ABSTRACT
Small noncoding RNAs (sRNAs) have been identified as important regulators of gene expression in various cellular processes. cia-dependent small RNAs (csRNAs), a group of sRNAs that are controlled by the two-component regulatory system CiaRH, are widely conserved in streptococci, but their targets have been identified only in Streptococcus pneumoniae. Streptococcus sanguinis, a pioneer colonizer of teeth and one of the most predominant bacteria in the early oral biofilm, has been shown to have six csRNAs. Using computational target prediction and the luciferase reporter assay, we identified pilT, a constituent of the type IV pilus operon, as a negative regulatory target for one of the csRNAs, namely, csRNA1-1, in S. sanguinis. RNA-RNA electrophoretic mobility shift assay using a nucleotide exchange mutant of csRNA1-1 revealed that csRNA1-1 binds directly to pilT mRNA. In addition, csRNA1-1 and csRNA1-2, a putative gene duplication product of csRNA1-1 that is tandemly located in the S. sanguinis genome, negatively regulated S. sanguinis biofilm formation. These results suggest the involvement of csRNAs in the colonization step of S. sanguinis.
KEYWORDS: CiaRH, PilT, Streptococcus sanguinis, csRNA, type IV pili
INTRODUCTION
Streptococcus sanguinis, a Gram-positive facultative anaerobe, is a commensal pioneer colonizer in oral biofilm formation and is also implicated in infective endocarditis. S. sanguinis adheres directly to saliva-coated teeth by a variety of mechanisms, including interaction with salivary components, e.g., α-amylase and secretory immunoglobulin A complex (1). After adherence, S. sanguinis provides a new scaffold for attachment and induces environmental changes, which include extracellular polysaccharide synthesis, H2O2 production, and DNA release (2, 3, 4). These changes affect the colonization of other oral microorganisms that constitute the oral biofilm. While various factors associated with the biofilm-forming ability of S. sanguinis have been identified, regulatory mechanisms of colonization and biofilm formation are extremely complex and still unclear.
In recent years, bacterial noncoding small RNAs (sRNAs) have received great attention because of their important roles in cellular processes, including adaptive responses and colonization (5). Bacterial sRNAs can control gene function using multiple molecular mechanisms to regulate the expression of target genes, including direct binding to complementary nucleotide sequences within target mRNA (6, 7). The best-characterized streptococcal sRNA is FasX of Streptococcus pyogenes. FasX positively or negatively regulates the expression of S. pyogenes virulence factors, including streptokinase, collagen-binding pili, and fibronectin-binding proteins (PrtF1 and PrtF2), through interaction with the 5′ untranslated region (UTR) of target mRNAs (8, 9, 10, 11). Since these posttranscriptional levels of regulation result in a quicker effect than transcriptional regulation, sRNAs are mainly associated with the fine-tuning of metabolic processes or stress adaptation.
Two-component systems (TCSs) are bacterial transcriptional regulatory systems that play important roles in the bacterial response to environmental changes (12, 13). A conventional TCS is composed of a transmembrane sensor histidine kinase and a cytoplasmic response regulator. When sensing an extracellular signal, the sensor kinase autophosphorylates on a histidine residue and then transfers the phosphate group to the response regulator, which in turn acts as a transcription factor of target genes. The first characterized TCS in streptococci, namely, the CiaRH system, which is composed of CiaH histidine kinase and a CiaR response regulator, was identified in a search for spontaneous cefotaxime-resistant mutants of Streptococcus pneumoniae (14). In addition to β-lactam antibiotic resistance, CiaRH has been implicated in genetic competence, host colonization, bacteriocin production, and virulence (15, 16, 17, 18). A search for CiaR target genes in S. pneumoniae identified 15 promoters, which control the expression of 24 genes. Five of these promoters control sRNA transcription and are designated cia-dependent small RNAs (csRNAs) (19). BLAST searches using the five csRNAs from S. pneumoniae as a query revealed that all streptococcal genomes sequenced thus far contain csRNA genes (20). Among the streptococcal genomes searched in the study, S. sanguinis has been shown to express six csRNAs, namely, csRNA1-1, csRNA1-2, csRNA1-3, csRNA2, csRNA7, and csRNA8. To date, the functions of these csRNAs have not been elucidated.
In the present study, we performed a computational target search for csRNAs in S. sanguinis. We identified pilT, a constituent of the type IV pilus gene cluster, as a target gene of csRNA1-1. Analysis of the translational reporter fusion of PilT and an RNA-RNA mobility shift assay revealed direct binding of csRNA1-1 to pilT mRNA to control PilT expression. We also report the involvement of csRNAs in S. sanguinis biofilm formation.
RESULTS
Expression of csRNAs is regulated by CiaRH.
To determine whether the expression of csRNAs is controlled by the CiaRH system in S. sanguinis ATCC 10556, we first constructed the following strains: wild-type (WT) ATCC 10556 containing an empty vector (WT mock), its ciaRH deletion mutant containing an empty vector (ΔciaRH mock), and the complemented mutant derivative containing a plasmid-borne WT ciaRH allele (ΔciaRH+pVACiaRH). Using these strains, real-time reverse transcription-PCR (RT-PCR) analysis of csRNA gene expression was performed. As shown in Table 1, transcription of all csRNAs, except for csRNA8, was almost completely lost in the ΔciaRH strain, and the expression levels were restored in the ΔciaRH+pVACiaRH complemented strain. These results confirm that the expression of csRNAs, except for csRNA8, is regulated by the CiaRH TCS in S. sanguinis ATCC 10556.
TABLE 1.
Transcriptional regulation of csRNAs by the CiaRH system
| Target | Fold change in expressiona |
||
|---|---|---|---|
| WT mock | ΔciaRH mock | ΔciaRH+pVACiaRH | |
| csRNA1-1 | 1 ± 0.060 | 0.001 ± 0.0003b | 0.64 ± 0.154 |
| csRNA1-2 | 1 ± 0.165 | 0.015 ± 0.017b | 1.67 ± 0.639 |
| csRNA1-3 | 1 ± 0.102 | 0.108 ± 0.060b | 1.47 ± 0.097 |
| csRNA2 | 1 ± 0.343 | 0.008 ± 0.027b | 1.90 ± 0.716 |
| csRNA7 | 1 ± 0.181 | 0.015 ± 0.027b | 1.47 ± 0.561 |
| csRNA8 | 1 ± 0.138 | 0.997 ± 0.342 | 1.34 ± 0.253 |
Mean values and standard deviations from three independent cultures are presented.
P < 0.01.
Computational prediction of csRNA targets.
