ABSTRACT
Treatment of intracellular bacterial pathogens with antibiotic therapy often requires a long course of multiple drugs. A barrier to developing strategies that enhance antibiotic efficacy against these pathogens is our poor understanding of the intracellular nutritional environment that maintains bacterial persistence. The intracellular pathogen Brucella abortus survives and replicates preferentially in alternatively activated macrophages (AAMs); however, knowledge of the metabolic adaptations promoting exploitation of this niche is limited. Here we show that one mechanism promoting enhanced survival in AAMs is a shift in macrophage arginine utilization from production of nitric oxide (NO) to biosynthesis of polyamines, induced by interleukin 4 (IL-4)/IL-13 treatment. Production of polyamines by infected AAMs promoted both intracellular survival of B. abortus and chronic infection in mice, as inhibition of macrophage polyamine synthesis or inactivation of the putative putrescine transporter encoded by potIHGF reduced both intracellular survival in AAMs and persistence in mice. These results demonstrate that increased intracellular availability of polyamines induced by arginase-1 expression in IL-4/IL-13-induced AAMs promotes chronic persistence of B. abortus within this niche and suggest that targeting of this pathway may aid in eradicating chronic infection.
KEYWORDS: nitrogen metabolism, putrescine
INTRODUCTION
Several intracellular pathogens that cause chronic infections can establish persistence within the mononuclear phagocyte system, including Salmonella enterica serovar Typhi (S. Typhi), Mycobacterium species, Leishmania, and Brucella species (1–4). Within infected tissues, these pathogens can be found in association with different populations of macrophages (5–7). Like other immune cells, macrophages are recognized to adopt a spectrum of different functional states that are influenced by the immune microenvironment (reviewed in references 8 and 9). Functional states that can be modeled in vitro include activation by gamma interferon (IFN-γ), which induces the classically activated macrophage (CAM; also known as M1) phenotype, characterized by production of nitric oxide (NO) and inflammatory cytokines such as tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6). During inflammation in vivo, cells with a similar phenotype can arise from Ly6Chigh inflammatory monocytes that leave the bone marrow in a C-C chemokine receptor type 2 (CCR2)-dependent manner (10). In contrast, in vitro, the Th2 cytokines IL-4 and IL-13 activate signal transducer and activator of transcription 6 (STAT6) to promote the development of alternatively activated macrophages (AAMs), which play important roles in allergic inflammation, helminth infection, and tissue repair (11–13).
In addition to roles in inflammation and immunity, these different macrophage populations differ in their metabolic capacity (reviewed in reference 14), with IFN-γ activation inducing a glycolytic energy metabolism while IL-4/IL-13 treatment promotes a shift to mitochondrial β-oxidation that is dependent on peroxisome proliferator-activated receptors (PPARs [15]). The resulting increase in intracellular glucose availability enhances intracellular replication of both S. Typhi and Brucella abortus (16, 17).
A second metabolic program with divergent functions in distinct macrophage subsets is arginine consumption. In IFN-γ-activated macrophages, arginine serves as a substrate of inducible nitric oxide synthase (iNOS), which converts it to NO and l-citrulline. In contrast, IL-4/IL-13 exposure induces expression of macrophage arginase-1 (18), which catalyzes the first step in biosynthesis of the polyamines putrescine, spermine, and spermidine. An IL-4/IL-13-mediated shift in macrophage arginine metabolism promotes infection with Leishmania spp.; however, the mechanism by which this growth promotion occurs is unclear (19).
Brucella abortus causes chronic intracellular infections in humans and animals (4). In mouse infection models, bacteria are found in association with phagocytic cells, most prominently macrophages, in which a subset of B. abortus can evade killing in phagolysosomes. These bacteria replicate within an endoplasmic reticulum-associated compartment and can interact subsequently with a modified autophagy pathway to promote their cell-to-cell spread (20, 21). While bacteria can be found in association with macrophages having characteristics of both classical (IFN-γ) and alternative (IL-4/IL-13 or IL-10) activation during the initial stages of infection in mice, as infection progresses bacteria are found preferentially within phagocytes that exhibit gene expression profiles characteristic of alternative activation states (16). Interestingly, the B. abortus genome encodes transporters for ornithine and polyamines, the products of macrophage arginase-1, suggesting that arginase-1 expression may provide nutrients for growth of B. abortus within arginase-expressing macrophages. The goal of this study was to determine whether activation of the arginase-1 pathway in AAMs contributes to the ability of this cell population to serve as a site for chronic infection with B. abortus.
RESULTS
B. abortus relies on putrescine uptake to boost bacterial growth in vitro.
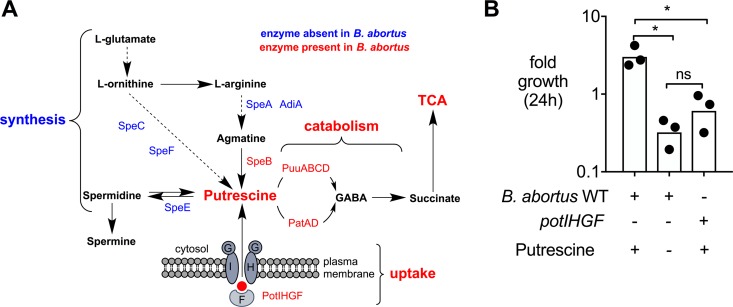
Polyamines such as putrescine are essential secondary metabolites that can be linked directly to basic cellular homeostasis in eukaryotic as well as prokaryotic cells. Interestingly, a survey of the B. abortus 2308 genome sequence (22) showed that homologs of the S. Typhimurium and Escherichia coli genes required for synthesis of putrescine from l-arginine and l-ornithine appear to be absent (Fig. 1A). However, the B. abortus genome does encode homologs of E. coli PatA, PatD, and PuuA, -B, -C, and -D, which mediate the catabolism of putrescine to succinate via γ-aminobutyraldehyde (GABA) (Fig. 1A) (23). Therefore, we investigated whether putrescine could be used as a nutrient source during B. abortus growth in vitro. For this purpose, we reformulated a chemically defined medium with and without putrescine based on the composition of Ham's F-12K (Kaighn's) medium from Invitrogen (USA). B. abortus 2308 exhibited modest growth over 24 h in medium containing putrescine; however, in the absence of putrescine no growth was observed (Fig. 1B; see also Fig. S1A in the supplemental material). To determine the contribution of putrescine uptake to the growth of B. abortus in this medium, we inactivated the potIHGF genes (BAB1_1624 to BAB1_1628; strain TOK13), which based on similarity to the pot genes in E. coli (24) were predicted to encode a putrescine-specific ATP binding cassette transporter. The potIHGF mutant grew normally in tryptic soy broth (TSB) but exhibited no growth in putrescine-containing F12/K medium (Fig. 1B and S1B). These results suggest that B. abortus PotIHGF contributes to growth in the presence of putrescine.
