ABSTRACT
Streptococcus suis is a bacterium that is commonly carried in the respiratory tract and that is also one of the most important invasive pathogens of swine, commonly causing meningitis, arthritis, and septicemia. Due to the existence of many serotypes and a wide range of immune evasion capabilities, efficacious vaccines are not readily available. The selection of S. suis protein candidates for inclusion in a vaccine was accomplished by identifying fitness genes through a functional genomics screen and selecting conserved predicted surface-associated proteins. Five candidate proteins were selected for evaluation in a vaccine trial and administered both intranasally and intramuscularly with one of two different adjuvant formulations. Clinical protection was evaluated by subsequent intranasal challenge with virulent S. suis. While subunit vaccination with the S. suis proteins induced IgG antibodies to each individual protein and a cellular immune response to the pool of proteins and provided substantial protection from challenge with virulent S. suis, the immune response elicited and the degree of protection were dependent on the parenteral adjuvant given. Subunit vaccination induced IgG reactive against different S. suis serotypes, indicating a potential for cross protection.
KEYWORDS: Streptococcus suis, adjuvant, functional genomics, subunit, vaccine
INTRODUCTION
Streptococcus suis is a Gram-positive bacterium commonly carried in the tonsil and nasal cavity of swine that can cause systemic disease and secondary pneumonia, especially in young pigs. Streptococcal disease is widespread wherever pig production occurs, and systemic invasion most commonly results in septicemia, meningitis, arthritis, and/or polyserositis, causing significant economic losses to the industry. S. suis is also a zoonotic agent capable of causing meningitis in humans, and although it is historically sporadic in nature, there have been recent larger outbreaks in China and Vietnam with high levels of mortality (1–3). There are at least 33 capsular serotypes (serotypes 1 to 31, 33, and 1/2) of S. suis, with serotypes 32 and 34 having been reassigned (4) and ongoing controversy over the appropriate species of serotypes 20, 22, 26, and 33 (5). In most countries, capsular serotype 2 is the most virulent and the most frequently isolated from both diseased swine and humans (6). However, depending on the geographic location, other serotypes, such as serotypes 1, 1/2, 3, 7, 8, 9, and 14, are commonly isolated from diseased pigs (7–10).
The mechanisms that enable S. suis to invade systemically from the respiratory tract are not well understood, though numerous potential virulence factors or virulence-related factors have been identified (reviewed by Segura et al. [11]). However, none of these factors individually appear to correlate completely with the ability to cause disease; thus, virulence is probably multifactorial, and to date, no highly effective vaccines have been developed to protect against S. suis disease. Genomic analysis of large numbers of isolates with known commensal or disease-associated provenance revealed a complex population structure with high levels of recombination and marked genomic differences between the two groups (12). The presence of multiple serotypes and the high genotypic variability may make it difficult to develop broadly protective vaccines.
A relatively new technique called transposon-directed insertion sequencing (TraDIS or TnSeq) is a method used to simultaneously identify bacterial fitness genes by the generation of a random transposon library disrupting individual gene expression and assessment of the effects of the disruption on survivability under selection conditions. The high-throughput sequencing technology is used to generate sequence reads spanning the transposon/chromosome boundaries of each insertion, allowing the en masse accurate mapping of transposon insertion sites (13–17). By identifying members of the library that are no longer present after the applied negative selection, disrupted genes that are important for fitness under the applied conditions can be readily identified. Prior to this study, we processed an S. suis strain P1/7 TraDIS library through an in vitro organ culture (IVOC) system using pig nasal epithelium to select genes encoding proteins that may be involved in colonization fitness. Using an in silico bioinformatics approaches, five S. suis proteins were further selected on the basis of a likely cell surface location and conservation. The five proteins were cloned, expressed, and purified in Escherichia coli and then tested as potential vaccine candidates in swine.
RESULTS
Characteristics of the five candidate vaccine proteins.
Five candidate vaccine proteins (SSU0185, SSU1215, SSU1355, SSU1773, SSU1915) were selected on the basis of the results of the experimental functional genomics screening and in silico bioinformatics approaches described in Materials and Methods (Table 1). Candidate genes resulting in a significant reduction in the fitness of transposon mutants in an IVOC system with swine respiratory epithelium were narrowed down to those encoding surface-associated proteins, excluding those containing transmembrane domains in the middle of the protein-coding sequence (Table 1). Homology searches were used to identify proteins highly conserved in 459 publically available S. suis genomes which cover all serotypes with the exception of serotypes 20, 22 and 33 and come from Argentina, Canada, China, Denmark, Germany, The Netherlands, the United Kingdom, and Vietnam (Tables 2 and 3). Of the five proteins chosen, SSU0185 and SSU1355 were found in the genomes of all 459 S. suis isolates, SSU1915 was found in >99% of the isolates, and SSU1215 and SSU1773 were found in >98% of the isolates (Table 2). The identities of the five subunit vaccine candidate proteins were compared to those of the proteins in S. suis strains for which complete genomes are available in GenBank (see Table S1 in the supplemental material) and disease-associated S. suis serotype representatives from the collection of 459 S. suis genomes (Table 3). These strains represent disease-associated S. suis serotypes isolated from diverse global geographic sources. Overall, the five candidate proteins in these strains had >91% protein sequence identity to those in P1/7. The immunoreactivity of the recombinant proteins was tested in a Western blot assay with serum collected from a convalescent-phase pig infected with a serotype 2 S. suis strain under experimental conditions (Fig. 1). Reactivity to four of the proteins (SSU1215, SSU1355, SSU1773, and SSU1915) was observed. The potential to apply the five candidate proteins as a pool of subunit vaccines has not been previously published, patented, or tested in pig protection studies.
TABLE 1.
Characteristics of the five candidate vaccine proteins
| Antigen-encoding gene | Function/ortholog | Range of TraDIS fitness scoresa | Size of full-length protein (no. of amino acids) | N-terminal signal peptide (amino acids)b | Protein subcellular localization predictionc | Conserved domain | No. of amino acids/molecular mass (kDa) of fusion proteind |
|---|---|---|---|---|---|---|---|
| SSU0185 | Putative tagatose-6-phosphate aldose/ketose isomerase (AgaS) | −4.66 to −8.58 (3/3) | 389 | Extracellular (literature mining) | 432/47.4 | ||
| SSU1215 | Putative surface-anchored dipeptidase | −0.90 to −10.22 (3/4) | 607 | 1–27 | Cell wall anchored (in silico) | LPSTG | 623/67.4 |
| SSU1355 | Putative surface-anchored 5′-nucleotidase | −0.81 to −8.23 (3/4) | 674 | 1–30 | Cell wall anchored (in silico) | LPNTG | 687/74.1 |
| SSU1773 | Putative surface-anchored serine protease | 1.00 to −8.74 (6/8) | 1,692 | 1–40 | Cell wall anchored (in silico) | LPQTG | 1695/187.4 |
| SSU1915 | Putative maltose/maltodextrin-binding protein precursor (Ma1X) | −5.03 to −5.05 (2/2) | 419 | Lipid anchored (in silico) | 462/49.0 |
TraDIS fitness scores are presented as the log2 fold change of the output/input determined by DESeq2 after normalization. The fraction of significantly attenuated mutants in each gene is shown in parentheses, using the parameters an input read of ≥500 and a P value of ≤0.05. Values in parentheses are the number of pigs with TraDIS fitness scores in the indicated range/total number of pigs tested.
Genes encoding the surface proteins were cloned without the N-terminal signal peptides.
In silico protein subcellular localization predictions by use of the PSORTb and LocateP databases.
The amino acid residues and molecular masses of pET-30 Ek/LIC fusion proteins were calculated by including the protein tag generated from the vector (43 amino acids, 4.8 kDa) and excluding the signal peptides, if present.
TABLE 2.
Presence of the five immunogenic antigens in 459 isolates of S. suis
| Protein | No. of isolates in which protein is present | % of isolates in S. suis isolate collection in which protein is presenta |
||
|---|---|---|---|---|
| Clinicalb (292 isolates) | Nonclinicalc (134 isolates) | Not knownd (33 isolates) | ||
| SSU0185 | 459 | 100 | 100 | 100 |
| SSU1215 | 452 | 99 | 97 | 94 |
| SSU1355 | 459 | 100 | 100 | 100 |
| SSU1773 | 450 | 98 | 97 | 100 |
| SSU1915 | 458 | 100 | 99 | 100 |
The presence of the protein was investigated by taking the sequence of the protein from P1/7 and comparing it with the sequences of 459 genomes using BLASTX analysis. If the protein had an 80% identity over 80% of the length, it was classified as present.
Isolates recovered from either systemic sites of pigs with clinical signs and/or a gross pathology consistent with S. suis infection (including meningitis, septicemia, and arthritis) or respiratory sites in the presence of gross lesions of pneumonia from the lung were classified as clinical.
Isolates from the tonsils or tracheobronchus of healthy pigs or pigs without any typical signs of S. suis infection but diagnosed with disease unrelated to S. suis (such as enteric disease or trauma) were classified as nonclinical.
Isolates for which there was insufficient information about the pigs sampled were classified as not known.
