Abstract
Shifting the balance away from tumor-mediated immune suppression toward tumor immune rejection is the conceptual foundation for a variety of immunotherapy efforts currently being tested. These efforts largely focus on activating antitumor immune responses but are confounded by multiple immune cell populations, including myeloid-derived suppressor cells (MDSCs), which serve to suppress immune system function. We have identified immune-suppressive MDSCs in the brains of GBM patients and found that they were in close proximity to self-renewing cancer stem cells (CSCs). MDSCs were selectively depleted using 5-flurouracil (5-FU) in a low-dose administration paradigm, which resulted in prolonged survival in a syngeneic mouse model of glioma. In coculture studies, patient-derived CSCs but not nonstem tumor cells selectively drove MDSC-mediated immune suppression. A cytokine screen revealed that CSCs secreted multiple factors that promoted this activity, including macrophage migration inhibitory factor (MIF), which was produced at high levels by CSCs. Addition of MIF increased production of the immune-suppressive enzyme arginase-1 in MDSCs in a CXCR2-dependent manner, whereas blocking MIF reduced arginase-1 production. Similarly to 5-FU, targeting tumor-derived MIF conferred a survival advantage to tumor-bearing animals and increased the cytotoxic T cell response within the tumor. Importantly, tumor cell proliferation, survival, and self-renewal were not impacted by MIF reduction, demonstrating that MIF is primarily an indirect promoter of GBM progression, working to suppress immune rejection by activating and protecting immune suppressive MDSCs within the GBM tumor microenvironment.
Keywords: Cancer stem cells, Myeloid-derived suppressor cells, Macrophage migration inhibitory factor, Glioblastoma, Tumor microenvironment, Tumor immune suppression, Immunotherapy, MIF
Introduction
Glioblastoma (GBM), the most prevalent primary malignant brain tumor, remains uniformly fatal despite aggressive therapies including surgery, radiation, and chemotherapy [1]. GBM growth and resistance are attributed to a series of integrated drivers including genetic mutations, deregulated signaling pathways, and the tumor microenvironment (TME) [2, 3]. Immune cells, including tumor-associated macrophages and microglia (TAMs) and T cell populations, are recruited to the GBM microenvironment [4–6], but their immune-rejecting activities are suppressed [7–10]. Additionally, self-renewing, tumorigenic cancer stem cells (CSCs) [11–15] challenge GBM treatment paradigms due to their inherent therapeutic resistance [16–18] and their ability to modulate the immune system. CSCs inhibit cytotoxic T cell proliferation and activation while stimulating T regulatory (TReg) cells [19] and immunosuppressive TAMs [20]. CSCs also directly impact TAMs by promoting their transition from the antitumor (M1) to the pro-tumor (M2) phenotype [21]. Therefore, reversing immune suppression has become an attractive next-generation GBM therapeutic strategy but will necessitate both an understanding of how the immune system is altered in GBM and the mechanisms by which CSCs modulate immune function [22].
Approaches aimed at reversing the immunosuppression imposed on cytotoxic immune cells and utilizing the immune system to attenuate tumor growth through vaccination and disruption of key immune checkpoints have shown promise in GBM preclinical models [23, 24]. However, there are other immune-suppressive cell types present within the GBM microenvironment, and the function of these in the context of tumor growth and therapeutic development has yet to be determined. Myeloid-derived suppressor cells (MDSCs) are a heterogeneous class of immature immunosuppressive cells that accumulate in multiple tumor types and suppress cytotoxic immune cells via cytokine secretion [25–27]. MDSCs are upregulated in the peripheral blood of GBM patients [28], but their presence in the microenvironment and role in tumor progression are uncharacterized. Based on these unresolved questions, we interrogated the GBM microenvironment and detected MDSCs. The activity of this immunosuppressive cell population was amplified by CSCs through secreted factors, including macrophage migration inhibitory factor (MIF).
Materials and Methods
GBM Cell Collection, Culture, and Usage
hGBM 4121 was obtained from Duke University as an established, subcutaneous engrafted tumor. Likewise, hGBM 10 was obtained from the Mayo Clinic as an established xenograft specimen. Both cell lines were transferred to the Cleveland Clinic under approved MTAs. hGBM 4121 and hGBM 10 were authenticated by short tandem repeat (STR) analysis performed by the DNA Analysis Facility at Duke University. GBM cells were dissociated from specimens maintained as subcutaneous xenografts as reported previously [16, 29–31] and briefly described as follows: 6-wk-old female nude mice (Charles River Laboratories, Wilmington, MA, www.criver.com) were injected subcutaneously with freshly dissociated bulk human glioma, and tumor cells were allowed to grow until tumor volume exceeded 5% of the animal’s body weight, after which the mice were sacrificed. Bulk tumors were dissected from the flank fat pad and mechanically dissociated using papain dissociation kits (Worthington Biochemical Co., Lake-wood, NJ, www.worthington-biochem.com). Bulk cells were then cultured overnight in neurobasal complete medium, which consisted of neurobasal medium (Life Technologies, Carlsbad, CA, www.thermofisher.com) supplemented with B27 (Life Technologies), 1% penicillin/streptomycin (Life Technologies), 1 mM sodium pyruvate, 2 mM L-glutamine, EGF (20 ng/ml, R&D Systems), and FGF (20 ng/ml, R&D Systems, Minneapolis, MN, www.rndsystems.com).
Separation of CSCs from bulk tumor cells was accomplished by CD133 affinity column purification. Bulk tumor cells were incubated with magnetic CD133 affinity beads (CD133 MicroBead Kit, Miltenyi Biotech, San Diego, CA, www.miltenyibiotec.com) for 1 hour and added to magnetic-activated cell sorting (MACS) columns (LS columns, Miltenyi Biotech). Nonstem tumor cells (NSTCs) were washed through the column using neurobasal null medium (neurobasal medium supplemented with 1% penicillin/streptomycin) under magnetization. Columns were removed from the influence of the MACS sorting magnet, and CSCs were extracted from the beads using neurobasal null medium. CSCs were subsequently cultured in neurobasal complete medium until use. NSTCs were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, www.sigmaaldrich.com) and 1% penicillin/streptomycin) until utilized.
CSCs and NSTCs were cultured in the appropriate complete medium until the day prior to usage. To isolate conditioned media, cells were washed once with null neurobasal medium and incubated in null neurobasal medium for 24 hours. Cell counts were normalized at the end of 24 hours, and the conditioned media were diluted in null media to normalize the numbers of CSCs and NSTCs from which the conditioned medium was derived. Conditioned medium was used immediately post normalization. GL261 cells were used as previously described [32] and cultured in complete RPMI 1640 medium (RPMI; supplemented with 10% FBS and 1% penicillin/streptomycin) until used. All cells were used at passage 5 or lower for all experiments.
Murine GL261 cells were engineered to contain MIF knockdown or a luciferase construct to facilitate in vivo bioluminescence imaging. MIF knockdown was achieved using lentivirus-delivered shRNA (Mission shRNA system, Sigma) as previously described [29–31, 33, 34]. Lentivirus containing the MIF shRNA was generated in 293T cells, and 1 × 107 viral particles were incubated with passage 1 native GL261 cells for 2 days to allow for infection. The following sequences were used:
Control Sequence (SHC002). CCGGGCAACTACAGTAAGCTGCTGTCTCGAGACAGCAGCTTACTGTAGTTGCTTTTTG
MIF shRNA 1 (TRCN0000067343). CCGGGCACCGCTGTTCTTTGAGC CTCTCGAGAGGCTCAAAGAACAGCGGTGCTTTTTG
MIF shRNA 2 (TRCN0000067344). CCGGCCGGGTCTACATCAACTATTACTCGAGTAATAGTTGATGTAGACCCGGTTTTTG
MIF shRNA 3 (TRCN0000067345). CCGGCCAGAACCGCAACTACA GTAACTCGAGTTACTGTAGTTGCGGTTCTGGTTTTTG
MIF shRNA 4 (TRCN0000067346). CCGGGCAACTACAGTAAGCTGCTGTCTC GAGACAGCAGCTTACTGTAGTTGCTTTTTG
MIF shRNA 5 (TRCN0000067347). CCGGCATCAACTATTACGACATGAACTCGAGTTCA TGTCGTAATAGTTGATGTTTTTG
Cells successfully infected with lentivirus were selected using puromycin (1 μg/ml, Life Technologies) and passaged as described above. GL261 luciferase-containing cells were generated in an identical fashion to MIF knockdown GL261 cells. After puromycin selection of luciferase-positive cell populations, luciferase expression was confirmed by incubating cells with luciferin and imaging using an IVIS in vivo imaging system.