FIG 1.
Putrescine is an essential nutrient source during B. abortus growth in vitro. (A) Schematic representation of differences in polyamine synthesis between S. Typhimurium and B. abortus based on Kusano and Suzuki (44) and the most-recent genomic and metabolomic data available in the MetaCyc/BioCyc (45) and KEGG (46) databases. (B) Fold growth of of B. abortus 2308 (wild type [WT]) in modified F12/k medium with and without putrescine as well as the potIHGF mutant (TOK13) at 24 h postinoculation. The inoculum was 0.5 × 105 CFU/ml. Bars represent geometric means, and circles represent results of independent replicates. The statistical significance of differences between groups was determined using a one-way ANOVA with the Sidak multiple-comparison test. Asterisks denote statistically significant differences between groups (P < 0.05); ns, not significant.
B. abortus persists in BALB/c mice within CD11b+ cells exhibiting features of alternate macrophage activation.
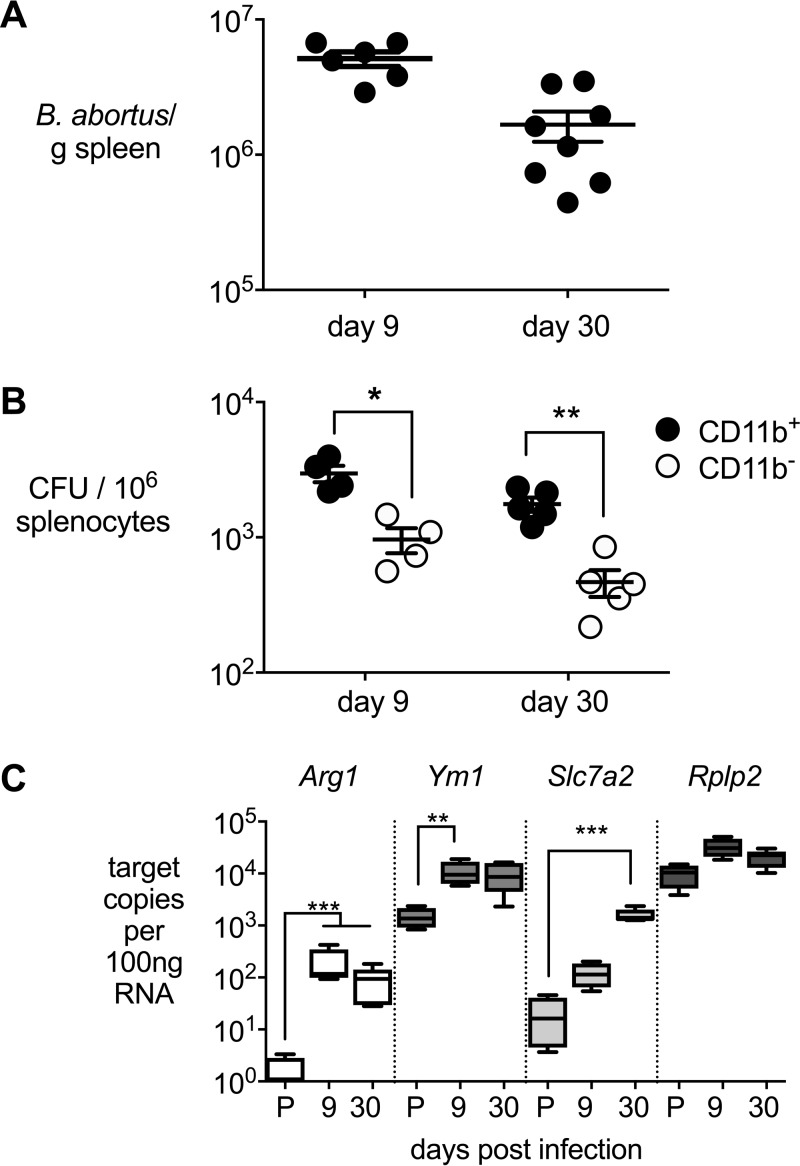
Our previous work showed that AAMs are a preferred replicative niche of B. abortus during chronic infections in C57BL/6J mice (16). However, for the current study, C57BL/6J mice proved not to be suitable, as this mouse strain is deficient for expression of the arginine transporter solute carrier family 7, member 2 (Slc7a2), which provides the substrate for arginase 1 during alternative activation of macrophages (25). Therefore, we used BALB/c mice for these studies. Since Il12p40−/− mice on the BALB/c background have been shown to harbor Brucella predominantly within the CD11c+ phagocyte subset (26), we first determined whether wild-type BALB/c mice harbor B. abortus within macrophages. To this end, we inoculated mice via the intraperitoneal (i.p.) route and recovered B. abortus from spleens at 9 and 30 days postinfection (Fig. 2). Spleens of mice were highly colonized at both time points (Fig. 2A). Enrichment of the CD11b+ population by immunomagnetic separation and plating of the CD11b+ cell-enriched and -depleted fractions revealed that while B. abortus could be isolated from both cellular subsets, greater numbers of bacteria were associated with the macrophage-containing CD11b+ fraction at both time points (Fig. 2B). Characterization of gene expression profiles revealed that compared to uninfected mice, the macrophage-containing CD11b+ splenocyte fraction from infected mice expressed higher levels of Arg1, encoding arginase, Ym1, encoding chitinase, and Slc7a2, encoding the arginine amino acid transporter, which are markers of alternate macrophage activation (Fig. 2C). In contrast to what was seen in C57BL/6 mice, expression of AAM markers in BALB/c mice at day 9 postinfection is higher, which could be attributed to a reduced production of gamma interferon in B. abortus-infected BALB/c mice compared to C57BL/6 mice (data not shown). These results show that in BALB/c mice B. abortus persists within macrophages in the spleen and that this cell population exhibits features of AAMs.
FIG 2.
B. abortus resides in CD11b+ splenocytes that express features of alternative activated macrophages. (A) B. abortus 2308 CFU in spleens from i.p. infected BALB/cJ mice enumerated 9 (n = 6) and 30 (n = 8) days postinfection. (B) Bacterial counts of B. abortus 2308 in CD11b+ and CD11b− splenocytes from BALB/cJ mice 9 (n = 4) and 30 (n = 5) days postinfection. (C) Absolute copy numbers of alternatively activated macrophage markers Arg-1, Ym1, and Slc7a2 of CD11b+ splenocytes from BALB/cJ mice infected with B. abortus 2308 or treated with D-PBS (Invitrogen) 9 days (n = 4) and 30 days (n = 5) postinfection. P, D-PBS; Rplp2, housekeeping control gene (encoding the large ribosomal subunit protein P2). Values represent means ± standard errors of the means (SEM). For panel B, the significance of differences between experimental groups at a single time point was determined using an unpaired t test on log-transformed data. For panel C, the significance of differences between each infection time point and uninfected mice was determined using an unpaired t test on log-transformed data. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Arginase-1 activity contributes to chronic infection by B. abortus.