TABLE 3.
Protein identities of the five subunit vaccine candidates in disease-associated S. suis serotype representativesa
| Strainb | Serotype | % protein identity |
||||
|---|---|---|---|---|---|---|
| SSU0185 | SSU1215 | SSU1355 | SSU1773 | SSU1915 | ||
| SS021 | 1 | 100 | 99 | 100 | 100 | 100 |
| SS045 | 1 | 100 | 100 | 100 | 100 | 100 |
| SS100 | 1/2 | 100 | 100 | 100 | 100 | 100 |
| SS043 | 1/2 | 98 | 99 | 99 | 98 | 100 |
| SS002 | 2 | 100 | 100 | 100 | 100 | 100 |
| SS008 | 2 | 98 | 99 | 99 | 98 | 100 |
| SS053 | 3 | 98 | 99 | 99 | 97 | 100 |
| SS084 | 3 | 98 | 99 | 99 | 97 | 100 |
| SS062 | 4 | 97 | 90 | 99 | 96 | 97 |
| SS079 | 4 | 88 | 43 | 75 | NPc | 85 |
| SS018 | 7 | 98 | 99 | 99 | 98 | 99 |
| SS024 | 7 | 98 | 99 | 99 | 98 | 99 |
| SS068 | 8 | 98 | 99 | 99 | 98 | 100 |
| SS091 | 8 | 98 | 99 | 99 | 97 | 100 |
| SS015 | 9 | 97 | 91 | 96 | 96 | 97 |
| SS088 | 9 | 97 | 90 | 96 | 96 | 97 |
| SS078 | 10 | 96 | 100 | 95 | 96 | 97 |
| SS063 | 14 | 100 | 100 | 100 | 100 | 100 |
| SS077 | 14 | 100 | 100 | 100 | 100 | 100 |
| SS097 | 16 | 98 | 98 | 97 | 97 | 98 |
| SS037 | 22 | 88 | 43 | 73 | NP | 85 |
| SS009 | 23 | 97 | 91 | 96 | 96 | 97 |
| SS082 | 31 | 98 | 92 | 96 | 97 | 97 |
The panel contains 2 representatives (where possible) of disease-associated serotypes. Respiratory isolates were selected when no other systemic isolate was available.
Strains in bold were also used in cross-reactive ELISAs, the results of which are shown in Fig. 6.
NP, not present. If the protein had less than an 80% identity over 80% of the length, it was classified as not present.
FIG 1.
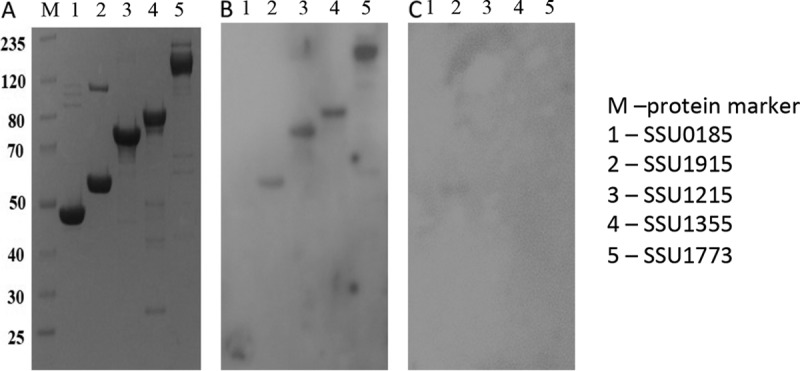
SDS-PAGE and Western blots of the five candidate vaccine proteins. The five candidate proteins were expressed in E. coli and purified as described in Materials and Methods. The purified proteins were run on an SDS-polyacrylamide gel (A) and also transferred to membranes and probed with either serum from a pig experimentally infected with S. suis serotype 2 (B) or sera from pigs raised in a pathogen-free environment as a negative control (C). The numbers on the left are molecular masses (in kilodaltons).
Parenteral adjuvant formulation and boosting significantly impact the serum IgG S. suis protein-specific response.
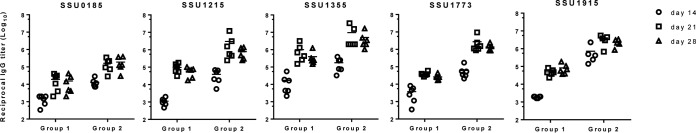
Two groups of pigs were vaccinated with the five proteins both intranasally (i.n.) with polyethyleneimine as the adjuvant and intramuscularly (i.m.) with one of two adjuvant combinations, Carbopol-AddaVax (group 1) or Emulsigen-D (group 2), as described in Materials and Methods (Table 4). Groups 3 to 5 were control groups given phosphate-buffered saline (PBS) mixed with the same adjuvants given to groups 1 and 2 (groups 3 and 4, respectively) or PBS only (group 5). Overall, serum IgG antibody reactive against all five proteins was detected in all vaccinated pigs, and there was an anamnestic response after administration of the boost vaccination (Fig. 2). No S. suis protein-specific IgG was detected in the pigs given adjuvant alone or PBS (data not shown), nor was there a response detected in serum collected at day 0. At 2 weeks following priming (day 14), the titers of IgG specific to individual S. suis proteins were significantly higher in serum from pigs in group 2 (Emulsigen-D adjuvant) than in serum from pigs in group 1 (Carbopol-AddaVax adjuvant), and this trend continued after the response was boosted (days 21 and 28). In fact, the titers of IgG to the proteins in group 2 pigs after a single injection were approximately equal to the titers in group 1 pigs after 2 injections.
TABLE 4.
Experimental groupsa
| Group | Vaccine | Adjuvant | Route | No. of pigs |
|---|---|---|---|---|
| 1 | S. suis proteins | Polyethyleneimine | i.n. | 6 |
| S. suis proteins | Carbopol-AddaVax | i.m. | ||
| 2 | S. suis proteins | Polyethyleneimine | i.n. | 6 |
| S. suis proteins | Emulsigen-D | i.m. | ||
| 3 | PBS | Polyethyleneimine | i.n. | 3 |
| PBS | Carbopol-AddaVax | i.m. | ||
| 4 | PBS | Polyethyleneimine | i.n. | 3 |
| PBS | Emulsigen-D | i.m. | ||
| 5 | PBS | None | i.n. | 4 |
| PBS | None | i.m. |
All groups were challenged with S. suis P1/7. i.n., intransal; i.m., intramuscular; PBS, phosphate-buffered saline.
FIG 2.
Titers of IgG antibodies to the individual subunit proteins among vaccinated pigs in groups 1 and 2 on day 14 (2 weeks after priming) and days 21 and 28 (1 and 2 weeks after the boost, respectively). Pigs in groups 1 and 2 (6 pigs each) were vaccinated with the 5 candidate proteins on days 0 and 14 of the experiment. Both groups were given the 5 proteins intranasally with polyethyleneimine as the adjuvant; in addition, group 1 pigs were given the 5 proteins intramuscularly with AddaVax and Carbopol as the adjuvant, while group 2 pigs were given the 5 proteins intramuscularly with Emulsigen-D as the adjuvant. Titers were determined via indirect ELISA with plates coated with the individual proteins using 2-fold serial dilutions of serum. The resulting OD data were modeled as a nonlinear function of the log10 dilution using the log (agonist)-versus-response variable slope four-parameter logistic model. Endpoints were interpolated by using 4 times the average OD of the day 0 sample for each respective pig as the cutoff.
The peripheral S. suis protein-specific IFN-γ recall response declines following boost immunization.
The number of peripheral blood mononuclear cells (PBMCs) producing gamma interferon (IFN-γ) following reexposure to the pool of S. suis proteins was used as a measure of vaccine-induced cell-mediated immunity. The number of IFN-γ-secreting cells (IFN-γ-SC) following restimulation with S. suis proteins was the greatest on day 14 postpriming, and the adjuvant formulation had a significant impact on the responses, with the pigs in group 2 (Emulsigen-D adjuvant) having significantly higher numbers of IFN-γ-SC than pigs in group 1 (Carbopol-AddaVax adjuvant) (Fig. 3). The number of IFN-γ-SC detected decreased over time, with an average of 263 and 32 IFN-γ-SC being detected in pigs in group 2 on days 14 and 28, respectively. PBMCs collected from pigs in groups 3, 4, and 5 (the no-antigen groups) did not have more than 13 IFN-γ-SC detected at any time point following stimulation with S. suis proteins. In addition, the number of IFN-γ-SC detected following stimulation with medium alone remained below 10 at each time point evaluated. While there was, on average, an increase in the number of IFN-γ-SC when PBMCs from pigs in group 1 were used at day 14 postpriming, it was not significantly increased over that in the control groups (groups 3 to 5).
FIG 3.
