GPRC5D, EVPL… |
0.023930017 | |
| GO:0052547 | Regulation of peptidase activity | 4 | CDH1, MYC… |
0.023930017 | |
|
| |||||
| Downregulated DEGs | GO:0070013 | Intracellular organelle lumen | 40 | SURF6, UTP18… |
4.57E − 04 |
| GO:0031974 | Membrane-enclosed lumen | 41 | HNRNPA2B1, SIRT4… |
4.70E − 04 | |
| GO:0043233 | Organelle lumen | 40 | SURF6, UTP18… |
4.81E − 04 | |
| GO:0031981 | Nuclear lumen | 31 | SURF6, UTP18… |
0.002647478 | |
| GO:0005730 | Nucleolus | 15 | TSEN54, TBL3… |
0.003596227 | |
| GO:0005643 | Nuclear pore | 6 | CSE1L, KPNA6… |
0.006174947 | |
| GO:0005739 | Mitochondrion | 39 | PGS1, PDP2… |
0.012109477 | |
| GO:0046930 | Pore complex | 6 | SIRT4, ACACA… |
0.014833322 | |
| GO:0005929 | Cilium | 8 | TTC30B, TTC30A1… |
0.018005139 | |
| GO:0005912 | Adherens junction | 7 | FMN1, ARHGAP31… |
0.01885244 | |
3.4. PPI Analysis of DEGs
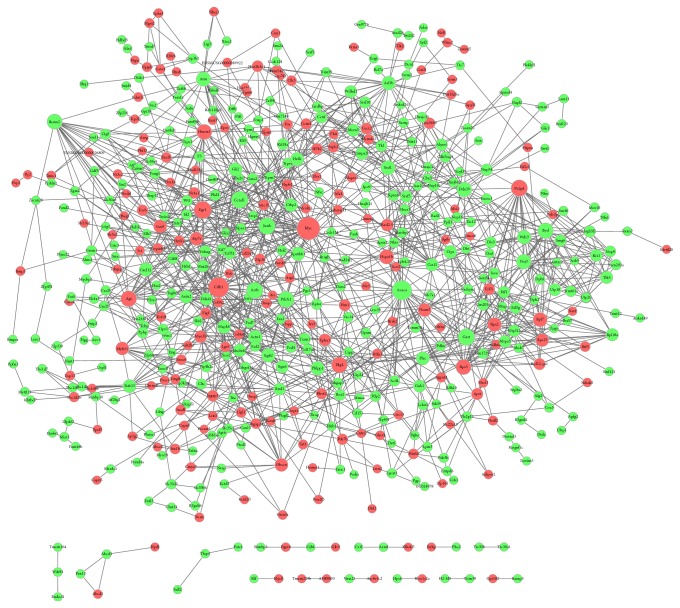
Totally, 552 DEGs including 162 upregulated DEGs and 360 downregulated DEGs were involved in 1337 interaction pairs (Figure 3). There were 7450 edges in PPI network for DEGs (Figure 3). MYC, Cdh1, Acaca, Egr1, and CCnd1 were located in the top 5 nodes with high degrees in PPI network.
Figure 3.
Protein-protein interaction network constructed for differentially expressed genes (DEGs). The red nodes stand for upregulated DEGs and the green nodes stand for downregulated DEGs.
4. Discussion
The present study found that CUR significantly promoted cell viability, reduced the expression of Beclin-1, and lowered the formation of autophagosomes in Aβ1–42 treated HT-22 cells. In addition, transcriptome sequencing results showed 324 upregulated DEGs and 558 downregulated DEGs, and PPI network showed that the pathogenesis of AD might be associated with MYC, Cdh1, and Acaca listed in the top 3 nodes with high degrees.
Previous study had shown a potential therapeutic role of CUR in the pathophysiology of AD [25]. Some in vivo studies demonstrated that oral administration of CUR could improve AD by removing Aβ deposition and improving behavioral impairment [10, 26]. It had been shown that CUR had an antiproliferation role in cancer cells [27]. However, this study found that CUR could promote cell proliferation. Similarly, Ma et al. [28] demonstrated that CUR could stimulate proliferation of rat neural stem cells. They found that low dose of CUR (0.1, 0.5, and 2.5 μM) increased the proliferation of neural stem cells, whereas high doles of CUR (12.5 and 62.5 μM) caused a decrease in the proliferation of neural stem cells [28], which was also consistent with our study. These results indicated the different role of CUR in cancer cells and neuronal cells. Autophagy had been reported to have contrary effect on Aβ aggregation in the different stage of AD [17]. In addition to antioxidant and anti-inflammatory effect, CUR could induce autophagy in various cancers, including human lung adenocarcinoma [29], colon cancer [30], glioblastomas [31], and oral cancer [32]. Furthermore, CUR was reported to induce autophagy and inhibit Aβ secretion in AD model mice [18]. Conversely, our study showed that CUR inhibited cell autophagy. This may explain that CUR removed intracellular Aβ depositions and then inhibited Aβ-induced toxicity, thereby exhibiting neuroprotective role by inhibiting cell autophagy [33]. However, our results showed only 10% inhibition of Beclin-1 expression caused by CUR treatment, so CUR-induced cell viability might be partly associated with cell autophagy in AD, while further study should be performed to confirm this ratiocination.