To identify genes that are controlled by csRNAs, we first searched the S. sanguinis SK36 genome with TargetRNA2 (25) using csRNA sequences as queries. The complete lists of genes identified by these searches are shown in Tables S1 to S6 in the supplemental material. The number of predicted target genes of the csRNAs ranged from 10 to 57, with some overlap. The predicted target genes of csRNA1-1 and csRNA1-2 showed a high degree of overlap (31 shared genes out of 52 or 56 candidate target genes, respectively), which suggested that these two csRNAs share some regulatory functions. In addition, Marx et al. (20) have shown that the expression levels of csRNA1-1 and csRNA1-2 in S. sanguinis were significantly higher than those of other csRNAs by Northern blot analyses. Thus, csRNA1-1 and csRNA1-2 were selected for further functional analyses. A list of the top 10-ranked candidate target genes for csRNA1-1 and csRNA1-2 is shown in Table 2. Predicted targets included several putative surface protein genes, e.g., SSA_0227, SSA_1632, SSA_2121, SSA_0906, and SSA_0905 (Table 2 and Table S1), which suggests an important role of csRNA1-1 and csRNA1-2 in S. sanguinis colonization. Among the predicted targets, type IV pilus retraction ATPase gene pilT (SSA_2317) was the most probable target, with the lowest thermodynamic energy (−15.87 kcal/mol) of hybridization between the two RNA molecules. The pilT gene is a constituent of the type IV pilus gene cluster (26, 27). Type IV pili have been shown to mediate many functions in bacteria, including adherence to host cells, twitching motility, genetic competence, and biofilm formation (28, 29). Thus, we chose pilT as a target gene for further analyses of csRNA function in S. sanguinis ATCC 10556.
TABLE 2.
Top 10-ranked putative target genes for csRNA1-1 and csRNA1-2
| Ranka | Energy | Locus tag | Gene description (gene) | Putative interaction between targets and csRNA1-1 or csRNA1-2b |
|---|---|---|---|---|
| 1 (1) | −15.87 | SSA_2317 | Tfp pilus assembly protein, pilus retraction ATPase PilT | csRNA1-1 47 ACUUUUUCAAAUCCUAA 31 |
| |:::||||||||||| | ||||
| SSA_2317 −14 AGGGGAAGUUUAGGAUG 3 | ||||
| 2 (2) | −14.36 | SSA_2345 | Hypothetical protein | csRNA1-1 60 ACCCUCUAAUAAUACU 45 |
| |||||||| ||||| | ||||
| SSA_2345 −11 AGGGAGAUU-UUAUGU 4 | ||||
| 3 (3) | −14.01 | SSA_2216 | Lipopolysaccharide biosynthesis protein (licD1) | csRNA1-1 52 UAAU-ACUUUUUCAAAUCCUA 32 |
| |||| |||||||||||:||| | ||||
| SSA_2216 −30 AUUAUUGAAAAAGUUUGGGAG −10 | ||||
| 4 | −13.14 | SSA_1763 | Molybdenum ABC transporter ATPase | csRNA1-1 35 CCUAAUAGUUUGU 23 |
| ||||||||||| | ||||
| SSA_1763 8 AGAUUAUCAAACU 20 | ||||
| (4) | −13.19 | SSA_1655 | Hypothetical protein | csRNA1-2 23 UUCAAUCCUCC 13 |
| ||||||||| | ||||
| SSA_1655 −78 CAGUUAGGAGA −68 | ||||
| 5 (5) | −12.43 | SSA_2217 | Cps9H | csRNA1-1 48 UACUUUUUCAAAUCCUA 32 |
| ||| |||||||||||| | ||||
| SSA_2217 −27 AUG-AAAAGUUUAGGAA −11 | ||||
| 6 (6) | −11.89 | SSA_1635 | Hypothetical protein | csRNA1-1 62 CCUCUAAUAAUACUUUU 46 |
| ||| |||||||||||| | ||||
| SSA_1635 2 GGAAAUUAUUAUGAAAU 19 | ||||
| 7 (7) | −11.64 | SSA_0227 | Collagen-binding surface protein | csRNA1-1 60 ACC-CUCUAAU-AAUACUUUU 42 |
| || |:||||| ||||| ||| | ||||
| SSA_0227 −14 CGGAGGGAUUAUUUAUG-AAA 6 | ||||
| 8 | −11.39 | SSA_1820 | Hypothetical protein | csRNA1-1 19 AAUCCCCCG 11 |
| ||||||| | ||||
| SSA_1820 4 AUAGGGGGA 12 | ||||
| 37 (8) | −11.59 | SSA_0718 | Hypothetical protein | csRNA1-2 23 UUCAAUCCUCCAGA 10 |
| |||||||| ||: | ||||
| SSA_0718 −78 CAGUUAGGAAGUUA −65 | ||||
| 9 (9) | −11.36 | SSA_1632 | Surface protein | csRNA1-1 59 CCCUCUAAUAAUACUU 44 |
| |||||||| |||| | ||||
| SSA_1632 −11 AGGAGAUUAAAAUG-A 4 | ||||
| 10 | −11.16 | SSA_0991 | DNase | csRNA1-1 51 UAAUACUUUUUCAAAUCCU 33 |
| ||||||||||||||||| | ||||
| SSA_0991 −3 UUUAUGAAAAAGUUUAGGU 16 | ||||
| (10) | −11.35 | SSA_2177 | Hypothetical protein | csRNA1-2 51 ACUUUUUCAA-AUCCU 37 |
| ||||||||| |||: | ||||
| SSA_1702 4 AGAAAAAGUUACAGGG 19 |
Rank for csRNA1-2 is indicated in parentheses.
Vertical lines and dots between nucleotides of mRNA and csRNA indicate standard (Watson-Crick) and nonstandard (G-U wobble) base-pairing interactions, respectively.
Expression of the pilT gene is negatively regulated by csRNA1-1 and csRNA1-2.