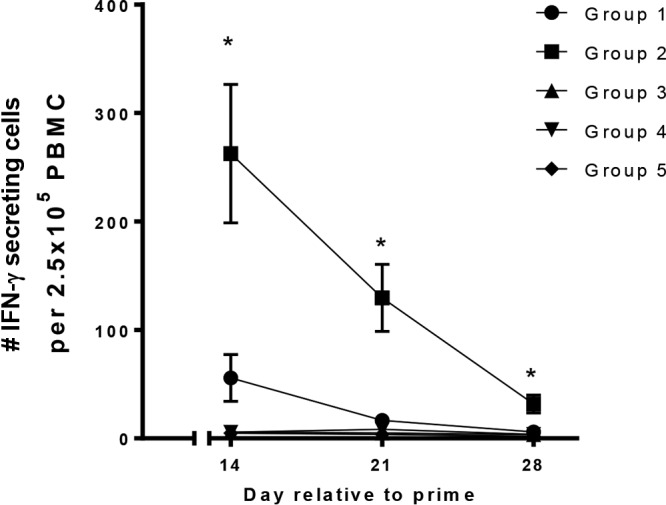
ELISpot assay data showing the number of IFN-γ-secreting cells detected in PBMCs isolated from pigs in the indicated groups on days 14 (2 weeks after priming), 21 (1 week after the boost), and 28 (2 weeks after the boost). Pigs in groups 1 and 2 (6 pigs each) were vaccinated with the 5 candidate proteins on days 0 and 14 of the experiment. Both groups were given the 5 proteins intranasally with polyethyleneimine as the adjuvant; in addition, group 1 pigs were given the 5 proteins intramuscularly with AddaVax and Carbopol as the adjuvant, while group 2 pigs were given the 5 proteins intramuscularly with Emulsigen-D as the adjuvant. Pigs in groups 3 to 5 were controls given the adjuvants alone (groups 3 and 4, 3 pigs each) or PBS (group 5, 4 pigs). PBMCs collected on days 14, 21, and 28 were seeded at 2.5 × 105 cells per well in duplicate and stimulated with a pool of the 5 candidate proteins. Control wells were stimulated with medium alone or pokeweed mitogen (data not shown). The means and standard errors of the means for each of the treatment groups are denoted. *, statistically significant differences between groups (P < 0.05).
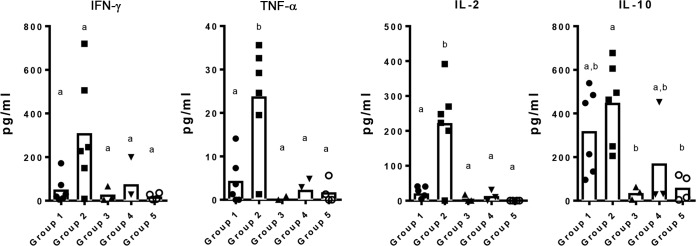
The levels of the cytokines produced by PBMCs following restimulation with the protein pool were the highest in pigs vaccinated with the Emulsigen-D adjuvant.
PBMCs collected on day 28, 2 weeks after the boost vaccination, were stimulated with the pool of five S. suis proteins as another measure of vaccine-induced cell-mediated immunity. Overall, the levels of cytokines produced by PBMCs following restimulation with the protein pool were the highest in pigs from group 2 (Emulsigen-D adjuvant) (Fig. 4). These levels were statistically significantly higher for group 2 than for all other groups for interleukin-2 (IL-2) and tumor necrosis factor alpha (TNF-α), whereas there was no statistically significant difference in the amount of these cytokines produced among groups 1 (Carbopol-AddaVax adjuvant) and 3 to 5 (the control groups).
FIG 4.
Cytokines produced by PBMCs isolated from pigs in the indicated groups on day 28 (2 weeks after the boost). Pigs in groups 1 and 2 (6 pigs each) were vaccinated with the 5 candidate proteins on days 0 and 14 of the experiment. Both groups were given the 5 proteins intranasally with polyethyleneimine as the adjuvant; in addition, group 1 pigs were given the 5 proteins intramuscularly with AddaVax and Carbopol as the adjuvant, while group 2 pigs were given the 5 proteins intramuscularly with Emulsigen-D as the adjuvant. Pigs in groups 3 to 5 were controls given the adjuvants (groups 3 and 4, 3 pigs each) alone or PBS (group 5, 4 pigs). PBMCs collected on day 28 were stimulated in vitro with a pool of the 5 candidate proteins, and the supernatants were collected to evaluate the levels of cytokines secreted by the cells by multiplex cytokine ELISA. Data are presented as box and dot plots representing the mean cytokine concentration for the group (box) and individual cytokine concentration for each pig (dots) (in picograms per milliliter). Significantly different cytokine concentrations among groups are identified with different letters (P < 0.05).
Subunit vaccination provides significant protection against lethal challenge with S. suis and is associated with the immune response and adjuvant given.
Following challenge with a virulent strain, 9 out of 10 pigs in nonvaccinated control groups 3 to 5 developed severe signs of systemic S. suis infection (lameness with swollen joints, anorexia, depression, dyspnea, and neurological signs) and had to be euthanized (Fig. 5). S. suis was cultured from systemic sites of these 9 pigs, including serosa (5/9), joint (9/9), cerebrospinal fluid (CSF) (9/9), and spleen (8/9), and macroscopic and microscopic lesions consistent with S. suis infection, including meningitis, polyserositis, and arthritis, were present. S. suis was readily isolated from the nasal cavities and tonsils of these pigs as well, but only small numbers of S. suis bacteria were isolated from the lung lavage fluid of 5 of them, and pneumonia was not a prominent lesion that was seen. There was one pig in group 5 that developed only intermittent mild lameness that began 1 day after challenge and that continued throughout the observation period but demonstrated no other clinical signs, and S. suis was isolated from the nasal wash and tonsil of this pig only at the termination of the experiment on day 15.
FIG 5.
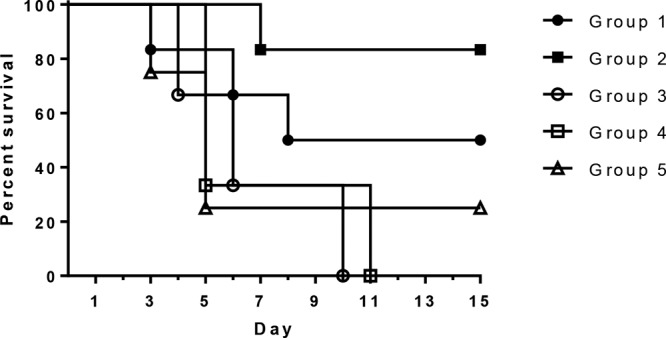
Survival rates of pigs vaccinated with 5 subunit proteins with different adjuvant formulations (groups 1 and 2) compared to those of pigs given adjuvant alone (groups 3 and 4) or PBS (group 5). Pigs in groups 1 and 2 (6 pigs each) were vaccinated with the 5 candidate proteins on days 0 and 14 of the experiment. Both groups were given the 5 proteins intranasally with polyethyleneimine as the adjuvant; in addition, group 1 pigs were given the 5 proteins intramuscularly with AddaVax and Carbopol as the adjuvant, while group 2 pigs were given the 5 proteins intramuscularly with Emulsigen-D as the adjuvant. Pigs in groups 3 to 5 were controls given the adjuvants alone (groups 3 and 4, 3 pigs each) or PBS (group 5, 4 pigs).
By comparison, in the two vaccinated groups, 3/6 pigs in group 1 (Carbopol-AddaVax adjuvant) and only 1/6 pigs in group 2 (Emulsigen-D adjuvant) developed severe systemic disease requiring euthanasia (Fig. 5). The rate of survival was significantly greater among pigs in group 2 than among the pigs in the nonimmunized control groups combined. Similar to the findings for the control groups, S. suis was isolated from systemic sites of the four pigs in the vaccinated groups that had to be euthanized (from the serosa in 4/4 pigs, joints in 4/4 pigs, CSF in 3/4 pigs, and the spleen in 3/4 pigs), and macroscopic and microscopic lesions consistent with S. suis infection were present. The nasal cavities and tonsils were heavily colonized in all the vaccinated pigs, but virtually no S. suis bacteria were isolated from the lung lavage fluid from any of these pigs. One pig from group 2 was lame for 2 days but showed no other clinical signs and recovered uneventfully; S. suis was isolated only from the nasal wash and tonsil and not from any systemic site of this pig, and no macroscopic or microscopic lesions consistent with S. suis infection were present at the end of the experiment, when all the remaining pigs were euthanized. In addition, S. suis was isolated from the spleen from one pig each in groups 1 and 2 that appeared clinically healthy throughout the experiment. Neither of these pigs had any macroscopic or microscopic lesions consistent with S. suis infection.
Subunit vaccination induces IgG reactive against whole S. suis bacteria.
An indirect enzyme-linked immunosorbent assay (ELISA) was performed to determine if serum IgG from vaccinated pigs collected on day 28 postvaccination reacted with whole S. suis P1/7 bacteria or other S. suis isolates representing serotypes commonly associated with disease (serotypes 1, 2, 1/2, 3, and 14). Although there were some differences in the degree of reactivity across the different isolates, there was an appreciable IgG response to all S. suis isolates tested, indicating a considerable amount of reactivity to different isolates of S. suis which vary in respect to serotype (Fig. 6). As with the other immune parameters measured, the S. suis-specific IgG response induced in group 2 pigs (Emulsigen-D adjuvant) was higher than that induced in group 1 pigs (Carbopol-AddaVax adjuvant).
