Abstract
Introduction: Infections due to antibiotic resistant bacteria have increased alarmingly in both developed and developing countries. Unrestrained and rapidly spreading bacterial growth has turned the management of wound infections into a serious challenge. This study aimed to determine the prevalence of different bacterial pathogens and their antibiotic susceptibility in various types of wound infections.
Methods: A cross-sectional study was conducted to collect 105 wound swabs. All isolated bacteria were identified based on colony characteristics, gram stain and standard biochemical tests, and antibiotic susceptibility testing (AST) with the disc diffusion method. Descriptive statistics were used to present the study findings, and all analyses were performed using Stata Version 13.
Results: The rate of isolation of bacteria was 92.3%. Staphylococcus aureus was found to be the most frequent isolate (55.7%), followed by Escherichia coli (23.7%), Pseudomonas spp. (8.2%), and Streptococcus pyogenes (7.2%). Gram-positive bacteria were mostly (60%) found sensitive to vancomycin, azithromycin, gentamicin, imipenem, cefixime, and ceftriaxone in this study. Among the Gram-negative bacteria, Escherichia coli (>60%) showed sensitivity to cefixime, azithromycin, cefuroxime, ceftriaxone, cefotaxime, gentamycin, and ceftazidime.
Conclusions: The diversity of isolated bacteria and their susceptibility patterns signify a need to implement a proper infection control strategy, which can be achieved by carrying out antibiotic sensitivity tests of the isolates.
Keywords: wound infection, bacterial pathogen, antibiotic susceptibility pattern
Introduction
Wounds follow the loss of skin integrity, which provides a moist, warm and nutritive environment that is known to be conducive to microbial colonization and proliferation 1. Wound infections are considered a major complication of surgery, and can be classified into three types: incisional surgical wounds, deep incisional wounds, and organ-specific infections 2. Despite maintaining the high standards of preoperative preparation, antibiotic prophylaxis, and operative procedures, the appearance of postoperative wound infections remains a grave threat among the clinicians 3. Some of the most frequent causative microorganisms are related to wound infections and include Staphylococcus aureus, Streptococcus pyogenes, Enterococci, Escherichia coli, Klebsiella pneumonia, Proteus species and Pseudomonas aeruginosa. However, the severity of complication is largely based on the virulence of the infecting pathogen and the site of infection 4. The reporting trend of infection varies depending on the surgeon’s ability, operative area, surgical procedures, patient characteristics, etc. For instance, approximately 5,00,000 infections per year take place in the United States among an estimated 27 million surgical procedures 5. The incidence of hospital-based postoperative infection varies from 10%–25% in India 6. Nosocomial infection is becoming a serious problem affecting hospitalized patients both in developed and developing countries. According to a study conducted in Bangladesh, it was reported that among 38% of nosocomial infections, more than 50% were due to wound infection 7. Moreover, wound infections were found to be higher (49%) among post-operative patients as compared to pre-operative patients (15.9%) in that study 7. Post-operative wound infections have emerged as one of the important causes of morbidity among the hospitalized patients 8. Emmerson et al. reported that surgical wound infections account for 12.3% of all hospital-acquired infections 9. Wound infection is becoming a major concern among patients and healthcare practitioners for its increased toll on morbidity and financial loss. It also generates demand for attaining expensive management within the public health system 5. Active and passive surveillance of surgical site infections in the hospital will help the surgeons and clinicians to know the antibiotic susceptibility pattern related to the surgical site, which can help reduce postoperative complications 10.
The present study aimed to collect data on the bacteriological profiles of wound infections and their antibiotic susceptibility patterns in a teaching hospital in Bangladesh.
Methods
Study design and study timeline
This cross-sectional study was conducted from the 10 th of July 2016 to the 30 th December 2016.