Isolation of Murine Cell Populations
To isolate bone marrow or MDSCs, mice bearing subcutaneous or intracranial tumors were sacrificed when symptomatic or when tumor volume exceeded 5% of body weight. Immediately after sacrifice, the bilateral femurs and tibias were dissected, and the epiphyses were removed bilaterally. Pushing neurobasal null media through the marrow-containing space using a 26-gauge needle mechanically extruded the marrow from within the long bones. Extruded cells were passed through a 70 μm filter, washed twice, and used as freshly isolated marrow. To isolate MDSCs, freshly isolated marrow was washed in FACS buffer (Neurobasal null medium supplemented with 5% bovine serum albumin) and blocked in FACS buffer with unlabeled antigoat IgG (1:250 dilution, Life Technologies) for 30 minutes. Cells were washed with Neurobasal null medium, stained with fluorescently labeled primary antibodies (CD11b, GR-1, CD244 [35]), and sorted using a BD FACSAria II flow cytometer (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). For visualization of MDSCs, the populations were gated first on live/dead cells, followed by aggregate correction using forward and side scatter, followed by a CD11b gate prior to being displayed as a CD244/GR-1 plot. Except for the immunohistochemistry (IHC) experiment presented in Fig. 1, we defined the MDSC population by the expression of the three cell-surface antigens: CD11b+, GR-1High, CD244+.
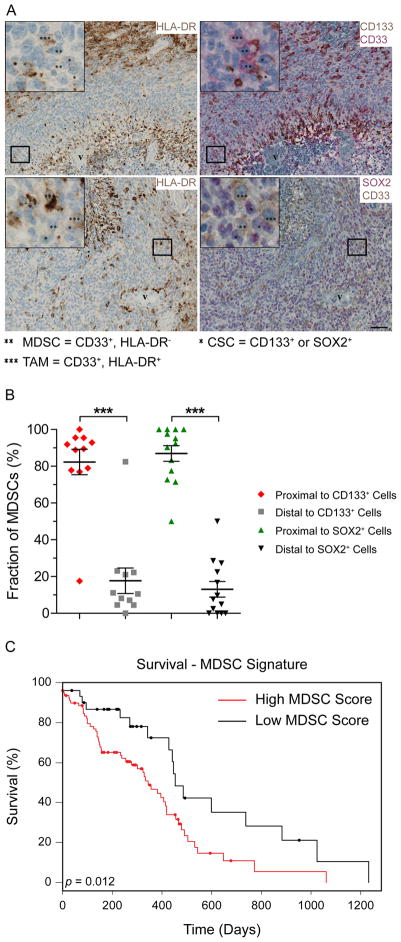
Figure 1.
MDSCs are present in glioblastoma tumors adjacent to cancer stem cells and predict poor survival. Virtual triple immunohistochemistry of GBM tumors (A) demonstrates the presence of MDSCs (** = HLA-DR−, CD33+) preferentially associated with CSCs (* = CD133+ or SOX2+). HLA-DR was stained in brown, CD33 was stained in red (upper right panel) or brown (lower right panel), CD133 was stained in brown, and SOX2 was stained in red. Scale bar = 50 μm. These results are quantified in (B) *** = p <.001 by Students’ unpaired t-test. Bioinformatics analysis of The Cancer Genome Atlas (C) indicated that an MDSC signature (CD11b+, CD33+, CD45+, CD244+, CXCR2+) negatively correlates with GBM patient prognosis. Statistical significance of survival was based on log-rank analysis. Abbreviations: CSCs, cancer stem cells; GBM, Glioblastoma; MDSC, Myeloid-derived suppressor cell.
To obtain T cells, spleens of nontumor-bearing wild-type C57BL/6 mice (Charles River Laboratories) were dissected from freshly sacrificed animals and mechanically dissociated in FACS buffer (5% serum in neurobasal medium). Isolation of native CD3 + T cells was accomplished by negative selection of splenic populations (Mouse T Cell Isolation Kit, StemCell Technologies, Vancouver, Canada, www.stemcell.com). For analysis of intratumoral cell populations, mice harboring intracranial tumors were sacrificed when they developed symptoms of tumor burden, and their tumors were dissected from the normal brain parenchyma. Tumors were mechanically dissociated, passed serially through 40 μm filters (single cell strainer, Fisher Scientific, Waltham, MA, www.fishersci.com), and analyzed without further dissociation. See Supporting Information Table S1 for information on antibodies used throughout this investigation.
Intracranial Injections
Human GBM cell populations or GL261 cells were injected as previously described [29–31]. Four-week-old female wild-type C57B/6 mice were purchased from Charles River Laboratories and used between 4 and 5 weeks of age for all experiments. A total of 20,000 GL261 cells were resuspended in 20 μl of RPMI null media and injected intracranially into the left hemisphere 2 mm caudal to the coronal suture, 3 mm lateral to the sagittal suture at a 90° angle with the murine skull to a depth of 2.5 mm. Mice were monitored twice daily for signs of tumor burden and sacrificed when symptomatic or when tumor volume exceeded 5% of body weight. 5-fluorouracil (5-FU, Sigma) was prepared fresh for every treatment by resuspending in DMSO and diluted in PBS to achieve working concentrations. 5-FU or equivalent concentrations of DMSO were injected intraperitoneally on a weekly basis for all treatment arms of the in vivo experiments. Mice were imaged every 2–3 days by injection of luciferin (50 μl) intraperitoneally after anesthetization by isoflurane and imaged using an IVIS in vivo imaging system (Brucker) to monitor tumor growth. Survival curves were generated and analyzed using SigmaStat (version 3.5.0.54, Dundas Software).
Proliferation, Sphere Formation, and Tumor Initiation Assays
Cell proliferation, self-renewal, and tumor initiation assays were performed as previously described [29–31]. For cell proliferation, self-renewal, and tumor initiation assays, freshly dissociated GBM xenografted cells were sorted by flow cytometry using a FACSAria II (BD Biosciences) into low and high populations for CD133. The 10% of cells with the lowest expression of the surface marker were classified as nonstem cells, and the 10% of cells with the highest expression were classified as stem cells.
For sphere formation assays, the FACSAria II was used to plate single, live cells into 96 well plates at increasing cell densities from 1 to 20 cells. Sorted cells were maintained for 10 days before assaying for sphere formation. Only spheres larger than 10 cells in diameter were counted. The Walter and Eliza Hall Institute Bioinformatics Division ELDA analyzer was used to analyze data and calculate stem cell frequency [36].
Proliferation of various cell populations in vitro was measured by seeding 2,000 cells per well of a 96 well tissue culture plate. Cells were cultured in complete medium, and Cell Titer analysis (CellTiter-Glo, Promega, Madison, WI, www.promega.com) of cell populations was assessed at 1, 3, 5, and 7 days post seeding.
Immunofluorescence and Immunohistochemistry
Immunostaining analysis of xenografted GBM specimens (generated by intracranial injection) or GL261 intracranial injections was performed as previously described [30]. For immunostaining analysis of transplanted mice, brains containing tumors were dissected from freshly sacrificed nude or C57B/6 mice, fixed in 4% formaldehyde, dehydrated using 30% sucrose, and embedded in optimum cutting temperature medium (OCT, Tissue-Tek). Ten micrometer tissue sections harboring tumors were mounted on glass slides (Superfrost microscope slides, Fisher Scientific), and cells were permeabilized using blocking buffer (PBS with 10% normal goat serum and 0.1% Triton X-100). Cells were washed, and primary and secondary antibodies were diluted in blocking buffer prior to staining. Cell nuclei were stained with Hoechst 33342 at a 1:10,000 dilution in PBS, coverslips were mounted using Fluorosave Mounting Reagent (Calbiochem, Billerica, MA, www.emdmillipore.com), and slides were stored at −20°C until imaging. Slides were imaged using the ×63 oil immersion lens of a Leica SP5 confocal microscope. All images are representative of four random fields within the stained sample and representative of three separate experiments.