FIG 6.
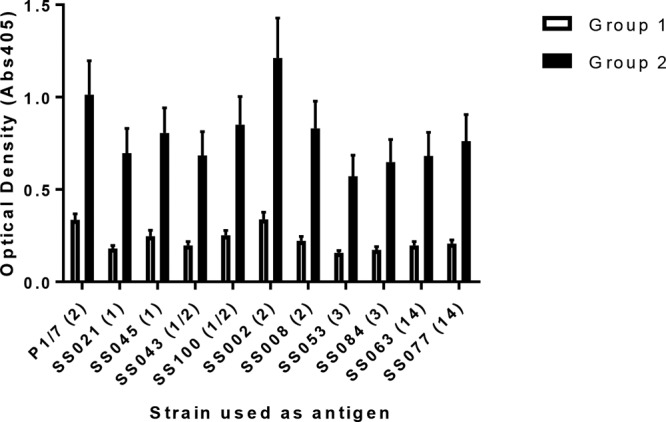
Cross-reactive IgG antibody to whole S. suis bacteria of serotypes that commonly cause systemic disease from group 1 and 2 pigs on day 28 (2 weeks after the boost). Pigs in groups 1 and 2 (6 pigs each) were vaccinated with the 5 candidate proteins on days 0 and 14 of the experiment. Both groups were given the 5 proteins intranasally with polyethyleneimine as the adjuvant; in addition, group 1 pigs were given the 5 proteins intramuscularly with AddaVax and Carbopol as the adjuvant, while group 2 pigs were given the 5 proteins intramuscularly with Emulsigen-D as the adjuvant. IgG reactivity was determined via indirect ELISA with plates coated with heat-inactivated whole bacteria. Serum samples collected on day 28 from each pig were diluted 1:500 and used in the assay. Data are reported as the mean ± SEM optical density at 405 nm. Bacterial strains are listed on the x axis, with the serotype being given in parentheses.
DISCUSSION
The five S. suis proteins in this study were chosen first on the basis of determination of the genes/proteins that were predicted to play a role in fitness during colonization of the respiratory tract, the initial stage in the establishment of infection, using a respiratory epithelium IVOC system and transposon mutant library. The identified proteins are predicted to have functions in several physiological processes, in particular, those associated with metabolism and nutrient acquisition, which might explain their role in survival on the respiratory epithelium.
SSU0185 was identified to be a putative tagatose-6-phosphate aldose/ketose isomerase. The ortholog of this protein, AgaS, is believed to be part of the pathway for utilization of the amino sugar N-acetyl-d-galactosamine in E. coli (18). The abundance of free sugars is scarce in the respiratory tract, and mucins, a major component of the mucus produced by respiratory surfaces, contain glycoproteins composed of sugars, amino sugars, and sulfated sugars commonly linked to a protein core via an N-acetylgalactosamine (19). Orthologs of agaS have been identified in other Streptococcus species, such as Streptococcus pneumoniae, where the ortholog of agaS was shown to be upregulated upon exposure to human macrophage-like cells and when it was grown in the presence of mucin, potentially explaining the importance of this protein for survival in the respiratory tract (20, 21).
SSU1915 was identified to be a putative maltose/maltodextrin-binding protein whose ortholog is MalX, a lipid-anchored solute binding protein of an ATP binding cassette (ABC) transporter. MalX has been reported to be a streptococcal virulence factor involved in carbohydrate metabolism, specifically, in polysaccharide degradation and synthesis (22). Members of the mal regulon of Streptococcus pyogenes have been shown to enhance colonization of the oropharynx through their niche-specific role in the utilization of dietary starch (23–25). Another study identified malX of S. pneumoniae to be one of the niche-specific virulence genes upregulated in the lung and confirmed attenuation of the virulence of a malX mutant during lung infection (26). In the same report, vaccination with MalX induced high antibody titers but not significant protection in an intraperitoneal challenge model (26). In contrast, Moffitt et al. demonstrated that intranasal vaccination with the S. pneumoniae protein SP2108, the MalX ortholog, was protective in a mouse model of pneumococcal nasopharyngeal colonization (27). Subsequently, they established that the lipid modification of this protein is critical to its immunogenicity in a Toll-like receptor 2-dependent manner, and there was an in trans effect of the lipoprotein that enhanced the immunogenicity of a coadministered nonlipidated antigen (28).
SSU1355 was identified to be a putative surface-anchored 5′-nucleotidase, a hydrolytic enzyme that catalyzes the hydrolysis of a nucleotide into a nucleoside and a phosphate. These enzymes have been identified to be virulence factors, purportedly by hydrolyzing extracellular nucleotides for purine salvage, by degrading nucleotide diphosphate sugars that can then be used by the cell, and/or by generating extracellular adenosine in the host, which is a powerful immunosuppressant-signaling molecule. Staphylococcus aureus produces extracellular adenosine to evade clearance by the host immune system, an activity attributed to the 5′-nucleotidase activity of adenosine synthase (AdsA) (29).
SSU1215 was identified to be a putative surface-anchored dipeptidase. These enzymes play roles in several physiological processes, such as the catabolism of exogenously supplied peptides and the final steps of protein turnover.
SS1773 was identified to be a putative surface-anchored serine protease. Prokaryotic serine proteases have roles in several physiological processes, such as those associated with metabolism, cell signaling, and the defense response and development; however, functional associations for a large number of prokaryotic serine proteases are relatively unknown.
Since the methods used to identify these proteins indicated that they are involved in respiratory colonization fitness, there was the possibility that a locally induced mucosal or parenterally induced systemic immune response, or both, would be important for protection. Since raising colostrum-deprived pigs delivered by cesarean section (CDCD pigs) is not a trivial matter and S. suis infection can have severe clinical consequences, it was decided to vaccinate the pigs with all five proteins by both routes to enhance the potential for success using the fewest number of pigs initially. Subsequently, further experiments could be conducted to determine the role of each of the proteins and the role of the route of delivery in protection and test protection against a heterologous challenge. Polyethyleneimine, an organic polycation, was chosen as the adjuvant for intranasal vaccination because it has previously been shown to be a potent mucosal adjuvant for the delivery of antigens of mucosal pathogens (30, 31). We chose a combination of AddaVax, a squalene-based oil-in-water adjuvant similar to MF-59 used in human influenza vaccines in Europe, and Carbopol, a polyanionic carbomer, as one choice for a parenteral adjuvant, based on previous work demonstrating that this type of combination yielded an additive or potentially synergistic adjuvant effect (32). In addition, we chose Emulsigen-D, an oil-in-water emulsion with dimethyldioctadecylammonium bromide, as the second parenteral adjuvant, which has also been shown to induce immune responses that are enhanced compared to those induced by some commonly used adjuvants (33). Both the magnitude of the systemic immune response and the degree of protection were dependent on the parenteral adjuvant administered with the proteins. This would suggest that parenteral vaccination is the delivery method important for protection; however, a role for mucosal immunization in protection or priming of the immune response cannot be ruled out, and additional studies separating the routes of administration will be needed to determine these roles.
Even though the proteins were identified to potentially contribute to fitness for respiratory colonization, all surviving vaccinated animals showed tonsil and nasal colonization by the challenge organism. A quantitative comparison of the colonizing bacterial load for immunized versus nonimmunized animals was beyond the scope of this preliminary study, so there could have been a reduction in the numbers of S. suis bacteria colonizing the respiratory sites that was not detected. In addition, since mucosal IgA was not measured, it is difficult to state whether there was a failure of induction of mucosal antibodies to these proteins or a failure of antibodies to prevent colonization. The impact of immunization on the reduction of the colonization load by pneumococcus in a mouse model was found to be dependent on individual host factors as well as vaccine-associated factors (34). There was a reduction in the incidence of systemic disease in vaccinated animals, which could have been due to reduced colonization and invasion or an increase in bactericidal/opsonic antibodies, or both. Streptococcus suis was also isolated from the spleens of two apparently healthy vaccinated pigs. These animals probably had an ongoing bacteremia that was being controlled and cleared by the immune response since, as indicated elsewhere in this article, the animals showed no antemortem, postmortem, or histopathological signs of streptococcal disease. It is possible that this represented a very recent bacteremia; however, in our infection model with this strain of S. suis, we rarely have pigs develop or succumb to disease past day 10 of exposure.
Peripheral IFN-γ recall responses were evaluated at various time points after vaccination, and there was a reduction in the number of peripheral S. suis-specific IFN-γ-SC after the boost (Fig. 3). However, there was an increase in peripheral S. suis-specific IgG levels after the second dose of vaccine, indicating a boost in immune responses following the second dosing. While the reduction in IFN-γ-SC was somewhat unexpected, it is important to note that IFN-γ-SC serve as a single measure of immune cell activation, and the cell-mediated immune responses after the prime-boost were likely skewed toward T-helper responses not involving IFN-γ production. Given the increased levels of S. suis-specific IgG after the boost, T cell responses were likely directed toward B cell affinity maturation and plasma cell generation, which would include the production of IL-13 and IL-5, though the levels of these cytokines were not measured in this study. Overall, subunit vaccination with the five S. suis proteins induced an immune response that provided substantial protection from lethal challenge with virulent S. suis bacteria, and specifics on the mechanism of protection warrant further investigation.