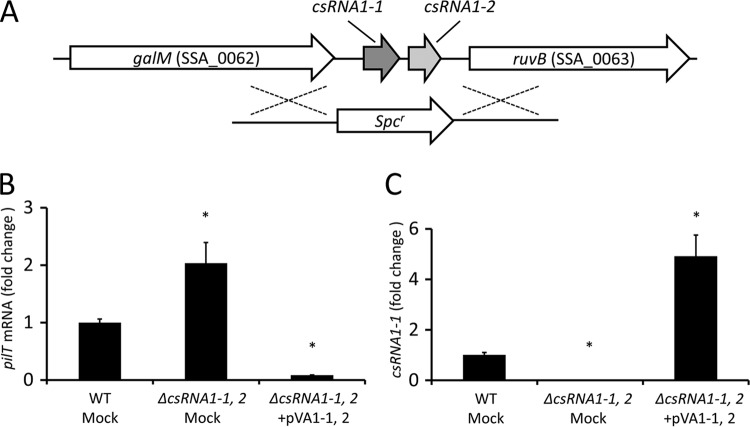
The csRNA1-1 (90 bp in size) and csRNA1-2 (94 bp in size) genes are tandemly located in the intergenic region between the galM (SSA_0062) and ruvB (SSA_0063) genes in the S. sanguinis SK36 genome (Fig. 1A) and have highly conserved nucleotide sequences (93% identity), suggesting they are gene duplications with similar physiological roles. Thus, to assess the physiological roles of csRNA1-1 and csRNA1-2, we generated a csRNA1-1 csRNA1-2 double-deletion mutant strain via double-crossover recombination (Fig. 1A) and its derivative containing an empty vector (ΔcsRNA1-1, 2 mock). The complemented mutant derivative containing plasmid-borne WT csRNA1-1 and csRNA1-2 alleles (ΔcsRNA1-1, 2+pVA1-1, 2) was also generated. Using these strains, we assessed whether the computationally identified target gene pilT is controlled by csRNA1-1 and csRNA1-2. As shown in Fig. 1B, real-time RT-PCR analyses revealed that the ΔcsRNA1-1, 2 strain contained about 2-fold increased levels of pilT mRNA, suggesting that csRNA1-1 and csRNA1-2 negatively regulate pilT mRNA levels. In addition, an approximately 10-fold reduction in pilT mRNA level was observed in the ΔcsRNA1-1, 2+pVA1-1, 2 strain compared to the level in the WT strain. This reduction seemed to be the result of increased expression (about 5-fold compared to that of the parent strain) of csRNA1-1 in the ΔcsRNA1-1, 2+pVA1-1, 2 strain (Fig. 1C). This increase in csRNA1-1 expression may be due to the increased copy number of the shuttle plasmid (pVA838) used for gene complementation. Using these strains, we also checked whether the other candidate target genes are controlled by csRNA1-1 and csRNA1-2. As shown in Fig. S1A, the level of SSA_2345 transcript (rank 2 in Table 2) was slightly decreased in the ΔcsRNA1-1, 2 mock strain, but the level was not changed in the ΔcsRNA1-1, 2+pVA1-1, 2 strain. In contrast, the expression of SSA_2216 (licD1; rank 3 in Table 2) was significantly decreased in the ΔcsRNA1-1, 2+pVA1-1, 2 strain but was not affected in the ΔcsRNA1-1, 2 mock strain (Fig. S1B). These results suggest that the mechanisms of gene regulation by csRNA1-1 and csRNA1-2 vary according to the target gene affected.
FIG 1.
Regulation of pilT mRNA expression by csRNA1-1 and csRNA1-2. (A) Schematic representation of S. sanguinis csRNA1-1 and csRNA1-2 genomic regions and the double-crossover event for generation of the csRNA1-1 csRNA1-2 double-deletion mutant strain (ΔcsRNA1-1, 2). (B and C) Quantitative RT-PCR analysis of pilT (B) and csRNA1-1 (C) expression in S. sanguinis. The ΔcsRNA1-1, 2 strain was transformed with either complementation plasmid pVA1-1, 2 or empty vector pVA838 (Mock). Strains were grown in BHI broth to the early exponential phase, and RNA was isolated and used in quantitative RT-PCR analysis. Data are expressed as means ± standard deviations (SD) from triplicate experiments. *, P < 0.01.
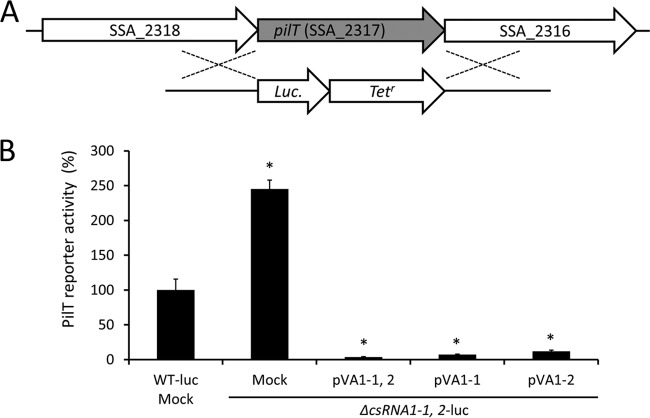
Expression of the pilT reporter gene is negatively regulated by csRNA1-1 and csRNA1-2.
To confirm the results of real-time RT-PCR analyses shown in Fig. 1, we created a luciferase translational reporter strain (WT-luc) in which the structural gene of pilT is replaced with a luciferase gene (Fig. 2A). Using this strain, we further constructed the following strains: WT-luc containing an empty vector (WT-luc mock), a csRNA1-1 csRNA1-2 double-deletion mutant containing an empty vector (ΔcsRNA1-1, 2-luc mock), and the complemented mutant (ΔcsRNA1-1, 2-luc+pVA1-1, 2). Luciferase assays using these strains indicated that the ΔcsRNA1-1, 2-luc mock strain showed a significant increase in luciferase activity, and complementation of both csRNA1-1 and csRNA1-2 completely reversed the elevated luciferase activity (Fig. 2B). These results are consistent with the results of real-time RT-PCR analyses shown in Fig. 1. We next used complementation plasmids that contained the WT allele of csRNA1-1 (pVA1-1) or csRNA1-2 (pVA1-2) to construct strains that express only csRNA1-1 (ΔcsRNA1-1, 2-luc+pVA1-1) or csRNA1-2 (ΔcsRNA1-1, 2-luc+pVA1-2). We then performed a luciferase assay to determine the effect of csRNA1-1 and csRNA1-2 on PilT reporter activity. As shown in Fig. 2B, the ΔcsRNA1-1, 2-luc+pVA1-1 and ΔcsRNA1-1, 2-luc+pVA1-2 strains both showed luciferase activity comparable to that of the ΔcsRNA1-1, 2-luc+pVA1-1, 2 strain, indicating that complementation with either csRNA1-1 or csRNA1-2 can sufficiently restore the inhibitory effect of csRNA1-1 and csRNA1-2 on PilT reporter activity. These results indicate that csRNA1-1 and csRNA1-2 have similar regulatory roles and can compensate for each other's functions, at least concerning the negative regulatory effect on PilT expression.
FIG 2.
(A) Schematic representation of S. sanguinis pilT genomic regions and the double-crossover event for generation of PilT-luciferase translational reporter strain. (B) Regulation of translational luciferase fusion of pilT by csRNA1-1 and csRNA1-2. PilT-luciferase reporter strains were grown in BHI broth to early exponential phase, and luciferase activity was measured. Data are normalized to the OD600 of corresponding cultures and converted to percent luciferase activity relative to that of the WT-luc strain. Data are expressed as means ± SD from triplicate experiments. *, P < 0.01.
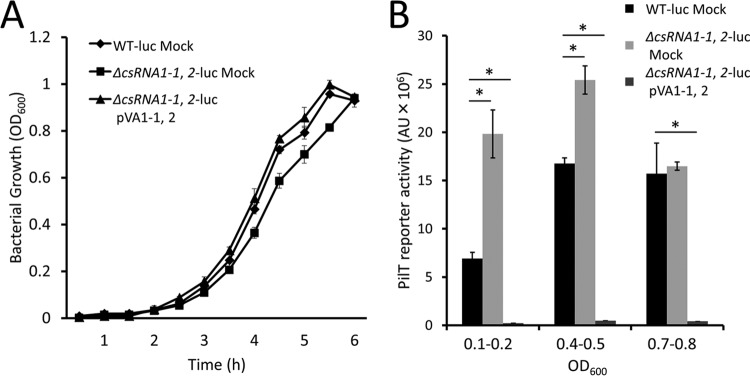
Time course of pilT gene expression.