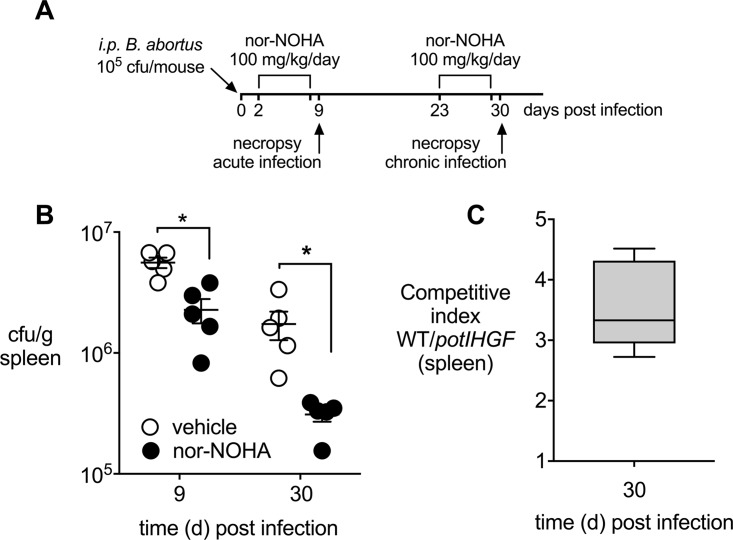
Our finding that Arg1 is upregulated in murine macrophages during B. abortus infection raised the question whether arginase activity promotes bacterial persistence. To determine the contribution of arginase-1 to survival of B. abortus in the mononuclear phagocyte system, we blocked arginase-1 activity in infected mice using the inhibitor N(omega)-hydroxy-nor-l-arginine (nor-NOHA) (27). For these experiments, mice inoculated via the i.p. route with B. abortus were treated daily with vehicle or nor-NOHA for 6 days before necropsy at either 9 days or 30 days postinfection (Fig. 3A and B). Treatment with nor-NOHA reduced the splenic bacterial burden 2-fold during the acute phase of infection (9 days) compared to the vehicle treatment (Fig. 3B). This effect was greater during chronic infection (30 days), when 6-fold-fewer bacteria were recovered from the nor-NOHA-treated group.
FIG 3.
Inhibition of arginase-1 activity reduces splenic B. abortus colonization. (A) Treatment regime to inhibit arginase-1 activity in vivo using 100 mg/kg/day nor-NOHA (Cayman Chemical, USA) for 6 consecutive days. (B) Survival of B. abortus 2308 in spleens from infected and nor-NOHA-treated BALB/cJ mice (n = 5) compared to infected but vehicle control (D-PBS)-treated mice (n = 5) at 6 and 30 days postinfection. (C) Competitive index, calculated for day 30 postinfection based on CFU from spleens of BALB/cJ mice infected with a 1:1 mixture of B. abortus 2308 and the potIHGF mutant. Values represent means ± SEM. The statistical significance of differences between groups was determined using an unpaired t test analysis on log-transformed data. *, P < 0.05.
The reduction in B. abortus infection levels by inhibition of arginase-1 raised the question of which downstream products B. abortus might utilize for its intracellular replication. The product of arginase-1, ornithine, can be decarboxylated by ornithine decarboxylase (ODC) into the polyamine putrescine. To determine whether uptake of putrescine by B. abortus contributes to its persistence in mice, we determined the effect of potIHGF inactivation on the fitness of B. abortus 30 days after i.p. infection of mice. The potIHGF mutant strain (TOK13) had a small but significant 3-fold competitive defect in the spleen compared to the parent strain at 30 days postinfection (Fig. 3C), suggesting that uptake of putrescine contributes to a higher pathogen burden in the spleen during chronic infection with B. abortus.
Inhibition of arginase-1 leads to reduced intracellular availability of l-ornithine and putrescine.
To determine if B. abortus replication in IL-4 and IL-13 (IL-4+IL-13)-polarized macrophages depends on products of the arginase-1 pathway, we measured the intracellular concentrations of l-ornithine, an arginase-1 product, and of putrescine, a product of the downstream ODC reaction. Treatment of bone marrow-derived macrophages (BMMs) with IL-4+IL-13 led to an increase in cellular ornithine (Fig. 4A). The elevated levels of ornithine were reversed by inhibition of arginase-1 with nor-NOHA (Fig. 4A) without significantly affecting the intracellular levels of other amino acids (Fig. 4B). Similar results were obtained when we measured intracellular putrescine concentrations by gas chromatography-mass spectrometry (GC-MS) (Fig. 4C), suggesting that inhibition of arginase-1 affects the abundance of polyamines that may promote intracellular replication of B. abortus. While treatment of macrophages with IFN-γ can shift their energy metabolism toward anaerobic glycolysis, inhibition of arginase-1 with nor-NOHA did not have this effect, since production of lactate, the end product of anaerobic glycolysis, was unaffected (Fig. 4D). Inhibition of arginase-1 also did not increase macrophage production of NO, as measured by the Griess assay (Fig. 4E). These observations imply that inhibition of arginase-1 limits the availability of l-ornithine and putrescine without shifting the phenotype of macrophages to that of IFN-γ-treated macrophages or inducing other pathways that could limit intracellular survival of B. abortus.
FIG 4.
