ABSTRACT
The host employs both cell-autonomous and system-level responses to limit pathogen replication in the initial stages of infection. Previously, we reported that the eukaryotic initiation factor 2α (eIF2α) kinases heme-regulated inhibitor (HRI) and protein kinase R (PKR) control distinct cellular and immune-related activities in response to diverse bacterial pathogens. Specifically for Listeria monocytogenes, there was reduced translocation of the pathogen to the cytosolic compartment in HRI-deficient cells and consequently reduced loading of pathogen-derived antigens on major histocompatibility complex class I (MHC-I) complexes. Here we show that Hri−/− mice, as well as wild-type mice treated with an HRI inhibitor, are more susceptible to listeriosis. In the first few hours of L. monocytogenes infection, there was much greater pathogen proliferation in the liver of Hri−/− mice than in the liver of Hri+/+ mice. Further, there was a rapid increase of serum interleukin-6 (IL-6) levels in Hri+/+ mice in the first few hours of infection whereas the increase in IL-6 levels in Hri−/− mice was notably delayed. Consistent with these in vivo findings, the rate of listeriolysin O (LLO)-dependent pathogen efflux from infected Hri−/− macrophages and fibroblasts was significantly higher than the rate seen with infected Hri+/+ cells. Treatment of cells with an eIF2α kinase activator enhanced both the HRI-dependent and PKR-dependent infection phenotypes, further indicating the pharmacologically malleability of this signaling pathway. Collectively, these results suggest that HRI mediates the cellular confinement and killing of virulent L. monocytogenes in addition to promoting a system-level cytokine response and that both are required to limit pathogen replication during the first few hours of infection.
KEYWORDS: stress response, eIF2α kinase, heme-regulated inhibitor, microbial pathogenesis, macrophages, Listeria monocytogenes, HRI
INTRODUCTION
Emerging evidence suggests that redirected mRNA translation plays an important role in immune responses to microbial pathogens. The translation rate of a substantial fraction of eukaryotic mRNAs is potentially regulated by the levels of phosphorylated eukaryotic initiation factor 2α (eIF2α-P) (1–3). In humans and in mice, the levels of eIF2α-P are controlled by eIF2α kinases (general control nonderepressible 2 [GCN2], heme-regulated inhibitor [HRI], protein kinase RNA-like endoplasmic reticulum kinase [PERK], and protein kinase R [PKR]), which are in turn activated by various ligands or stimuli. The heme-regulated inhibitor (HRI) was first isolated in reticulocytes, where it regulates the translation of β-globin-encoding mRNAs during erythropoiesis, and was later shown to play a protective role against oxidative stress during erythroid differentiation (4, 5). HRI also mediates protective responses to a variety of stresses in nonerythroid cells as well as in yeast (Saccharomyces cerevisiae) (6–10). Protein kinase R (PKR) is activated by double-stranded DNA (dsRNA) and acts to inhibit translation of virus-derived mRNAs primarily through its eIF2α kinase activity (11).
We showed using a yeast-based screen that HRI genetically interacts with two different bacterial virulence factors (12). We went on to demonstrate that eIF2α phosphorylation in mammalian cells occurs following infection and that this activation is required for the infection-induced activation of NF-κB signaling and expression of proinflammatory cytokines (13), similarly to what others reported for lipopolysaccharide (LPS) stimulation (14). Beyond cytokine expression, both HRI and PKR regulate key events upon cellular infection by a variety of bacterial pathogens. Specifically, cells lacking PKR are more susceptible to invasion by the bacterial pathogens Yersinia, Chlamydia, and Listeria whereas the functioning of the type III secretion system (Yersinia), intracellular growth (Chlamydia), and trafficking to the cytosol (Listeria) are greatly reduced in cells lacking HRI (15). Cytosolic invasion is considered a critical component of the virulence strategy of Listeria. Here we show the unanticipated consequences of HRI deficiency in mice following infection with Listeria and provide a description of a cellular mechanism that may contribute to their heightened susceptibility.
RESULTS
HRI-deficient mice are susceptible to listeriosis.
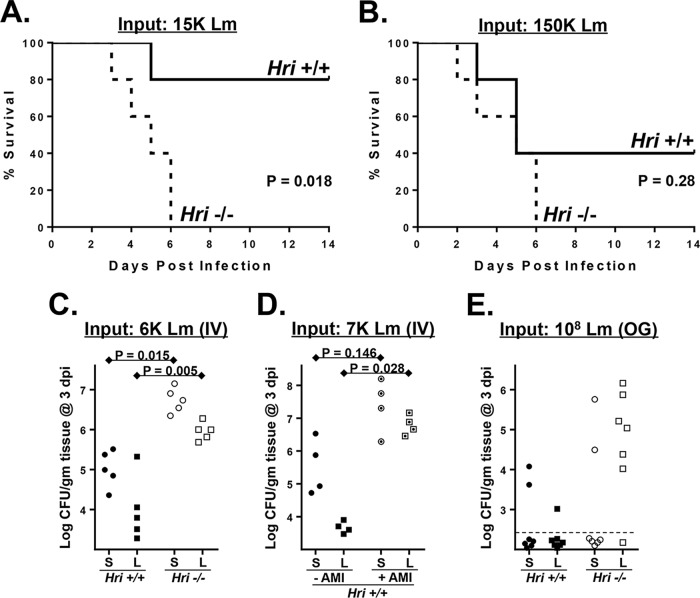
Hri+/+ and Hri−/− mice (16) were infected intravenously (i.v.) with various doses of L. monocytogenes. At the lowest dose tested (3,000 CFU), neither strain of mice displayed any obvious external signs of disease (not shown). At the intermediate dose tested (15,000 CFU), Hri+/+ mice presented with mildly ruffled coats by day 3 but were otherwise normal (with one exception) in their movements and were healthy looking by day 14 (Fig. 1A). In contrast, Hri−/− mice infected with a dose of 15,000 CFU started to present with ruffled coats and retarded movements by day 2 and all succumbed between day 3 and day 6 (Fig. 1A). At the highest dose tested (150,000 CFU; approximately the 50% lethal dose [LD50]), the two strains of mice displayed comparable levels of disease symptoms by day 2; however, all of the Hri−/− mice succumbed to this dose whereas half of the Hri+/+ mice survived (Fig. 1B). These data indicate that HRI plays a protective role in the early phases of L. monocytogenes infection.
FIG 1.