To assess whether S. sanguinis pilT gene expression varies according to growth phase, we measured the growth of the WT-luc mock, ΔcsRNA1-1, 2-luc mock, and ΔcsRNA1-1, 2-luc+pVA1-1, 2 strains over time (Fig. 3A). Luciferase activity of the strains then was measured at early log (optical density at 600 nm [OD600] of 0.1 to 0.2), mid-log (0.4 to 0.5), and late log (0.7 to 0.8) phases (Fig. 3B). At early log phase, the luciferase activity of the ΔcsRNA1-1, 2-luc mock strain was about three times higher than that of the WT-luc mock strain. The reporter activities increased to maximal levels at mid-log phase and decreased to similar levels at late-log phase. The PilT reporter activity did not directly correlate with the csRNA1-1 transcription level (Fig. S2) in the WT-luc mock strain, and growth phase-dependent changes in reporter activity were still observed in the ΔcsRNA1-1, 2-luc mock strain, suggesting that PilT expression is regulated by additional factors. In contrast to the ΔcsRNA1-1, 2-luc mock strain, the PilT reporter activity was scarcely detectable in the ΔcsRNA1-1, 2-luc+pVA1-1, 2 strain throughout the growth phase. These results indicate that the observed difference in Fig. 2 is not caused by a difference in the timing of maximal expression of PilT reporter activity.
FIG 3.
Growth phase-dependent expression of PilT. PilT-luciferase reporter strains were grown over a 6-h period, and samples were collected for luciferase assay at an OD600 of 0.1 to 0.2 (early log phase), 0.4 to 0.5 (mid-log phase), and 0.7 to 0.8 (late-log phase). (A) Growth curves of the strains over the measurement period. (B) PilT reporter activities of the strains. Luciferase activity was measured and divided by the OD600 of corresponding cultures. Data are expressed as means ± SD of arbitrary units (AU) from three independent cultures. *, P < 0.01.
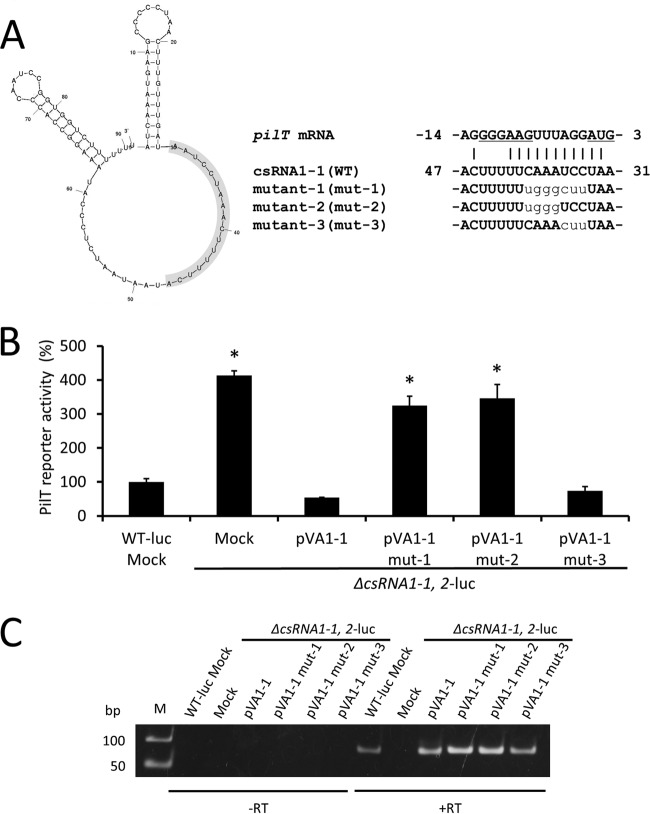
A specific region of csRNA1-1 is important for regulation of pilT gene expression.
To confirm the importance of nucleotides in csRNA1-1 predicted to base pair with pilT mRNA in the TargetRNA2 search (Fig. 4A), nucleotide substitution mutations (mut-1, mut-2, and mut-3) were introduced into the plasmid containing the csRNA1-1 gene (pVA1-1) to obtain pVA1-1 mut-1, pVA1-1 mut-2, and pVA1-1 mut-3. These plasmids then were transformed into the ΔcsRNA1-1, 2-luc mock strain, and the luciferase assay was performed to evaluate their ability to regulate PilT expression. As shown in Fig. 4B, strains expressing mut-1 or mut-2 exhibited elevated levels of luciferase activity, similar to the ΔcsRNA1-1, 2-luc mock strain, indicating that these mutants have lost their inhibitory effect on PilT expression. In contrast, the strain expressing mut-3 retained csRNA1-1 activity comparable to the WT level (Fig. 4B). The expression of all csRNA1-1 versions used was confirmed by RT-PCR (Fig. 4C). Collectively, these results indicate that the AAAC sequence within csRNA1-1, which was mutated in both mut-1 and mut-2, is essential for the inhibitory effect of csRNA1-1 on PilT expression.
FIG 4.
Effect of csRNA1-1 mutation on PilT expression. (A, left) Putative secondary structure of csRNA1-1, as predicted by Mfold (41). The nucleotides involved in the putative region for hybridization with pilT mRNA are highlighted in gray background. (Right) Putative interactions between the 5′ end of pilT mRNA and csRNA1-1 WT or mutant strains (mut-1, mut-2, and mut-3). The ribosome binding site and start codon are underlined. Substituted nucleotides in csRNA1-1 mutants are shown in plain lowercase. (B) Luciferase activity of S. sanguinis csRNA1-1 mutant strains (mut-1, mut-2, and mut-3). Strains were grown in BHI broth to early exponential phase and luciferase activity was measured. Data are presented as percent luciferase activity relative to that of the WT-luc strain. Data are expressed as means ± SD from triplicate experiments. *, P < 0.01. (C) RT-PCR analysis of csRNA1-1 expression in S. sanguinis csRNA1-1 mutant strains. Strains were grown to early exponential phase, and RNA was isolated and used in RT-PCR analysis. The image shown is representative of three independent experiments.
csRNA1-1 directly binds to pilT mRNA.