In order to further investigate the mechanism of CUR, transcriptome sequencing and bioinformation analysis were performed. The results found some important genes, such as MYC, Cdh1, and Acaca in PPI network. MYC oncogenes, containing C-myc, N-myc, and L-myc, had been proved to be overexpressed in tumor cells and closely associated with tumorigenesis by regulating cell proliferation, apoptosis, and differentiation [34]. In normal hematopoietic cells and hepatocytes, upregulated MYC expression could induce cell cycle progression [35, 36]. MYC was also overexpressed in AD and traumatic brain, which led to cognitive deficits and neurodegeneration [37, 38]. Cdh1 gene was cell cycle-related gene and could activate anaphase-promoting complex (APC) [39]. Cdh1-APC had been demonstrated to control the G0 and G1 phases of the cell cycle and regulate axonal growth during the neuronal differentiation of the mammalian brain [40]. Cdh1 could promote neuronal survival and lead to apoptotic cell death by inhibiting cyclin B1 accumulation in primary cortical neurons, indicating that upregulated Cdh1 prevented neuron damage induced by the neurotoxicity of Aβ [41]. Similarly, the present study revealed that, in Aβ1–42 treated HT-22 cells, CUR increased the expression of MYC and promoted cell growth. Acetyl-CoA carboxylase α (ACC-α) protein, encoded by Acaca gene, was a key enzyme in fatty acid synthesis pathway and expressed in various cells especially in lipogenic tissues [42]. ACC-α had been reported to be a potential target in metabolic syndromes and cancers because of the roles in fatty acid metabolism [43]. Some studies had shown overexpressed ACC-α in some cancers, including breast cancer and prostate cancer, indicating the protective role for cancer cell survival [44–46]. Effective interventions against ACC-α had been reported to inhibit tumor growth by regulating cell fate, transformation, and differentiation [47]. However, some studies should be performed to investigate the effect of ACC-α on neurodegenerative disease. In addition, our study only suggested preliminary results and further experiments for the validation of DEGs expression were still needed.
5. Conclusions
The current study revealed that CUR might have a potential protective effect by promoting cell viability in AD, which might be associated with cell autophagy. Furthermore, MYC, Cdh1, and Acaca might be involved in the early stage of AD, which should be further confirmed.
Acknowledgments
This study was supported by Zhengzhou Science & Technology Basic Research Program (no. 131PPTGG409-21).
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article and regarding funding.
Authors' Contributions
Lu Zhang and Yu Fang contributed equally to this work.
References
- 1.Vinters H. V. Emerging concepts in alzheimer's disease. Annual Review of Pathology: Mechanisms of Disease. 2015;10:291–319. doi: 10.1146/annurev-pathol-020712-163927. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer's Association. 2013 Alzheimer's disease facts and figures. Alzheimer's & Dementia. 2013;9(2):208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Hatcher H., Planalp R., Cho J., Torti F. M., Torti S. V. Curcumin: from ancient medicine to current clinical trials. Cellular and Molecular Life Sciences. 2008;65(11):1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai C., Ciccotosto G. D., Cappai R., et al. Curcumin Attenuates Colistin-Induced Neurotoxicity in N2a Cells via Anti-inflammatory Activity, Suppression of Oxidative Stress, and Apoptosis. Molecular Neurobiology. 2016:1–14. doi: 10.1007/s12035-016-0276-6. [DOI] [PubMed] [Google Scholar]
- 5.Yu S., Wang X., He X., et al. Curcumin exerts anti-inflammatory and antioxidative properties in 1-methyl-4-phenylpyridinium ion (MPP+)-stimulated mesencephalic astrocytes by interference with TLR4 and downstream signaling pathway. Cell Stress and Chaperones. 2016;21(4):697–705. doi: 10.1007/s12192-016-0695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernd A. Visible light and/or UVA offer a strong amplification of the anti-tumor effect of curcumin. Phytochemistry Reviews. 2014;13(1):183–189. doi: 10.1007/s11101-013-9296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra V., Pandav R., Dodge H. H., et al. Incidence of Alzheimer's disease in a rural community in India: the Indo-US study. Neurology. 2001;57(6):985–989. doi: 10.1212/wnl.57.6.985. [DOI] [PubMed] [Google Scholar]
- 8.Vas C. J., Pinto C., Panikker D., et al. Prevalence of dementia in an urban Indian population. International Psychogeriatrics. 2001;13(4):439–450. doi: 10.1017/S1041610201007852. [DOI] [PubMed] [Google Scholar]