S. suis is a diverse species of multiple serotypes, each of which is represented by immunologically different capsule types and displays a wide range of immune-evading features that, to date, have challenged the development of efficacious vaccines (35). In particular, although opsonizing antibody is believed to be key to the killing of S. suis in infected animals (36), the antibody response to the S. suis capsule has been shown to be limited in infected animals (37). Although much effort has already focused on subunit candidates, especially surface-associated targets (reviewed by Baums and Valentin-Weigand [38]), recent reports emphasize the ongoing challenges of matching candidates with promising measures of protection in mouse models and in vitro assays with in vivo survival outcomes in live challenged pigs (39).
The five proteins identified are highly conserved and present in almost all strains of S. suis tested, including probable nonvirulent strains. Since these strains are normal colonizers of pigs, one might expect that antibodies against these proteins are already present in pigs on farms. There was reactivity to four of the proteins in serum collected from a convalescent-phase pig infected with virulent S. suis (Fig. 1); however, nonvirulent strains are commensal microbes that could colonize without triggering a significant immune response. The diversity of antibody responses to these proteins in pigs naturally exposed to S. suis, with or without disease, might shed further light on their respective contribution to immune protection. Further studies will also be needed to evaluate the optimum approach to the field application of these subunits as protective immunogens, including the potential for sow versus piglet immunization and the possibility of prior passive or active antibody interference. In addition, the reactivity of the sera from vaccinated pigs against several diverse S. suis strains commonly associated with disease in pigs may indicate a potential for cross protection that will have to be confirmed through further challenge studies.
MATERIALS AND METHODS
Bacterial strains, vectors, media, and antibiotics used in the study.
The bacterial strains and the vector used in this study are listed in Table 5. S. suis strains were routinely grown at 37°C in Todd-Hewitt broth (Oxoid) supplemented with 0.2% yeast extract (Sigma) (THY) or on Columbia agar (Oxoid) containing 5% (vol/vol) defibrinated horse blood (TCS Bioscience) (CBA). E. coli strains were routinely grown at 37°C on Luria-Bertani (LB) agar plates or cultured in LB broth (Oxoid). E. coli strains expressing recombinant proteins were grown at 37°C in 2YT broth (Life Technologies). Kanamycin (Sigma) at a concentration of 100 μg/ml was used to select E. coli transformants. All the strains were stored at −80°C in 20% glycerol.
TABLE 5.
Bacterial strains and vector used in this study
| Strain or plasmid | Serotype | Clinical associationa or application | Tissue origin |
|---|---|---|---|
| S. suis pig isolates | |||
| P1/7 | 2 | SYS-brain | Blood |
| SS021 | 1 | SYS-other | Joint/skin |
| SS045 | 1 | SYS-brain | Meninges |
| SS100 | 1/2 | SYS-brain | Brain |
| SS043 | 1/2 | RESP | Lung |
| SS002 | 2 | SYS-brain | Brain |
| SS008 | 2 | SYS-other | Pericardial swab |
| SS053 | 3 | SYS-brain | Brain |
| SS084 | 3 | RESP | Lung |
| SS062 | 4 | SYS-brain | Brain |
| SS079 | 4 | SYS-brain | Brain |
| SS018 | 7 | SYS-other | Lung/pericardium |
| SS024 | 7 | SYS-brain | Brain |
| SS068 | 8 | SYS-brain | Brain |
| SS091 | 8 | RESP-SD | Lung |
| SS015 | 9 | SYS-brain | Brain |
| SS088 | 9 | SYS-other | Joint |
| SS078 | 10 | SYS-other | Joint |
| SS063 | 14 | SYS-other | Joint |
| SS077 | 14 | SYS-brain | Brain |
| SS097 | 16 | SYS-other | Spleen |
| SS037 | 22 | RESP | Lung |
| SS009 | 23 | RESP | Lung |
| SS082 | 31 | RESP-SD | Lung |
| E. coli strains | |||
| NovaBlue | E. coli host for cloning | ||
| BL21(DE3) | E. coli host for expressing recombinant protein | ||
| pET-30 Ek/LICb vector | Vector for cloning, expression, and purification of target proteins |
Isolates recovered from systemic sites in pigs with clinical signs and/or a gross pathology consistent with S. suis infection (including meningitis, septicemia, and arthritis) were classified as systemic (SYS), whereas those recovered from the lung in the presence of gross lesions of pneumonia were classified as respiratory (RESP). Isolates recovered from the lung of pigs with pneumonia but also with gross signs of systemic streptococcus-type disease were classified as RESP-SD.
The pET-30 Ek/LIC (ligation-independent cloning) vector is designed for cloning and high-level expression of target proteins fused with the His tag and STag coding sequences that are cleavable with enterokinase (Ek) protease. The plasmid contains a strong T7 lac promoter, an optimized ribosome binding site, the coding sequence for the Ek protease cleavage site (Asp Asp Asp Asp Lys↓), and a multiple-cloning site that contains restriction enzyme sites found in many other Novagen expression vectors to facilitate insert transfer. An optional C-terminal His tag-coding sequence is compatible with purification, detection, and quantification.
S. suis (P1/7), a serotype 2 isolate from the blood of a pig with meningitis (40), was used for challenge and was grown on tryptic soy agar containing 5% sheep blood (Becton, Dickinson and Company) at 37°C overnight, scraped from the plates, and resuspended in phosphate-buffered saline (PBS) to an optical density at 600 nm (OD600) of 0.42 to give an inoculum dose of 1 × 109 CFU/ml. Each challenged pig received 1 ml per nostril (2 ml total).
General molecular biology techniques.
The genomic DNA of the S. suis strains was isolated using a MasterPure Gram-positive bacterial DNA purification kit (Epicentre Biotechnologies). Bacterial lysates of S. suis were prepared using the InstaGene matrix, a Chelex-based resin (Bio-Rad Laboratories Ltd.), according to the manufacturer's instructions. The plasmid DNA samples were prepared using a QIAprep spin miniprep kit (Qiagen) or a HiSpeed plasmid maxi kit (Qiagen). Plasmids and genomic DNA were stored at −20°C.
The PCRs for screening bacterial colonies were set up with Go Taq Green master mix (Promega Ltd.) according to the manufacturer's instructions. The amplification conditions used were as follows: initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for a period determined by the size of the PCR product (1 min/kb), with a final extension step at 72°C for 7 min.
The PCR products used for cloning were amplified using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific) according to the manufacturer's instructions. The reaction mixtures contained 100 ng of template DNA or 1 to 5 μl bacterial lysate, 200 μM each deoxynucleoside triphosphate (Bioline Ltd.), 0.5 to 1 μM each primer (Sigma-Aldrich Ltd.), 1× PCR buffer, 1 unit of DNA polymerase, and dimethyl sulfoxide at a final concentration of 3% when required. The initial denaturation was done at 98°C for 30 s, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at appropriate temperatures for 30 s, and extension at 72°C for a period determined by the size of the PCR product (10 to 30 s/kb). The final extension was done at 72°C for 7 min.
The primers used in this study are listed in Table 6. The primers were designed using Primer3web software (version 4.0.0; http://primer3.ut.ee) and synthesized by Sigma-Aldrich Ltd. The primers were rehydrated with deionized water to a concentration of 100 μM on arrival, and working stocks of 10 μM concentration were prepared. All primers were stored at −20°C.
TABLE 6.
Primers used for protein cloning in this study
| Primer | Primer function | Sequencea (5′-3′) |
|---|---|---|
| 0185-4F | Cloning primers for SSU0185 | GACGACGACAAGATGTTCCGTTTAGCAAAAGAAGAAC |
| 0185-1167R | GAGGAGAAGCCCGGTTATTTTTCTAAAGGATGGATGA | |
| 1915-4F | Cloning primers for SSU1915 | GACGACGACAAGATGAAACACAATCTCCTTAAGAGCG |
| 1915-1257R | GAGGAGAAGCCCGGTTAGTTGCTGTGTTTTTGAGCAA | |
| 1215-82F | Cloning primers for SSU1215 | GACGACGACAAGATGGGCTTTATTATTGGGAAAGG |
| 1215-1831R | GAGGAGAAGCCCGGTTATTCTTTACTGGATTTTTTTC | |
| 1355-91F | Cloning primers for SSU1355 | GACGACGACAAGATGTTAGCTGTCCAAATTATGGGAG |
| 1355-2022R | GAGGAGAAGCCCGGTTACTCCCCTTCCTTACGTCTCA | |
| 1773-121F | Cloning primers for SSU1773 | GACGACGACAAGATGGATACTAGTGGAGAAGGATTGG |
| 1773-5076R | GAGGAGAAGCCCGGTTATTCTTTTCGCTTCAAATTTC |
Underlined nucleotides correspond to the sequence extensions required for LIC compatibility with the pET-30 Ek/LIC cloning vector.