Study participants
105 samples of pus or wound swab were collected from the Microbiology Department of the Enam Medical College Hospital, Dhaka, which is a teaching hospital located in Bangladesh. The Microbiology department collected the samples from the outpatient and inpatient department of Surgery, Medicine, Gynaecology, and Orthopaedic.
Data collection
105 swab samples were collected from patients with various wound infections including post-operative surgical wounds, burn wounds and superficial and soft tissue infections (SSTI) by paramedics. Selective criteria were considered: infected wound, adult patients, and before administration of antibiotics. Specimens were collected aseptically by nurses or technicians before the wound cleaning and before application of an antiseptic solution. At the time of swab collection, standard care was taken to avoid contamination by the normal flora of the surrounding skin. Then the specimens were transported within one hour to the Microbiology laboratory of the hospital to perform the culture and susceptibility tests. Subsequently, each specimen was inoculated on appropriate agar media: blood agar, MacConkey’s agar, nutrient agar, and mannitol salt agar media. Finally, the cultures were incubated aerobically at 37°C for 24–48 hours with proper care. All the plates were regularly inspected for growth, and identification of the isolated bacteria was done by colony morphology, gram-staining and standard biochemical tests by microbiologists 11. Antimicrobial susceptibility patterns of the isolated bacterial pathogens were tested by using commonly used antibiotics such as amoxicillin (10 µg), penicillin (10 µg), vancomycin (30 µg), azithromycin (15 µg), cephradine (30 µg), tetracycline (30 µg), cloxacillin (5 µg), co-trimoxazole (23.75 µg), ciprofloxacin (5 µg), cefixime (5 µg), cefuroxime (30 µg), imipenem (10 µg), ceftriaxone (30 µg), and nitrofurantoin (300 µg) using the Kirby Bauer disc diffusion method according to the guidelines of Clinical Laboratory Standards Institute 12.
Statistical analysis
Errors in data were revised after cross-checking the laboratory records and clinical case recording forms. Descriptive statistics were used to interpret the data. Frequency and proportions were used to present categorical variables while mean and standard deviation (SD) were given to describe continuous variables. Stata (v.13) was utilized to analyze the data.
Ethical statement
Written informed consent was obtained from each participant. All study participants were informed verbally about the objective of the study. The research team paid the costs related to patient sample collection. The study was conducted under the clearance of the Ethical Review Committee (approval# 2017/218) of Enam Medical College Hospital, Dhaka, Bangladesh.
Results
Characteristics of study participants
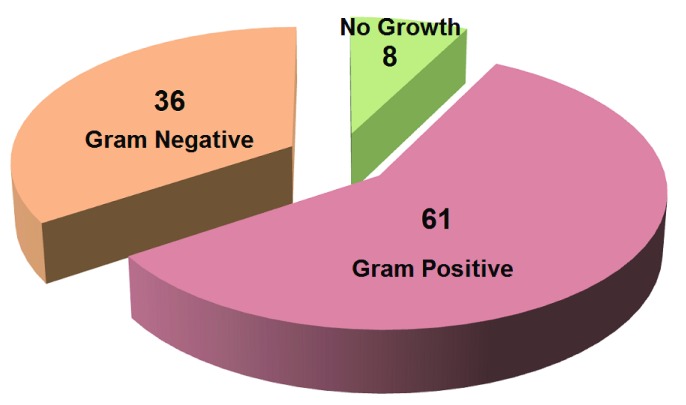
The mean (±standard deviation) age of the study participants was 37 (±08) years, and 57.1% of participants were male. The rate of isolation of bacteria was 92.3%. Figure 1 shows the frequency of bacterial growth. Around 62.9% of culture positive plates turned out to be Gram-positive organisms, and 37.1% Gram-negative (n=97). Only 7.6% did not yield any growth in a culture plate.
Figure 1. Pattern of bacterial growth among total samples (N=105).
In this figure, red, magenta, and green portion indicates the Gram Positive, Gram Negative, No growth, respectively and indicates the percentage of bacterial growth.