HRI is protective against listeriosis. (A and B) Hri+/+ and Hri−/− mice (5 per group) were infected intravenously with 15,000 (A) or 150,000 (B) CFU of L. monocytogenes and monitored for 14 days. (P values were determined by a Mantel-Cox log rank test of a single representative experiment performed two times with similar results). (C) Hri+/+ mice (closed symbols) and Hri−/− mice (open symbols) were infected intravenously (IV) with L. monocytogenes and were humanely euthanized at 3 days postinfection (dpi). Data represent CFU in spleen (S; circles) and liver (L; squares) homogenates. (D) Hri+/+ mice were infected with L. monocytogenes and treated with either vehicle (closed symbols) or HRI AMI (dotted open symbols) on days 0, 1, and 2 postinfection. The load of L. monocytogenes in the spleen and liver was determined at 3 dpi. (E) Mice were inoculated orogastrically (OG) with the indicated dose of L. monocytogenes, and the bacterial loads in the spleen and liver were determined at 3 dpi by CFU assay. The dotted lines indicate detection limits. (P values were calculated using a Student t test of results of a single representative experiment performed two times.)
To determine whether the increased disease susceptibility of HRI null mice was associated with enhanced pathogen colonization, bacterial loads in the spleen and liver were assessed in Hri+/+ and Hri−/− mice infected for 3 days with an intermediate dose of L. monocytogenes. The levels of viable L. monocytogenes recovered in the spleens (44-fold) and livers (21-fold) of Hri−/− mice were significantly higher than those seen with Hri+/+ mice (Fig. 1C).
An HRI inhibitor was recently identified in a high-throughput screen and was further modified to increase its specificity and bioavailability (17, 18). The resulting compound, here referred to as AMI {N-(2,6-dimethylbenzyl)-6,7-dimethoxy-2H-[1]benzofuro[3,2-c]pyrazol-3-amine hydrochloride}, disrupts HRI-dependent memory retention in the rat hippocampus as well as HRI-mediated activation of the β-site APP cleaving enzyme 1 (BACE1) in synaptic spines (19, 20). After 3 days of infection with L. monocytogenes, there were greatly increased levels of spleen (55-fold) and liver (1,478-fold) colonization among AMI-treated mice compared to untreated mice (Fig. 1D). These data show that an HRI null-like infection phenotype can be achieved pharmacologically in infected wild-type mice.
To further compare the innate responses of Hri+/+ and Hri−/− mice, an orogastric route of infection was used that mimics that of foodborne pathogens. At 3 days following inoculation with L. monocytogenes, viable bacteria were recovered from the livers of only 1 of 7 Hri+/+ mice; in contrast, the livers of 6 of 7 Hri−/− mice had substantial bacterial loads following this infection period (Fig. 1E). Since the blood supply of the liver is primarily derived from the hepatic portal vein that drains the gastrointestinal tract, these data could indicate that HRI is important in front-line responses occurring in intestinal tissue. Collectively, these findings indicate that HRI plays a protective role against L. monocytogenes in a variety of infection scenarios.
HRI restricts postseeding proliferation of L. monocytogenes in the liver.
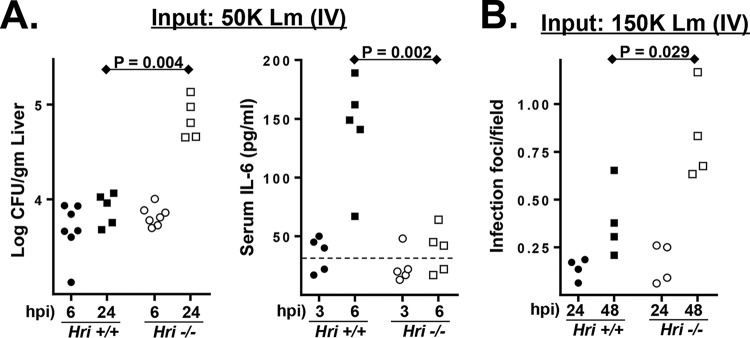
Relatively brief infection periods were used to distinguish between initial pathogen seeding of the liver by blood-borne L. monocytogenes and the subsequent intraliver proliferation of the pathogen. Following i.v. infection of L. monocytogenes, the initial colonization of the spleen and liver occurred with comparable kinetics in Hri+/+ and Hri−/− mice. In both mouse strains, ∼10% of the inoculum was recovered in the spleen and liver 6 h postinfection (hpi) (Fig. 2A). However, by 24 hpi the levels of recovery of L. monocytogenes from livers were 10-fold higher in Hri−/− mice than in Hri+/+ mice (Fig. 2A).
FIG 2.
Enhanced postseeding proliferation of L. monocytogenes in the liver is associated with delayed cytokine response. (A) (Left panel) Hri+/+ and Hri−/− mice were infected with L. monocytogenes, and the bacterial load of the liver was determined at either 6 or 24 hpi by CFU assay. (Right panel) At 6 hpi, blood was collected and analyzed for IL-6 levels. The dotted line indicates the detection limit. (B) Histological analysis of livers from infected Hri+/+ and Hri−/− mice. Inflammatory microabscesses were enumerated by examining >20 microscopic fields for each of 4 individual mice per test condition. (P values were calculated using a Student t test of results of a single representative experiment performed three times.)
Serum cytokine levels were measured in Hri+/+ and Hri−/− mice to determine whether the levels of immune activation differed in these strains. In both uninfected Hri+/+ and uninfected Hri−/− mice, the levels of interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), monocyte chemoattractant protein 1 (MCP-1), IL-10, and IL-12p70 were below the level of detection. IL-6 was detected in the majority (3/5) of the Hri+/+ mice after 3 h of infection, and the levels had increased 3-fold by 6 hpi (levels of the other cytokines remained undetected at 6 hpi) (Fig. 2A). In contrast, IL-6 was detected in only 1 of 5 Hri−/− mice after 3 h of infection and was minimally detected in 3 of 5 mice at 6 hpi (Fig. 2A). Histological examination of livers from infected mice showed that there were comparable levels of inflammatory foci in Hri+/+ and Hri−/− mice at 24 hpi whereas there were considerably greater numbers of these foci in the livers of Hri−/− mice by 48 hpi (Fig. 2B). Collectively, these data suggest that a delay in cytokine expression in Hri−/− mice may contribute to allowing the pathogen to gain a foothold in the liver. These data are consistent with our previous findings showing defective cytokine expression in peritoneal and splenic Hri−/− macrophages infected in vitro with L. monocytogenes (15). Collectively, these data suggest that events occurring subsequently to the initial pathogen seeding of these organs account for the differences observed at ≥24 hpi.