To confirm whether csRNA1-1 can bind directly to target mRNA to regulate its expression, we performed an RNA-RNA electrophoretic mobility shift assay (EMSA) using csRNA1-1 and pilT mRNA. To show the specificity of RNA-RNA interaction, the mutant version of csRNA1-1 (mut-2 in Fig. 4) and the pilT mRNA mutant (mut) with the respective compensatory nucleotide substitutions were also used. Putative interactions between pilT mRNA (WT) and csRNA1-1 (WT) or pilT mRNA (mut) and csRNA1-1 (mut-2) are shown in Fig. 5A. Incubation of in vitro-transcribed pilT mRNA (WT) with digoxigenin (DIG)-labeled csRNA1-1 (WT) resulted in the emergence of a higher-molecular-weight complex, and the band intensity decreased significantly when DIG-labeled csRNA1-1 (mut-2) was used as the probe (P < 0.01) (Fig. 5B and C, lanes 5 and 6). In contrast, incubation of pilT mRNA (mut) with DIG-labeled csRNA1-1 (WT) resulted in a faint band, and a significantly higher-intensity band was observed after incubation with csRNA1-1 (mut-2) (P < 0.01) (Fig. 5B and C, lanes 7 and 8). An approximately 2-fold increase in the intensity of the shifted band was observed in lane 8 compared with lane 5 (Fig. 5B and C). This difference may be caused by the increased GC content within the putative binding region of pilT mRNA (mut) and csRNA1-1 (mut-2) (Fig. 5A). These results clearly indicate that csRNA1-1 directly binds to pilT mRNA, and the AAAC sequence within csRNA1-1 is important for the binding. In addition, together with the results shown in Fig. 4, the inhibitory effect of csRNA1-1 on PilT expression appears to be dependent on its ability to bind pilT mRNA.
FIG 5.
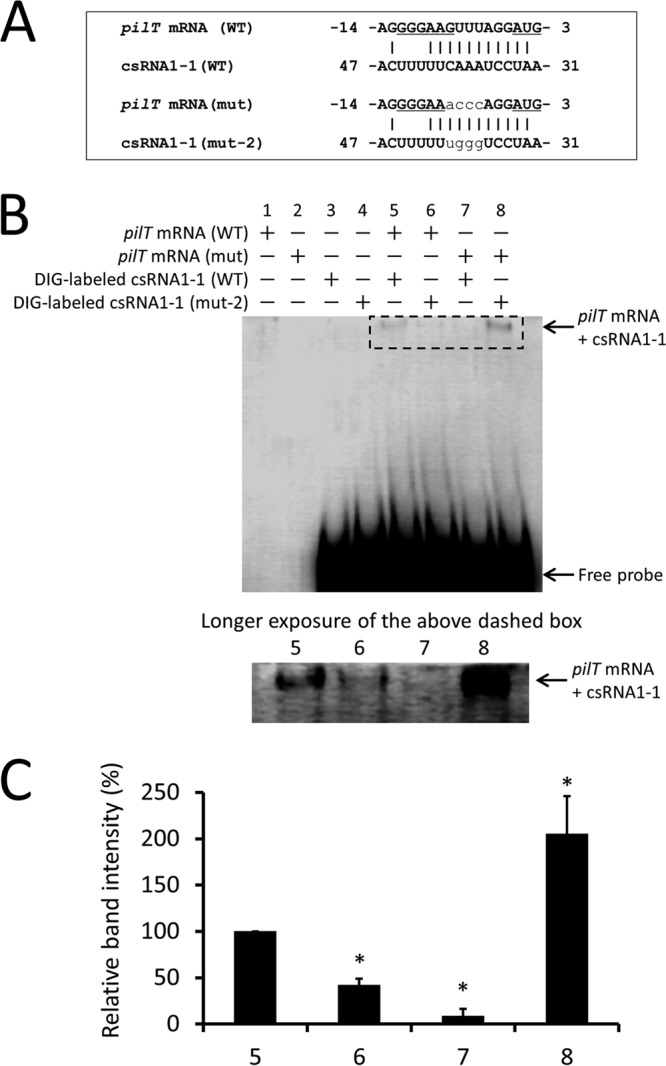
RNA-RNA electrophoretic mobility shift assay of csRNA1-1 with pilT mRNA. (A) Putative interactions between the 5′ end of pilT mRNA (WT) and csRNA1-1 (WT) or pilT mRNA (mut) and csRNA1-1 (mut-2). The ribosome binding site and start codon are underlined. Substituted nucleotides in mutants are shown in plain lowercase. (B) DIG-labeled csRNA1-1 WT or csRNA1-1 mut-2 was incubated with pilT mRNA (WT) or pilT mRNA mutant (mut). Reaction products were separated on 5% native polyacrylamide gel and transferred to a membrane. DIG-labeled RNA signals on the membrane were detected using anti-DIG antibody. The image shown is representative of three independent experiments. The shifted bands in lanes 5 to 8 (the region surrounded by a dashed-line rectangle) are shown with a longer exposure time in the lower panel. (C) Quantification of the intensity of shifted bands in lanes 5 to 8. Data are expressed as means ± SD from three independent experiments. *, P < 0.01.
csRNA1-1 and csRNA1-2 inhibit biofilm formation in S. sanguinis.
Type IV pili are known to mediate attachment and colonization of various surfaces (29). In addition, some of the predicted target genes of csRNA1-1 (Table 2), such as SSA_0227, SSA_1632, SSA_2121, SSA_0906, and SSA_0905, are putative surface proteins that may be involved in S. sanguinis biofilm formation. Thus, we hypothesized that csRNA1-1 and csRNA1-2 play a regulatory role in S. sanguinis biofilm formation. To test this hypothesis, we performed biofilm formation assays using WT mock, ΔcsRNA1-1, 2 mock, and ΔcsRNA1-1, 2+pVA1-1, 2 strains. As shown in Fig. 6, the ΔcsRNA1-1, 2 mock strain showed a significant increase in biofilm formation compared with the WT mock strain, and this increase was reversed in the ΔcsRNA1-1, 2+pVA1-1, 2 strain. On the basis of these results, csRNA1-1 and csRNA1-2 appear to play inhibitory roles in S. sanguinis biofilm formation.
FIG 6.
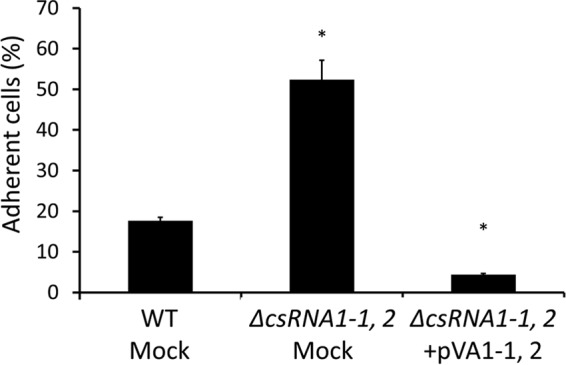
Effect of csRNA1-1 and csRNA1-2 deletion on biofilm formation. S. sanguinis strains were grown in BHI broth containing 1% sucrose on saliva-coated 24-well polystyrene plates. Adherent and nonadherent cells were separately collected and quantified by measuring OD620. Percentages of adherent cells were calculated by the following equation: 100 × OD620 of adherent cells/OD620 of total (adherent and nonadherent) cells. *, P < 0.01.