Isolation of different types of bacteria
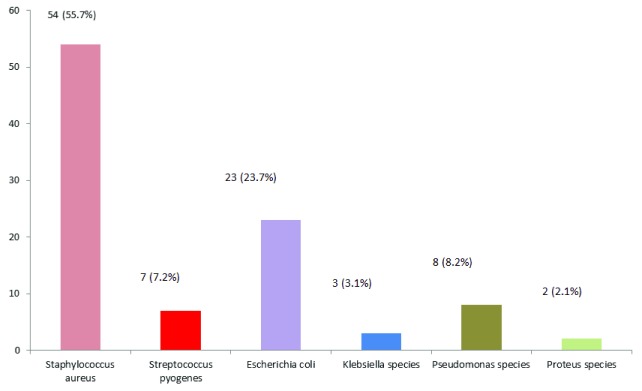
Staphylococcus aureus (n=54; 55.7%) was predominantly found to be isolated among all the presenting bacteria. The frequency of Escherichia coli and Pseudomonas species was 23.7% and 8.2%, respectively ( Figure 2).
Figure 2. Rate of isolation of different bacteria (N=97).
Rate of isolation of different bacteria are mentioned here based on number and their corresponding percentage.
Sensitivity pattern of isolated Gram-positive and Gram-negative bacteria
The susceptibility pattern of Gram-positive bacteria was mostly isolated to imipenem (90%), followed by ceftriaxone (85.5%), gentamycin (81.8%), vancomycin (80.8%), azithromycin (76.5%) and other antibiotics (<75.0%) ( Table 1).
Table 1. Sensitivity pattern of isolated Gram-positive bacteria (N = 61).
| Antimicrobial agents |
Staphylococcus
aureus (n=54) |
Streptococcus
pyogenes (n=7) |
|---|---|---|
| Amoxicillin (10 µg) | 32 (59.3%) | 4 (57.1%) |
| Penicillin (10 µg) | 30 (55.6%) | 4 (57.1%) |
| Vancomycin (30 µg) | 41 (75.9%) | 6 (85.7%) |
| Azithromycin (15 µg) | 44 (81.5%) | 5 (71.5%) |
| Cephradine (30 µg) | 32 (59.3%) | 4 (57.1%) |
| Tetracycline (30 µg) | 32 (59.3%) | 4 (57.1%) |
| Cloxacillin 5( µg) | 31 (57.4%) | 4 (57.1%) |
| Co-trimoxazole (23.7 µg) | 31 (57.4%) | 3 (42.9%) |
| Gentamicin (10 µg) | 42 (77.8%) | 6 (85.7%) |
| Ciprofloxacin (5 µg) | 32 (59.3%) | 4 (57.1%) |
| Cefixime (5 µg) | 40 (74.1%) | 5 (71.5%) |
| Cefuroxime (30 µg) | 32 (59.3%) | 4 (57.1%) |
| Imipenem (10 µg) | 51 (94.4%) | 6 (85.7%) |
| Ceftriaxone (30 µg) | 46 (85.2%) | 6 (85.7%) |
Most of the Gram-negative isolates were sensitive to ceftazidime (79.0%), ceftriaxone (71.8%), gentamicin (70.7%) and other antibiotics (<70.0%) ( Table 2). Most of the Pseudomonas spp. (>50%) were sensitive to ceftriaxone, imipenem, and gentamycin.
Table 2. Sensitivity pattern of isolated Gram-negative bacteria (N = 36).
| Antimicrobial Agents | Escherichia
coli (n=23) |
Klebsiella spp.