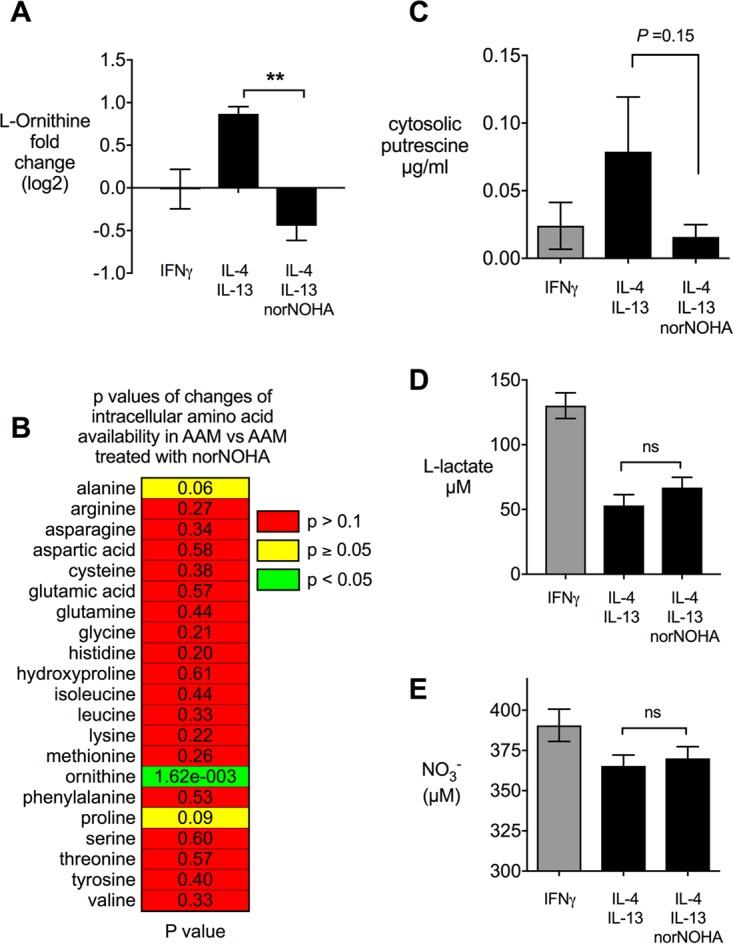
Inhibition of arginase-1 in alternatively (IL-4+IL-13) activated macrophages leads to reduced intracellular availability of l-ornithine and putrescine without changing the activation state of the macrophages. (A) Intracellular l-ornithine levels measured in BMMs after treatment with inducers. (B) Influence of the arginase-1 inhibitor nor-NOHA on the amino acid pool in IL-4+IL-13-polarized macrophages. (C) Concentration of putrescine in IFN-γ-polarized and IL-4+IL-13-polarized BMMs and effect of nor-NOHA treatment. (D and E) Effects of arginase-1 inhibition on production of lactate (D) and NO (E) by IL-4+IL-13-polarized macrophages. Results from IFN-γ-polarized macrophages are shown as a reference. Values represent means ± SEM. Differences between IL-4+IL-13-treated groups were analyzed using an unpaired t test analysis on log-transformed data. **, P < 0.01; ns, not significant.
Increased abundance of polyamines supports growth of B. abortus in IL-4+IL-13-treated macrophages.
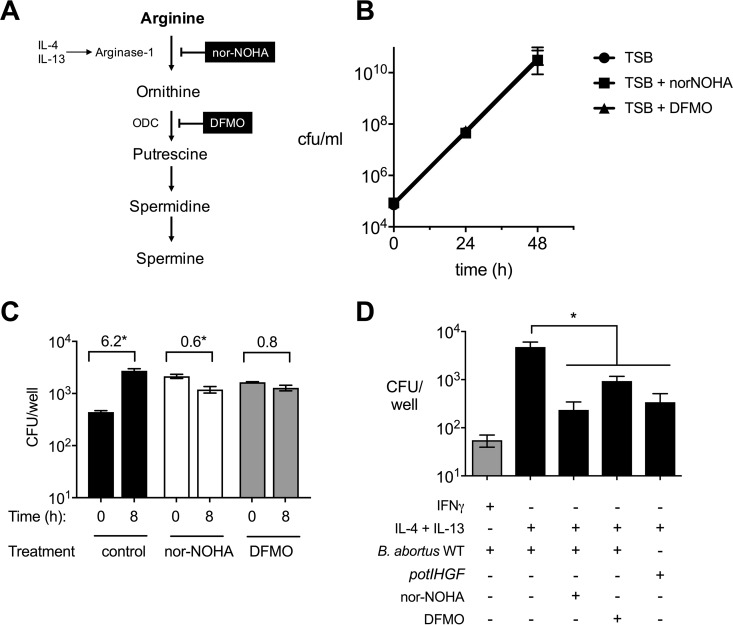
The above-described observations in mice suggested a role for metabolites of the arginase-1 pathway in supporting chronic B. abortus infection, but since arginase-1 inhibition in the mouse could affect other cells in addition to macrophages, we asked whether IL-4+IL-13 treatment of macrophages promotes intracellular replication of B. abortus via increased availability of polyamines. To test this idea, BMMs from BALB/c mice were differentiated with IL-4+IL-13 and treated either with nor-NOHA or with the ODC inhibitor difluoromethylornithine (DFMO), which prevents production of putrescine (Fig. 5A). In addition, two control experiments were performed to assess the effects of the inhibitors on extracellular growth of B. abortus (Fig. 5B) and on uptake of bacteria by treated macrophages (Fig. 5C). While nor-NOHA and DFMO had no effect on B. abortus growth in TSB (Fig. 5B), treatment of IL-4+IL-13-treated J774 macrophages with either inhibitor increased uptake of B. abortus but blocked intracellular replication of the internalized bacteria compared to untreated cells (Fig. 5C). In BMMs, blockade of either arginase-1 or ODC also significantly reduced intracellular replication of B. abortus, suggesting that the arginase-1 pathway contributes significantly to intracellular replication of B. abortus within IL-4+IL-13-treated macrophages (Fig. 5D). We used IFN-γ-treated macrophages as a control for a condition that restricts intracellular replication of B. abortus. Interestingly, the effect of inhibiting arginase-1 was similar to that of inactivating the putrescine uptake system encoded by potIHGF, suggesting putrescine as a potential nutrient that fuels replication within this population of macrophages.
FIG 5.
Intracellular de novo synthesis of polyamines is beneficial for B. abortus replication in IL-4+IL-13-activated BMMs. (A) Simplified model of the polyamine synthesis pathway in macrophages treated with IL-4 and IL-13. Targets of the inhibitors nor-NOHA and DFMO are shown. ODC, ornithine decarboxylase. (B) Growth of B. abortus in TSB supplemented with inhibitors of arginase-1 (nor-NOHA) or ornithine decarboxylase (DFMO). (C) Effects of nor-NOHA and DFMO treatments on uptake and early replication of B. abortus in IL-4+IL-13-treated J774 macrophages. Numbers above each experimental group represent the fold increase in intracellular B. abortus between 0 h and 8 h. Asterisks represent significant differences between time points determined using Student's t test on logarithmically transformed data. (D) Intracellular survival of B. abortus 2308 WT and the potIHGF mutant at 48 h postinfection of BMMs. IFN-γ-treated macrophages are shown as a reference. Values represent means ± SEM. The statistical significance of differences between IL-4+IL-13-treated macrophages and macrophages treated with inhibitors or infected with the potIHGF mutant was determined using a one-way ANOVA with a Tukey's multiple-comparison test. Asterisks represent significant differences between groups (P < 0.05).
DISCUSSION
B. abortus persists within the cells of the mononuclear phagocyte system to cause chronic infection. One niche for persisting bacteria is within macrophages expressing phenotypic markers of alternate activation (16). Considering the metabolic differences between classically (M1, IFN-γ-) activated macrophages and other, less inflammatory macrophage populations, it is likely that the metabolism of B. abortus is adapted to take advantage of this niche by utilizing other metabolites that are more abundant in AAMs. Previous work has shown that glucose uptake in this niche supports the chronic stages of brucellosis (16, 28). To further define this important replicative niche especially during chronic Brucella infection, we investigated the role of polyamines, mainly putrescine, in the survival of B. abortus inside AAMs since it was described that the arginase-1-driven polyamine synthesis is considered to be a hallmark of M2 or AAMs (29).