The PCR products and DNA samples were analyzed by agarose gel electrophoresis. The agarose gels were visualized and photographed using the Gel Doc XR+ imaging system with Image Lab image acquisition and analysis software (Bio-Rad Laboratories Ltd.).
SDS-PAGE analyses were performed with whole-cell lysates or purified proteins. Samples were diluted in equal volumes of 2× SDS sample buffer, heated at 70°C for 10 min, and run on 4 to 12% (vol/vol) bis-Tris gels (Life Technologies) to confirm protein expression.
Selection of candidate vaccine proteins.
A strategy of combining experimental functional genomics screening (in an IVOC system with TraDIS) with in silico bioinformatics approaches was applied for selection of candidate vaccine proteins using a library generated in S. suis strain P1/7 (13–17, 41). The selection consists of the following steps: (i) candidate fitness genes (defined as genes that harbored at least one transposon insertion mutant with a significant reduction in fitness in a swine respiratory epithelium IVOC system) were determined through previous functional genomics screening; (ii) protein subcellular localization was predicted in silico with bioinformatics approaches using the PSORTb (http://db.psort.org/) and LocateP (http://www.cmbi.ru.nl/locatep-db/cgi-bin/locatepdb.py) databases or on the basis of literature mining to shortlist fitness genes encoding surface-associated proteins (cell wall-anchored or extracellular [lipid-anchored or secretory] proteins); (iii) proteins containing transmembrane domains in the middle of protein-coding sequence were excluded; (iv) in silico protein homology-based searches were used to identify proteins with cross-protection potential; i.e., the protein from the S. suis P1/7 genome was used as a query in a BLASTX search, and we identified the proteins present (80% identity over 80% of the length) in 459 publically available strains or in the majority of disease-associated strains (12); and (v) a final pool with five potential candidate vaccine proteins whose potential to be applied as a cassette of the subunit vaccine and that have not been previously published, patented, or tested in pig protection studies was chosen.
Cloning and expression of candidate vaccine proteins.
The genes of interest were cloned from the genome of S. suis strain P1/7, with the signal sequences being excluded when they were present. The signal peptide cleavage sites of the open reading frames (ORFs) were predicted using the SignalP server (http://www.cbs.dtu.dk/services/SignalP). The PCR products of candidate genes were cloned into the pET-30 Ek/LIC vector (Merck Millipore), and fusion plasmids were transformed into E. coli NovaBlue (Merck Millipore) according to the manufacturer's instructions. The positive recombinants were confirmed by PCR and DNA sequencing and then transformed into E. coli BL21(DE3) (Merck Millipore) for expression. Overnight cultures of E. coli BL21(DE3) strains carrying the recombinant plasmids were used to inoculate 1 to 6 liters fresh 2YT broth, grown to an OD595 of 0.6 at 37°C in broth supplemented with 100 μg/ml kanamycin, and then induced with 1 mM IPTG (isopropyl-β-d-1-thiogalactopyranoside; Sigma) at 37°C for 2, 4, and 24 h. Protein expression was checked by SDS-PAGE using whole-cell lysates.
Purification of recombinant vaccine proteins.
Recombinant proteins were purified from 1- to 6-liter cultures grown in 2YT broth and induced with 1 mM IPTG for 2 to 4 h. Cell pellets were washed once in PBS and centrifuged at 3,000 × g for 15 min. The cell pellets were resuspended in binding buffer (10 mM imidazole, 300 mM NaCl, 50 mM phosphate, pH 8.0) and sonicated on ice for 6 min. Appropriate amounts of Benzonase and rLysozyme (Novagen, Merck Millipore) were added to reduce the viscosity of the lysate and improve the protein extraction efficiency. The lysates were first centrifuged at 3,000 × g for 10 min at 4°C to pellet the debris, and the supernatants were subjected to further centrifugation at 75,000 × g for 1.5 h at 4°C. Recombinant proteins were subjected to purifications by nickel His tag affinity chromatography, anion-exchange chromatography, CHAP {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} chromatography, and gel filtration, when appropriate. Target proteins were confirmed by peptide mass fingerprinting. The protein concentration was determined using spectrophotometry, and purified proteins were stored at −80°C.
Immunoreactivity of the recombinant proteins with convalescent-phase pig serum.
Immunoreactivity against the purified recombinant proteins was tested using serum from a conventionally reared pig experimentally infected with S. suis serotype 2. As a control, naive serum samples were collected from Gottingen minipigs (Serolabs Ltd.), which were reared in a pathogen-free environment and not expected to have any antibodies against S. suis, and pooled. The purified recombinant proteins were separated on 4 to 12% (vol/vol) bis-Tris gels under denaturing conditions and transferred to polyvinylidene difluoride membranes. The membranes were rinsed in Tris-buffered saline (30 mM Tris base, 138 mM NaCl, 2.7 mM KCl, pH 8.0) with 0.05% Tween 20 (TBST) and then blocked with 2% casein in TBST overnight at 4°C. The pig serum samples (1:2,000) were used as the primary antibody, and horseradish peroxidase (HRP)-conjugated goat anti-pig immunoglobulin (1:10,000; Sigma) was used as the secondary antibody. The primary and secondary antibodies were diluted in 1% casein in TBST, and the membranes were probed at room temperature (RT) for 1 to 1.5 h. The blots were then washed three times in TBST for 10 min at room temperature. The membranes were developed with chemiluminescent substrate (Novex ECL substrate reagent kit; Life Technologies) according to the manufacturer's instructions. The enhanced chemiluminescent (ECL) substrate-treated membranes were exposed to X-ray film (Amersham Hyperfilm ECL; GE Healthcare Life Sciences) for a suitable duration and developed in an X-ray film developer.
Vaccine protection study.
The USDA-ARS-National Animal Disease Institutional Animal Care and Use Committee approved all animal work. Twenty-two 5-week-old colostrum-deprived pigs delivered by cesarean section (CDCD pigs) were distributed into groups as follows (Table 3): group 1 pigs (6 pigs) were given a 2-ml dose of vaccine containing 250 μg protein (50 μg per subunit) with 1 ml of an AddaVax emulsion (a squalene-based oil-in-water adjuvant; InvivoGen) and 5 mg of Carbopol (Carbopol-971; Lubrizol Corporation) intramuscularly (i.m.) in the neck and a 2-ml dose of vaccine containing 500 μg protein (100 μg per subunit) and 500 μg of polyethyleneimine (Sigma) intranasally (i.n.; 1 ml per nostril); group 2 pigs (6 pigs) were vaccinated i.n. similarly to pigs in group 1, but in the 2-ml i.m. dose, the proteins were mixed with Emulsigen-D (an oil-in-water emulsion with dimethyldioctadecylammonium bromide; MVP Technologies) at a 1:5 (vol/vol) mix; group 3 and 4 pigs (3 pigs each) were controls given PBS mixed with the same adjuvants given to groups 1 and 2, respectively; and group 5 pigs (4 pigs) were given PBS only. The pigs received a booster dose of the same respective formulation 2 weeks after priming, and 2 weeks after the boost the pigs were challenged with 2 ml of 109 CFU/ml S. suis P1/7 i.n. (1 ml per nostril). Blood was collected on day 0 (prime) for serum and days 14 (boost), 21 (one week postboost), and 28 (challenge) for serum and peripheral blood mononuclear cells (PBMCs) to evaluate vaccine immunogenicity. After challenge, the pigs were observed for clinical signs of disease (approximately every 4 to 5 h, except for an 8-h overnight period), including lameness, lethargy, and neurological symptoms. If the presentation was severe (such as neurological involvement, severe lameness, or depression that resulted in recumbency with a reluctance to stand) the pig was euthanized. Pigs not showing signs of disease or only transitory or mild signs of disease were euthanized at 15 days postchallenge. At necropsy, nasal wash specimens, swabs of serosa and the hock joint (or other affected joint), cerebrospinal fluid (CSF) specimens, lung lavage fluid specimens, and sections of tonsil and spleen tissues were collected for culture. Nasal turbinate, tonsil, lung, heart, kidney, liver, spleen, retropharyngeal lymph node, brain, and synovium tissues were collected for microscopic pathological examination.
Evaluation of the humoral immune response to vaccination.