(n=3) |
Pseudomonas
spp. (n=8) |
Proteus
spp. (n=2) |
|---|---|---|---|---|
| Cephradine (30 µg) | 10 (43.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Co-trimoxazole (23.7 µg) | 12 (52.2%) | 1 (33.3%) | 1 (12.5%) | 1 (50.0%) |
| Cefixime (5 µg) | 19 (82.6%) | 1 (33.3%) | 1 (12.5%) | 1 (50.0%) |
| Penicillin (10 µg) | 8 (34.8%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Aztreonam (30 µg) | 17 (73.9%) | 1 (33.3%) | 1 (12.5%) | 2 (50.0%) |
| Cloxacillin (5 µg) | 11 (47.8%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Cefuroxime (30 µg) | 18 (78.3%) | 0 (0.0%) | 0 (0.0%) | 2 (100%) |
| Tetracycline (30 µg) | 14 (60.9%) | 0 (0.0%) | 3 (37.5%) | 1 (50.0%) |
| Imipenem (10 µg) | 21 (91.3%) | 1 (33.3%) | 4 (50.0%) | 2 (100%) |
| Ceftriaxone (30 µg) | 21 (91.3%) | 1 (33.3%) | 5 (62.5%) | 2 (100%) |
| Ciprofloxacin (5 µg) | 5 (21.7%) | 0 (0.0%) | 3 (37.5%) | 1 (50%) |
| Azithromycin (15 µg) | 8 (34.8%) | 2 (66.7%) | 3 (37.5%) | 2 (100%) |
| Amoxicillin (10 µg) | 1 (4.3%) | 1 (33.3%) | 0 (0.0%) | 0 (0.0%) |
| Cefotaxime (30 µg) | 20 (86.9%) | 1 (33.3%) | 0 (0.0%) | 1 (50%) |
| Gentamycin (10 µg) | 19 (82.6%) | 3 (100%) | 4 (50%) | 1 (50.0%) |
| Ceftazidime (30 µg) | 18 (78.3%) | 3 (100%) | 3 (37.5%) | 2 (100%) |
| Nitrofurantoin (300 µg) | 18 (78.3%) | 1 (33.3%) | 1 (12.5%) | 1 (50.0%) |
Age, sex, residence, occupation, blood pressure, and diabetic mellitus status are given.
Copyright: © 2017 Roy S et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Includes data on S. aureus, S. pyogenes, E. coli, Klebsiella spp, Pseudomonas, and Proteus.
Copyright: © 2017 Roy S et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Discussion
Management of post-operative wound infection remains a significant concern for physicians globally 13. The problem has magnified due to the rapidly spreading resistance to the available array of antimicrobial agents 14, 15. We found that Gram-positive organisms accounted for 62.9% of isolates, compared to Gram-negative isolates that accounted for 37.1%. Staphylococcus aureus (55.7%) was the major microbial pathogen responsible for the wound infections. According to Centre for Disease Control and Prevention (CDC), Staphylococcus aureus is the most common organism associated with surgical wound infections. This study supports the results reported by Nwachukwu et al. 16, where 42.3% of infections were found to be caused by Staphylococcus aureus. Among the Gram-negative organisms, Escherichia coli were frequently isolated (23.7%) in our study. This finding is in line with a previous study which identified Escherichia coli as the major pathogen in the wound infection, followed by Staphylococcus aureus in a different setup 17. A previous survey conducted in Lahore supported our findings demonstrating that Staphylococcus aureus was the main causative organism of surgical infection 18.
In our study, we found imipenem as the most active antibiotic, with a susceptibility of 94.4% against Staphylococcus aureus. This study showed high sensitivity of Staphylococcus aureus against imipenem, vancomycin, and gentamycin. This finding corresponds to a previous study that also found that Staphylococcus aureus was susceptible to higher generation of antibiotics 19. The high sensitivity to gentamycin has also been reported by other authors as well 20. We found that Staphylococcus aureus is usually resistant to various antibiotics and the infection might be acquired in the hospital.