Immunohistochemical analysis of human GBM specimens was performed as previously described [37]. To examine the proximity between GSCs and MDSCs, virtual triple immunohistochemistry was performed with antibodies detecting CD133 or SOX-2, CD33, and HLA-DR. CD133 and CD33 as well as SOX-2 and CD33 were established as double immunostainings, while HLA-DR was established as a single staining. Stainings were performed on serial sections of two TMAs. Briefly, formalin-fixed, paraffin-embedded human glioblastoma samples were mounted onto slides, deparaffinized, rehydrated, subjected to epitope retrieval, and incubated with the respective antibodies. Double staining with CD33/SOX-2 was carried out on an automated immunostainer (Benchmark Ultra IHC/ISH staining system, Ventana Medical Systems, Tucson, AZ, U.S.) using the UltraView Universal DAB detection kit containing five prediluted and ready-to-use dispensers plus amplification (Ventana). Sections were dried at 75°C for 4 minutes and deparaffinized at 72°C. The staining procedure included pre-treatment with cell conditioner 1 at 100°C for 32 minutes and quenching of endogenous peroxidases with H2O2. Slides were then incubated with primary antibody to CD33 (NCL-L, Novocastra, Newcastle, UK, 1:200) at 36°C for 16 minutes, and UltraView-DAB with amplification was used for visualization. Next, sections were reheated in a reaction buffer at 94°C for 4 minutes followed by incubation of the primary antibody to SOX2 (Clone #245610, R&D systems, 1:200) for 32 minutes at 36°C and visualized using UltraView Red Universal Alkaline Phosphatase detection kit with amplification (Ventana). Double staining with CD133/CD33 was performed on the AutostainerPlus and the BenchMark Ultra; sections were deparaffinized, and HIER was performed in TEG buffer. After blocking of endogenous peroxidase activity by incubation in 1.5% H2O2, sections were incubated with protein block to suppress nonspecific binding of subsequent reagents, followed by sequential incubation with mouse antihuman CD133 antibody (W6B3C1, Miltenyi Biotec, Germany, 1:40) for 60 minutes, antimouse immunoglobulin-HRP, fluorescyl-tyramide H2O2, and antifluorescein-HRP. After a second round of HIER and endogenous peroxidase blocking, slides were incubated with anti-CD33 for 32 minutes at 36°C on the BenchMark Ultra and visualized using UltraView Red Universal Alkaline Phosphatase detection kit with amplification (Ventana). HLA-DR staining was executed using the BenchMark Ultra. The standard protocol includes epitope retrieval in cell conditioner 1 buffer (Ventana) for 32 minutes at 100°C. Endogenous tissue peroxidase activity was inactivated using OptiView peroxidase inhibitor (Ventana). Primary antibody against HLA-DR (CR3/43, Dako, 1:200) was incubated for 32 minutes at 36°C, and OptiView-DAB (Ventana) was used according to the manufacturer’s recommendations for visualization of the bound primary antibody. All slides were scanned on a Hamamatsu Digital Slide Scanner (Hamamatsu, Japan). Omission of primary antibodies served as negative controls and controls for nonspecific staining related to the detection systems.
The stained sections were evaluated using the Visiopharm software (Visiopharm, Hørsholm, Denmark). Each core in the CD33/SOX2 stainings (n = 13) and in the CD33/CD133 stainings (n = 11) were aligned individually with their respective core in the HLA-DR stainings using the TISSUEalign module (Visiopharm). Sample images were then collected for each core using systematic uniform random sampling (meander fraction-based) at a sampling fraction that resulted in approximately 20 images per core. To ensure optimal alignment, sample images were reviewed, and the six best aligned images for each core were included for further analysis. Using the newCAST module (Visiopharm), the cells were counted manually to estimate the number of 1) CD133+/SOX2+ cells, 2) CD33+/HLA-DR− cells, 3) CD33+/HLA-DR+ cells, 4) CD33−/HLA-DR+ cells, and 5) negative cells (i.e., CD133−/SOX-2−/CD33−/HLA-DR− cells). For the CD33+/HLA-DR− cells, it was assessed whether they interacted directly with CD133+/SOX2+ cells (i.e., no other nucleus in between the two cells) or not. Cells were considered proximal when they were directly adjacent with no other nuclei in between. Cells were considered distal when one or more nuclei were located between the CD133+/SOX2+ cell and the CD33+/HLA-DR− cell. Cell fractions were calculated based on the total cell number and the total number CD33+ cells.
Immunoblotting
Immunoblotting of cell and marrow populations isolated as described above was performed as previously described [29–31]. Cell and marrow populations were generated as described above. Cell populations were incubated with RIPA lysis buffer (RIPA buffer supplemented with PMSF, protease inhibitor cocktail, and sodium orthovanadate; Santa Cruz Biotechnology, Dallas, TX, http://www.scbt.com), and protein concentrations were normalized using a BCA protein assay (Pierce Biotechnology, Rockford, IL, www.thermofisher.com). Proteins were denatured using Laemmli denaturing buffer (Bio-Rad Laboratories, Hercules, CA, www.bio-rad.com) supplemented with β-mercaptoethanol and separated using 12% or 15% polyacrylamide SDS-PAGE gels. Proteins were transferred to PVDF membranes and immunoblotted using the antibodies described. Membranes were developed using ECL-2 reagent (Pierce Biotechnology).
Flow Cytometry Analysis
Peripheral blood analysis to determine MDSC populations in GBM patients versus age-matched controls was performed in accordance with an approved Cleveland Clinic Foundation IRB protocol and analyzed as previously described [28]. To assess MDSC/CSC interactions, MDSC marrow populations were generated as described above. Marrow containing MDSCs was incubated with conditioned media generated from CSCs or NSTCs for 24 hours, after which analysis was performed.
Cell preparation and analysis was performed as previously described [29–31]. For analysis, cells were assessed post coincubation. Cells were blocked in FACS buffer, and live cells were stained with the listed antibodies to cell surface receptors. To stain for intracellular proteins, live cells were incubated with a Golgi plug prior to fixation (4% PFA) and permeabilization (FACS buffer supplemented with 0.1% Triton X-100). Fixed, permeabilized cells were subsequently stained using antibodies to the intracellular proteins of interest. Quantification of apoptosis was performed by staining live cell populations using Annexin V apoptosis kits (Life Technologies). Cell populations were analyzed using an LSRFortessa flow cytometer (BD Biosciences), and populations were separated and quantified using FlowJo imaging software (Tree Star Inc.). To determine the effect of MDSCs on T cells, freshly sorted MDSCs (CD244+/GR-1+) were incubated with freshly isolated splenic T cells from nontumor-bearing mice in CSC or NSTC conditioned media. T cells were activated by CD3/CD28 microbeads (CD3/CD28 Dynabeads, Life Technologies) and murine recombinant IL-2 (R&D systems) for 72 hours, after which flow cytometry analysis was performed as described above. MIF rescue experiments and competitive inhibition by small molecule inhibitors or neutralizing antibodies were accomplished by addition of the factors into the conditioned media immediately prior to incubation with MDSC populations. Recombinant MIF and G-CSF were purchased from R&D systems.
Cytokine Array
Conditioned media from CSCs and NSTCs were generated as described above and incubated with PVDF membranes containing antibodies to the cytokine array library (RayBiotech, Inc, Norcross, GA, www.raybiotech.com, Human Cytokine Antibody Array C Series 1000). Membranes were washed and developed in a manner similar to the previously described immunoblotting procedures, and pixel intensity was calculated by ImageJ.
Patient Dataset Analysis
Overall Myeloid Signature was calculated for 493 TCGA [38] patients from the glioblastoma (GBM) dataset for whom survival and mRNA expression data were available, based on CD11b (ITGAM), CD33 (CD33), and CD45 (PTPRC) expression levels. Survival was calculated using a Kaplan–Meier estimator and a logrank test with patients classified into high or low cohorts based on expression above or below the mean score, respectively. Overall Myeloid Signatures of 112 TCGA patients with RNA-sequencing data were subdivided into contributions from myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), and neutrophils based on the following gene sets: MDSC: CD11b, CD33, CD45, CD244, and CXCR2, TAM: CD11b, CD33, CD45, and IBA1 (AIF1), Neutrophil: CD11b, CD33, CD45, CD54 (ICAM1), CCR5, and CCR7 [39]. Additional analysis was performed using GlioVis (https://gliovis.bio-info.cnio.es) and exported directly from the program.