HRI limits the emergence of virulent L. monocytogenes from macrophages.
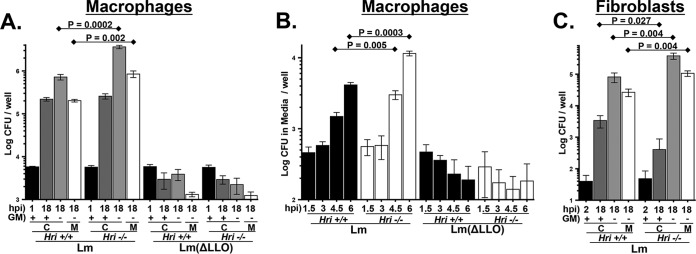
The L. monocytogenes infection cycle consists of four distinct steps: cellular invasion and initial enclosure within a Listeria-containing vacuole (LCV); virulence factor-mediated disintegration of the LCV membrane; replication within the cytosol; and, finally, penetration of the plasma membrane resulting in infection of adjacent cells (as in an epithelial sheet) or, in solitary cells, release of the pathogen into the extracellular media that gives rise to secondary infections. Peritoneal exudate macrophages (PEMs) derived from Hri+/+ and Hri−/− mice were used to determine whether HRI impacts the intracellular replication of L. monocytogenes. Freshly isolated Hri+/+ and Hri−/− PEMs were infected in vitro for 30 min, noninternalized bacteria were killed with gentamicin, and then viable intracellular bacteria were enumerated in cells infected for a total of 1 or 18 h. Comparable levels of L. monocytogenes were recovered from Hri+/+ and Hri−/− PEMs following 1 h of infection, indicating that HRI does not play an apparent role in infectivity (Fig. 3A, black bars). Similarly, there were comparable levels of recovery of L. monocytogenes from Hri+/+ and Hri−/− PEMs following 18 h of infection in the presence of gentamicin (Fig. 3A, dark gray bars).
FIG 3.
HRI restricts emergence of L. monocytogenes from infected macrophages. (A) Peritoneal exudate macrophages (PEMs) isolated from Hri+/+ and Hri−/− mice were infected in vitro with either wild-type L. monocytogenes (Lm) or a derivative mutant strain lacking the virulence factor listeriolysin O (ΔLLO). After 30 min of infection, macrophages were treated with gentamicin for an additional 30 min to kill extracellular bacteria. Levels of viable intracellular L. monocytogenes (“C”) were determined by CFU assay at either 1 hpi (black bars) or 18 hpi in either the presence or absence of gentamicin (dark gray or light gray bars, respectively). For 18-h infections occurring in the absence of gentamicin (light gray bars), the levels of extracellular L. monocytogenes were determined in the overlying media (“M”; unfilled bars). (B) PEMs were infected as described above and, following the removal of gentamicin at 1 hpi, the levels of extracellular L. monocytogenes in the overlying media were determined at the indicated time points. (C) Mouse embryonic fibroblasts (MEFs) derived from from Hri+/+ and Hri−/− mice were infected with L. monocytogenes (Lm). After 90 min of infection, cells were treated with gentamicin for an additional 30 min. Levels of viable intracellular L. monocytogenes (“C”) were determined by CFU assay at either 2 hpi (black bars) or 18 hpi in either the presence or absence of gentamicin (dark gray or light gray bars, respectively). For 18-h infections occurring in the absence of gentamicin (light gray bars), the levels of extracellular L. monocytogenes were determined in the overlying media (“M”; unfilled bars). (The means and standard deviations of results of 3 separate samples per condition are plotted; P values were calculated using a Student t test of results of a single representative experiment performed multiple times.)
However, when gentamicin was removed from the infected cultures following the 30-min killing phase, there was a 5-fold reduction in the level of L. monocytogenes recovered from Hri+/+ PEMs compared to Hri−/− PEMs following 18 h of infection (Fig. 3A, light gray bars) as well as a similar reduction in the bacterial levels in the overlying media (Fig. 3A, unfilled bars), suggesting that HRI impacts an extracellular phase of the L. monocytogenes infection cycle. A shorter-term kinetic analysis was performed to further characterize L. monocytogenes emergence from infected PEMs. The emergence of L. monocytogenes from infected Hri+/+ and Hri−/− PEMs occurred with similar kinetics (beginning >3 hpi); however, the magnitude of the increase was significantly greater in Hri−/− PEMs than in Hri+/+ PEMs (Fig. 3B). Control experiments showed that the appearance of L. monocytogenes in the overlaying media was dependent on the presence of cells and was not due to incomplete killing of the pathogen during the 30-min extracellular killing phase (not shown). These data further indicate that the cellular efflux rate of L. monocytogenes is enhanced in the absence of HRI.
L. monocytogenes lacking the virulence factor listeriolysin O (LLO), which is required for disintegration of the LCV membrane (discussed above), displayed similar infection profiles in Hri+/+ and Hri−/− PEMs. The infectivity (i.e., the initial invasion) of the L. monocytogenes (ΔLLO) strain was comparable to that of the LLO-expressing L. monocytogenes strain; however, the ΔLLO strain was subsequently efficiently killed in both Hri+/+ and Hri−/− PEMs and very few ΔLLO bacteria were detected in the overlaying media (Fig. 3A and B). These data show that HRI is not required for the cellular defense against attenuated L. monocytogenes.
To determine whether HRI also acts to confine L. monocytogenes within fibroblastic cells, Hri+/+ and Hri−/− mouse embryonic fibroblasts (MEFs) were infected with L. monocytogenes and analyzed in a manner similar to that described above for macrophages. As was previously shown (see reference 15), Hri+/+ and Hri−/− MEFs were similarly infected by L. monocytogenes (Fig. 3C, black bars) and, with prolonged infection in the presence of gentamicin, bacterial recovery from Hri+/+ MEFs actually exceeded (>8-fold) that from Hri−/− MEFs (Fig. 3C, dark gray bars). In that earlier study (15), the reduced bacterial recovery from Hri−/− MEFs was interpreted (as it turned out, erroneously) to indicate that HRI was required for optimal L. monocytogenes intracellular proliferation. However, in the absence of gentamicin, bacterial recovery from Hri+/+ MEFs was reduced 5-fold compared to that from Hri−/− MEFs (Fig. 3C, light gray bars). Similarly to the macrophage results, the increased levels of L. monocytogenes recovered from Hri−/− MEFs under antibiotic-free conditions were likely due to secondary infections since the level of L. monocytogenes in the overlaying media of Hri−/− MEFs was 4-fold higher than that seen with Hri+/+ MEFs (Fig. 3C, unfilled bars). Collectively, these findings show that, in both macrophages and fibroblastic cells, HRI promotes the cellular confinement of virulent L. monocytogenes, which may account for the enhanced susceptibility of the Hri−/− mouse to this pathogen.
