Abstract
Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) has a specific antitumour activity against many malignant tumours. However, more than half of lung cancer cells are resistant to TRAIL-relevant drugs. Trichosanthin (TCS) is a traditional Chinese medicine with strong inhibitive effects on various malignancies. Nevertheless, its function on TRAIL resistance has not been revealed in non-small cell lung cancer (NSCLC). To examine the molecular mechanisms of TCS-induced TRAIL sensitivity, we administrated TCS to TRAIL-resistance NSCLC cells, and found that the combination treatment of TCS and TRAIL inhibited cancer cell proliferation and invasion, and induced cell apoptosis and S-phase arrest. This combined therapeutic method regulated the expression levels of extrinsic apoptosis-associated proteins Caspase 3/8 and PARP; intrinsic apoptosis-associated proteins BCL-2 and BAX; invasion-associated proteins E-cadherin, N-cadherin, Vimentin, ICAM-1, MMP-2 and MMP-9; and cell cycle-associated proteins P27, CCNE1 and CDK2. Up-expression and redistribution of death receptors (DRs) on the cell surface were also observed in combined treatment. In conclusion, our results indicated that TCS rendered NSCLC cells sensitivity to TRAIL via upregulating and redistributing DR4 and DR5, inducing apoptosis, and regulating invasion and cell cycle related proteins. Our results provided a potential therapeutic method to enhance TRAIL-sensitivity.
Keywords: TRAIL, NSCLC, Trichosanthin, TRAIL-resistance, death receptor
Introduction
Lung cancer has been the leading cause of cancer-related death across the world over the last 30 years. Up to 85% of lung cancer cases are non-small cell lung cancer (NSCLC), almost 75% of which are diagnosed as advanced cancer at the first visit 1. Chemotherapy and radiotherapy are still the major therapies to treat advanced cancer 2. Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) can selectively induce cancer cell apoptosis and keep normal cells alive 3-5. There are five different TRAIL receptors, including death receptor 4 (DR4), DR5, decoy receptor 1 (DcR1), DcR2, and the soluble receptor osteoprotegerin (OPG) 6. The interaction between TRAIL and DRs initiates TRAIL-induced apoptosis via aggregating Fas-associated death domain (FADD) and Caspase 8 7-8. Activated Caspase 8 directly induces apoptosis via extrinsic or intrinsic apoptotic pathway, also called the DR-mediated or mitochondrial-mediated apoptotic pathway. In the extrinsic apoptotic pathway, the activation of Caspase 3 by Caspase 8 cleaves a broad range of apoptosis-related protein substrates and induces programmed cell death. In the intrinsic apoptotic pathway, the cleavage of BH3 interacting domain death agonist (Bid) by Caspase 8 induces tBid 9 and upregulates BCL-2 associated X protein (BAX). Clinical trials on TRAIL-related drugs were carried out in lung cancer, pancreatic cancer, colorectal cancer, breast cancer, lymphoma, soft tissue sarcoma, and head and neck tumours 10-12. However, cancer cells can escape TRAIL-induced apoptosis and gradually acquire TRAIL resistance during the process of treatment 13-15. Therefore, further investigation on the mechanisms of TRAIL resistance and enhancement of TRAIL-sensitivity will substantially improve its therapeutic potential.
Trichosanthin (TCS) is a type I ribosome inactivating protein (RIP), which is used in choriocarcinoma therapy and midterm pregnancy termination in China 16. Its primary sequence and 3 dimensional structure were reported in 1990 17, 18. TCS is a traditional Chinese medicine extracted from Trichosanthes root. TCS has various pharmacological effects including induction of abortion, inhibition of protein synthesis, anti-HIV, and anti-tumour 19-21. TCS was also reported to kill tumour cells specifically without damaging normal cells 22. We hypothesized that TCS could promote TRAIL sensitivity in NSCLC cells. We administrated TCS individually or combined it with TRAIL to TRAIL-resistance NSCLC cells, and examined its effects on cell proliferation, cell apoptosis, invasion ability, cell cycle distribution and the regulation of DRs. Our studies revealed the molecular mechanisms of TRAIL resistance and broadened the application of TRAIL.
Materials and methods
Reagents and cell culture
TRAIL was purchased from Shanghai TRAIL Bio-Technical Co., Ltd. TCS was obtained from the Chengdu Institute of Biological Products Co., Ltd. Purified TCS was prepared as previously reported 23, 24. Briefly, TCS was purified from the grinded and extracted root tuber of Trichosanthes kirilowii by ammonium sulphate fractionation followed by Blue-sepharose CL-B and SP-sepharose column. The homogeneity of purified TCS was assessed by SDS-PAGE and PAGE.
NSCLC cells (NCI-H1299, NCI-H1975 and A549) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and authenticated by Guangzhou Cellcook Biotech Ltd (Identification certificates listed in Supplemental Fig. S1-3). Cells were cultured in RPMI-1640 medium (Hyclone Ltd., USA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin, and maintained at 37°C in 5% CO2.