Inhibition of host-derived putrescine production impaired the intracellular bacterial survival of B. abortus ex vivo in AAMs as well as in vivo in BALB/c mice. Further, a mutant in a locus encoding a predicted putrescine transporter, potIHGF, had a competitive fitness defect during chronic infection, suggesting that putrescine is a likely nutrient for B. abortus during this phase of infection. While no predicted bacterium-type ODC that could serve as a source of putrescine was identified in the B. abortus genome, the protein encoded by ORF BAB2_0098 shares similarity with eukaryotic ODC (KEGG), suggesting that B. abortus might be able to produce putrescine under some conditions. However, if this is the case, it appears to be unable to compensate for the blockade of host-derived putrescine production in macrophages (Fig. 5C and D). It should also be noted that potIHGF is not the only predicted transporter gene for arginine metabolites in the B. abortus genome, as BAB2_0876-0879 and BAB2_1062-1064/BAB2_1129 encode putative transporters for ornithine and putrescine/spermidine, respectively. Interestingly, the Brucella melitensis homolog of BAB2_0876-0879 appears also to contribute to chronic infection (30), suggesting that Brucella spp. have multiple mechanisms for acquiring metabolites of the arginase-1 pathway in vivo.
Our observation that survival of B. abortus during chronic infection is promoted by arginase-1 activity is consistent with the findings of Hanot Mambres et al., who found that in IL-12p40−/− mice, which are defective in IFN-γ production and express elevated levels of IL-4 (31), B. abortus was associated with cells expressing arginase-1 (26). In this previous study, in situ analysis showed that the arginase-expressing cells had features of dendritic cells, suggesting that in our in vivo experiments, inhibition of arginase-1 could affect other cell types in addition to macrophages. During its intracellular survival, access of B. abortus to polyamines such as putrescine can be explained by its route of intracellular trafficking. Polyamines have been localized within acidified compartments in the host cell that have features of late endocytic compartments (32). Upon phagocytic uptake, vacuoles containing Brucella mature along the endocytic pathway and fuse with lysosomes (21). After exclusion of lysosomal markers, B. abortus replicates within a compartment that is associated with the endoplasmic reticulum (21, 33). To complete its intracellular cycle and spread to the next cell, B. abortus subsequently subverts a subset of autophagy proteins to form a compartment that regains features of the endosome, including its acidified nature (34). Therefore, during at least two stages of its intracellular life cycle, B. abortus should be exposed to a polyamine-containing environment.
The contribution of polyamines to the survival of B. abortus in the host may shed light on the differing susceptibilities of specific genetic mouse backgrounds to Brucella infections. It is generally accepted that mice of the C57BL/6 genetic background are more resistant to Brucella infections than are BALB/c mice (35), with a difference of about 1 order of magnitude in splenic colonization levels after i.p. inoculation. In contrast to what is observed in BALB/c mice, the enhanced clearance of Brucella in C57BL/6 mice was attributed to the expression of the proinflammatory cytokine IFN-γ during the acute phase of the infection, which further translated to the diminished ability of Brucella to establish chronic infections (36). The less-permissive C57BL/6 mice produce the proinflammatory cytokine IFN-γ, which shifts the balance in the macrophage pool toward classic or M1 activation and thus would be expected to contain lower levels of intracellular polyamines as a nutrient source for Brucella. In contrast, the macrophages of BALB/c mice, which have a reduced and delayed expression of IFN-γ during infection, expressed markers of alternative activation earlier during infection than what was previously described for C57BL/6 mice (16). These findings are consistent with a report on Leishmania infection showing that macrophages from C57BL/6 mice have impaired arginine uptake as a result of a mutation in the promoter of the arginine solute carrier SLC7a2 (25). Expression of this solute carrier is induced by IL-4+IL-13 and is essential to import sufficient arginine for arginase-1 activity and downstream polyamine production. These polyamines can then can be exploited by B. abortus as a nutrient source to promote the intracellular chronicity of the pathogen. In conclusion, our results show that products of the arginase-1/polyamine pathway in IL-4+IL-13-activated macrophages support intracellular replication and chronic infection by B. abortus. This finding identifies this pathway as a potential target for strategies to limit or eradicate chronic infection, an idea that merits further investigation.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Brucella abortus 2308 was used as the wild-type strain for this work. All work with live B. abortus was performed at biosafety level 3 and was approved by the Institutional Biosafety Committee at the University of California, Davis. To generate the putrescine uptake-negative potIHGF mutant strain (TOK13), we inserted the plasmid pTK411 by homologous recombination to disrupt the operon encoding the potIHGF locus in B. abortus 2308. The plasmid pTK411 is a pBlueScript II SK(+)-based plasmid carrying two amplified 500-bp fragments flanking the potIHGF operon using the primer combinations (i) pBl-KS-3-A-Fwd (5′-gcggtggcggccgctGACCGGCATCGGCTACAA-3′) and H2-0414-Rev (5′-ccagttgttCGAATAGCCGATCTCCACC-3′) for fragment H1-0411-A and (ii) H1-0411-Fwd (5′-gctattcgAACAACTGGTCCCGCTTC-3′) and pBl-KS-3-B-Rev (5′-ggccccccctcgaggGGTCGAAATTCATCGCCAC-3′) for fragment H2-0414-B (where lowercase bases represent overlapping regions for Gibson assembly of the plasmid). The fragments were fused together and cloned into the vector backbone by using the Gibson cloning (NEB, USA). Successful cloning steps were verified by PCR and sequencing. The resulting construct, designated pTK411t, was introduced into B. abortus 2308 by electroporation, and ampicillin resistance was used to identify recombinants. The phenotype of the potIHGF mutant was confirmed by loss of growth promotion by putrescine in modified F-12/K medium. Strains were cultured on tryptic soy agar (TSA; Difco/Becton Dickinson, Sparks, MD) or tryptic soy broth at 37°C on a rotary shaker. For mouse infection, bacteria were cultured on TSA plates containing 5% sheep blood for 3 days. Ampicillin (250 mg/ml) or kanamycin (100 mg/ml) was added to the culture media to select resistant B. abortus strains. To address the importance of putrescine to growth of B. abortus in vitro, we formulated a modified version (modified F-12/K; see Table S1 in the supplemental material) of the commercially available F-12/K medium (Invitrogen, USA) according to the manufacturer's formulation, which included all inorganic salts, d-glucose, hypoxanthine Na, lipoic acid, sodium pyruvate, and thymidine, with the addition of minimum essential medium (MEM) amino acids solution (50×) liquid and MEM vitamin solution (100×) liquid (both from Invitrogen, USA) to complete the medium. When needed, putrescine (Santa Cruz Biotechnology, USA) was added at the manufacturer's suggested concentration of 0.32 mg/liter.