Serum IgG titers to individual S. suis proteins and reactivity to inactivated P1/7 were determined using an indirect ELISA. Blood was collected into a BD Vacutainer serum separator tube (SST), and the serum was isolated according to the manufacturer's recommendation (BD Pharmingen) and stored at −80°C until it was used in assays. For evaluation of the titers of antibodies to individual S. suis proteins, Immulon-2 plates were coated with 0.1 ml of each individual protein in 100 mM carbonate-bicarbonate buffer (pH 9.6) overnight at 4°C at the following concentrations: 1 μg/ml for SSU1773, 2 μg/ml for SSU1355, 1 μg/ml for SSU1915, 1 μg/ml for SSU0185, and 0.5 μg/ml for SSU1215. On the next day, the plates were blocked with 0.2 ml of blocking buffer (2% bovine serum albumin [BSA] in PBS–0.05% Tween 20 [PBS-T]) for 2 h at RT and then washed three times with PBS-T. Eleven 2-fold serial dilutions of serum (starting at 1:2,000) collected from each pig were made in 1% BSA–PBS-T, transferred to the ELISA plate in duplicate, and incubated at RT for 2 h. The plates were washed, S. suis-specific IgG was detected by adding 0.1 ml of anti-porcine IgG conjugated to horseradish peroxidase (dilution, 1:10,000; catalog number 14-14-06; KPL), and the mixture was incubated at RT for 1 h. The plates were washed, and tetramethylbenzidine substrate was added according to the manufacturer's recommendations (Life Technologies). After 15 min with substrate, 0.05 ml of stop solution (2 N H2SO4) was added and the optical density at 450 nm with correction at 655 nm was read. The resulting OD data were modeled as a nonlinear function of the log10 dilution using GraphPad Prism software (La Jolla, CA) and the log (agonist)-versus-response variable slope four-parameter logistic model. Endpoints were interpolated by using 4 times the average OD of the day 0 sample of each respective pig serum sample as the cutoff.
To determine whether serum IgG reacted with whole S. suis P1/7 bacteria, heat-inactivated (HI) P1/7 was used as the antigen in an indirect ELISA. To make the antigen, a single P1/7 colony was inoculated into 5 ml Todd-Hewitt broth and incubated at 37°C in 5% CO2 at 200 rpm for approximately 6 h, at which time it had reached an OD600 of 0.6. The bacteria were centrifuged at 4,000 × g to pellet them, the medium was decanted, and the bacteria were resuspended in 5 ml PBS. The bacteria were heat inactivated by incubating the suspension in a water bath at 85°C for 20 min. Inactivation was confirmed by plating 0.1 ml of the heat-inactivated preparation on blood agar plates and incubating the plates at 37°C in 5% CO2. No growth was observed on the plate after 2 days. Aliquots were stored frozen at −80°C. The protein concentration of HI P1/7 was determined using a bicinchoninic acid protein microtiter assay according to the manufacturer's recommendations (Pierce). Immulon-2 plates were coated with 0.1 ml of 7.5 μg/ml of HI P1/7 diluted in 100 mM carbonate-bicarbonate buffer (pH 9.6). Serum samples collected on day 0 and day 28 from each pig were diluted 1:500 and used in the assay. P1/7-specific IgG was detected, and the ELISA was completed as described above for individual proteins. Data are reported as the OD at 450 nm with correction at 655 nm. A checkerboard of different HI P1/7 concentrations and a pool of sera from day 0 and day 28 was used to determine the optimal ELISA conditions (data not shown). Similar techniques were used to evaluate IgG reactivity with a collection of other HI S. suis strains comprised of two randomly selected representatives of those serotypes most commonly associated with disease (serotypes 1, 2, 1/2, 3, and 14) (Table 1), with bacteria reaching OD600s of 0.6 to 1.1 in the 6- to 8-h culture period prior to heat inactivation (data not shown), and plate wells were coated with 0.1 ml containing 7.5 μg/ml of the HI S. suis preparations for the ELISA.
Evaluation of the cell-mediated immune response to vaccination.
To evaluate the induction of cell-mediated immunity following vaccination, enzyme-linked immunosorbent spot (ELISpot) assays were performed to enumerate the IFN-γ-secreting cells following in vitro stimulation with a pool of the vaccine proteins. Blood was collected by venipuncture and placed into BD Vacutainer cell preparation tubes (CPT) with sodium citrate for the isolation of PBMCs using culture medium, as previously described (42). PBMCs were enumerated and seeded at 2.5 × 105 cells per well in the IFN-γ ELISpot assay plates in duplicate for each treatment. The PBMCs were stimulated with a protein pool in a final volume of 0.25 ml (1 μg/ml of each individual protein per well). Control wells received medium alone or pokeweed mitogen (0.5 μg/ml). At approximately 18 h after stimulation, the ELISpot assay was completed according to the manufacturer's recommendations (R&D Systems, Minneapolis, MN). Spots were enumerated using an S5UV ImmunoSpot instrument and software (Cellular Technology Ltd., Shaker Heights, OH), and the data were analyzed using GraphPad Prism software (La Jolla, CA). The count for duplicate wells for each treatment for each pig was determined and used to calculate the mean for each group.
The levels of the cytokines produced by PBMCs collected on day 28 following restimulation with the protein pool were also measured. PBMC culture supernatants were collected 72 h after restimulation with the protein pool or medium only and used to evaluate the levels of cytokines secreted by the cells. The amounts of IFN-γ, TNF-α, IL-2, and IL-10 in the medium were determined by a multiplex cytokine ELISA according to the manufacturer's recommendations using the recombinant proteins provided by the manufacturer as standards to determine the concentrations in the supernatants (Aushon Biosystems).
Statistical analysis.
Survival analysis was performed using the product limit method of Kaplan and Meier and comparing survival curves using the log-rank test (Prism software; GraphPad, La Jolla, CA). Antibody titers were converted to log10 values, and a two-tailed Student's t test was used to evaluate whether statistically significant differences existed between groups 1 and 2 for the comparisons indicated above and in the figures, with a P value of <0.05 being considered significant. One-way analysis of variance (ANOVA) with Tukey's multiple-comparison posttest was performed to evaluate whether statistically significant differences in the number of IFN-γ-secreting cells and cytokine production exited between groups (P < 0.05). GraphPad Prism software (version 6.0) was used for statistical analysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steven Kellner, Sarah Shore, Zahra Olson, and Lilia Walther for their excellent technical assistance; Dhaarini Raghunathan and Shaowen Li for their contribution to protein purification; and Nate Horman, Jason Huegel, and Tyler Standley for their excellent animal care.
This work was supported by a Longer and Larger (LoLa) grant from the Biotechnology and Biological Sciences Research Council (BBSRC grant numbers BB/G020744/1, BB/G019177/1, BB/G019274/1, and BB/G018553/1), the UK Department for Environment, Food and Rural Affairs, and Zoetis (formerly Pfizer Animal Health), awarded to the Bacterial Respiratory Diseases of Pigs-1 Technology (BRaDP1T) consortium.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00559-17.
REFERENCES
- 1.Gottschalk M, Segura M. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol 76:259–272. doi: 10.1016/S0378-1135(00)00250-9. [DOI] [PubMed] [Google Scholar]
- 2.Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NT, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ, Parkhill J. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4:e6072. doi: 10.1371/journal.pone.0006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngo TH, Tran TB, Tran TT, Nguyen VD, Campbell J, Pham HA, Huynh HT, Nguyen VV, Bryant JE, Tran TH, Farrar J, Schultsz C. 2011. Slaughterhouse pigs are a major reservoir of Streptococcus suis serotype 2 capable of causing human infection in southern Vietnam. PLoS One 6:e17943. doi: 10.1371/journal.pone.0017943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. 2005. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet Microbiol 107:63–69. doi: 10.1016/j.vetmic.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Okura M, Osaki M, Nomoto R, Arai S, Osawa R, Sekizaki T, Takamatsu D. 2016. Current taxonomical situation of Streptococcus suis. Pathogens 5:E45. doi: 10.3390/pathogens5030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottschalk M, Segura M, Xu J. 2007. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev 8:29–45. doi: 10.1017/S1466252307001247. [DOI] [PubMed] [Google Scholar]
- 7.Berthelot-Herault F, Morvan H, Keribin AM, Gottschalk M, Kobisch M. 2000. Production of muraminidase-released protein (MRP), extracellular factor (EF) and suilysin by field isolates of Streptococcus suis capsular types 2, 1/2, 9, 7 and 3 isolated from swine in France. Vet Res 31:473–479. doi: 10.1051/vetres:2000133. [DOI] [PubMed] [Google Scholar]
- 8.Wisselink HJ, Smith HE, Stockhofe-Zurwieden N, Peperkamp K, Vecht U. 2000. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet Microbiol 74:237–248. doi: 10.1016/S0378-1135(00)00188-7. [DOI] [PubMed] [Google Scholar]