Cell viability and clonogenic assays
Cell viability was detected by the cell counting kit (CCK-8, Dojindo Ltd., Japan). Cells with confluence of 70-80% were treated with different concentrations of TCS (0, 10, 20, 40, 60, 80, 100 μg/ml) and TRAIL (0, 5, 10, 25, 50, 100, 250, 500, 750, 1000 ng/ml) for 48 h. After mixing with basal medium containing 10% (v/v) CCK-8 for 1 h, the optical density (OD) at 450 nm was measured by a microplate reader (Rayto Ltd., China). Cell viability was calculated by the formula: cell viability (100%) = (OD value of the treated wells - OD value of the blank control wells) / (OD value of the negative control wells - OD value of the blank control wells). Each group had at least five wells and the whole assay was repeated 3 times.
For the clonogenic assay, cells were seeded into 6-well plates (100 cells/well), and then treated with 50 ng/ml TRAIL or/and the IC20 dose of TCS for 14 days. Colonies were fixed with 4% paraformaldehyde (Sangon Ltd., China) and stained with 0.5% crystal violet (Beyotime Ltd., China). The colony number was quantified under a microscope, and the assay was performed in triplicate.
Immunofluorescence
Cell climbing sheets were treated with 50 ng/ml TRAIL or/and the IC20 dose of TCS for 48 h. After fixing with 4% paraformaldehyde (Sangon Ltd., China) and permeabilizing with 0.5% Triton X-100 (Beyotime Ltd., China), the cells were blocked with concentrated normal goat serum for 30 min and incubated with primary antibodies (listed in Supplemental Table S1) at 4°C overnight. After incubating with fluorescein isothiocyanate (FITC) /Cy3-conjugated secondary antibody (listed in Supplemental Table S2) for 45 min and washing with PBS, the slides were stained with DAPI (Sigma Ltd., Germany). Images were taken by microscope (Olympus Ltd., Japan) or confocal laser scanning microscopy (Nikon Ltd., Japan).
Flow cytometry
The cells were treated with 50 ng/ml TRAIL or/and the IC20 dose of TCS for 48 h. For the detection of apoptotic cells, the cells were digested with EDTA-free trypsin (Corning Ltd., USA) and harvested. After washing twice and dissolving in 400 μl binding buffer (BestBio Ltd., China), the single cell suspension was stained with 5 μl Annexin V-FITC for 30 min and then with 7 μl propidium iodide (PI) for 5 min at room temperature. For the detection of cell cycle, the cells were digested with trypsin and harvested. After washing with PBS and fixing in ice-cold 70% ethanol overnight, cells (1 × 106) were washed twice with PBS, and then resuspended in PI staining buffer (50 μg/ml PI; 100 μg/ml RNase and 0.2% Triton X-100 in PBS) for 30 min. For the detection of DR cytomembrane expression, cells were trypsinized with 0.25% trypsin (Corning Ltd., USA) and washed twice with ice-cold PBS. Cells were then incubated in 50 μl PBS containing 1% goat serum at room temperature for 40 min. After washing with PBS 3 times, cells were incubated with the primary antibody (listed in Supplemental Table S1) at 4°C overnight. After washing 3 times with PBS and incubating with FITC-labelled secondary antibody (listed in Supplemental Table S2) in dark at room temperature for 45 min, the cells were washed with PBS once and resuspended in 500 μl PBS. For the negative control, cells were processed as described above without using primary antibody. The samples were analyzed using flow cytometer (Becton-Dickinson Ltd., USA).
In situ cell death detected by terminal deoxynucleotidyl transferase (TdT) ‑ mediated dUTP nick end‑labelling (TUNEL) assay
Cell climbing sheets were treated with 50 ng/ml TRAIL or/and 40 μg/ml TCS for 48 h. The in situ cell death was detected by a TUNEL Kit (Roche Ltd., Switzerland). Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. The positive control group was treated with 100 µl DNase I at 25°C for 10 min. After incubating with 50 µl TUNEL reaction solution in the dark for 1 h and washing with PBS, the slides were mounted with DAPI, and images were taken. Five visual fields of view were randomly selected to count the positive cells from per 100 cells under the microscope at 100X magnification, and the positive rate was calculated as apoptosis index (AI).
Invasion assay
The H1299 cells were resuspended in basal medium after treatment. Cells (1×104) were seeded into the upper chamber with 8 µm pore inserts (Corning Ltd., USA) pretreated with Matrigel (Becton-Dickinson Ltd., USA), and 600 µl RPMI-1640 medium containing 20% FBS was added into the lower chamber. After 24 h, the cells on the upper surface of the membrane were removed, whereas the cells on the lower surface were fixed with 4% paraformaldehyde (Sangon Ltd., China) for 30 min at room temperature and stained with 0.5% crystal violet solution (Beyotime Ltd., China) for 15 min. The numbers of invasive cells were counted under the microscope at 200X magnification. The images were analyzed using Image-Pro Plus software (version 6.0).
RNA isolation, RT-PCR and qRT-PCR
Total RNA was extracted with TRIzol (Sangon Ltd., China). RNA concentration was detected by a Nanodrop spectrophotometer (Thermo Scientific Ltd., USA). Total RNA (500 ng) was used for the synthesis of first-strand cDNA using HiScript® II Q RT SuperMix for qPCR kit (Vazyme Ltd., China). The following primers were used:
DR4:
forward 5'-ACCTTCAAGTTTGTCGTCGTC-3' and reverse 5'-CCAAAGGGCTATGTTCCCATT-3';
DR5:
forward 5'-ACAGTTGCAGCCGTAGTCTTG -3' and reverse 5'- CCAGGTCGTTGTGAGCTTCT -3';
GAPDH:
forward 5'-TGGAAGGACTCATGACCACA-3' and reverse 5'- TCAGCTCAGGGATGACCTT -3'.