To investigate the contribution of putrescine uptake to growth of B. abortus 2308 and of the potIHGF mutant strain (TOK13) in vitro, bacterial pellets of fresh overnight cultures were washed three times with sterile Dulbecco's phosphate-buffered saline (D-PBS; Invitrogen) and suspended in modified F-12/K with and without putrescine at 0.5 × 105 to 1 × 105 CFU/ml. Bacterial cultures (5 ml) were incubated for 24 h at 37°C in 50-ml conical tubes with tightly closed lids on a rotary shaker. Serial dilutions of the bacterial culture in sterile PBS were plated to enumerate CFU/ml and to calculate bacterial growth over a 24-h period.
Animal experiments.
Female BALB/cJ mice obtained from the Jackson Laboratory (USA) were used for this study. Mice were held in individually ventilated microisolator cages (Tecniplast, West Chester, PA) with sterile bedding and irradiated feed in a biosafety level 3 laboratory. All animal experiments were approved by the University of California Laboratory Animal Care and Use Committee and were conducted in accordance with institutional guidelines. Groups of four to six mice were inoculated intraperitoneally (i.p.) with 0.1 ml of PBS containing 1 × 105 CFU of B. abortus 2308 or a 1:1 mixture of B. abortus 2308 and the potIHGF mutant as previously described (37). When needed, mice were treated daily for 7 days with i.p. injection of 100 mg/kg of body weight/day of the arginase inhibitor nor-NOHA (Cayman Chemical, USA) diluted in 0.1 ml of sterile D-PBS. Sterile D-PBS was used as a vehicle control. At various time points after infection, mice were euthanized via CO2 inhalation, and spleens and serum from the animals were collected aseptically at necropsy. Spleens were weighed and homogenized in 4 ml of PBS to enumerate CFU per organ using serial dilutions of the homogenate plated on TSA alone or TSA with ampicillin (250 ng/ml). Spleen samples were also collected for gene expression, flow cytometry, and immunohistochemistry analysis.
BMM infection.
BMMs were differentiated as previously described by Murray et al. (29) and Rolan et al. (37) from femora and tibiae of female 6- to 8-week-old BALB/c mice. To determine the contribution of de novo polyamine synthesis to the cellular survival of B. abortus strains, BMMs were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated and dialyzed (3,500 molecular weight cutoff [MWCO]) fetal calf serum (FCS), 0.03% l-glutamine, 1 mM nonessential amino acids, 1 mM sodium pyruvate (all from Invitrogen, USA) according to the method of Van den Bossche et al. (38). For infection of BMMs with different B. abortus strains, 24-well microtiter plates were seeded with 5 × 105 macrophages per well in 0.5 ml of medium and incubated for 48 h at 37°C in 5% CO2. Bacterial inoculation and infection of BMMs were done using a gentamicin protection assay as described in reference 16 with a multiplicity of infection (MOI) of 100 in RPMI that was formulated as described above. Bacterial survival was enumerated by plating serial dilutions on TSA or on TSA plus ampicillin (250 μg/ml) 48 h after infection using BMMs that were lysed with 0.5 ml of 0.5% Tween 20. For purification of proteins and RNA, BMMs were directly resuspended in 0.5 ml of TRIzol (Invitrogen, USA) and stored at −80°C until further use. To block de novo synthesis of polyamines, the specific arginase inhibitors nor-NOHA (Cayman Chemicals, USA) and DFMO (Sigma) to block ornithine decarboxylase were used according to the recommendation of Van den Bossche et al. (38). To induce classical activation of BMMs (CAMs), 10 ng/ml of mouse recombinant IFN-γ (rIFN-γ; BD Bioscience, San Jose, CA) was used. AAMs were generated using 10 ng/ml of mouse rIL-4 (mrIL-4; R&D Systems, USA) and 5 ng/ml rIL-13 (R&D Systems, USA). All treatments were added to the cells either 48 h (nor-NOHA, DFMO) or 24 h (mrIL-4/-13) prior to experiments and maintained throughout the experiments. Experiments were performed independently in duplicate at least four times, and the standard error for each time point was calculated. To control for any effect of treatments on macrophage viability, lactate dehydrogenase (LDH) release was assayed using the CytoTox 96 nonradioactive cytotoxicity (Promega). At the concentrations used, no significant difference in LDH release was observed between groups treated with IFN-γ, IL-4/IL-13, DFMO, or NOHA and untreated controls (data not shown).
CD11b+ cell isolation.
Macrophages (CD11b+ cells) were isolated from the spleens of B. abortus-infected BALB/cJ mice using a MACS CD11b MicroBeads magnetic cell sorting kit from Miltenyi Biotech (Auburn, CA) according to the manufacturer's instructions as previously described (39). To enumerate viable B. abortus in CD11b− and CD11b+ splenocyte fractions, 10-fold serial dilutions of each fraction in sterile PBS were plated on TSA plates and the numbers of viable bacteria were normalized to CFU per 106 splenocytes.
Gene expression analysis.
Eukaryotic gene expression was determined by quantitative real-time PCR (qRT-PCR) as previously described (40). RNA was isolated using TRIzol (Invitrogen, USA) according to the manufacturer's instructions. To synthesize cDNA, a reverse transcriptase reaction was performed using 500 ng of total RNA, random hexamers (Applied Biosystems, USA), and TaqMan reverse transcription reagents (Applied Biosystems, Carlsbad, CA). Real-time PCR was performed using SYBR green (Applied Biosystems) with the primers listed in Table S2 in the supplemental material. Absolute copy numbers for Arg1, Ym1, and Slc7a2 were calculated based on individually cloned plasmid standards and were normalized to 100 ng input RNA used for the cDNA synthesis. The housekeeping gene Rplp2, encoding the large ribosomal protein 2, was included as a control.
Intracellular amino acid content measurement.
Measurement of total intracellular amino acid contents of differentially activated and/or nor-NOHA-treated BALB/c BMMs was performed by the UC Davis Genome Center Proteomics core. For sample preparation, BMMs treated for 48 h were intensively washed 4 times with sterile D-PBS (Invitrogen, USA). Three wells of a 6-well plate were combined in a total of 1 ml of sterile PBS, and cells were detached by scraping. All samples were stored at −80°C until further use. For amino acid analysis, 50 μl of 10% sulfosalicylic acid (Sigma-Aldrich, USA) was added to 200 μl of sample and mixed at room temperature for 15 min, and the mixture was stored overnight at −20°C. The thawed samples were vortexed and centrifuged, and 100 μl of supernatant was spiked with 3.75 nmol AE-Cys as an internal standard. The amount of free amino acids was analyzed using the Hitachi L-8900 amino acid analyzer.
Intracellular putrescine measurements by GC-MS.