- 9.Ng T.-P., Chiam P.-C., Lee T., Chua H.-C., Lim L., Kua E.-H. Curry consumption and cognitive function in the elderly. American Journal of Epidemiology. 2006;164(9):898–906. doi: 10.1093/aje/kwj267. [DOI] [PubMed] [Google Scholar]
- 10.Begum A. N., Jones M. R., Lim G. P., et al. Curcumin structure-function, bioavailability, and efficacy in models of neuroinflammation and Alzheimer's disease. The Journal of Pharmacology and Experimental Therapeutics. 2008;326(1):196–208. doi: 10.1124/jpet.108.137455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Alloza M., Borrelli L. A., Rozkalne A., Hyman B. T., Bacskai B. J. Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. Journal of Neurochemistry. 2007;102(4):1095–1104. doi: 10.1111/j.1471-4159.2007.04613.x. [DOI] [PubMed] [Google Scholar]
- 12.Lim G. P., Chu T., Yang F., Beech W., Frautschy S. A., Cole G. M. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. The Journal of Neuroscience. 2001;21(21):8370–8377. doi: 10.1523/JNEUROSCI.21-21-08370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Strooper B., Annaert W. Novel research horizons for presenilins and γ-secretases in cell biology and disease. Annual Review of Cell and Developmental Biology. 2010;26:235–260. doi: 10.1146/annurev-cellbio-100109-104117. [DOI] [PubMed] [Google Scholar]
- 14.LaFerla F. M., Green K. N., Oddo S. Intracellular amyloid-β in Alzheimer's disease. Nature Reviews Neuroscience. 2007;8(7):499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 15.Nixon R. A., Yang D.-S. Autophagy failure in Alzheimer's disease-locating the primary defect. Neurobiology of Disease. 2011;43(1):38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nasica-Labouze J., Nguyen P. H., Sterpone F., et al. Amyloid β Protein and Alzheimer's Disease: When Computer Simulations Complement Experimental Studies. Chemical Reviews. 2015;115(9):3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipinski M. M., Zheng B., Lu T., et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proceedings of the National Acadamy of Sciences of the United States of America. 2010;107(32):14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C., Zhang X., Teng Z., Zhang T., Li Y. Downregulation of PI3K/Akt/mTOR signaling pathway in curcumin-induced autophagy in APP/PS1 double transgenic mice. European Journal of Pharmacology. 2014;740:312–320. doi: 10.1016/j.ejphar.2014.06.051. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Zhang X.-G., Zhou T.-T., et al. Elevated neuronal excitability due to modulation of the voltage-gated sodium channel Nav1.6 by Aβ1-42. Frontiers in Neuroscience. 2016;10, article no. 94 doi: 10.3389/fnins.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., Li Z., Qiu D., Gu Q., Lei Q., Mao L. The inhibitory effects of different curcuminoids on β-amyloid protein, β-amyloid precursor protein and β-site amyloid precursor protein cleaving enzyme 1 in swAPP HEK293 cells. Neuroscience Letters. 2010;485(2):83–88. doi: 10.1016/j.neulet.2010.08.035. [DOI] [PubMed] [Google Scholar]
- 21.Smyth G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1, article 3) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 22.Dennis Jr. G., Sherman B. T., Hosack D. A., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(5):p. P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 23.Franceschini A., Szklarczyk D., Frankild S., et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013;41(1):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohl M., Wiese S., Warscheid B. Data Mining in Proteomics. Springer; 2011. Cytoscape: software for visualization and analysis of biological networks; pp. 291–303. [DOI] [PubMed] [Google Scholar]
- 25.Ringman J. M., Frautschy S. A., Cole G. M., Masterman D. L., Cummings J. L. A potential role of the curry spice curcumin in Alzheimer's disease. Current Alzheimer Research. 2005;2(2):131–136. doi: 10.2174/1567205053585882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frydman-Marom A., Levin A., Farfara D., et al. Orally administrated cinnamon extract reduces β-amyloid oligomerization and corrects cognitive impairment in Alzheimer's disease animal models. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0016564.e16564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunnumakkara A. B., Anand P., Aggarwal B. B. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Letters. 2008;269(2):199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Ma X. X., Wang C. M., Zhang G. L., et al. Curcumin stimulates proliferation of rat neural stem cells by inhibiting glucocorticoid receptors. Chinese Journal of Pharmacology Toxicology. 2015;29:202–207. [Google Scholar]