The qRT-PCR reactions were performed using a CFX96 qRT-PCR system (Applied Biosystems Ltd., USA) according to the manufacturer's instruction. The 2-ΔΔCT method was used to calculate the fold changes. GAPDH was used as an internal control for the normalization of target gene expression.
Western blot analysis
H1299 Cells were treated with 50 ng/ml TRAIL or/and 40 μg/ml TCS for 48 h. Whole cell lysate was extracted using RIPA buffer (Beyotime Ltd., China) supplemented with protease and phosphatase inhibitors cocktails (Sigma Chemical Ltd., USA). Cell membrane proteins DR4 and DR5 were extracted following the membrane protein extraction kit instruction (Merck Ltd., Germany). Protein concentration was measured by bicinchoninic acid system (Beyotime Ltd., China) with bovine serum albumin as a standard control. Aliquots of 40 μg protein per lane were separated by 10% SDS-PAGE, and the proteins were then transferred to polyvinylidene fluoride (PVDF) membranes. Primary and secondary antibodies used for detection were listed in Supplemental Table S1 and S2 for 90 min. Then, the PVDF membranes were visualized with an enhanced chemiluminescence kit (Bio-Rad Ltd., USA) and exposed on a gel imaging analyzer (Bio-Rad Ltd., USA). The total protein levels were related to GAPDH and the membrane protein levels were related to ATP1A1.
Statistical analysis
Results were presented as the mean ± standard deviation (SD). The difference between 2 measurements was analyzed by the unpaired Student's T-test using GraphPad Prism 5.0 Software. A p value of < 0.05 was defined as a significant difference. IC20 and IC50 values were calculated using SPSS 17.0 software.
Results
Combination of TCS and TRAIL inhibited the proliferation of TRAIL-resistant cells
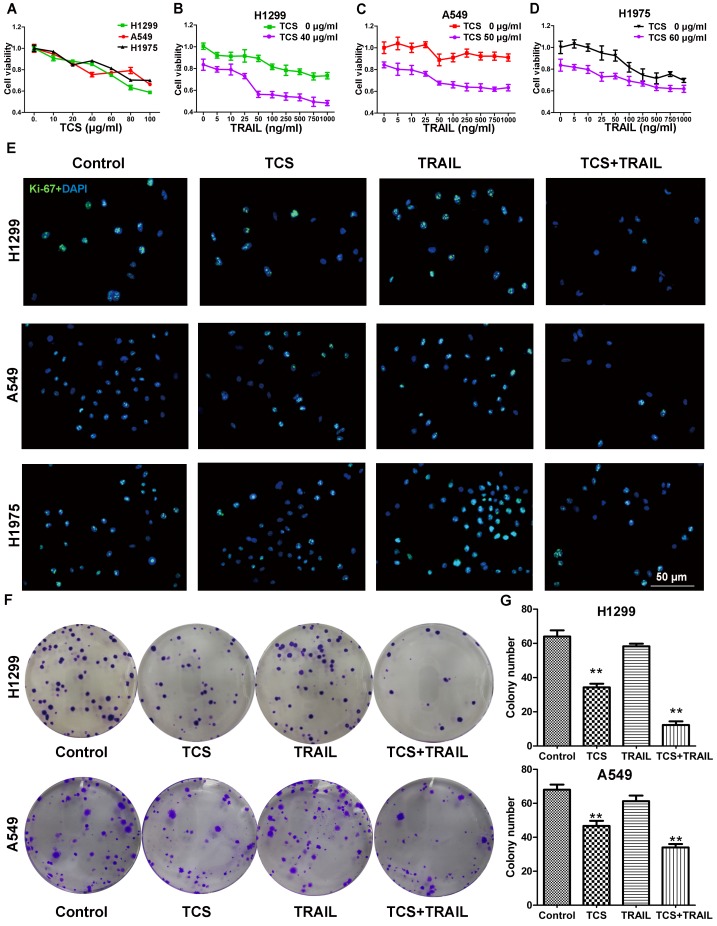
H1299, A549 and H1975 cells were reported to be resistant to TRAIL 26. Our CCK8 results also showed that, even when treated with 1,000 ng/ml TRAIL, these cells still maintained approximately 70% cell viability (Fig. 1B-D, the upper curve). As the combination index was less than 1 at every drug concentration with the combined treatments, TCS and TRAIL had synergistic effects on these cells (Supplemental Fig. S4). We next investigated whether or not TCS enhanced the TRAIL sensitivity of H1299, A549 and H1975 cells. We found that TCS inhibited the cell viability in a dose-dependent manner. The IC20 values of TCS were 39.5 ± 1.1, 49.3 ± 4.2 and 61.1 ± 6.8 μg/ml in H1299, A549 and H1975 cells respectively (Fig. 1A). Cell viability was significantly inhibited by the combined therapy with 50 ng/ml TRAIL and the IC20 dose of TCS (Fig. 1B-D, the lower curve). The levels of Ki-67 in all these 3 cell lines were also decreased by combined treatment (Fig. 1E). As expected, the colony formation of H1299 and A549 cells were significantly decreased in combined treatment groups (Fig. 1F). The average colony numbers decreased from 64 ± 3.61% to 12.3 ± 2.08% for H1299, and from 57.67 ± 2.05% to 26 ± 1.63% for A549 with combined treatment (Fig. 1G). Those results indicated that TCS enhanced the sensitivity to TRAIL by inhibiting cell proliferation and colony formation in TRAIL-resistance cells.