Statistical Analysis
All experiments were performed in triplicate. Values reported in the results represent mean values ±standard deviation. Unless otherwise stated, one-way ANOVA was used to calculate statistical significance; p-values are detailed in the text and figure legends. In vivo survival analysis was calculated by log-rank analysis.
Results
MDSCs are Present in the GBM Microenvironment Proximal to CSCs
To examine MDSCs in GBM, we interrogated 13 patient-derived specimens for the presence of MDSCs as detected by the expression of CD33 and the absence of HLA-DR. By omitting HLA-DR+ cells, we remove contaminating TAMs from our IHC analysis. We were readily able to detect MDSCs within the GBM microenvironment (Fig. 1A). This extends previous findings in which MDSCs were found in GBM patient blood [28]. We further validated that the MDSCs in GBM patient blood were skewed toward the more immunosuppressive, granulocytic subtype (Supporting Information Fig. S1). Based on the previously reported role of CSCs in immune modulation [19–21], we assessed the proximity of MDSCs to CSCs. Using the CSC markers CD133 and SOX2, we found that a significant fraction of MDSCs were located directly adjacent to CSCs (Fig. 1B). To assess whether the MDSC content in a given tumor was informative for patient survival, we constructed a gene signature based on the combined positive expression of CD11b, CD33, CD45, CD244, and CXCR2. We evaluated the TCGA dataset [38] and found that GBM patients with a higher MDSC gene signature had a significantly poorer prognosis (Fig. 1C). We additionally established that a high overall myeloid gene signature (CD11b+, CD33+, CD45+) was prognostic of poorer survival, but not those presenting with high neutrophil (CD11b+, CD33+, CD45+, CD54+, CCR5+, and CCR7+) [39] or TAM-specific (CD11b+, CD33+, CD45+, Iba1+) gene signatures (Supporting Information Fig. S6). These results indicate that MDSCs are present in patient tumors, they are associated with aggressive disease progression, and suggest that there may be an interaction between MDSCs and CSCs.
MDSCs Promote Tumor Development
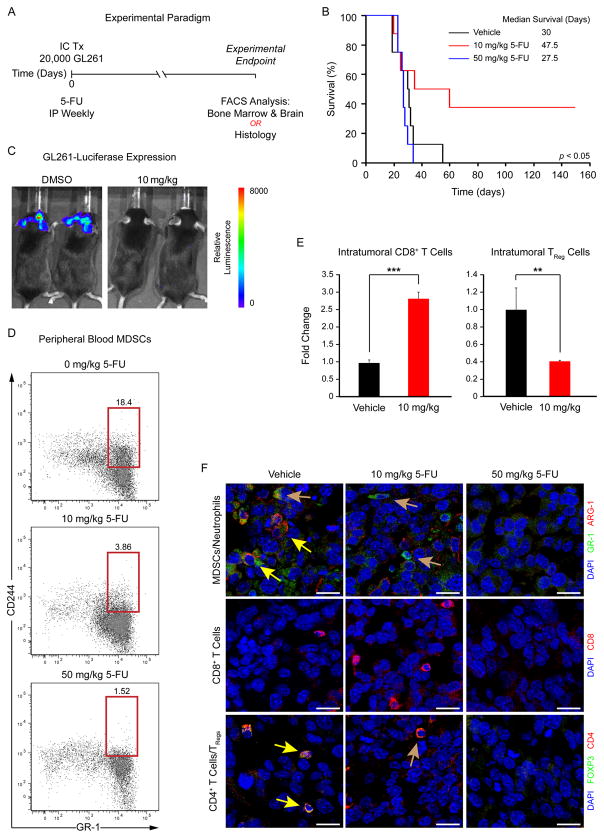
To test the hypothesis that MDSCs impact tumor development, we leveraged previous findings that low-dose 5-fluorouracil (5-FU) is selectively toxic to MDSCs [40]. This pharmacological approach was chosen due to the lack of a specific genetic strategy to selectively eliminate MDSCs [40]. Using the GL261 syngeneic mouse model, we initiated tumors and treated with varying doses of 5-FU weekly (Fig. 2A). A 10 mg/kg dose selectively targeted MDSCs, whereas a 50 mg/kg dose nonspecifically ablated immune cells. We found that 10 mg/kg 5-FU significantly increased tumor latency and reduced tumor growth compared with 50 mg/kg or vehicle treatments (Fig. 2B, 2C). We evaluated the peripheral blood of tumor-bearing mice and found a reduction in MDSCs as reported by coexpression of CD11b, CD244, and GR-1 [35] in mice treated with 10 mg/kg and 50 mg/kg 5-FU compared with vehicle control (Fig. 2D). After depleting MDSCs with 10 mg/kg 5-FU, we observed an increase in the number of cytotoxic CD8-positive T cells and a reduction in the number of immune-suppressive CD4/CD25-positive T regulatory cells (TRegs) in the tumor (Fig. 2E, Supporting Information Table S2). We verified the changes in the immune cell composition within tumors after 5-FU treatment using immunofluorescence (Fig. 2F). In the vehicle control, we observed MDSCs, limited CD8-positive T cells, and a moderate number of TRegs. In the tumors from 10 mg/kg-treated mice, we were not able to detect MDSCs and found increased CD8-positive T cells and reduced TRegs. In tumors from 50 mg/kg-treated mice, we were not able to reliably detect any immune cell types. These results indicate that specifically targeting MDSCs alters tumor growth and immune composition within the tumor microenvironment (TME).
Figure 2.
MDSCs promote tumor development. Schematic depicting treatment paradigm (A), in which MDSCs were targeted using a low-dose 5-fluorouracil (5-FU) treatment strategy during tumor engraftment and growth in the GL261 syngeneic glioma model. Kaplan–Meier survival curves (B) demonstrate that tumor-bearing mouse survival was increased with low-dose 5-FU treatment (10 mg/kg, red) compared with higher-dose (50 mg/kg, blue) and vehicle control (black) groups. Examples of tumor burden as measured by bioluminescence (C) show differences in low-dose 5-FU compared with the control treatment group. Analysis of peripheral blood MDSC levels (based on CD244.2 and GR-1 positivity) in tumor-bearing mice using flow cytometry (D) demonstrates that 5-FU treatment reduces MDSCs. Analysis of brain tissue in tumor-bearing mice using flow cytometry (E) reveals that 10 mg/kg 5-FU increased intratumoral CD8-positive T cells (CD3+/CD8+) and reduced T regulatory (TReg) cells (CD3+/CD4+/CD25+/Foxp3+). Micrographs of tumor-bearing mice (F) confirm a reduction in MDSCs (GR-1+/ARG-1+, yellow arrows in top panels), neutrophils (GR-1+/ARG-1−, yellow arrows in top panels), TRegs (FOXP3+/CD4+, yellow arrows in bottom panels), and CD4+ activated T cells (FOXP3−/CD4+, brown arrow bottom panel) with a concomitant increase in CD8+ T cells with 10 mg/kg 5-FU treatment compared with vehicle control. These immune cell populations were not detected after treatment with 50 mg/kg 5-FU. Values shown are means ±standard deviation, scale bar = 20 μm, and nuclei were counterstained with DAPI. *, p <.05, **, p <.01, and ***, p <.001 by one-way ANOVA, and statistical significance for in vivo studies was calculated using log-rank analysis. Abbreviations: IC Tx, intracranial transplantation; IP, intraperitoneal; MDSC, Myeloid-derived suppressor cell.
CSCs Promote MDSC Function and Survival
Evaluating interactions between MDSCs and human CSCs is confounded by limitations in model systems. Although a population of CD133-expressing cells has been identified in the GL261 system [41], CSCs have been historically defined in human patient-derived xenograft models [11, 16, 42]. However, these models are generated in severely immunocompromised mice that lack key immune cell populations including MDSCs. To allow for the evaluation of MDSCs and human CSCs, nude mice were required. We identified MDSCs in the marrow and brains of mice engrafted with CSCs in contrast to those engrafted with nonstem tumor cells (NSTCs, Supporting Information Fig. S2A, S2B). Arginase-1, a marker of MDSC function, was elevated as assessed by flow cytometry in the myeloid cells of CSC tumor-bearing mice, specifically in the MDSCs (Supporting Information Fig. S2C).