An eIF2 activator enhances HRI and PKR infection phenotypes.
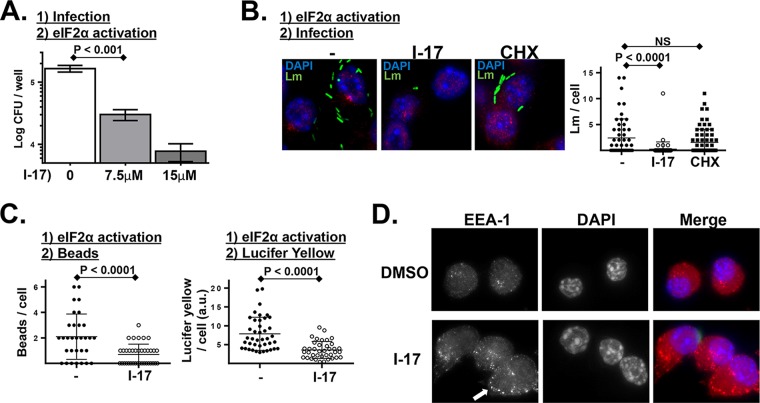
The findings described above were drawn from HRI deficiency-based models using either genetic knockouts or inhibitors. To determine whether HRI-regulated cellular activities could be hyperactivated, we employed a recently identified small-molecule activator of eIF2α phosphorylation (21). To determine whether this activator acted on postinternalization infection dynamics, macrophages were first infected and then treated with this eIF2α phosphorylation activator (I-17). This treatment regimen resulted in a significant reduction in the intracellular proliferation of L. monocytogenes (Fig. 4A). When, alternatively, macrophages were first treated with I-17 and then infected, the result was a notable reduction in the infectivity (i.e., internalization) of L. monocytogenes (Fig. 4B). Macrophages pretreated with cycloheximide (CHX) had levels of infectivity comparable to those seen with untreated cells (Fig. 4B). The latter finding suggests that the I-17 effect is not due to general translation inhibition but instead, as discussed in the introduction, may indicate that the I-17 effect is due to its enhancing the expression of upstream open reading frame (uORF)-regulated cellular factors. Additionally, I-17-treated macrophages were similarly resistant to internalizing inert beads and the soluble dye Lucifer yellow (Fig. 4C). Macrophages treated with I-17 exhibited a highly punctate distribution of the early endosomal marker EEA1 (Fig. 4D), further indicating that I-17 disrupts early trafficking events. These findings are consistent with our previously published findings (13, 15) that HRI and PKR impact L. monocytogenes intracellular replication and pathogen invasion, respectively. Importantly, these data indicate that these separable cellular processes (invasion and postinvasion trafficking) can both be targeted by a small-molecule activator.
FIG 4.
L. monocytogenes infection is inhibited in cells treated with a small-molecule activator of eIF2α phosphorylation. (A) PEMs were infected with L. monocytogenes for 30 min, noninternalized bacteria were killed with gentamicin, and then cells were treated with the indicated concentration of eIF2α phosphorylation activator I-17. Following 6 h of infection, intracellular L. monocytogenes levels were determined by CFU assay. (P values were calculated using a Student t test of results of a single representative experiment performed three times.) (B) Cultured murine macrophage-like cells (RAW 267.4) were left untreated or treated with I-17 or treated with cycloheximide (CHX) for 60 min. Cells were then infected with GFP-expressing L. monocytogenes (Lm) for 45 min, washed extensively, and analyzed by fluorescence microscopy. Plotted data represent the number of L. monocytogenes bacteria per cell from a representative experiment performed multiple times with similar results. NS, not significant. (C) Similar experiment in which untreated and I-17-treated macrophages were pulsed with fluorescent beads (left plot) for 5 min or with the soluble dye Lucifer yellow (LY) for 20 min (right plot), washed extensively, and then analyzed by fluorescence microscopy. Plotted data represent the number of beads or level of fluorescence per cell from a representative experiment performed three times with similar results. a.u., arbitrary units. (D) Similar experiment in which cells were treated with either vehicle alone or I-17 (10 μM) for 60 min and then were fixed and stained for the early endosomal marker EEA1. The arrow indicates the cumulative early EEA1-positive endosomes in I-17-treated cells.
HRI regulates intracellular trafficking dynamics.
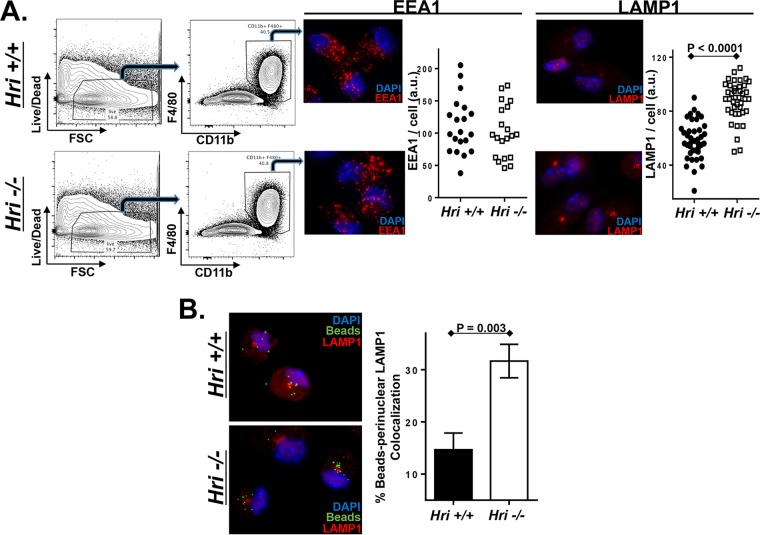
To determine whether HRI regulates intracellular trafficking in a noninfection setting, Hri+/+ and Hri−/− macrophages were analyzed to determine whether they differed in their endosomal components or dynamics. PEMs derived from Hri+/+ and Hri−/− mice were phenotypically similar in terms of surface expression levels of macrophage-specific markers as well as the levels of intracellular distribution of the early endosome marker EEA1 (Fig. 5A). In contrast, levels of the integral lysosomal factor LAMP-1 were notably higher in Hri−/− PEMs and were particularly concentrated in the perinuclear region (Fig. 5A). To compare dynamic responses, Hri+/+ and Hri−/− PEMs were pulsed with fluorescently labeled beads. There were comparable levels of internalization of beads in Hri+/+ and Hri−/− PEMs; however, following their internalization, the beads were much more rapidly trafficked to perinuclear regions in Hri−/− PEMs than in Hri+/+ PEMs (Fig. 5B). These data reveal that HRI regulates the composition and dynamics of postinternalization trafficking and may account for the differential L. monocytogenes infection profiles of Hri+/+ and Hri−/− macrophages.