Figure 1.
The combination of TCS and TRAIL inhibited the proliferation of TRAIL-resistance cells. (A) Sensitivity of H1299, A549 and H1975 cells to TCS was determined by CCK8. The cell lines showed different responsivity to 0-100 μg/ml TCS. All assays were performed in 5 replicates and repeated 3 times. (B-D) Sensitivity of H1299, A549 and H1975 cells to TRAIL or the combined treatment of TRAIL and TCS was determined by CCK8. Cells were resistant to TRAIL but more sensitive to the combined treatment. (E) Proliferation of H1299, A549 and H1975 cells were detected by Ki-67 immunofluorescence. Cells showed decreased proliferation rate after the combination treatment of 50 ng/ml TRAIL and the IC20 dose of TCS. The magnification was 400X. The scale bar: 50 μm. (F-G) Colony forming assay for H1299 and A549 cells after treatment with 50 ng/ml TRAIL or/and IC20 dose of TCS for 14 days. Less colony numbers in the combination group compared with individual drug groups. All assays were repeated 3 times. **, p < 0.01.
Combination of TCS and TRAIL promoted the apoptosis of TRAIL-resistant cells
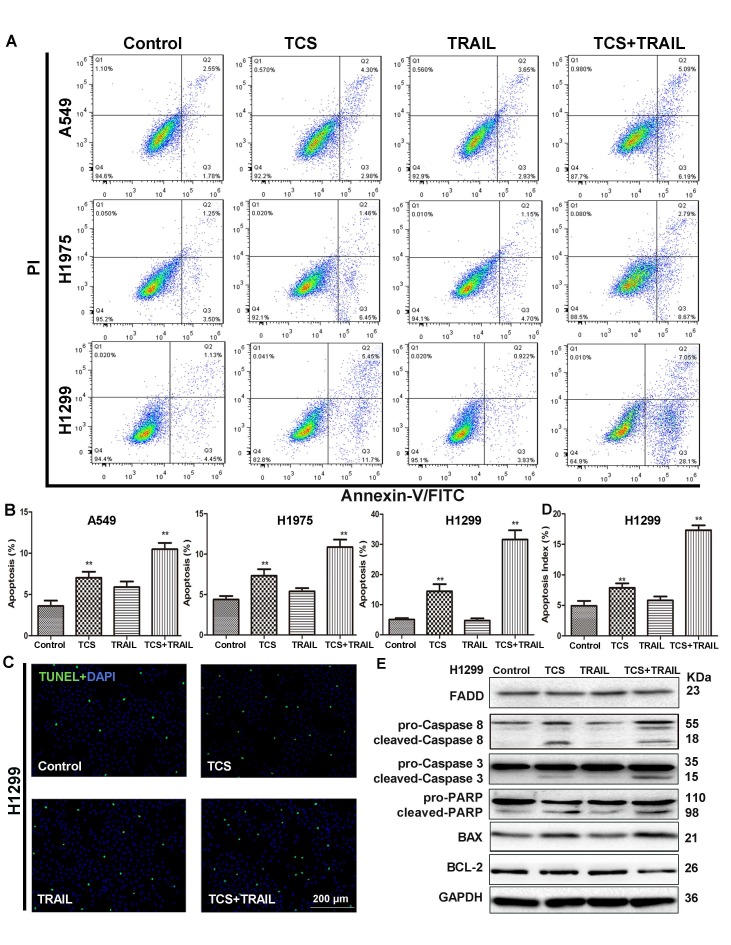
The apoptosis of A549, H1975 and H1299 cells treated with TCS or/and TRAIL was detected by an Annexin V-FITC/PI apoptosis detection kit. While individual treatment of TRAIL had no effect on the apoptotic rate, the combined use of TCS and TRAIL significantly induced Annexin V expression, suggesting more cell apoptosis (Fig. 2A-B). The number of TUNEL positive cells was also significantly increased in H1299 cells treated with both TCS and TRAIL (Fig. 2C-D). Compared with the control group or the individual treatment groups, the levels of cleaved-Caspase 3/8, cleaved- poly ADP-ribose polymerase (PARP) and BAX of the combined group were substantially increased in H1299 cells, while the levels of BCL-2 were decreased (Fig. 2E). These results suggested that TCS and TRAIL dual treatment induced apoptosis in TRAIL-resistance cells via promoting both the extrinsic and intrinsic apoptotic pathways.
Figure 2.