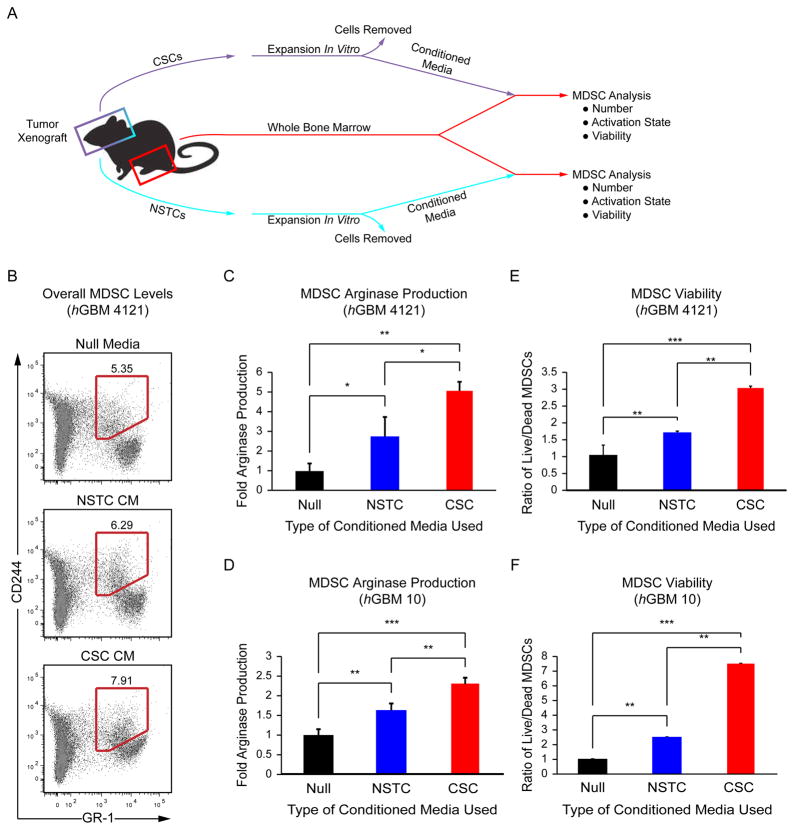
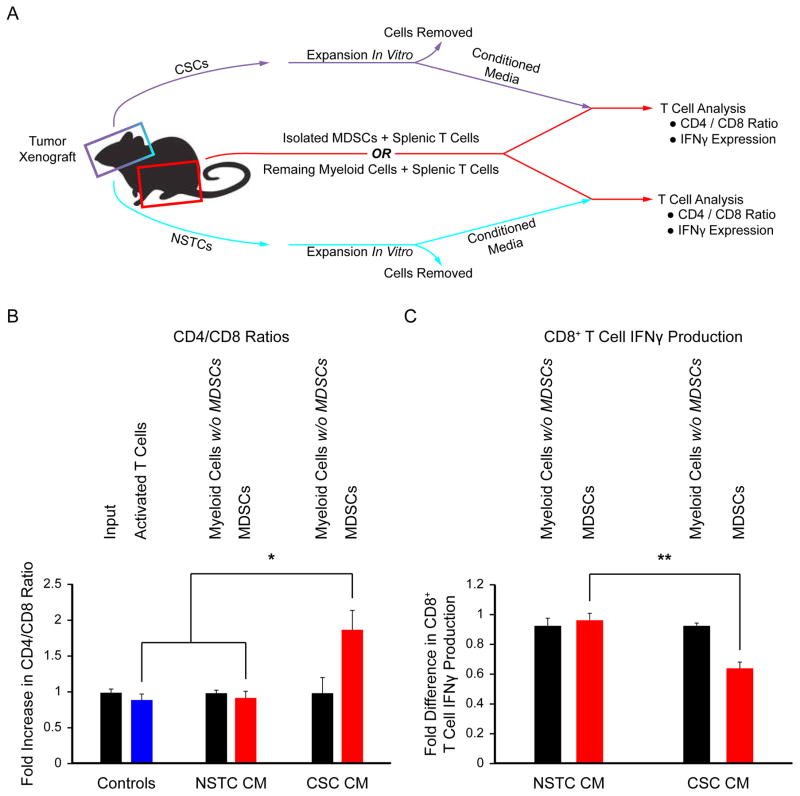
Based on our observation that MDSCs were adjacent to CSCs, we hypothesized that there was a functional interaction between these two cell types. Whole bone marrow from tumor-bearing nude mice was cultured with conditioned media from CSCs or NSTCs and compared with a null media control (Fig. 3A). While MDSCs were present in all media conditions (Fig. 3B), incubation in CSC conditioned media resulted in the largest increase in arginase-1 expression (Fig. 3C, 3D; Supporting Information Table S3) and granulocytic MDSC production (Supporting Information Fig. S3). CSC conditioned media also increased the survival of MDSCs (Fig. 3E, 3F, Supporting Information Table S3). We introduced immature splenic T cells into this coculture system (Fig. 4A) and found that MDSCs primed with CSC conditioned media increased the ratio of CD4-positive to CD8-positive T cells (Fig. 4B, Supporting Information Table S4) and decreased interferon-γ (IFNγ) production (Fig. 4C, Supporting Information Table S5), indicating the emergence of an immunosuppressive phenotype. These results demonstrate that MDSC-mediated immune suppression is enhanced by CSCs and suggests that the enrichment in MDSCs in the GBM microenvironment may require this interaction.
Figure 3.
CSC conditioned media increases arginase-1 production and survival in MDSCs. Schematic (A) depicting the coculture strategy employed to determine the effect of CSC and NSTC conditioned media on MDSCs. Flow cytometry plots (B) of overall MDSC levels (red region) with control (unconditioned) media (null), NSTC conditioned media, and CSC conditioned media. Arginase-1 expression in MDSCs derived from the marrow of tumor-bearing mice was elevated as assessed by flow cytometry with CSC conditioned media compared with NSTC conditioned and control media. These results were repeated using mice implanted with two separate patient-derived human GBM specimens, hGBM 4121 (C) and hGBM 10 (D). The ratio of live to dead cells as assessed by Annexin V in MDSCs derived from the marrow of tumor-bearing mice was elevated with CSC conditioned media and compared with NSTC conditioned and control media. Experiments were repeated with conditioned media generated from the CSCs and NSTCs isolated from hGBM 4121 (E) and hGBM 10 (F) xenografts. Values shown are means ±standard deviation, *, p <.05, **, p <.01, and ***, p <.001 by one-way ANOVA. Abbreviations: CM, conditioned media; CSCs, cancer stem cells; MDSC, Myeloid-derived suppressor cell; NSTC, Nonstem tumor cell.
Figure 4.
MDSCs exposed to CSC conditioned media promote immune suppression. Schematic (A) depicting the coculture strategy employed to determine the effect of MDSCs exposed to CSCs and NSTC conditioned media on T cell function. The ratio of CD4+ to CD8+ cells (B) was altered by MDSCs exposed to CSC conditioned media compared with NSTC conditioned media. The activation of T cells as reported by interferon-γ (IFNγ) expression (C) was suppressed when MDSCs were exposed to CSC conditioned media compared with NSTC conditioned media. Values shown are means ±standard deviation, *, p <.05, **, p <.01, and ***, p <.001 by one-way ANOVA. Activated T cells and neutrophils were used as controls. Abbreviations: CSC, cancer stem cell; MDSCs, Myeloid-derived suppressor cells; NSTC, Nonstem tumor cell.