DISCUSSION
Bacterial gene regulation by sRNAs has attracted increasing attention in recent years because sRNAs are implicated in many biological processes, including environmental adaptation and pathogenesis. In streptococci, a substantial number of sRNAs, mainly from the two major human pathogens S. pyogenes and S. pneumoniae, have been predicted through computational approaches or experimentally identified using next-generation sequencing (RNA-seq) (30, 31). However, only a few of these sRNAs have known functions thus far. csRNAs of S. pneumoniae are a well-studied example of streptococcal sRNAs, which are involved in competence control (32, 33, 34). Because the CiaRH TCS is conserved in all streptococci and controls a variety of physiological processes, including environmental stress tolerance, it is conceivable that csRNAs play important roles in the physiology of streptococci. Using a target prediction program (TargetRNA2), we searched for genes that are regulated by csRNA1-1 and csRNA1-2 in the S. sanguinis genome. Among various candidate target genes, pilT, a constituent of the type IV pilus gene cluster, was the most probable target, and indeed pilT gene expression was shown to be negatively regulated by csRNA1-1 and csRNA1-2 in S. sanguinis (Fig. 1 and 2).
Type IV pili are surface-exposed filaments that promote properties such as adhesion, protein secretion, DNA uptake, and motility (29). Although type IV pili have been studied extensively in a few Gram-negative species, including Pseudomonas and Neisseria, little is known about type IV pili of Gram-positive species, including streptococci. It is only recently that the functional analysis of type IV pili in S. sanguinis has been reported (27). The 22-kb-long type IV pilus gene cluster of S. sanguinis is conserved in almost all S. sanguinis strains and absent from all other streptococcal species (27). This species-specific conservation of the gene cluster raises the possibility that type IV pili play important roles in the physiology of S. sanguinis.
Twitching motility is a form of solid-surface translocation that occurs in a wide range of bacteria (35). The movement is mediated by type IV pilus retraction, which is powered by the ATPase PilT (36). Twitching motility has been observed in a limited number of S. sanguinis strains other than SK36 or ATCC 10556 (27). We first hypothesized that the twitching motility-negative phenotype of S. sanguinis strains such as SK36 and ATCC 10556 is a consequence of a negative regulatory effect of csRNA1-1 and csRNA1-2 on PilT expression. However, we did not observe twitching motility in either the ATCC 10556 WT or ΔcsRNA1-1, 2 strain (data not shown). Because twitching motility can be influenced by many factors, including substrate topography, fluidity, stiffness, surface coating, secretion of biological molecules, and oxygen levels (37), it is possible that under some currently unknown physiological conditions, twitching motility-negative strains such as SK36 and ATCC 10556 can exhibit twitching motility.
csRNAs show a high degree of similarity to each other, particularly in the unpaired region between the two stem-loop structures. Complementarity to the ribosome binding site and the AUG start codon within this unpaired region has been suggested to allow csRNA to bind to target mRNA and inhibit translational initiation (20). Consistent with these predictions, we showed that csRNA1-1 binds directly to its target, pilT mRNA, in RNA-RNA EMSA (Fig. 5). To our knowledge, this is the first experimental observation of direct binding between a csRNA and a target mRNA. The nucleotide sequence that was important for base pairing with pilT mRNA, which resided in proximity of the ribosome binding site and AUG (Fig. 4), was not identical to the previously defined complemental sequence to the start codon and ribosome binding site (20). This discrepancy suggests the diversity of mechanisms by which csRNAs control target mRNA expression.
Biofilm formation by S. sanguinis is a complex process involving many factors, such as adhesins, signaling systems, glucosyltransferases, and extracellular DNA (1, 2, 4, 38, 39). In our in vitro biofilm assay using sucrose-containing medium, a saliva coating on culture plates was also essential for biofilm formation of S. sanguinis WT and mutant strains, because all strains tested formed almost no biofilm on uncoated plates (data not shown). In addition, the inhibitory effect of csRNA1-1 and csRNA1-2 on biofilm formation was observed only in cultures grown under gentle agitation (Fig. 6). No inhibitory effect was observed with static culture, in which all S. sanguinis strains tested readily formed biofilms and showed no significant difference in biofilm-forming ability (data not shown). In static culture, S. sanguinis cells that have settled to the bottom of the well can synthesize water-soluble glucan and may adhere to the polystyrene surface regardless of the presence or absence of initial attachment. Conversely, during culture with agitation, in which S. sanguinis cannot readily settle to the bottom of the well, initial attachment to saliva-coated surfaces with adhesins may be essential for biofilm formation. Thus, the observed difference between S. sanguinis WT and ΔcsRNA1-1, 2 strains in biofilm formation with agitated culture seemed to be caused by the difference in the ability of initial attachment.
Although S. sanguinis SK36 has pili that contribute to cell adhesion and biofilm formation (38, 40), strain ATCC 10556 does not express pili (40). It has not been assessed whether type IV pili of S. sanguinis contribute to initial attachment; however, it is conceivable that increased PilT expression in the ΔcsRNA1-1, 2 strain contributed, at least in part, to elevated biofilm formation in this strain. In addition, although we did not assess the expression of other putative adhesin genes in the ΔcsRNA1-1, 2 strain, it is possible that the regulation of these genes by csRNA1-1 and csRNA1-2 also could affect biofilm-forming ability. Although extracellular signals that are sensed by the CiaRH system in S. sanguinis are yet to be determined, the regulation of PilT expression by csRNA1-1 and csRNA1-2 provides new insight into molecular mechanisms underlying csRNA-mediated regulation in S. sanguinis colonization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. sanguinis strains used in this study are listed in Table S7 in the supplemental material. Strains were routinely grown in brain heart infusion (BHI) broth (BD) at 37°C under anaerobic conditions (10% H2, 10% CO2, and 80% N2). For antibiotic selection, 250 μg/ml of spectinomycin (Spec), 12.5 μg/ml of tetracycline (Tet), or 5 μg/ml of erythromycin was used. Escherichia coli strain DH5α was aerobically grown in Lennox L broth (Invitrogen), and 100 μg/ml of ampicillin or 17 μg/ml of chloramphenicol was used for antibiotic selection. Transformation of S. sanguinis strains was performed as described previously (21), with some modifications. Briefly, cultures in 3 ml of Todd-Hewitt (TH) broth (Difco) adjusted to pH 7.6 were incubated at 37°C overnight under anaerobic conditions. One microliter of each culture was transferred to 5 ml of TH broth (pH 7.6) containing 10% horse serum and statically incubated at 37°C under aerobic conditions for 2 h. After incubation, aliquots of the cultures were mixed with transforming DNA (1 μg/ml) in a 1.5-ml microtube and further incubated for 5 h. The cells then were plated on BHI agar containing appropriate antibiotics and grown under anaerobic conditions at 37°C for 48 h.
Construction of S. sanguinis mutant and complemented strains.
The csRNA1-1 csRNA1-2 double-deletion mutant and ciaRH deletion mutant strains of S. sanguinis were created by replacement of the target genes with a Spec resistance cassette (Spr). Spr was cloned into the BamHI site of pUC18 to obtain pUC18 Spr. The upstream and downstream fragments of the target genes (400 to 500 bp in size) were cloned into pUC18 Spr at sites SphI/SalI and SmaI/EcoRI, respectively, using primers listed in Table S8. The obtained plasmids were linearized with EcoRI digestion and transformed into S. sanguinis ATCC 10556, and deletion of the target genes was confirmed by PCR. For complementation studies, wild-type (WT) csRNA1-1 and/or csRNA1-2 or ciaRH alleles were cloned into the E. coli-streptococci shuttle vector pVA838 (22) via SalI/SphI sites for csRNA1-1 and csRNA1-2 or SalI site for ciaRH to obtain pVA1-1, 2, pVA1-1, pVA1-2, and pVAciaRH plasmids, respectively. These plasmids were transformed into the respective mutant strains, and genetic complementation of the genes was confirmed by real-time PCR.