The combination of TCS and TRAIL promoted apoptosis of TRAIL-resistance cells. (A) Apoptosis was detected by Annexin V-FITC/PI double-staining. Cells were treated with 50 ng/ml TRAIL or/and the IC20 dose of TCS for 48 h. (B) The percentage of apoptotic cells to total cells. Data were presented as column graph of 3 independent experiments with the mean value. *, p < 0.05; **, p < 0.01. (C) After H1299 cells were incubated with 50 ng/ml TRAIL or/and 40 μg/ml TCS for 48 h, the apoptosis of H1299 cells was measured by the TUNEL assay. The magnification was 100X. The scale bar: 200 μm. (D) A graphical representation of the data presented the apoptotic index of 100 cells from 3 independent experiments. *, p < 0.05; **, p < 0.01. (E) The protein levels of apoptosis-related proteins FADD, Caspase 3/8, PARP, BCL-2 and BAX were analyzed by Western blot. GAPDH was used as an internal control to verify equal protein loading.
Combination of TCS and TRAIL inhibited invasion and induces S-phase arrest in H1299 cells
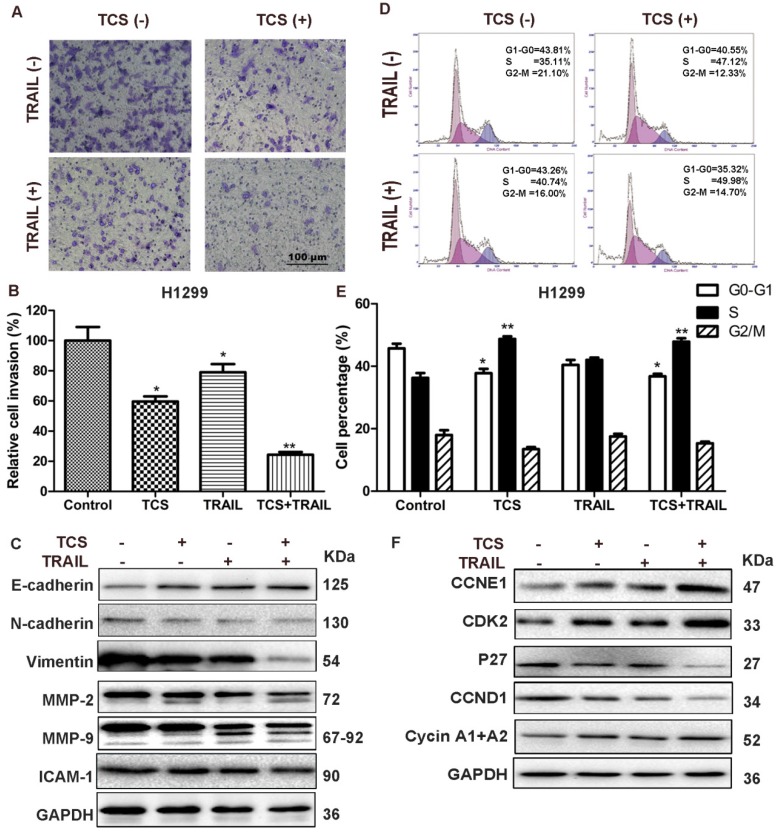
The results of Matrigel invasion assay showed that combined treatment of TCS and TRAIL significantly decreased cell invasion compared with the control and the individual drug groups (Fig. 3A-B). The protein levels of E-cadherin were increased (Fig. 3C and Supplemental Fig. S5), while the protein levels of N-cadherin, Vimentin, Intercellular Adhesion Molecule 1 (ICAM-1), Matrix Metallopeptidase-2 (MMP-2) and MMP-9 were decreased in the combination group (Fig. 3C). The results of flow cytometry indicated that combination of TCS and TRAIL increased the proportion of cells in S phase and decreased the proportion of cells in G1 phase compared with the control group and the individual drug groups (Fig. 3D-E). Western blot results showed that the protein levels of CCNE1, CDK2 and Cyclin A1+A2 were up-regulated and those of CCND1 and P27 were decreased in combination group (Fig. 3F).
Figure 3.
The combination of TCS and TRAIL inhibited cell invasion and induced cell cycle redistribution. (A) H1299 cells were incubated with 50 ng/ml TRAIL or/and 40 μg/ml TCS for 48 h, and the cell invasion was detected by a transwell invasion assay. The magnification was 200X. The scale bar: 100 μm. (B) A graphical representation of the data from 3 independent experiments. *, p < 0.05; **, p < 0.01. (C) The protein levels of invasion-related proteins E-cadherin, N-cadherin, Vimentin, ICAM-1, MMP-2 and MMP-9 were analyzed by Western blot. GAPDH was used as an internal control to verify equal protein loading. (D) Cell cycles were detected by flow cytometry. H1299 cells were treated with 50 ng/ml TRAIL or/and 40 μg/ml TCS for 48 h. Cells were then stained with PI and analyzed using Flow cytometry. (E) The percentage of G0-G1, S and G2-M phase cells were presented as histogram from 3 independent experiments with the mean value. *, p < 0.05 vs G0-G1 control, **, p < 0.01 vs S control. (F) The protein levels of cell cycle-related proteins CCNE1, CDK2, CCND1, Cyclin A1+A2 and P27 were analyzed by Western blot. GAPDH was used as an internal control to verify equal protein loading.