FIG 5.
HRI regulates intracellular trafficking and lysosomal dynamics in macrophages. (A) PEMs isolated from Hri+/+ and Hri−/− mice were evaluated for viability (first column), and the gated population was assessed for the macrophage markers F4/80+ and CD11b+ (second column). PEMs were stained for either EEA1 or LAMP1, and plotted data represent the total EEA1- and LAMP1-associated signal of individual cells. (B) Hri+/+ and Hri−/− PEMs were pulsed in vitro for 5 min with fluorescently labeled beads and then analyzed 90 min later for bead localization and LAMP1 colocalization. Plotted data represent the percentage of beads localized to the LAMP1+ perinuclear region determined in three independent experiments in which >75 beads/experiment were tabulated. (P values were calculated using a Student t test of results of a single representative experiment performed multiple times.)
Susceptibility of HRI-deficient mice to infection is not due to developmental defects in lymphoid and myeloid cells.
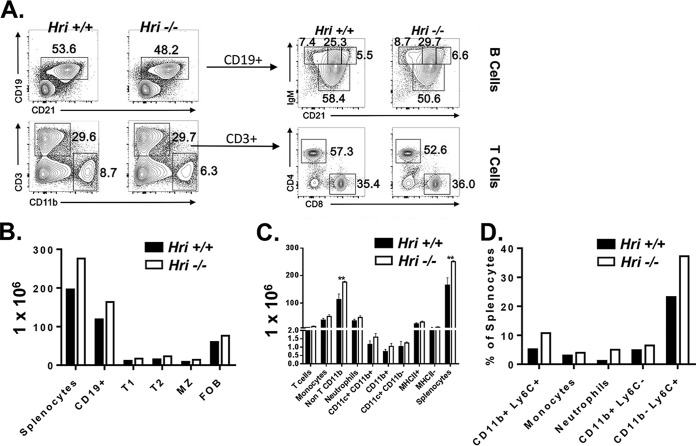
The findings presented above and previously (15) suggest that altered cellular pathogen trafficking, as well as a delayed cytokine response, may account for the enhanced susceptibility of HRI-deficient mice to L. monocytogenes. These early phases of immune responses to infection are largely regulated by innate immune cells. Therefore, whether HRI impacted the development of the immune system was investigated. A comparative phenotypic analysis of immune cells isolated from the spleens of Hri+/+ and Hri−/− mice was performed by flow cytometry. There were no major differences between 21-, 27-, and 34-day-old Hri+/+ and Hri−/− mice in splenic cellularity or composition; the results from both T and B lymphocytes and myeloid cells were comparable (data for 27-day-old mice are shown in Fig. 6).
FIG 6.
Largely intact immune cell development in HRI-deficient mice. Splenocytes isolated from 27-day-old Hri+/+ and Hri−/− mice (n = 4 each) were analyzed for levels of lymphocytes and myeloid-derived cells by flow cytometry. (A) Data at the left represent results from a representative mouse showing B cells, T cells, and macrophages; data at the right represent a higher-resolution representation of B and T lineage subpopulations. (B to D) Relative numbers of total splenocytes, total B cells, and B cell subpopulations (T1, transition 1 B cells; T2, transition 2 B cells; MZ, marginal zone B cells; FOB, follicular B cells) (B) and splenic T and innate cells resolved by separate cell surface markers (C, numbers of cells; D, percentages of cells) to reveal specific cell populations. Data are from a representative experiment repeated multiple times with similar results.
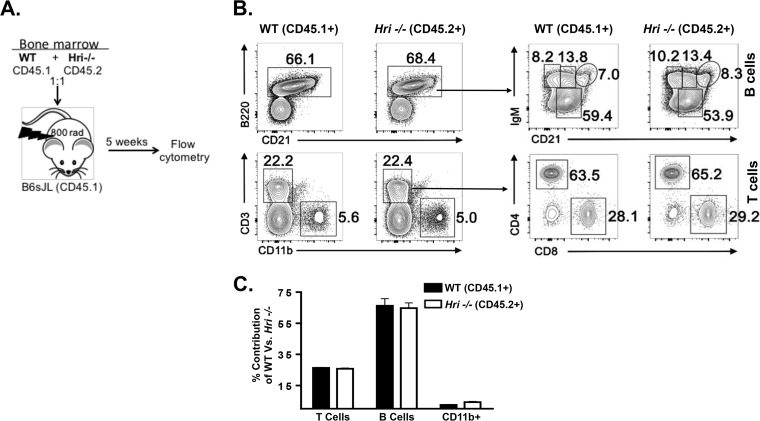
To reveal any possible cell-autonomous effects of HRI deficiency on hematopoietic stem cell (HSC) development, we performed a bone marrow competitive chimera experiment in which a 1:1 ratio of Hri+/+ and Hri−/− bone marrow cells (“donors”) was transplanted into lethally irradiated Hri+/+ recipient mice (Fig. 7A). After 5 weeks, the reconstituted immune systems of the recipient mice were analyzed. There were no discernible differences in the ability of Hri+/+ and Hri−/− bone marrow-derived HSCs to contribute to the total number of B cells (B220+), to B cell maturation (CD21), or to the number of CD3+ T cells or CD11b+ myeloid cells (Fig. 7B). Higher-resolution analysis of B and T cells showed that all major subsets of splenic B cells, including transitional B cells (T1 and T2) and mature follicular and mature innate-like marginal zone B cells (FoB and MZ B, respectively), were present in the two genotypes in similar proportions (Fig. 7B). Likewise, Hri+/+ and Hri−/− CD4+ and CD8+ T cells were seen in comparable proportions (Fig. 7B). In addition to the ratios, the numbers of B, T, and myeloid cells were also comparable (Fig. 7C). These data show that HRI does not play an apparent role in the development of immune cells. Further, Hri−/− immune cells can compete with Hri+/+ cells in the development of the immune system, indicating that HRI is dispensable for the development of all major hematopoietic cell populations.