- 29.Xiao K., Jiang J., Guan C., et al. Curcumin induces autophagy via activating the AMPK signaling pathway in lung adenocarcinoma cells. Journal of Pharmacological Sciences. 2013;123(2):102–109. doi: 10.1254/jphs.13085FP. [DOI] [PubMed] [Google Scholar]
- 30.Kantara C., O'Connell M., Sarkar S., Moya S., Ullrich R., Singh P. Curcumin promotes autophagic survival of a subset of colon cancer stem cells, which are ablated by DCLK1-siRNA. Cancer Research. 2014;74(9):2487–2498. doi: 10.1158/0008-5472.CAN-13-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanotto-Filho A., Braganhol E., Klafke K., et al. Autophagy inhibition improves the efficacy of curcumin/temozolomide combination therapy in glioblastomas. Cancer Letters. 2015;358(2):220–231. doi: 10.1016/j.canlet.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 32.Kim J. Y., Cho T. J., Woo B. H., et al. Curcumin-induced autophagy contributes to the decreased survival of oral cancer cells. Archives of Oral Biolog. 2012;57(8):1018–1025. doi: 10.1016/j.archoralbio.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Goozee K. G., Shah T. M., Sohrabi H. R., et al. Examining the potential clinical value of curcumin in the prevention and diagnosis of Alzheimer's disease. British Journal of Nutrition. 2015;115(3):449–465. doi: 10.1017/S0007114515004687. [DOI] [PubMed] [Google Scholar]
- 34.Vita M., Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Seminars in Cancer Biology. 2006;16(4):318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 35.Shachaf C. M., Kopelman A. M., Arvanitis C., et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 36.Felsher D. W., Bishop J. M. Reversible tumorigenesis by MYC in hematopoietic lineages. Molecular Cell. 1999;4(2):199–207. doi: 10.1016/S1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 37.Lee H.-G., Casadesus G., Nunomura A., et al. The neuronal expression of MYC causes a neurodegenerative phenotype in a novel transgenic mouse. The American Journal of Pathology. 2009;174(3):891–897. doi: 10.2353/ajpath.2009.080583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Giovanni S., Movsesyan V., Ahmed F., et al. Cell cycle inhibition provides neuroprotection and reduces glial proliferation and scar formation after traumatic brain injury. Proceedings of the National Acadamy of Sciences of the United States of America. 2005;102(23):8333–8338. doi: 10.1073/pnas.0500989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M., Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Division. 2009;4(1):p. 1. doi: 10.1186/1747-1028-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konishi Y., Stegmüller J., Matsuda T., Bonni S., Bonni A. Cdh1-APC Controls Axonal Growth and Patterning in the Mammalian Brain. Science. 2004;303(5660):1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- 41.Almeida A., Bolaños J. P., Moreno S. Cdh1/Hct1-APC is essential for the survival of postmitotic neurons. The Journal of Neuroscience. 2005;25(36):8115–8121. doi: 10.1523/JNEUROSCI.1143-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abu-Elheiga L., Matzuk M. M., Kordari P., et al. Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal. Proceedings of the National Acadamy of Sciences of the United States of America. 2005;102(34):12011–12016. doi: 10.1073/pnas.0505714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J. E., Walsh M. C., Hoehn K. L., James D. E., Wherry E. J., Choi Y. Regulator of fatty acid metabolism, acetyl coenzyme a carboxylase 1, controls T cell immunity. The Journal of Immunology. 2014;192(7):3190–3199. doi: 10.4049/jimmunol.1302985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhajda F. P. Fatty-acid synthase and human cancer: New perspectives on its role in tumor biology. Nutrition Journal . 2000;16(3):202–208. doi: 10.1016/S0899-9007(99)00266-X. [DOI] [PubMed] [Google Scholar]
- 45.Chajès V., Cambot M., Moreau K., Lenoir G. M., Joulin V. Acetyl-CoA carboxylase α is essential to breast cancer cell survival. Cancer Research. 2006;66(10):5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- 46.Brusselmans K., de Schrijver E., Verhoeven G., Swinnen J. V. RNA interference—mediated silencing of the acetyl-Coa-carboxylase-α gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Research. 2005;65(15):6719–6725. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- 47.Alli P. M., Finn M. L., Jaffee E. M., McFadden J. M., Kuhajda F. P. Fatty acid synthase inhibitors are chemopreventive for mammary cancer in neu-N transgenic mice. Oncogene. 2005;24(1):39–46. doi: 10.1038/sj.onc.1208174. [DOI] [PubMed] [Google Scholar]