Combination of TCS and TRAIL up-regulated DRs expression and accumulation on the cell surface in H1299 cells
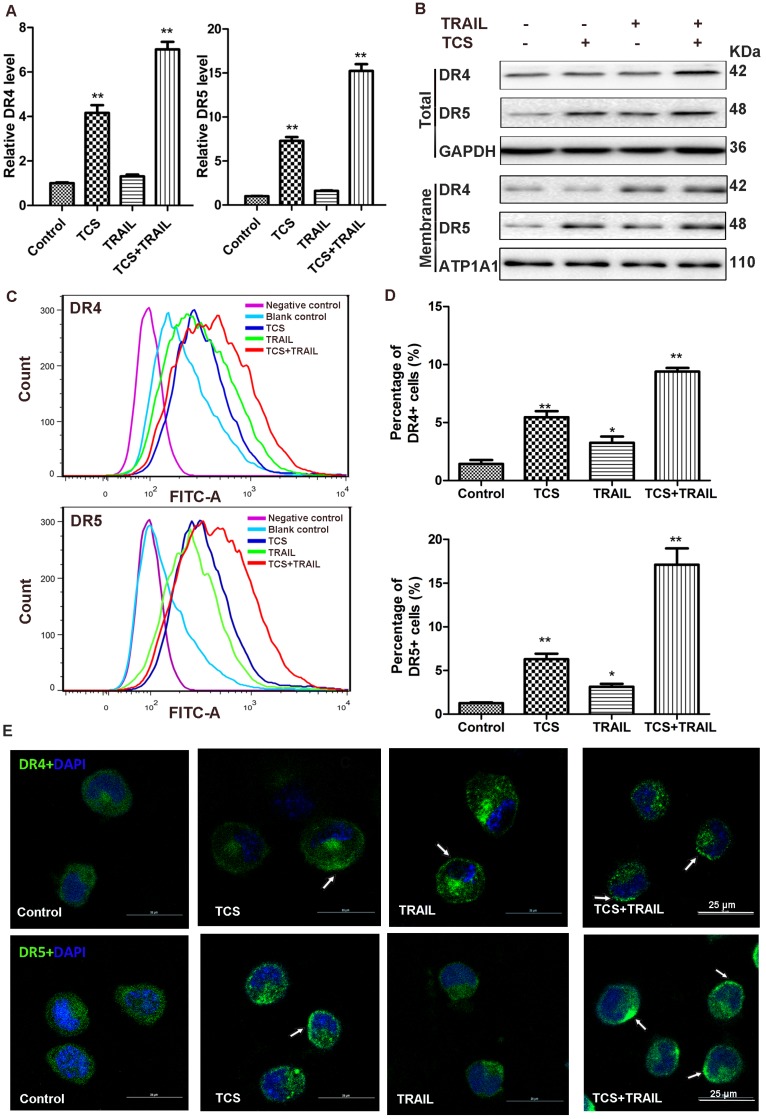
The transcriptional levels of DR4 and DR5 in H1299 cells were significantly increased in the combination group (Fig. 4A and Supplemental Fig. S6). The results of Western blot analysis also suggested that the protein levels of DR4 and DR5 were up-regulated after combined treatment, in both whole cell lysate and the membrane compartment (Fig. 4B). The results of flow cytometry indicated that combination of TCS and TRAIL significantly increased the levels of DR4 and DR5 on the cell surface (Fig. 4C-D). These results suggested that TCS improved TRAIL-sensitivity by increasing the expression levels of DR4 and DR5 and leading them to accumulate onto the cell membrane. Immunofluorescence also confirmed that higher expression and induced accumulation of DRs on the cell surface in H1299 cells treated with both TCS and TRAIL (Fig. 4E, the white arrow).
Figure 4.
The combination of TCS and TRAIL upregulated the expression and cell surface accumulation of DR4 and DR5. (A) The transcriptional levels of DR4 and DR5 (relative to GAPDH) were analyzed by qRT-PCR in H1299 cells treated with 50 ng/ml TRAIL or / and 40 μg/ml TCS for 48 h. All assays were performed in triplicates and repeated 3 times. *, p < 0.05; **, p < 0.01. (B) The protein levels of DR4 and DR5 were analyzed by Western blot. GAPDH and ATP1A1 were used as internal controls to verify equal protein loading. (C) The cell surface levels of DR4 and DR5 were detected by indirect immunofluorescence staining using Flow cytometry. (D) A graphical representation of the data from 3 independent experiments. *, p < 0.05; **, p < 0.01. (E) The accumulation of DR4 and DR5 on the surface of the cell membrane was revealed by confocal microscopy, marked by white arrow. The magnification was 800X. The scale bar: 25 μm.
Discussion
The clinical application of TRAIL is limited due to the drug resistance. As described in previous researches, cells with an IC50 < 100 ng/ml were classified as sensitive, those with an IC50 between 100 and 500 ng/ml were classified as moderately sensitive, and those with an IC50 > 500 ng/ml were defined as resistant 26. One study found that 32 of the 39 tumour cells including colon, lung, breast, kidney, and skin cancers were sensitive to the cytotoxic effects of recombinant TRAIL 27. Among 13 detected glioma cell lines, while 3 cell lines were sensitive, and 4 cell lines were moderately sensitive, the rest and normal astrocytes were resistant to TRAIL 28. In vitro studies showed that more than half of the lung cancer cell lines were resistant to either recombinant human TRAIL or TRAIL DR agonists 26. Our CCK8 results showed that H1299, A549 and H1975 cells still preserved at least 70% cell viability when treated with the maximum concentration of TRAIL (1,000 ng/ml) (Fig. 1B-D), and they were considered as TRAIL-resistance cells. Compared with A549 and H1975 cell lines, the H1299 cell line was more sensitive to the combined treatment in both proliferation and apoptosis assays. Therefore, the cell function assays were only preformed in H1299 cells.