FIG 7.
Hri−/− lymphoid and myeloid cells develop comparably in competition with Hri+/+ in vivo. (A) Bone marrow cells from wild-type (WT) and Hri−/− mice were cotransferred into congenic hosts. Splenocytes isolated from the recipient mice were analyzed after 5 weeks posttransfer to determine the percentages of the wild-type and Hri−/−-derived B cells, T cells, and macrophages. (B) (Left) Plots of data from a representative mouse showing B cell, T cell, and macrophage populations. (Right) Further resolved B and T cell subpopulations. (C) Bar graph representing percentages of T cells, B cells, and macrophages in recipient mice that were derived from wild-type or HRI-deficient mice analyzed as described for panel B. Data are from a representative experiment repeated twice with similar results.
DISCUSSION
The increased proliferation of L. monocytogenes in in vitro-infected Hri−/− macrophages (∼18 hpi), which occurred with kinetics similar to the results seen with enhanced proliferation of this pathogen in the liver of Hri−/− mice, may indicate that the increased susceptibility of Hri−/− mice to L. monocytogenes infection results from altered pathogen dynamics in initially infected host cells. Our findings reported here and previously (15) indicate that HRI impacts the trafficking of L. monocytogenes in such a way that it limits the emergence or efflux of the pathogen from infected cells. Consistently, screens to identify host genes that affect L. monocytogenes intracellular trafficking have found that factors involved in endosomal maturation can, when their expression levels are reduced, greatly impact infection (22–24). Other studies have identified host factors that either neutralize the activity of Listeria-encoded virulence factors or can be exploited by the pathogen to promote infection. For example, IFN-γ-inducible lysosomal thiol reductase (GILT) is required for the intracellular activation of listeriolysin O (LLO) (25). Furthermore, we have recently shown that Perforin-2 inhibits the rapid acidification of Listeria-containing vacuoles, preventing virulence gene expression and activity (26).
Mechanistically, the findings presented here and earlier (in a study in which it was directly shown that there is markedly reduced trafficking of L. monocytogenes to the cytosol in HRI-deficient cells [15]) indicate that HRI and GILT are broadly similar at the cellular level in terms of their positive roles in promoting the vacuole-to-cytosol transitioning of L. monocytogenes. However, where these two host factors differ is in regard to the consequences that occur in their absence. The killing of L. monocytogenes is enhanced in GILT-deficient cells, and GILT knockout mice are relatively protected against infection from this pathogen (25). In contrast, we have shown here that the replication of L. monocytogenes is actually increased in HRI-deficient cells and that HRI knockout mice are relatively more susceptible to infection by this pathogen. The intracellular infection cycle in HRI-deficient cells, bypassing a cytosolic phase, is characterized by a greatly enhanced rate of cellular efflux of the pathogen. Reduced levels of cytosolic L. monocytogenes are usually correlated with reduced intracellular proliferation and disease, as exemplified in the case of GILT. However, in the absence of a functioning immune system (e.g., in the SCID mouse), vacuole-bound L. monocytogenes can cause chronic infections, showing that, at least under certain conditions, cytosolic invasion is not invariably linked to disease (27). More recently, it was shown that L. monocytogenes is capable of long-term survival and persistence in lysosome-like vacuoles (28). Collectively, our findings indicate that modulation of intracellular pathogen trafficking pathways by the eIF2a kinases HRI and PKR suggests an important role for protein translational control in host response to infection and that these processes can be targeted with small-molecule inhibitors and activators.
MATERIALS AND METHODS
Mice and mouse infections.
Heme-regulated inhibitor (Hri)−/− mice (C57BL/6), as well mice of the derivative Hri−/− mouse embryonic fibroblast (MEF) cell line, were generously provided by Jane-Jane Chen and Randal J. Kaufman, respectively (9, 16). A breeding colony of Hri−/− mice is maintained at the University of Miami Miller School of Medicine and is frequently backcrossed to wild-type, Hri+/+, C57BL/6 mice. The green fluorescent protein (GFP)-expressing wild-type Listeria monocytogenes 10403S strain and the corresponding ΔLLO derivative mutant strain were provided by Daniel Portnoy. Mice were treated humanely in accordance with all appropriate government guidelines for the care and use of laboratory animals of the National Institutes of Health, and their use was approved for this entire study by the University of Miami institutional animal care and use committee (protocols 14-134 and 16-024). Seven-week-old mice were infected intravenously with exponentially growing L. monocytogenes (optical density [OD], 0.2) that had been propagated in brain heart infusion media at 37°C. Alternatively, mice were infected orogastrically with a 1 × 108 CFU of L. monocytogenes as described previously (29). In HRI inhibition infections, mice were treated with either vehicle only (phosphate-buffered saline [PBS]) or AMI {N-(2,6-dimethylbenzyl)-6,7-dimethoxy-2H-[1]benzofuro[3,2-c]pyrazol-3-amine hydrochloride} (17, 18) at a dose of 1.2 mg/kg of body weight administered intraperitoneally on days 0, 1, and 2 days postinfection (dpi) and harvested at 3 dpi. In all infection experiments, mice were monitored for signs of distress daily and animals that presented reduced movements and/or ruffled coats were humanely euthanized. Livers were removed and homogenized in sterile water containing 0.05% Triton X-100 by grinding through a fine wire mesh. Resulting homogenates were diluted further with water to fully lyse individual cells to release intracellular L. monocytogenes and the resulting dilutions plated to determine CFU. For histological analysis, intact frontal lobes were fixed in 10% neutral buffered formalin for 24 h and then embedded, sectioned, and stained with hematoxylin and eosin (H&E) by a licensed histotechnologist (Pathology Research Resources Histology Laboratory, University of Miami Miller School of Medicine). Slides were examined microscopically at ×60 magnification for inflammatory microabscesses characterized by neutrophilic infiltrates.