- 9.Vela AI, Goyache J, Tarradas C, Luque I, Mateos A, Moreno MA, Borge C, Perea JA, Dominguez L, Fernandez-Garayzabal JF. 2003. Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J Clin Microbiol 41:2498–2502. doi: 10.1128/JCM.41.6.2498-2502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa AT, Lobato FC, Abreu VL, Assis RA, Reis R, Uzal FA. 2005. Serotyping and evaluation of the virulence in mice of Streptococcus suis strains isolated from diseased pigs. Rev Inst Med Trop Sao Paulo 47:113–115. doi: 10.1590/S0036-46652005000200012. [DOI] [PubMed] [Google Scholar]
- 11.Segura M, Fittipaldi N, Calzas C, Gottschalk M. 2017. Critical Streptococcus suis virulence factors: are they all really critical? Trends Microbiol 25:585–599. doi: 10.1016/j.tim.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Weinert LA, Chaudhuri RR, Wang J, Peters SE, Corander J, Jombart T, Baig A, Howell KJ, Vehkala M, Valimaki N, Harris D, Chieu TT, Van Vinh Chau N, Campbell J, Schultsz C, Parkhill J, Bentley SD, Langford PR, Rycroft AN, Wren BW, Farrar J, Baker S, Hoa NT, Holden MT, Tucker AW, Maskell DJ, BRaDP1T Consortium. 2015. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat Commun 6:6740. doi: 10.1038/ncomms7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. 2013. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weerdenburg EM, Abdallah AM, Rangkuti F, Abd El Ghany M, Otto TD, Adroub SA, Molenaar D, Ummels R, Ter Veen K, van Stempvoort G, van der Sar AM, Ali S, Langridge GC, Thomson NR, Pain A, Bitter W. 2015. Genome-wide transposon mutagenesis indicates that Mycobacterium marinum customizes its virulence mechanisms for survival and replication in different hosts. Infect Immun 83:1778–1788. doi: 10.1128/IAI.03050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barquist L, Mayho M, Cummins C, Cain AK, Boinett CJ, Page AJ, Langridge GC, Quail MA, Keane JA, Parkhill J. 2016. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics 32:1109–1111. doi: 10.1093/bioinformatics/btw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan SL, Chaudhuri RR, Peters SE, Mayho M, Weinert LA, Crowther SA, Wang J, Langford PR, Rycroft A, Wren BW, Tucker AW, Maskell DJ, BRaDP1T Consortium. 2013. Generation of a Tn5 transposon library in Haemophilus parasuis and analysis by transposon-directed insertion-site sequencing (TraDIS). Vet Microbiol 166:558–566. doi: 10.1016/j.vetmic.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Patel IR, Mukherjee A. 2013. Genetic analysis of the roles of agaA, agaI, and agaS genes in the N-acetyl-d-galactosamine and d-galactosamine catabolic pathways in Escherichia coli strains O157:H7 and C. BMC Microbiol 13:94. doi: 10.1186/1471-2180-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose MC, Voynow JA. 2006. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev 86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 20.Song XM, Connor W, Hokamp K, Babiuk LA, Potter AA. 2009. Transcriptome studies on Streptococcus pneumoniae, illustration of early response genes to THP-1 human macrophages. Genomics 93:72–82. doi: 10.1016/j.ygeno.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Paixao L, Oliveira J, Verissimo A, Vinga S, Lourenco EC, Ventura MR, Kjos M, Veening JW, Fernandes VE, Andrew PW, Yesilkaya H, Neves AR. 2015. Host glycan sugar-specific pathways in Streptococcus pneumoniae: galactose as a key sugar in colonisation and infection [corrected]. PLoS One 10:e0121042. doi: 10.1371/journal.pone.0121042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott DW, Higgins MA, Hyrnuik S, Pluvinage B, Lammerts van Bueren A, Boraston AB. 2010. The molecular basis of glycogen breakdown and transport in Streptococcus pneumoniae. Mol Microbiol 77:183–199. doi: 10.1111/j.1365-2958.2010.07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shelburne SA III, Sumby P, Sitkiewicz I, Okorafor N, Granville C, Patel P, Voyich J, Hull R, DeLeo FR, Musser JM. 2006. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect Immun 74:4605–4614. doi: 10.1128/IAI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shelburne SA III, Okorafor N, Sitkiewicz I, Sumby P, Keith D, Patel P, Austin C, Graviss EA, Musser JM. 2007. Regulation of polysaccharide utilization contributes to the persistence of group A Streptococcus in the oropharynx. Infect Immun 75:2981–2990. doi: 10.1128/IAI.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shelburne SA III, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A 105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogunniyi AD, Mahdi LK, Trappetti C, Verhoeven N, Mermans D, Van der Hoek MB, Plumptre CD, Paton JC. 2012. Identification of genes that contribute to the pathogenesis of invasive pneumococcal disease by in vivo transcriptomic analysis. Infect Immun 80:3268–3278. doi: 10.1128/IAI.00295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffitt KL, Gierahn TM, Lu YJ, Gouveia P, Alderson M, Flechtner JB, Higgins DE, Malley R. 2011. T(H)17-based vaccine design for prevention of Streptococcus pneumoniae colonization. Cell Host Microbe 9:158–165. doi: 10.1016/j.chom.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffitt K, Skoberne M, Howard A, Gavrilescu LC, Gierahn T, Munzer S, Dixit B, Giannasca P, Flechtner JB, Malley R. 2014. Toll-like receptor 2-dependent protection against pneumococcal carriage by immunization with lipidated pneumococcal proteins. Infect Immun 82:2079–2086. doi: 10.1128/IAI.01632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thammavongsa V, Schneewind O, Missiakas DM. 2011. Enzymatic properties of Staphylococcus aureus adenosine synthase (AdsA). BMC Biochem 12:56. doi: 10.1186/1471-2091-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wegmann F, Gartlan KH, Harandi AM, Brinckmann SA, Coccia M, Hillson WR, Kok WL, Cole S, Ho LP, Lambe T, Puthia M, Svanborg C, Scherer EM, Krashias G, Williams A, Blattman JN, Greenberg PD, Flavell RA, Moghaddam AE, Sheppard NC, Sattentau QJ. 2012. Polyethyleneimine is a potent mucosal adjuvant for viral glycoprotein antigens. Nat Biotechnol 30:883–888. doi: 10.1038/nbt.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin T, Yin Y, Huang L, Yu Q, Yang Q. 2015. H9N2 influenza whole inactivated virus combined with polyethyleneimine strongly enhances mucosal and systemic immunity after intranasal immunization in mice. Clin Vaccine Immunol 22:421–429. doi: 10.1128/CVI.00778-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai RP, Seaman MS, Tonks P, Wegmann F, Seilly DJ, Frost SD, LaBranche CC, Montefiori DC, Dey AK, Srivastava IK, Sattentau Q, Barnett SW, Heeney JL. 2012. Mixed adjuvant formulations reveal a new combination that elicit antibody response comparable to Freund's adjuvants. PLoS One 7:e35083. doi: 10.1371/journal.pone.0035083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park ME, Lee SY, Kim RH, Ko MK, Lee KN, Kim SM, Kim BK, Lee JS, Kim B, Park JH. 2014. Enhanced immune responses of foot-and-mouth disease vaccine using new oil/gel adjuvant mixtures in pigs and goats. Vaccine 32:5221–5227. doi: 10.1016/j.vaccine.2014.07.040. [DOI] [PubMed] [Google Scholar]
- 34.Kuipers K, van Selm S, van Opzeeland F, Langereis JD, Verhagen LM, Diavatopoulos DA, de Jonge MI. 2017. Genetic background impacts vaccine-induced reduction of pneumococcal colonization. Vaccine 35:5235–5241. doi: 10.1016/j.vaccine.2017.08.023. [DOI] [PubMed] [Google Scholar]
- 35.Segura M. 2015. Streptococcus suis vaccines: candidate antigens and progress. Expert Rev Vaccines 14:1587–1608. doi: 10.1586/14760584.2015.1101349. [DOI] [PubMed] [Google Scholar]
- 36.Chabot-Roy G, Willson P, Segura M, Lacouture S, Gottschalk M. 2006. Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb Pathog 41:21–32. doi: 10.1016/j.micpath.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Calzas C, Lemire P, Auray G, Gerdts V, Gottschalk M, Segura M. 2015. Antibody response specific to the capsular polysaccharide is impaired in Streptococcus suis serotype 2-infected animals. Infect Immun 83:441–453. doi: 10.1128/IAI.02427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baums CG, Valentin-Weigand P. 2009. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim Health Res Rev 10:65–83. doi: 10.1017/S146625230999003X. [DOI] [PubMed] [Google Scholar]
- 39.Gómez-Gascón L, Cardoso-Toset F, Tarradas C, Gómez-Laguna J, Maldonado A, Nielsen J, Olaya-Abril A, Rodríguez-Ortega MJ, Luque I. 2016. Characterization of the immune response and evaluation of the protective capacity of rSsnA against Streptococcus suis infection in pigs. Comp Immunol Microbiol Infect Dis 31: 47:52–59. doi: 10.1016/j.cimid.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Li P, Liu J, Zhu L, Qi C, Bei W, Cai X, Sun Y, Feng S. 2010. VirA: a virulence-related gene of Streptococcus suis serotype 2. Microb Pathog 49:305–310. doi: 10.1016/j.micpath.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Nunes SF, Murcia PR, Tiley LS, Brown IH, Tucker AW, Maskell DJ, Wood JL. 2010. An ex vivo swine tracheal organ culture for the study of influenza infection. Influenza Other Respir Viruses 4:7–15. doi: 10.1111/j.1750-2659.2009.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braucher DR, Henningson JN, Loving CL, Vincent AL, Kim E, Steitz J, Gambotto AA, Kehrli ME Jr. 2012. Intranasal vaccination with replication-defective adenovirus type 5 encoding influenza virus hemagglutinin elicits protective immunity to homologous challenge and partial protection to heterologous challenge in pigs. Clin Vaccine Immunol 19:1722–1729. doi: 10.1128/CVI.00315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.