There are various factors that cause TRAIL resistance, such as endoplasmic reticulum stress 29, protein synthesis disorders 30, DR expression decreased, and anti-apoptotic protein over-expression 31. Many therapeutic ways been developed to increase TRAIL-sensitivity. Previous researches proved that the cell surface levels of DR4 were significantly reduced in acquired TRAIL-resistance SW480 cells, and that TRAIL resistance was reversed by increased cytomembrane expression of DRs 32. However, the mechanism of TRAIL resistance is complex and unclear at present. The causes of TRAIL resistance and the methods to overcome resistance are still to be investigated.
Many researchers have reported that TCS induced cell apoptosis and inhibited cell proliferation in many malignant tumours, including human choriocarcinoma 33, 34, breast cancer 35, nasopharyngeal carcinoma 36, leukemia and lymphoma 37 and cervical cancer 38. TCS might promote demethylation of antioncogenes such as adenomatous polyposis coli (APC) and tumour suppressor in lung cancer 1 (TSLC1) in cervical cancer cells 38. One study reported that TCS was cytotoxic towards A549 cells, which could lead to redistribution of cell cycle and induce apoptosis39. Another study demonstrated that TCS up-regulated the expression of TSLC1 and class I-restricted T cell-associated molecule (CRTAM), and inhibited tumour growth via promoting the conjunction of TSLC1 and CRTAM in mouse Lewis lung cancer cells 40. These studies suggested the potential role of TCS as an anti-cancer drug.
In this study, we investigated the effects and molecular mechanisms of the combined treatment of TCS and TRAIL on cell proliferation, cell apoptosis, invasion ability, cell cycle distribution and the expression of DRs in TRAIL-resistance cells. The cell viability and cell colony formation were significantly reduced by the combined treatment (Fig. 1B-F and Suppmental Fig. S4). Apoptosis was remarkably increased in the combination groups (Fig. 2A), especially in H1299 cells (Fig. 2D). The combined treatment with TCS and TRAIL also inhibited cell invasion and induced S-phase arrest in H1299 cells via regulating the expression of invasion- and apoptosis-associated proteins (Fig. 3C, F). These results indicated that TCS increased the TRAIL-sensitivity and suggested a new strategy to overcome TRAIL resistance.
As the initiators of apoptosis, the DRs significantly restrict the effectiveness of apoptotic signal transduction. Wang et al. 41 constructed a colon cancer cell line which was deficient in DR5 using RNA interference technology, and transplanted these cells into nude mice. It was found that the mice acquired resistance to 5-Fluoracil, suggesting that DRs played an important role in the process of drug resistance. There are many potential mechanisms involved in the regulation of DR4 and DR5. One study reported that bufalin promoted the clustering of DR4 and DR5 in aggregated lipid rafts in breast cancer cells MCF-7 and MDA-MB-231, and that bufalin enhanced TRAIL-induced apoptosis by activating the extrinsic apoptotic pathway. The cholesterol-sequestering agent methyl-β-cyclodextrin reversed the DR4 and DR5 clustering and reduced bufalin+TRAIL-induced apoptosis 42. In colon cancer cells, resveratrol induced DR redistribution into lipid rafts and facilitated the Caspase cascade activation in response to DR stimulation without increased levels of DRs on the cell surface 43. In glioblastomas, targeting of receptor-interacting protein (RIP), cellular Fas-associated death domain Like IL-1 beta converting enzyme (c-FLIP) or phosphoprotein enriched in diabetes/phosphoprotein enriched in astrocytes (PED/PEA-15) with siRNA led to the redistribution of death inducing signaling complex (DISC) from non-rafts to lipid rafts and eliminated the inhibition of Caspase 8 cleavage 44. Another study stated that TRAIL induced gastric cancer cell apoptosis through the casitas B-lineage lymphoma (Cbl)-regulated DR redistribution in lipid rafts in MGC803, BGC823, and SGC7901 cells partially 45. We found that DR4 and DR5 were significantly increased by combined treatment with TCS and TRAIL (Fig. 4 and Supplemental Fig. S6). The cell surface levels of DR4 and DR5 were increased by the combined treatment (Fig. 4B-F). These results suggested that TCS promoted the TRAIL-sensitivity via increasing the cytomembrane levels of DR4 and DR5 in lung cancer cells.
Our study indicated that the combined treatment of TCS and TRAIL in TRAIL-resistance cells increased TRAIL sensitivity via inhibiting cell proliferation and invasion, inducing apoptosis and S phrase arrest, and upregulating the cytomembrane levels of DR4 and DR5. These results provided a new strategy to enhance TRAIL-sensitivity. Further research on the effects of this combined treatment on both primary and acquired TRAIL-resistance using animal models is on the way. The molecular mechanism, such as DNA methylation and cell autophagy, is to be investigated. Taken together, our study suggests that TCS might act as a potential therapeutic candidate to enhance TRAIL-sensitivity in NSCLC cells.