For cytokine analysis, serum was collected as indicated in the figure legends and IL-6, TNF-α, IFN-γ, MCP-1, IL-10, and IL-12p70 levels were determined by a cytometric bead array assay (BD Biosciences). For developmental analyses, Hri+/+ and Hri−/− mice were sacrificed on day 21, 27, or 34. A single cell suspension of splenocytes was stained with CD19, CD11b, CD11c, CD4, CD8, CD3, and LIVE/DEAD and analyzed by flow cytometry using a LSR II cytometer (BD Biosciences). Chimera mice were generated using recipient B6.SJL (congenic wild-type) mice expressing the CD45.1 allele which were lethally irradiated with 1,100 rads in a Gammacell 40 137Cs irradiator (Best Theratronics). At 16 h later, recipient mice were injected intravenously with 3.5 × 106 bone marrow cells consisting of equal numbers of cells isolated from B6.SJL and Hri−/− donor mice. Six weeks after injection, mice were sacrificed and splenic leukocytes were analyzed as described above. Surface markers CD45.1 on B6.SJL cells and CD45.2 on Hri−/− cells were used to distinguish donor bone marrow cells.
Cells and in vitro infections.
Peritoneal exudate macrophages (PEMs) were isolated from Hri+/+ and Hri−/− mice that had previously been injected with thioglycolate, assessed for viability using LIVE/DEAD cell stain (Life Technologies), and analyzed as described previously (26). Live cells were stained with antibodies allophycocyanin (APC) clone M1/70 (Tonbo Bioscience) and phycoerythrin (PE)/Cy7 clone BM8 (eBiosciences) to assess surface expression of macrophage-specific markers CD11b and F4/80. PEMs were seeded either at low density on glass coverslips (for microscopy) or into 48-well tissue culture dishes for CFU assays (2 × 105 cells per well). In some experiments, cells were treated with a freshly prepared solution of the eIF2α activator I-17 (reference 21) (solubilized in dimethyl sulfoxide [DMSO] and diluted in PBS) 1 h after seeding. Two hours following their seeding, adherent PEMs were infected as described below. Hri+/+ and Hri−/− MEFs were seeded in 48-well tissue culture dishes (2 × 105 cells per well) and infected the next day. Cultured cells were infected with exponentially growing L. monocytogenes (OD, ∼0.2) at a multiplicity of infection (MOI) of 5. We showed previously that these infection conditions resulted in detectable levels of viable L. monocytogenes for highly bactericidal PEMs after 60 min of infection (26). At either 30 min (PEMs) or 60 min (MEFs) postinfection, gentamicin (Gibco 15750-060) (2 μg/ml) was added to kill extracellular L. monocytogenes. It was determined in control experiments that this concentration of gentamicin kills >99.992% of extracellular L. monocytogenes within 5 min and that the killing of L. monocytogenes was essentially complete after 30 min (the exposure time used in macrophage infections). Our unpublished findings, as well as the published findings of others (28), show that higher (∼ 25 μg/ml) concentrations of gentamicin penetrate the eukaryotic plasma membrane and kill intracellular L. monocytogenes. Thirty minutes after the addition of gentamicin, cells were either lysed immediately (for early time points) or left undisturbed (for later time points) or gentamicin-containing media were removed and replaced with antibiotic-free media (to measure cellular L. monocytogenes efflux). Intracellular L. monocytogenes levels were determined by removing media from wells, adding 0.5 ml distilled sterile water for 30 s, and plating the resulting water lysate (or dilutions thereof) on semisolid LB media to enumerate CFU following 24 h incubation at 37°C.
Microscopy.
Following infections, macrophages bound to glass coverslips (see above) were fixed with 3.7% paraformaldehyde for 12 min and then stained for EEA1 (Abcam 2900) or LAMP1 (Abcam 24170) for 60 min at room temperature. Macrophages were then washed and stained with Alexa Fluor 555 for 60 min prior to being washed and mounted onto precleaned glass slides. DAPI (4′,6-diamidino-2-phenylindole) was included in the Pro-Long Gold mounting medium (Invitrogen). Imaging was performed using a Olympus fluorescence BX61 microscope equipped with Nomarski differential interference contrast (DIC) optics, a Uplan S Apo 100× objective (numerical aperture [NA], 1.4), a Roper CoolSnap HQ camera, Sutter Lambda 10-2 excitation and emission filters, and a 175-W xenon remote source. Intelligent Imaging Innovations Slidebook 4.01 was used for image capture. A series of optical Z-sections (0.35 μm) were imaged, and individual stacks were deconvolved using a nearest-neighbor algorithm prior to analysis. Representative projected images were chosen to be included in the figures. ImageJ software was used to quantify the level of fluorescence signal per infected cell or per bacterium. The fluorescence signal was divided by the area of the cell or bacterium to generate a signal/area ratio that was termed “fluorescence intensity in arbitrary units.”
ACKNOWLEDGMENTS
We declare that we have no conflict of interest with regard to the contents of this article.
This work was supported in whole or part by National Institutes of Health grants R01 AI101041 and DK084246 (W.N.K.) and R01 AI53459 (K.S.).
W. Bahnan and K. Schesser conceived the project. W. Bahnan, J. C. Boucher, and K. Schesser conducted the majority of the experiments and, together with W. N. Khan, analyzed and interpreted the results. P. Gayle and N. Shrestha assisted in performing experiments. M. Rosen provided the HRI inhibitor, and B. Aktas provided the eIF2α kinase activator. B. Adkins and A. Ager provided technical and material assistance. K. Schesser, with assistance from W. N. Khan, wrote the paper.