RNA isolation and quantitative RT-PCR.
S. sanguinis cultures were grown to early exponential phase (OD600 of 0.1 to 0.2), and RNA was isolated from cells subjected to bead beating using the RNeasy minikit (Qiagen) and treated with Turbo DNase (Ambion). cDNA was synthesized from RNA using the ReverTra Ace quantitative PCR RT kit (Toyobo) according to the manufacturer's instructions. Quantitative PCR was performed using the StepOnePlus real-time PCR system (Life Technology) with Fast SYBR green master mix (Thermo Fisher). Primers used in the experiments are listed in Table S8. Results were normalized to S. sanguinis 16S rRNA gene expression. Experiments were performed in triplicate with at least three independent RNA samples.
Construction of plasmid-borne mutant csRNA1-1 alleles.
Nucleotide substitution mutants of plasmid-borne csRNA1-1 were constructed with the QuikChange II site-directed mutagenesis kit (Agilent) according to the manufacturer's instructions, with slight modifications. The pVA1-1 plasmid, which contains the WT allele of csRNA1-1, and primers containing the desired mutation (listed in Table S8) were used for PCR amplification. After PCR, template DNA was digested with DpnI, and the resultant reaction product containing mutant plasmid was transformed into E. coli DH5α cells. The generation of mutations was confirmed by DNA sequencing. The obtained plasmids that contained the desired mutations are designated pVA1-1 mut-1, pVA1-1 mut-2, and pVA1-1 mut-3, respectively.
Construction of PilT-luciferase reporter strain and luciferase assay.
A single-copy reporter fusion of pilT in the native chromosomal context was generated as follows. The Tet resistance cassette (Tetr) was cloned into pUC18 at the BamHI site to obtain pUC18 Tetr. Using the two-step overlap PCR method (23), the upstream region of pilT was fused to the open reading frame of the luciferase gene NanoLuc, amplified from the pNL1.1 vector (Promega), and cloned into pUC18 Tetr at SphI/SalI sites. The downstream PCR fragment of the pilT gene then was cloned into SmaI/EcoRI sites, resulting in generation of the pilT-NanoLuc-Tetr fusion plasmid, which was linearized with EcoRI digestion and transformed into S. sanguinis ATCC 10556. A schematic illustration of the double-crossover event is shown in Fig. 2A. The obtained PilT-luciferase reporter strain, which has the promoterless luciferase gene replaced with the pilT gene in the chromosome, is designated WT-luc. Using WT-luc as a parent strain, csRNA1-1 and csRNA1-2 deletion mutants and complemented strains were also generated (Table S7). The luciferase assay was performed using the Nano-Glo Dual-Luciferase reporter assay kit (Promega) according to the manufacturer's instructions, with some modifications. Briefly, S. sanguinis luc strains were grown in BHI broth to early exponential phase (OD600 of 0.1 to 0.2). Subsequently, 50 μl of the culture was transferred to a 96-well plate, and 25 μl of NanoDLR Stop and Glo reagent (Promega) was added to the wells. After incubation at room temperature for 10 min, luminescence was measured with the GloMax-Multi detection system (Promega). Relative luminescence units were normalized to the OD600 of corresponding cultures. Experiments were performed in triplicate with at least three independent cultures.
In vitro transcription reactions.
In vitro-transcribed RNAs for use in the RNA-RNA electrophoretic mobility shift assay (EMSA) were created using the CUGA7 in vitro transcription kit (Nippon Gene), and digoxigenin (DIG)-labeled csRNA1-1 and csRNA1-1 mut-2, which were used as RNA probes, were synthesized using the DIG Northern Starter kit (Roche) according to the manufacturer's instructions. Template DNA for pilT mRNA (WT) and pilT mRNA (mut) or csRNA1-1 transcription was PCR amplified or chemically synthesized, respectively. The T7 promoter sequence was fused to the 5′ end of DNA templates to allow transcription using T7 RNA polymerase. The primers used are listed in Table S8. After the transcription reaction, template DNA was removed by DNase treatment, and RNA products were purified with the RNA Clean & Concentrator kit (Zymo Research).
RNA-RNA EMSA.
Direct interaction between csRNA1-1 and pilT mRNA was examined by RNA-RNA EMSA analysis as described previously (24), with some modifications. RNAs were heated at 90°C for 2 min in 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 50 mM KCl, 0.5 mM EDTA, and 10% glycerol. RNAs then were cooled on ice for 5 min to allow them to fold. After the folding reaction, the DIG-labeled RNA probe, csRNA1-1 WT or csRNA1-1 mut-2 (15 ng each), and pilT mRNA (WT) or pilT mRNA (mut) (500 ng each) were mixed and incubated at 37°C for 15 min. The reaction products were then separated on 5% native polyacrylamide gel in 1× Tris-borate-EDTA buffer and electrically transferred onto a Hybond N+ membrane (GE). After UV cross-linking, DIG-labeled RNA signals on the membrane were detected using an alkaline phosphatase-conjugated anti-DIG antibody with CSPD-Star detection reagent (Roche) according to the manufacturer's instructions. The signal intensity for each band was quantified using LI-COR Image Studio software.
Biofilm formation assay.
Overnight cultures of S. sanguinis strains were diluted 1:100 into fresh BHI broth containing 1% sucrose, transferred (500 μl/well) to a 24-well polystyrene plate (Iwaki) coated with saliva, and incubated anaerobically at 37°C with rotary agitation at 80 rpm for 24 h. After incubation, culture medium containing nonadherent cells was transferred to a 1.5-ml microtube and the cells were collected by centrifugation (12,000 × g, 5 min). The precipitated cells then were suspended with 200 μl of 0.1 M NaOH. Adherent cells were detached and suspended by adding 200 μl of 0.1 M NaOH to the wells. The suspensions of adherent or nonadherent cells were transferred to a 96-well plate and quantified by measuring the OD620 with a microplate reader (Tecan). The percentages of adherent cells were calculated by the following equation: 100 × OD620 of adherent cells/OD620 of total (adherent and nonadherent) cells.
Statistical analysis.
The statistical significance of data obtained from quantitative RT-PCR, luciferase assay, and biofilm formation assay was determined by Student's t tests. P values of <0.01 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina Croney from Edanz Group for editing a draft of the manuscript.
This work was supported in part by JSPS KAKENHI grant number JP17K11627. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
C.O., H.M., M.N., N.O., and H. Kuwata designed the study. C.O. and H.M. performed experiments and wrote the initial draft of the manuscript. T.A., H.F., H. Kataoka, and H. Kuwata contributed to analysis and interpretation of data and assisted in the preparation of the manuscript. All other authors contributed to data collection and interpretation and critically reviewed the manuscript. All authors approved the final version of the manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00894-17.