CSCs Secrete MIF to Promote MDSC-Mediated Immune Suppression
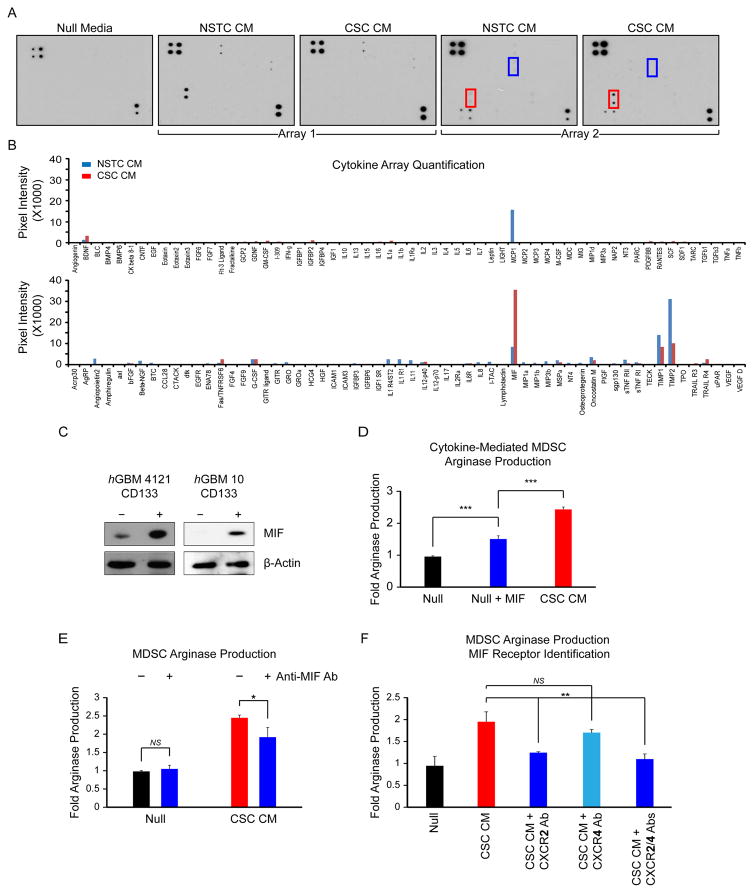
Based on the ability of CSCs to amplify MDSC function, we hypothesized that CSCs secrete one or more factors that promote MDSC-mediated immune suppression. To identify these factors, we interrogated the secreted cytokines present in CSC, NSTC, and null conditioned media. Using a cytokine array, we observed a robust difference between the secretion of macrophage migratory inhibitory factor (MIF) in CSC and NSTC conditioned media (Fig. 5A, 5B). It is worth noting that because the array data was acquired based on an HRP-ECL reaction similar to western analysis, we used an exposure time appropriate to prevent signal saturation from MIF, the most highly-expressed species on the array. Allowing for a longer exposure could have accentuated the weaker signals from other cytokines, however we would then lose the ability to quantify the expression of these additional cytokines relative to the dominant species. Thus, other cytokines of interest were not absent; rather, the intensity of the MIF signal dwarfed the signals from most of the other species on the array. For example, TGFβ, which has been linked to the CSC phenotype and to tumor progression, presented a twofold enrichment in CSC conditioned media compared with NSTC media [43, 44]. We validated the differential expression of MIF between CSCs and NSTCs using immunoblotting (Fig. 5C). Additionally, to add support to the differential expression of MIF between CSCs and NSTCs, we conducted a targeted qRT-PCR screen for the top candidates identified in our cytokine array screen comparing CSC enriched cultures to nontransformed astrocytes. In accordance with our cytokine array findings, we identified MIF expression across 8 additional hGBM CSC-enriched specimens compared with control astrocytes. In contrast, levels of CCL2 (MCP1), TIMP1 and TIMP2 were markedly lower than in the astrocyte control (Supporting Information Fig. S7A). Furthermore, in order to control for the complication of the different culture conditions used to cultivate CSCs and NSTCs, we interrogated MIF expression levels from CSCs and NSTCs cultured in Neurobasal complete media (NBMC), Neurobasal null media (NBM−EF), and null media supplemented with 10% FCS (NBM−EF+FCS). Both cell types were conditioned in each of the divergent media types for 24 hours prior to collection and analysis. Regardless of culture condition, we observed increased MIF expression in the CSC pool compared with the NSTC pool, suggesting a cell-intrinsic higher level of MIF-expression in the CSC population (Supporting Information Fig. S7B). The addition of MIF to null media potentiated MDSC arginase-1 production (Fig. 5D; Supporting Information Table S6), and the effect of CSC conditioned media on arginase-1 production was blunted by the addition of a MIF blocking antibody (Fig. 5E; Supporting Information Table S6). To identify the MDSC MIF receptor, we evaluated two known receptors that bind MIF, CXCR2 and CXCR4. Using blocking antibodies against each receptor, we found that blockade of CXCR2 but not CXCR4 inhibited arginase-1 production (Fig. 5F; Supporting Information Table S6). These results substantiate that MIF is a factor secreted by CSCs that mediates the MDSC immune suppressive phenotype.
Figure 5.
CSC conditioned media is enriched in macrophage MIF, which increases arginase-1 production in MDSCs in a CXCR2-dependent manner. Dot blots (A) and quantitation (B) of cytokine arrays. Red box: MIF; blue box: G-CSF. Immunoblots of MIF in GBM CSCs (CD133+) and NSTCs (CD133−) in human GBM cells from two patient specimens (C). MDSC ARG-1 production as assessed by flow cytometry after the addition of exogenous MIF (D) or MIF receptor neutralizing antibodies (E) to conditioned media. (F) MDSC ARG-1 production in the presence of neutralizing antibodies to the CXCR2 and CXCR4 receptors. Values shown are means ±standard deviation, *, p <.05, **, p <.01, and ***, p <.001 by one-way ANOVA. Abbreviations: CM, conditioned media; CSC, cancer stem cell; MDSCs, Myeloid-derived suppressor cells; MIF, migration inhibitory factor; NSTC, Nonstem tumor cell.
MIF Depletion Increases Tumor Latency and Alters Tumor Immune Cell Composition
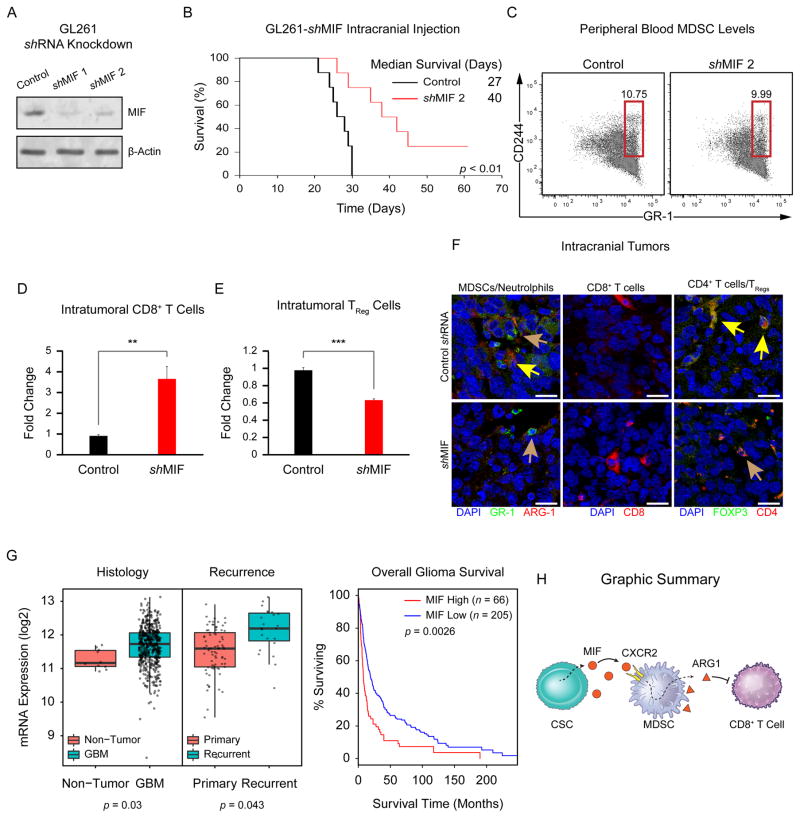
Based on the robust expression of MIF in CSCs, we hypothesized that silencing MIF would attenuate immune suppression and prolong survival. To interrogate this hypothesis in an immune-competent context, we utilized the GL261 system. We attenuated MIF expression using short hairpin RNA (Fig. 6A) and found an increase in tumor latency with MIF knockdown (Fig. 6B). While we did not observe an appreciable difference in the number of MDSCs in the peripheral blood (Fig. 6C), we did note a significant increase in CD8-positive cytotoxic T cells and a significant decrease in TRegs in the brains of these animals (Fig. 6D–6F; Supporting Information Table S7). As it was critical to establish that MIF knockdown did not directly impair the ability of these cells to generate tumors, we evaluated proliferation, apoptosis, and self-renewal in GL261 cells depleted of MIF. Indeed, we found that MIF knockdown did not alter these behaviors in GL261 cells (Supporting Information Fig. S4). To further support the role of MIF in the TME, we evaluated a series of bioinformatics databases and found that MIF was significantly elevated in GBM compared with nontumor tissue and elevated in recurrent GBM compared with primary tumors (Fig. 6G). Further, high MIF levels correlated with poorer prognosis of glioma patients (Fig. 6G), especially in the proneural GBM subclass (Supporting Information Fig. S5A–S5D). These results indicate that MIF alters tumor growth by dampening immune suppression in the TME.