REFERENCES
- 1.Barbosa C, Peixeiro I, Romão L. 2013. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 3.Young SK, Wek RC. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291:16927–16935. doi: 10.1074/jbc.R116.733899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen JJ, Pal JK, Petryshyn R, Kuo I, Yang JM, Throop MS, Gehrke L, London IM. 1991. Amino acid microsequencing of internal tryptic peptides of heme-regulated eukaryotic initiation factor 2 alpha subunit kinase: homology to protein kinases. Proc Natl Acad Sci U S A 88:315–319. doi: 10.1073/pnas.88.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suragani RN, Zachariah RS, Velazquez JG, Liu S, Sun CW, Townes TM, Chen JJ. 2012. Heme-regulated eIF2α kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood 119:5276–5284. doi: 10.1182/blood-2011-10-388132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlanga JJ, Herrero S, de Haro C. 1998. Characterization of the hemin-sensitive eukaryotic initiation factor 2alpha kinase from mouse nonerythroid cells. J Biol Chem 273:32340–32346. doi: 10.1074/jbc.273.48.32340. [DOI] [PubMed] [Google Scholar]
- 7.Lu L, Han AP, Chen JJ. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol Cell Biol 21:7971–7980. doi: 10.1128/MCB.21.23.7971-7980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhan K, Vattem KM, Bauer BN, Dever TE, Chen JJ, Wek RC. 2002. Phosphorylation of eukaryotic initiation factor 2 by heme-regulated inhibitor kinase-related protein kinases in Schizosaccharomyces pombe is important for resistance to environmental stresses. Mol Cell Biol 22:7134–7146. doi: 10.1128/MCB.22.20.7134-7146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 10.Mittal SP, Kulkarni AP, Mathai J, Chattopadhyay S, Pal JK. 2014. Dose-dependent differential response of mammalian cells to cytoplasmic stress is mediated through the heme-regulated eIF2α kinase. Int J Biochem Cell Biol 54:186–197. doi: 10.1016/j.biocel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129–141. doi: 10.1016/S1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 12.Wiley DJ, Shrestha N, Yang J, Atis N, Dayton K, Schesser K. 2009. The activities of the Yersinia protein kinase A (YpkA) and outer protein J (YopJ) virulence factors converge on an eIF2alpha kinase. J Biol Chem 284:24744–24753. doi: 10.1074/jbc.M109.010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha N, Bahnan W, Wiley DJ, Barber G, Fields KA, Schesser K. 2012. Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J Biol Chem 287:28738–28744. doi: 10.1074/jbc.M112.375915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Suragani RN, Wang F, Han A, Zhao W, Andrews NC, Chen JJ. 2007. The function of heme-regulated eIF2alpha kinase in murine iron homeostasis and macrophage maturation. J Clin Invest 117:3296–3305. doi: 10.1172/JCI32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrestha N, Boucher J, Bahnan W, Clark ES, Rosqvist R, Fields KA, Schesser K. 2013. The host-encoded Heme Regulated Inhibitor (HRI) facilitates virulence-associated activities of bacterial pathogens. PLoS One 8:e68754. doi: 10.1371/journal.pone.0068754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH, Chen JJ. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J 20:6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanelakis KC, Palomino HL, Li L, Wu J, Yan W, Rosen MD, Rizzolio MC, Trivedi M, Morton MF, Yang Y, Venkatesan H, Rabinowitz MH, Shankley NP, Barrett TD. 2009. Characterization of a robust enzymatic assay for inhibitors of 2-oxoglutarate-dependent hydroxylases. J Biomol Screen 14:627–635. doi: 10.1177/1087057109333976. [DOI] [PubMed] [Google Scholar]
- 18.Rosen MD, Woods CR, Goldberg SD, Hack MD, Bounds AD, Yang Y, Wagaman PC, Phuong VK, Ameriks AP, Barrett TD, Kanelakis KC, Chuang JC, Shankley NP, Rabinowitz MH. 2009. Discovery of the first known small-molecule inhibitors of heme-regulated eukaryotic initiation factor 2alpha (HRI) kinase. Bioorg Med Chem Lett 19:6548–6551. doi: 10.1016/j.bmcl.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 19.ILL-Raga G, Köhler C, Radiske A, Lima RH, Rosen MD, Muñoz FJ, Cammarota M. 2013. Consolidation of object recognition memory requires HRI kinase-dependent phosphorylation of eIF2α in the hippocampus. Hippocampus 23:431–436. doi: 10.1002/hipo.22113. [DOI] [PubMed] [Google Scholar]
- 20.Ill-Raga G, Tajes M, Busquets-García A, Ramos-Fernández E, Vargas LM, Bosch-Morató M, Guivernau B, Valls-Comamala V, Eraso-Pichot A, Guix FX, Fandos C, Rosen MD, Rabinowitz MH, Maldonado R, Alvarez AR, Ozaita A, Muñoz FJ. 2015. Physiological control of nitric oxide in neuronal BACE1 translation by heme-regulated eIF2α kinase HRI induces synaptogenesis. Antioxid Redox Signal 22:1295–1307. doi: 10.1089/ars.2014.6080. [DOI] [PubMed] [Google Scholar]
- 21.Chen T, Takrouri K, Hee-Hwang S, Rana S, Yefidoff-Freedman R, Halperin J, Natarajan A, Morisseau C, Hammock B, Chorev M, Aktas BH. 2013. Explorations of substituted urea functionality for the discovery of new activators of the heme-regulated inhibitor kinase. J Med Chem 56:9457–9470. doi: 10.1021/jm400793v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 23.Cheng LW, Viala JP, Stuurman N, Wiedemann U, Vale RD, Portnoy DA. 2005. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc Natl Acad Sci U S A 102:13646–13651. doi: 10.1073/pnas.0506461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burrack LS, Harper JW, Higgins DE. 2009. Perturbation of vacuolar maturation promotes listeriolysin O-independent vacuolar escape during Listeria monocytogenes infection of human cells. Cell Microbiol 11:1382–1398. doi: 10.1111/j.1462-5822.2009.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R, Jamieson A, Cresswell P. 2008. GILT is a critical host factor for Listeria monocytogenes infection. Nature 455:1244–1247. doi: 10.1038/nature07344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormack R, Bahnan W, Shrestha N, Boucher J, Barreto M, Barrera CM, Dauer EA, Freitag NE, Khan WN, Podack ER, Schesser K. 2016. Perforin-2 protects host cells and mice by restricting the vacuole to cytosol transitioning of a bacterial pathogen. Infect Immun 84:1083–1091. doi: 10.1128/IAI.01434-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. 2008. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature 451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 28.Kortebi M, Milohanic E, Mitchell G, Péchoux C, Prevost MC, Cossart P, Bierne H. 2017. Listeria monocytogenes switches from dissemination to persistence by adopting a vacuolar lifestyle in epithelial cells. PLoS Pathog 13:e1006734. doi: 10.1371/journal.ppat.1006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Echeverry A, Schesser K, Adkins B. 2007. Murine neonates are highly resistant to Yersinia enterocolitica following orogastric exposure. Infect Immun 75:2234–2243. doi: 10.1128/IAI.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]