Intracellular levels of the polyamine putrescine were measured according to the protocol for sample preparation, derivatization chemistry, and GC-MS conditions that was previously published by Chen et al. (41, 42). BMMs were cultured as described for the amino acid analysis, and 2 wells of a 6-well culture plate were combined and detached by scraping in 1 ml of 10% NaCl (pH = 10) according to the method of Chen et al. (42). One microgram of Put-D8 (CDN Isotopes, Canada) was used as an internal standard. A TQ8040 GC-MS/MS instrument (Shimadzu) was employed to detect intracellular levels of putrescine in differentially activated bone marrow-derived macrophages. All mass spectra were acquired in electron ionization (EI) for all measurements in the selected-ion monitoring (SIM) mode for putrescine-d8 and the multiple-reaction monitoring (MRM) mode for putrescine. Helium carrier gas was set to a column head pressure of 149.5 kPa at a flow rate of 1.75 ml/min. Sample aliquots of 2 μl were injected with a split ratio of 5 with a 5-min solvent delay. GC analyses were done with an Rtx-5Sil MS capillary column (30-m length, 0.25-mm inner diameter [i.d.], and 0.25-μm thickness; Restek, USA) programmed from 140°C to 210°C at 8°C/min, followed by a 2-min hold, and then to 300°C at 20°C/min, followed by a 4-min hold. A final temperature increase to 320°C at 20°C/min was held as bake out for 4 min. MS conditions were the following: source, 200°C; interface, 300°C; injector, 260°C; electron energy, 70 eV; typical source pressure, 1.8 × 10−5 torr. Full scans were over the mass range m/z 10 to 700 at 2.0 scans/s for the SIM mode. A total of 3 ions from putrescine and its fragmentation products were used to monitor Put. SIM and full-scan acquisitions were started at 5.20 min and ended at 17.73 min. The first ion group consisted of three ions with m/z 166, 283, and 355 for putrescine-d8 running for 5 to 8 min with a dwell time of 100 ms. Detection and quantification of the abundance of the internal standard putrescine by MRM was done using the ions 283 > 142 m/z and 355 > 211 in the above-mentioned time frame. While all ions in each group were monitored for peak verification, the ions used for quantification were those in italics above. Instrument run, peak measurements, and data analysis were done with the GCMS solution software (Shimadzu, USA). To normalize the data, we used the detected concentration of the putrescine-d8 in each individual sample as a calibrator to calculate a factor that normalizes all samples to the same putrescine-d8 level as the one that was detected in the untreated BMM within each mouse. These factors were multiplied with the detected putrescine areas for each sample based on the corresponding individual calibration factor calculated for each sample in each mouse to normalize the data. Prism (GraphPad, USA) was used to graph the normalized areas and to compute statistical analysis based on the log of normalized areas using a paired, one-tailed Student's t test.
Metabolite measurements.
For determination of secreted l-lactate levels, BALB/c BMMs were grown in 24-well plates and infected as described above. At 24 and 48 h postinfection, the cell culture supernatant was harvested and deproteinized using perchloric acid (Abcam, USA). l-lactate was measured using the L-lactate assay kit II (Eton Bioscience, USA). To follow the production of reactive nitrogen species by macrophages ex vivo, we measured the levels of nitrate and nitrite in cell culture supernatants of activated and infected macrophages 48 h postinfection using an adaptation of the Griess assay as described by Lopez et al. (43).
Statistical analysis.
Fold changes of ratios (bacterial numbers or mRNA levels) and percentages (flow cytometry and fluorescence microscopy) were transformed logarithmically prior to statistical analysis. An unpaired Student's t test was used on the transformed data to determine whether differences between groups were statistically significant (P < 0.05). When more than two treatments were used, statistically significant differences between groups were determined by an ordinary one-way analysis of variance (ANOVA) and the Sidak test for multiple comparisons. All statistical analyses were performed using GraphPad Prism software (GraphPad Software, USA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by PHS grants AI112258 and AI109799 to R.M.T. and AI118807 and AI128151 to S.E.W. Work in S.E.W.'s laboratory is supported by a grant from the Welch Foundation (I-1858).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00458-17.
REFERENCES
- 1.Antoine JC, Prina E, Jouanne C, Bongrand P. 1990. Parasitophorous vacuoles of Leishmania amazonensis-infected macrophages maintain an acidic pH. Infect Immun 58:779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss G, Schaible UE. 2015. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev 264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruby T, McLaughlin L, Gopinath S, Monack D. 2012. Salmonella's long-term relationship with its host. FEMS Microbiol Rev 36:600–615. doi: 10.1111/j.1574-6976.2012.00332.x. [DOI] [PubMed] [Google Scholar]
- 4.Byndloss MX, Tsolis RM. 2016. Chronic bacterial pathogens: mechanisms of persistence. Microbiol Spectr 4(2). doi: 10.1128/microbiolspec.VMBF-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byndloss MX, Tsolis RM. 2016. Brucella spp. virulence factors and immunity. Annu Rev Anim Biosci 4:111–127. doi: 10.1146/annurev-animal-021815-111326. [DOI] [PubMed] [Google Scholar]
- 6.Scott P, Novais FO. 2016. Cutaneous leishmaniasis: immune responses in protection and pathogenesis. Nat Rev Immunol 16:581–592. doi: 10.1038/nri.2016.72. [DOI] [PubMed] [Google Scholar]
- 7.Tan S, Russell DG. 2015. Trans-species communication in the Mycobacterium tuberculosis-infected macrophage. Immunol Rev 264:233–248. doi: 10.1111/imr.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Dyken SJ, Locksley RM. 2013. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol 31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon S, Martinez FO. 2010. Alternative activation of macrophages: mechanism and functions. Immunity 32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Shi C, Pamer EG. 2011. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes JL, Terrazas LI. 2007. The divergent roles of alternatively activated macrophages in helminthic infections. Parasite Immunol 29:609–619. doi: 10.1111/j.1365-3024.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 12.Shirey KA, Cole LE, Keegan AD, Vogel SN. 2008. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J Immunol 181:4159–4167. doi: 10.4049/jimmunol.181.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence T, Natoli G. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 14.Chawla A. 2010. Control of macrophage activation and function by PPARs. Circ Res 106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, Chawla A. 2007. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xavier MN, Winter MG, Spees AM, den Hartigh AB, Nguyen K, Roux CM, Silva TM, Atluri VL, Kerrinnes T, Keestra AM, Monack DM, Luciw PA, Eigenheer RA, Baumler AJ, Santos RL, Tsolis RM. 2013. PPARgamma-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe 14:159–170. doi: 10.1016/j.chom.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisele NA, Ruby T, Jacobson A, Manzanillo PS, Cox JS, Lam L, Mukundan L, Chawla A, Monack DM. 2013. Salmonella require the fatty acid regulator PPARdelta for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 14:171–182. doi: 10.1016/j.chom.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corraliza IM, Soler G, Eichmann K, Modolell M. 1995. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochem Biophys Res Commun 206:667–673. doi: 10.1006/bbrc.1995.1094. [DOI] [PubMed] [Google Scholar]