Figure 6.
MIF depletion attenuates tumor immune suppression and confers a survival advantage in vivo. Immunoblot for MIF after MIF shRNA treatment of GL261 cells (A). Survival of mice intracranially injected with control (black) or MIF KD (red) GL261 cells, p <.01 by log-rank test (B). Median survival is indicated. MDSCs in the peripheral blood of mice bearing control and MIF KD GL261 intracranial xenografts (C). Intratumoral CD8+ T cells (CD3+/CD8+), **, p <.01 by pairwise t-test (D) and TReg cells (CD3+/CD4+/CD25+/FOXP3+), ***, p <.001 by pairwise t-test (E) in MIF KD GL261-injected mice vs. controls. Immunofluorescence of intracranial mouse xenografts (F). (Left) Yellow arrows: MDSCs (GR-1+/ARG-1+); brown arrows: neutrophils (GR-1+/ARG-1−). (Right) Yellow arrows: TRegs (FOXP3+/CD4+); brown arrows: CD4+ activated T cells (FOXP3−/CD4+). Bioinformatics analysis of MIF mRNA expression levels in human GBM pathological specimens (G) compared with nonmalignant control tissues (Left Panel, p = .03 by pairwise t-test). MIF mRNA expression levels in primary versus recurrent human GBM (Central Panel, p = .043 by pairwise t-test). Kaplan–Meier analysis of MIF expression levels informs human GBM patient survival (Right Panel, p = .0026 by log-rank test). Cartoon summary of the CSC/MDSC/T Cell interaction that takes place in the GBM TME (H). Scale bar = 20 μm. Abbreviations: GBM, glioblastoma; MDSCs, myeloid-derived suppressor cells; MIF, migration inhibitory factor.
Discussion
The success of immunotherapy for GBM is predicated on a thorough understanding of the immune cell composition within the TME. This requires delineating the function of individual immune cell types as well as their interaction with cancer cells and other stromal populations. Building on observations that GBM patients exhibited elevated numbers of MDSCs in their blood [28], we determined that MDSCs infiltrate the GBM-bearing brain, take up residence adjacent to CSCs and contribute to immune suppression. Previously published works substantiate that CSCs alter the function of various immune cell populations in service of the developing tumor [19–21]. Here, we extend the scope of this understanding, demonstrating the critical importance of CSC-secreted MIF as a potent activator of the MDSC immune-suppressive phenotype (Fig. 6H). While MIF has been implicated in multiple cancers outside of the central nervous system [45, 46, 55, 56] and its activation of tumor immune suppression is well documented [47–49], its role as a mediator of a GBM CSC/tumor-infiltrating MDSC interaction was previously unknown. Further, the finding that CXCR2, but not CXCR4, facilitated MIF-driven MDSC function (Fig. 6H) is concordant with recent reports demonstrating the importance of CXCR2-expressing MDSCs in the suppression of chronic inflammation and the progression of colon cancer [50]. Because MIF knockdown did not alter the growth characteristics of tumor cells themselves, we reason that MIF is primarily an indirect promoter of GBM progression that works to suppress immune rejection by activating and protecting immune-suppressive MDSCs within the GBM TME.
Interestingly, Zhai and colleagues observed a survival advantage in GL261-injected animals treated with MIF [51], which directly contrasts with our observations of prolonged survival upon MIF knockdown. We speculate that the discrepancy between our data and the work published by the Tsirka group may be due to MIF dosage and the relative contributions to tumor progression from TAMs versus MDSCs. The Tsirka groups in vivo experiment involved inhibition of TAM activity via continuous delivery of high concentrations (250 μg/ml) of MIF over 28 days. At such exaggerated levels, we speculate that TAM-mediated tumor inhibition may dominate over the tumor promoting effects of CSCs interacting with MDSCs.
The mechanisms responsible for the differential expression of MIF between CSCs and NSTCs remain unknown. Recently, O-GlcNAcylation [52] of MIF and binding to heat shock protein 90 [53] were identified as mechanisms of MIF stabilization. We speculate that one or both of these mechanisms may operate within CSCs. While our data indicate a functional role for MIF in MDSC activation by CSCs, additional factors that mediate this activation likely also exist. Future investigations along these lines may reveal additional opportunities to compromise CSC-driven immune suppression and enhance tumor immune rejection. In total, our data support a CSC/MIF/immunosuppressive axis that serves as a critical mediator of GBM immune evasion and one that deserves further inquiry.
Conclusion
At a basic level, our results reveal even greater complexity within the immune compartment of the GBM microenvironment than had previously been appreciated. With accumulating evidence that targeting immune-suppressive cell types can result in complete immune rejection of established tumors [54], identifying all cell types responsible for immune suppression is critical. We anticipate that MDSC-targeted therapies will ultimately be used in conjunction with other immunotherapies intended to compromise immune checkpoints and enhance immune-mediated tumor rejection. With the limited efficacy of targeted therapies based on molecular genetic interrogation of tumors, immunotherapies represent a promising alternative. We envision the targeted eradication of the tumor-associated MDSC population becoming a critical component of future immunotherapeutic management of GBM.
Supplementary Material
Supplemental Table 1. Primary and Secondary Antibody Information
Significance Statement.
Cancer stem cells drive immune suppression by secreting macrophage migration inhibitor factor that enhances the immune-suppressive properties of myeloid-derived suppressor cells.
Acknowledgments
We thank the members of the Lathia laboratory for insightful discussion and constructive comments on the manuscript. We thank Cathy Shemo, Patrick Barrett, and Sage O’Bryant for flow cytometry assistance and the Cleveland Clinic Foundation Tissue Procurement Service and Mary McGraw. A Distinguished Scientist Award from the Sontag Foundation supports the work on CSCs and the immune system in the Lathia Laboratory. Blast GBM and the Cleveland Clinic VeloSano Bike Race fund work on CSCs and the immune system in the Lathia and Vogelbaum laboratories. The Lathia laboratory also receives funding from the National Institutes of Health grants NS089641, NS083629, CA157948, and CA191263; a Research Scholar Award from the American Cancer Society; and the Case Comprehensive Cancer Center. Work in the Vogelbaum laboratory is also supported by the Wolf Family Foundation, and work in the Finke laboratory is supported by National Institutes of Health grant CA150959. This work was supported by Sontag Foundation Distinguished Scientist Award (JDL), Blast GBM (JDL, MAV), and the Cleveland Clinic VeloSano Bike Race funds (JDL, MAV). R.M.R. is currently affiliated with the Biogen IDEC, Cambridge, Massachusetts.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interests.