Among the Gram-negative bacteria, Escherichia coli was found to be susceptible to ceftriaxone, cefotaxime, gentamycin, cefixime, ceftazidime, and cefuroxime. Furthermore, we found that Escherichia coli were less sensitive to cloxacillin with a frequency of 47.8%. Among three isolated Klebsiella spp., all organisms were resistant to cephradine, penicillin, cloxacillin, cefuroxime, tetracycline, and ciprofloxacin. Similarly, Okonko et al. 21 had observed a high level of resistance by Klebsiella spp. to most antibiotics. However, they noticed that all three Klebsiella spp. Isolates were susceptible to gentamycin and ceftazidime. This high susceptibility pattern might support gentamycin as a suitable antibiotics to treat Klebsiella infection 22. Among eight isolated Pseudomonas spp., all were resistant to cephradine, penicillin, cloxacillin, cefuroxime, amoxicillin, and cefotaxime in this study. We found that five Pseudomonas isolates were susceptible to ceftriaxone, four were susceptible to imipenem and gentamycin, and three were susceptible to tetracycline, ciprofloxacin, azithromycin, and ceftazidime. Only one Pseudomonas spp. isolate was susceptible to co-trimoxazole, cefixime, and nitrofurantoin.
The susceptibility pattern that we found indicates that most of the isolated strains were multi-drug resistant. Similarly, a study conducted in European setting reported a high resistance of Pseudomonas spp., mostly isolated from surgical wounds 23. Several previous studies carried out in different settings also support the multi-drug resistance pattern of Pseudomonas spp. 24– 26. The mechanisms of intrinsic resistance of Pseudomonas spp. over most of the antimicrobial agents has emerged because of the low permeability of its outer membrane and the naturally occurring chromosomal Amp β-lactamase 27, 28.
The control of wound infections is becoming difficult due to widespread bacterial resistance to antibiotics. Previous studies also notified an increased incidence of bacterial infections by methicillin-resistant Staphylococcus aureus, polymicrobial flora and different fungi 29. As wound infections are found to be common in this study, prior knowledge of the causative agents of can be a helpful tool in selecting the empiric antimicrobial therapy to control infection
In developing countries, physicians generally do not wait for the culture reports and sometimes, there may be a delay in conducting or reporting of a culture sensitivity test. Hence, with our study, we would like to urge the physicians to start an empirical therapy with a combination of either of the following as an empirical treatment regime:
1) Azithromycin/Imipenem and Ceftriaxone;
2) Gentamycin and Imipenem/Ceftriaxone
3) Ceftazidime and Imipenem.
After application of the above mentioned combination regime, culture sensitivity is advised to be performed in next step. Irrespective of the report, the entire course should be completed and if the condition still remains has not improved, urgent change of treatment plan according to the culture sensitivity report should be carried out.
We would discourage the use of penicillin and amoxicillin, since the resistance towards them has been on the rise. We would also urge physicians to not to prescribe the last resort drugs like vancomycin and linezolid, since they should be used as only in high resistance cases.
Study limitations
The susceptibility patterns of bacterial isolates to commonly prescribed antibiotics like ceftriaxone, cefuroxime, ciprofloxacin, and azithromycin might not be generalized globally. The fact that our research was a single center study and had a small sample size were other drawbacks. However, our result might represent the scenario of a developing country. Moreover, high-quality data and laboratory support were the particular strengths of this study.
Conclusions
The most common isolate in wound infection was Staphylococcus aureus, followed by Escherichia coli, Pseudomonas spp., Klebsiella spp., and Streptococcus pyogenes. Gram-negative bacteria were sensitive to fewer than thirty percent of the commonly prescribed antibiotics, which can be a matter of great concern when treating wound infections. The judicious use of antibiotic prophylaxis and reporting can be the most effective means to reduce the wound infection rate.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2017 Roy S et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
Dataset 1: Patient characteristics. Age, sex, residence, occupation, blood pressure, and diabetic mellitus status are given. DOI, 10.5256/f1000research.12887.d185740 30.