- 19.Iniesta V, Gomez-Nieto LC, Corraliza I. 2001. The inhibition of arginase by N(omega)-hydroxy-l-arginine controls the growth of Leishmania inside macrophages. J Exp Med 193:777–784. doi: 10.1084/jem.193.6.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorvel JP, Moreno E. 2002. Brucella intracellular life: from invasion to intracellular replication. Vet Microbiol 90:281–297. doi: 10.1016/S0378-1135(02)00214-6. [DOI] [PubMed] [Google Scholar]
- 21.Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J. 2008. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic 9:678–694. doi: 10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 22.Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, Aguero F, Land ML, Ugalde RA, Garcia E. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect Immun 73:8353–8361. doi: 10.1128/IAI.73.12.8353-8361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki H, Kurihara S. 2015. Polyamine catabolism in prokaryotes. In Kusano T, Suzuki H (ed), Polyamines. Springer, Tokyo, Japan. [Google Scholar]
- 24.Terui Y, Saroj SD, Sakamoto A, Yoshida T, Higashi K, Kurihara S, Suzuki H, Toida T, Kashiwagi K, Igarashi K. 2014. Properties of putrescine uptake by PotFGHI and PuuP and their physiological significance in Escherichia coli. Amino Acids 46:661–670. doi: 10.1007/s00726-013-1517-x. [DOI] [PubMed] [Google Scholar]
- 25.Sans-Fons MG, Yeramian A, Pereira-Lopes S, Santamaria-Babi LF, Modolell M, Lloberas J, Celada A. 2013. Arginine transport is impaired in C57Bl/6 mouse macrophages as a result of a deletion in the promoter of Slc7a2 (CAT2), and susceptibility to Leishmania infection is reduced. J Infect Dis 207:1684–1693. doi: 10.1093/infdis/jit084. [DOI] [PubMed] [Google Scholar]
- 26.Hanot Mambres D, Machelart A, Vanderwinden JM, De Trez C, Ryffel B, Letesson JJ, Muraille E. 2015. In situ characterization of splenic Brucella melitensis reservoir cells during the chronic phase of infection in susceptible mice. PLoS One 10:e0137835. doi: 10.1371/journal.pone.0137835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenu JP, Lepoivre M, Moali C, Brollo M, Mansuy D, Boucher JL. 1999. Effects of the new arginase inhibitor N(omega)-hydroxy-nor-L-arginine on NO synthase activity in murine macrophages. Nitric Oxide 3:427–438. doi: 10.1006/niox.1999.0255. [DOI] [PubMed] [Google Scholar]
- 28.Hong PC, Tsolis RM, Ficht TA. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect Immun 68:4102–4107. doi: 10.1128/IAI.68.7.4102-4107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronneau S, Moussa S, Barbier T, Conde-Alvarez R, Zuniga-Ripa A, Moriyon I, Letesson JJ. 2016. Brucella, nitrogen and virulence. Crit Rev Microbiol 42:507–525. doi: 10.3109/1040841X.2014.962480. [DOI] [PubMed] [Google Scholar]
- 31.Magram J, Connaughton SE, Warrier RR, Carvajal DM, Wu CY, Ferrante J, Stewart C, Sarmiento U, Faherty DA, Gately MK. 1996. IL-12-deficient mice are defective in IFN gamma production and type 1 cytokine responses. Immunity 4:471–481. doi: 10.1016/S1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- 32.Soulet D, Gagnon B, Rivest S, Audette M, Poulin R. 2004. A fluorescent probe of polyamine transport accumulates into intracellular acidic vesicles via a two-step mechanism. J Biol Chem 279:49355–49366. doi: 10.1074/jbc.M401287200. [DOI] [PubMed] [Google Scholar]
- 33.Celli J, Salcedo SP, Gorvel JP. 2005. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc Natl Acad Sci U S A 102:1673–1678. doi: 10.1073/pnas.0406873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, Celli J. 2012. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grillo M-J, Blasco J-M, Gorvel J-P, Moriyon I, Moreno E. 2012. What have we learned from brucellosis in the mouse model? Vet Res 43:29. doi: 10.1186/1297-9716-43-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy EA, Sathiyaseelan J, Parent MA, Zou B, Baldwin CL. 2001. Interferon-gamma is crucial for surviving a Brucella abortus infection in both resistant C57BL/6 and susceptible BALB/c mice. Immunology 103:511–518. doi: 10.1046/j.1365-2567.2001.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolan HG, Tsolis RM. 2008. Inactivation of the type IV secretion system reduces the Th1 polarization of the immune response to Brucella abortus infection. Infect Immun 76:3207–3213. doi: 10.1128/IAI.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van den Bossche J, Lamers WH, Koehler ES, Geuns JMC, Alhonen L, Uimari A, Pirnes-Karhu S, Van Overmeire E, Morias Y, Brys L, Vereecke L, De Baetselier P, Van Ginderachter JA. 2012. Pivotal advance: arginase-1-independent polyamine production stimulates the expression of IL-4-induced alternatively activated macrophage markers while inhibiting LPS-induced expression of inflammatory genes. J Leukoc Biol 91:685–699. doi: 10.1189/jlb.0911453. [DOI] [PubMed] [Google Scholar]
- 39.Rolan HG, Xavier MN, Santos RL, Tsolis RM. 2009. Natural antibody contributes to host defense against an attenuated Brucella abortus virB mutant. Infect Immun 77:3004–3013. doi: 10.1128/IAI.01114-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rolán HG, Tsolis RM. 2007. Mice lacking components of adaptive immunity show increased Brucella abortus virB mutant colonization. Infect Immun 75:2965–2973. doi: 10.1128/IAI.01896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen GG, Fiori LM, Mamer OA, Turecki G. 2011. High-resolution capillary gas chromatography in combination with mass spectrometry for quantification of three major polyamines in postmortem brain cortex. Methods Mol Biol 720:427–436. doi: 10.1007/978-1-61779-034-8_27. [DOI] [PubMed] [Google Scholar]
- 42.Chen GG, Turecki G, Mamer OA. 2009. A quantitative GC-MS method for three major polyamines in postmortem brain cortex. J Mass Spectrom 44:1203–1210. doi: 10.1002/jms.1597. [DOI] [PubMed] [Google Scholar]
- 43.Lopez CA, Rivera-Chavez F, Byndloss MX, Baumler AJ. 2015. The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar Typhimurium during colitis. Infect Immun 83:3470–3478. doi: 10.1128/IAI.00351-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusano T, Suzuki H. 2015. Polyamines: a universal molecular nexus for growth, survival, and specialized metabolism. Springer, Tokyo, Japan. [Google Scholar]
- 45.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD. 2016. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.