Author Contributions
B.O.: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. D.J.S.: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. E.E.M-H.: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. A.G.A.: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript. S.M.T.: Collection and/or assembly of data, final approval of manuscript. M.D.S.: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript. P.R.: Collection and/or assembly of data, final approval of manuscript. W.A.F.: Collection and/or assembly of data, final approval of manuscript. J.S.H.: Collection and/or assembly of data, final approval of manuscript. K.S.: Collection and/or assembly of data, final approval of manuscript. M.S.: Collection and/or assembly of data, final approval of manuscript. Q.W.: Collection and/or assembly of data, final approval of manuscript. A.J.: Collection and/or assembly of data, final approval of manuscript. S-H.K.: Collection and/or assembly of data, final approval of manuscript. P.L.F.: Data analysis and interpretation, final approval of manuscript. I.N.: Data analysis and interpretation, final approval of manuscript. J.N.R.: Data analysis and interpretation, final approval of manuscript. R.M.R.: Data analysis and interpretation, final approval of manuscript. J.F.: Data analysis and interpretation, financial support, final approval of manuscript. B.W.K.: Data analysis and interpretation, provision of study materials or patients, final approval of manuscript. M.A.V.: Conception and design, data analysis and interpretation, financial support, final approval of manuscript. J.D.L.: Conception and design, data analysis and interpretation, manuscript writing, financial support, administrative support, final approval of manuscript.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Gene Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 4.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohanbash G, McKaveney K, Sakaki M, et al. GM-CSF promotes the immunosuppressive activity of glioma-infiltrating myeloid cells through Interleukin-4 receptor. Cancer Res. 2013;73:6413–6423. doi: 10.1158/0008-5472.CAN-12-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gielen PR, Schulte BM, Kers-Rebel ED, et al. Increase in both CD14-positive and CD15-positive myeloid-derived suppressor cell subpopulations in the blood of patients with glioma but predominance of CD15-positive myeloid-derived suppressor cells in glioma tissue. J Neuropathol Exp Neurol. 2015;74:390–400. doi: 10.1097/NEN.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 7.Platten M, Kretz A, Naumann U, et al. Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas. Ann Neurol. 2003;54:388–392. doi: 10.1002/ana.10679. [DOI] [PubMed] [Google Scholar]
- 8.Fecci PE, Mitchell DA, Whitesides JF, et al. Increased regulatory T-Cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–3302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- 9.Lewis CE. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 10.Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 12.Singh SK, Clarke ID, Hide T, et al. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 13.Ignatova TN, Kukekov VG, Laywell ED, et al. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 14.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 16.Bao S, Wu Q, Mclendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133 +cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Li Y, Yu T-S, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2013;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei J, Barr J, Kong LY, et al. Glioblastoma cancer-initiating cells inhibit T-Cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar S, Doring A, Zemp FJ, et al. Therapeutic activation of macrophages and microglia to suppress brain tumor-initiating cells. Nat Neurosci. 2013;17:46–55. doi: 10.1038/nn.3597. [DOI] [PubMed] [Google Scholar]
- 21.Zhou W, Ke SQ, Huang Z, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol. 2015;17:170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silver DJ, Sinyuk M, Vogelbaum MA, et al. The intersection of cancer, cancer stem cells, and the immune system: Therapeutic opportunities. Neuro Oncol. 2016;18:153. doi: 10.1093/neuonc/nov157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufmann JK, Chiocca EA. Glioma virus therapies between bench and bedside. Neuro Oncol. 2014;16:334–351. doi: 10.1093/neuonc/not310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wainwright DA, Chang AL, Dey M, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin Cancer Res. 2014;20:5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko JS, Zea AH, Rini BI, et al. Sunitinib mediates reversal of myeloid-derived suppressor cell accumulation in renal cell carcinoma patients. Clin Cancer Res. 2009;15:2148–2157. doi: 10.1158/1078-0432.CCR-08-1332. [DOI] [PubMed] [Google Scholar]
- 26.Ostrand-Rosenberg S. Myeloid-derived suppressor cells: More mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother. 2010;59:1593–1600. doi: 10.1007/s00262-010-0855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozao-Choy J, Ma G, Kao J, et al. The novel role of tyrosine kinase inhibitor in the reversal of immune suppression and modulation of tumor microenvironment for immune-based cancer therapies. Cancer Res. 2009;69:2514–2522. doi: 10.1158/0008-5472.CAN-08-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raychaudhuri B, Rayman P, Ireland J, et al. Myeloid-derived suppressor cell accumulation and function in patients with newly diagnosed glioblastoma. Neuro Oncol. 2011;13:591–599. doi: 10.1093/neuonc/nor042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyler CE, Wu Q, Yan K, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lathia JD, Gallagher J, Heddleston JM, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: Development of an experimental model. Cancer Res. 1970;30:2394–2400. [PubMed] [Google Scholar]
- 33.Hitomi M, Deleyrolle LP, Mulkearns-Hubert EE, et al. Differential connexin function enhances self-renewal in glioblastoma. Cell Rep. 2015;11:1031–1042. doi: 10.1016/j.celrep.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lathia JD, Li M, Sinyuk M, et al. High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep. 2014;6:117–129. doi: 10.1016/j.celrep.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youn JI, Collazo M, Shalova IN, et al. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc Biol. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Christensen K, Schrøder HD, Kristensen BW. CD133 +niches and single cells in glioblastoma have different phenotypes. J Neurooncol. 2010;104:129–143. doi: 10.1007/s11060-010-0488-y. [DOI] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest. 2014;124:5466–5480. doi: 10.1172/JCI77053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vincent J, Mignot G, Chalmin F, et al. 5-fluorouracil selectively kills tumor-associated myeloid-derived suppressor cells resulting in enhanced T cell-dependent antitumor immunity. Cancer Res. 2010;70:3052–3061. doi: 10.1158/0008-5472.CAN-09-3690. [DOI] [PubMed] [Google Scholar]
- 41.Wu A, Oh S, Wiesner SM, et al. Persistence of CD133 +cells in human and mouse glioma cell lines: Detailed characterization of GL261 glioma cells with cancer stem cell-like properties. Stem Cell Dev. 2008;17:173–184. doi: 10.1089/scd.2007.0133. [DOI] [PubMed] [Google Scholar]
- 42.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 43.PeNuelas S, Anido J, Prieto-SAnchez RM, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, et al. TGF-beta receptor inhibitors target the CD44high/Id1high glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 45.Simpson KD, Templeton DJ, Cross JV. Macrophage migration inhibitory factor promotes tumor growth and metastasis by inducing myeloid-derived suppressor cells in the tumor microenvironment. J Immunol. 2012;189:5533–5540. doi: 10.4049/jimmunol.1201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lippitz BE. Review Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol. 2013;14:e218–e228. doi: 10.1016/S1470-2045(12)70582-X. [DOI] [PubMed] [Google Scholar]
- 47.Mittelbronn M, Platten M, Zeiner P, et al. Macrophage migration inhibitory factor (MIF) expression in human malignant gliomas contributes to immune escape and tumour progression. Acta Neuropathol. 2011;122:353–365. doi: 10.1007/s00401-011-0858-3. [DOI] [PubMed] [Google Scholar]
- 48.Hussain F, Freissmuth M, Volkel D, et al. Human anti-macrophage migration inhibitory factor antibodies inhibit growth of human prostate cancer cells in vitro and in vivo. Mol Cancer Ther. 2013;12:1223–1234. doi: 10.1158/1535-7163.MCT-12-0988. [DOI] [PubMed] [Google Scholar]
- 49.Põlajeva J, Bergstrom T, Edqvist P-H, et al. Glioma-derived macrophage migration inhibitory factor (MIF) promotes mast cell recruitment in a STAT5-dependent manner. Mol Oncol. 2014;8:50–58. doi: 10.1016/j.molonc.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh H, Wang D, Daikoku T, et al. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell. 2013;24:631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhai H, Heppner FL, Tsirka SE. Microglia/macrophages promote glioma progression. Glia. 2010;59:472–485. doi: 10.1002/glia.21117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Y, Li X, Qian X, et al. Secreted and O-GlcNAcylated MIF binds to the human EGF receptor and inhibits its activation. Nat Cell Biol. 2015;17:1348–1355. doi: 10.1038/ncb3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz R, Marchenko ND, Holembowski L, et al. Inhibiting the HSP90 chaperone destabilizes macrophage migration inhibitory factor and thereby inhibits breast tumor progression. J Exp Med. 2012;209:275–289. doi: 10.1084/jem.20111117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu P, Lee Y, Liu W, et al. Intratumor depletion of CD4 +cells unmasks tumor immunogenicity leading to the rejection of late-stage tumors. J Exp Med. 2005;201:779–791. doi: 10.1084/jem.20041684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaddanapudi K, et al. MIF Is Necessary for Late-Stage Melanoma Patient MDSC Immune Suppression and Differentiation. Cancer Immunology Research. 2016;4:101–112. doi: 10.1158/2326-6066.CIR-15-0070-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta Y, Pasupuleti V, Du W, Welford SM. Macrophage Migration Inhibitory Factor Secretion Is Induced by Ionizing Radiation and Oxidative Stress in Cancer Cells. PLoS ONE. 2016;11:e0146482–11. doi: 10.1371/journal.pone.0146482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Primary and Secondary Antibody Information