Dataset 2: Antibiotic susceptibility of bacterial cultures. Includes data on S. aureus, S. pyogenes, E. coli, Klebsiella spp, Pseudomonas, and Proteus. DOI, 10.5256/f1000research.12887.d185741 31.
Acknowledgements
We are grateful to the director of the hospital for giving us the permission to collect data.
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 2 approved]
References
- 1. Insan NG, Payal N, Singh M, et al. : Post operative wound infection: Bacteriology and antibiotic sensitivity pattern. International Journal of Current Research and Review. 2013;5(13):74–79. Reference Source [Google Scholar]
- 2. Howard R, Lee J: Surgical wound infections: epidemiology, surveillance, and clinical management. Surgical Infectious Diseases. 1995;401–12. [Google Scholar]
- 3. Bowler PG, Duerden BI, Armstrong DG: Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14(2):244–69. 10.1128/CMR.14.2.244-269.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lilani S, Jangale N, Chowdhary A, et al. : Surgical site infection in clean and clean-contaminated cases. Indian J Med Microbiol. 2005;23(4):249–52. [PubMed] [Google Scholar]
- 5. Haley RW, Culver DH, White JW, et al. : The nationwide nosocomial infection rate. A new need for vital statistics. Am J Epidemiol. 1985;121(2):159–67. 10.1093/oxfordjournals.aje.a113988 [DOI] [PubMed] [Google Scholar]
- 6. Mustafa A: Incidence of nosocomial wound infection in postoperative patients at a teaching hospital in Kashmir. JK— Practitioner. 2004;2(1):38–4. [Google Scholar]
- 7. Hussain T, Fazal M, Ahmed A: Nosocomial infection-A cross-sectional study in the surgical wards of Dhaka Medical College Hospital. J Prev Soc Med. 1991;10:70–3. [Google Scholar]
- 8. Koontz FP: Trends in post-operative infections by Gram-positive bacteria. Int J Antimicrob Agents. 2000;16 Suppl 1:S35–7. 10.1016/S0924-8579(00)00304-6 [DOI] [PubMed] [Google Scholar]
- 9. Emmerson AM, Enstone JE, Griffin M, et al. : The Second National Prevalence Survey of infection in hospitals--overview of the results. J Hosp Infect. 1996;32(3):175–90. 10.1016/S0195-6701(96)90144-9 [DOI] [PubMed] [Google Scholar]
- 10. Zaman SB, Hussain MA, Nye R, et al. : A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus. 2017;9(6):e1403. 10.7759/cureus.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheesbrough M: District laboratory practice in tropical countries. Cambridge university press;2006. Reference Source [Google Scholar]
- 12. CLSI C: Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement. CLSI Document M100-S20, Clinical and Laboratory Standards Institute, Wayne, Pa USA;2010;30. [Google Scholar]
- 13. Zaman SB, Hussain MA, Hossain N, et al. : Antibiotic Resistance: A Tragedy of the Common. International Journal of Research Studies. 2017;1(2):7–9. Reference Source [Google Scholar]
- 14. Raza MS, Chander A, Ranabhat A: Antimicrobial susceptibility patterns of the bacterial isolates in post-operative wound infections in a tertiary care hospital, Kathmandu, Nepal. Open Journal of Medical Microbiology. 2013;3(3):159 10.4236/ojmm.2013.33024 [DOI] [Google Scholar]
- 15. Dionigi R, Rovera F, Dionigi G, et al. : Risk factors in surgery. J Chemother. 2001;13(Spec No 1(1)):6–11. 10.1179/joc.2001.13.Supplement-2.6 [DOI] [PubMed] [Google Scholar]
- 16. Nwachukwu NC, Orji FA, Okike UM: Antibiotic susceptibility patterns of bacterial isolates from surgical wounds in Abia State University Teaching Hospital (ABSUTH), Aba–Nigeria. Research Journal of Medicine and Medical Sciences. 2009;4(2):575–9. Reference Source [Google Scholar]