REFERENCES
- 1.Gong K, Mailloux L, Herzberg MC. 2000. Salivary film expresses a complex, macromolecular binding site for Streptococcus sanguis. J Biol Chem 275:8970–8974. doi: 10.1074/jbc.275.12.8970. [DOI] [PubMed] [Google Scholar]
- 2.Tamesada M, Kawabata S, Fujiwara T, Hamada S. 2004. Synergistic effects of streptococcal glucosyltransferases on adhesive biofilm formation. J Dent Res 83:874–879. doi: 10.1177/154405910408301110. [DOI] [PubMed] [Google Scholar]
- 3.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreth J, Vu H, Zhang Y, Herzberg MC. 2009. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J Bacteriol 191:6281–6291. doi: 10.1128/JB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner EG, Romby P. 2015. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet 90:133–208. [DOI] [PubMed] [Google Scholar]
- 6.Miller EW, Cao TN, Pflughoeft KJ, Sumby P. 2014. RNA-mediated regulation in Gram-positive pathogens: an overview punctuated with examples from the group A Streptococcus. Mol Microbiol 94:9–20. doi: 10.1111/mmi.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brantl S, Brückner R. 2014. Small regulatory RNAs from low-GC Gram-positive bacteria. RNA Biol 11:443–456. doi: 10.4161/rna.28036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol 39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez-Peña E, Treviño J, Liu Z, Perez N, Sumby P. 2010. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol 78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Treviño J, Ramirez-Peña E, Sumby P. 2012. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol Microbiol 86:140–154. doi: 10.1111/j.1365-2958.2012.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danger JL, Makthal N, Kumaraswami M, Sumby P. 2015. The FasX small regulatory RNA negatively regulates the expression of two fibronectin-binding proteins in group A Streptococcus. J Bacteriol 197:3720–3730. doi: 10.1128/JB.00530-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26:369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 14.Guenzi E, Gasc AM, Sicard MA, Hakenbeck R. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol Microbiol 12:505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 15.Giammarinaro P, Sicard M, Gasc AM. 1999. Genetic and physiological studies of the CiaH-CiaR two-component signal-transducing system involved in cefotaxime resistance and competence of Streptococcus pneumoniae. Microbiology 145(Part 8):1859–1869. [DOI] [PubMed] [Google Scholar]
- 16.Sebert ME, Palmer LM, Rosenberg M, Weiser JN. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect Immun 70:4059–4067. doi: 10.1128/IAI.70.8.4059-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawid S, Sebert ME, Weiser JN. 2009. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J Bacteriol 191:1509–1518. doi: 10.1128/JB.01213-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. 2004. Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J Bacteriol 186:5258–5266. doi: 10.1128/JB.186.16.5258-5266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halfmann A, Kovács M, Hakenbeck R, Brückner R. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol 66:110–126. doi: 10.1111/j.1365-2958.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 20.Marx P, Nuhn M, Kovács M, Hakenbeck R, Brückner R. 2010. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics 11:661. doi: 10.1186/1471-2164-11-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paik S, Senty L, Das S, Noe JC, Munro CL, Kitten T. 2005. Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect Immun 73:6064–6074. doi: 10.1128/IAI.73.9.6064-6074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macrina FL, Tobian JA, Jones KR, Evans RP, Clewell DB. 1982. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19:345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- 23.Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res 30:E2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson CA, Vincent HA, Stone CM, Phillips JO, Cary PD, Gowers DM, Callaghan AJ. 2013. Characterization of MicA interactions suggests a potential novel means of gene regulation by small non-coding RNAs. Nucleic Acids Res 41:3386–3397. doi: 10.1093/nar/gkt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kery MB, Feldman M, Livny J, Tjaden B. 2014. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res 42:W124–W129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, Manque P, Ge X, Serrano MG, Puiu D, Hendricks S, Wang Y, Chaplin MD, Akan D, Paik S, Peterson DL, Macrina FL, Buck GA. 2007. Genome of the opportunistic pathogen Streptococcus sanguinis. J Bacteriol 189:3166–3175. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurung I, Spielman I, Davies MR, Lala R, Gaustad P, Biais N, Pelicic V. 2016. Functional analysis of an unusual type IV pilus in the Gram-positive Streptococcus sanguinis. Mol Microbiol 99:380–392. doi: 10.1111/mmi.13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melville S, Craig L. 2013. Type IV pili in Gram-positive bacteria. Microbiol Mol Biol Rev 77:323–341. doi: 10.1128/MMBR.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berry JL, Pelicic V. 2015. Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 39:134–154. doi: 10.1093/femsre/fuu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Rhun A, Charpentier E. 2012. Small RNAs in streptococci. RNA Biol 9:414–426. doi: 10.4161/rna.20104. [DOI] [PubMed] [Google Scholar]
- 31.Patenge N, Pappesch R, Khani A, Kreikemeyer B. 2015. Genome-wide analyses of small non-coding RNAs in streptococci. Front Genet 6:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsui HC, Mukherjee D, Ray VA, Sham LT, Feig AL, Winkler ME. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol 192:264–279. doi: 10.1128/JB.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laux A, Sexauer A, Sivaselvarajah D, Kaysen A, Brückner R. 2015. Control of competence by related non-coding csRNAs in Streptococcus pneumoniae R6. Front Genet 6:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnorpfeil A, Kranz M, Kovács M, Kirsch C, Gartmann J, Brunner I, Bittmann S, Brückner R. 2013. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol 89:334–349. doi: 10.1111/mmi.12277. [DOI] [PubMed] [Google Scholar]
- 35.Mattick JS. 2002. Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 36.Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 37.Maier B, Wong GC. 2015. How bacteria use type IV pili machinery on surfaces. Trends Microbiol 23:775–788. doi: 10.1016/j.tim.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Okahashi N, Nakata M, Terao Y, Isoda R, Sakurai A, Sumitomo T, Yamaguchi M, Kimura RK, Oiki E, Kawabata S, Ooshima T. 2011. Pili of oral Streptococcus sanguinis bind to salivary amylase and promote the biofilm formation. Microb Pathog 50:148–154. doi: 10.1016/j.micpath.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Moraes JJ, Stipp RN, Harth-Chu EN, Camargo TM, Höfling JF, Mattos-Graner RO. 2014. Two-component system VicRK regulates functions associated with establishment of Streptococcus sanguinis in biofilms. Infect Immun 82:4941–4951. doi: 10.1128/IAI.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okahashi N, Nakata M, Sakurai A, Terao Y, Hoshino T, Yamaguchi M, Isoda R, Sumitomo T, Nakano K, Kawabata S, Ooshima T. 2010. Pili of oral Streptococcus sanguinis bind to fibronectin and contribute to cell adhesion. Biochem Biophys Res Commun 391:1192–1196. doi: 10.1016/j.bbrc.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 41.Zuker M. 2003. Mfold web server for Nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.