- 17. Afroz H, Fakruddin M, Masud MR, et al. : Incidence of and risk factors for Hospital Acquired Infection in a Tertiary Care Hospital of Dhaka, Bangladesh. Bangladesh Journal of Medical Science. 2017;16(3):358–69. 10.3329/bjms.v16i3.32847 [DOI] [Google Scholar]
- 18. Aman S: Bacteriological analysis of wound infection in Mayo hospital, Lahore. J Pak Med Assoc. 1982;32(3):66–68. [PubMed] [Google Scholar]
- 19. Mengesha RE, Kasa BG, Saravanan M, et al. : Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder Teaching and Referral Hospital, Mekelle, Ethiopia. BMC Res Notes. 2014;7(1):575. 10.1186/1756-0500-7-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zaman SB, Hossain N, Yasir Arafat SM, et al. : Management of Newborn Infection: Knowledge and attitude among health care providers of selected sub-district hospitals in Bangladesh. International Journal of Perceptions in Public Health. 2017;1(2):127–32. Reference Source [Google Scholar]
- 21. Okonko IO, Soleye FA, Amusan TA, et al. : Incidence of multi-drug resistance (MDR) organisms in Abeokuta, Southwestern Nigeria. Global Journal of Pharmacology. 2009;3(2):69–80. Reference Source [Google Scholar]
- 22. Abe-Aibinu IE, Ohaegbulam V, Odugbemi TO: A comparative study on the antimicrobial susceptibility patterns of Klebsiella and Enterobacter species from the Lagos university teaching hospital. Journal of the Nigerian Infection Control Association. 2000;3(2):14–7. 10.4314/jnica.v3i2.10720 [DOI] [Google Scholar]
- 23. Fluit AC, Jones ME, Schmitz FJ, et al. : Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance proGram, 1997 and 1998. Clin Infect Dis. 2000;30(3):454–60. 10.1086/313710 [DOI] [PubMed] [Google Scholar]
- 24. Daini OA, Effiong MJ, Ogbolu OD: Quinolones Resistance and R-Plasmids of clinical isolates of Pseudomonas species. Sudan JM Sci. 2008;3(2):139–46. 10.4314/sjms.v3i2.38528 [DOI] [Google Scholar]
- 25. Sekiguchi J, Asagi T, Miyoshi-Akiyama T, et al. : Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6')-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob Agents Chemother. 2005;49(9):3734–42. 10.1128/AAC.49.9.3734-3742.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nordmann P, Guibert M: Extended-spectrum beta-lactamases in Pseudomonas aeruginosa. J Antimicrob Chemother. 1998;42(2):128–31. [DOI] [PubMed] [Google Scholar]
- 27. Sexton DJ: The impact of antimicrobial resistance on empiric antibiotic selection and antimicrobial use in clinical practice. J Med Liban. 2000;48(4):215–20. [PubMed] [Google Scholar]
- 28. Olayinka AT, Olayinka BO, Onile BA: Antibiotic susceptibility and plasmid pattern of Pseudomonas aeruginosa from the surgical unit of a university teaching hospital in north central Nigeria. International Journal of Medicine and Medical Sciences. 2009;1(3):079–83. Reference Source [Google Scholar]
- 29. Shittu AO, Kolawole DO, Oyedepo EA: A study of wound infections in two health institutions in Ile-Ife, Nigeria. Afr J Biomed Res. 2002;5(3):97–102. 10.4314/ajbr.v5i3.53994 [DOI] [PubMed] [Google Scholar]
- 30. Roy S, Ahmed MU, Uddin BMM, et al. : Dataset 1 in: Evaluation of antibiotic susceptibility in wound infections: A pilot study from Bangladesh. F1000Research. 2017. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roy S, Ahmed MU, Uddin BMM, et al. : Dataset 2 in: Evaluation of antibiotic susceptibility in wound infections: A pilot study from Bangladesh. F1000Research. 2017. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]