Supplementary Material
Supplementary figures and tables.
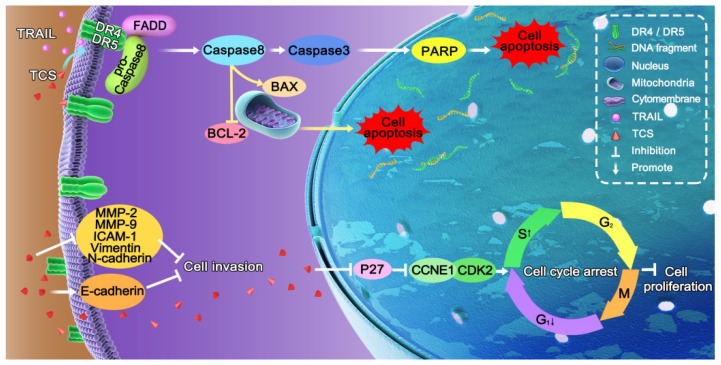
Figure 5.
The molecular mechanism of TCS enhanced TRAIL-sensitivity. TCS promoted the redistribution of DR4 and DR5 to the cytomembrane, and induced both the extrinsic and intrinsic apoptotic pathways mediated by TRAIL. TCS also induced S phrase arrest and inhibited cell proliferation and invasion of TRAIL-resistance cancer cells.
Acknowledgments
The excellent technical assistance of Ms. Xiaohua Leng is gratefully acknowledged.
Grant support
This study was supported in part by grants from the Chinese National Natural Science Foundation (grant No. 81572967 and No. 81372498), Hubei Natural Science Foundation (grant No. 2013CFA006), Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund (grant No. znpy2016050), Wuhan City Huanghe Talents Plan and Chinese National Key Clinical Specialty Construction Program.
Author Contribution
Conceived and designed the study: C.Y., J.Z., and C.X. Performed the study: C.Y., Y.S., S.Z., G.T., N.Z., C.L., X.T., S.M., and Y.L. Analyzed the results: C.Y., Y.S., W.S., and F.W. Contributed reagents/ materials/analysis tools: X.L. Wrote the manuscript: C.Y., Y.G., Y.X., and C.X. All authors reviewed and agreed to the publication of the manuscript.
Abbreviations
- TRAIL
Tumour necrosis factor related apoptosis inducing ligand
- NSCLC
non-small cell lung cancer
- Caspase
cysteineaspartic acid proteasess
- CCK-8
cell counting kit 8
- DD
death domain
- DR
death receptors
- DcR
decoy receptor
- FADD
Fas associated death domain
- PBS
phosphate buffered solution
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TBST
tris buffered saline tween
- PVDF
polyvinylidene fluoride
- PCR
polymearse chain reaction
- FITC
fluorescein isothiocyanate
- PI
propidium iodide
- BCL-2
B cell lymphoma-2
- BAX
BCL-2 associated X protein
- Bid
BH3 interacting domain death agonist
- BAK
BCL2-antagonist/killer 1
- RIP
Ribosome inactivating protein
- PARP
poly ADP-ribose polymerase
- ICAM-1
Intercellular Adhesion Molecule 1
- MMP
Matrix Metallopeptidase.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Cao C, D'Amico T, Demmy T, Dunning J, Gossot D, Hansen H. et al. Surgery versus SABR for resectable non-small-cell lung cancer. The Lancet Oncology. 2015;16:e370–e1. doi: 10.1016/S1470-2045(15)00036-4. [DOI] [PubMed] [Google Scholar]
- 3.Steven R. Wiley, Ken Schooley, Pamela J. Smolak, Din; WS, Huang; C-P, Jillian K. Nicholl, et al. Identification and Characterization of a New Member of the TNF Family that Induces Apoptosis. Immunity; 1995. p. 3. [DOI] [PubMed] [Google Scholar]
- 4.Pitti RM, Marster SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–90. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 5.Hao C, Song JH, Hsi B, Lewis J, Song DK, Petruk KC. et al. TRAIL inhibits tumor growth but is nontoxic to human hepatocytes in chimeric mice. Cancer Res. 2004;64:8502–6. doi: 10.1158/0008-5472.CAN-04-2599. [DOI] [PubMed] [Google Scholar]
- 6.Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov. 2008;7:1001–12. doi: 10.1038/nrd2637. [DOI] [PubMed] [Google Scholar]
- 7.Kischkel F, Lawrence D, Chuntharapai A, Schow P, Kim K, Ashkenazi A. Apo2L/ TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–20. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 8.Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782–98. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- 9.de Miguel D, Lemke J, Anel A, Walczak H, Martinez-Lostao L. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016;23:733–47. doi: 10.1038/cdd.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greco FA, Bonomi P, Crawford J, Kelly K, Oh Y, Halpern W. et al. Phase 2 study of mapatumumab, a fully human agonistic monoclonal antibody which targets and activates the TRAIL receptor-1, in patients with advanced non-small cell lung cancer. Lung Cancer. 2008;61:82–90. doi: 10.1016/j.lungcan.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Hotte SJ, Hirte HW, Chen EX, Siu LL, Le LH, Corey A. et al. A phase 1 study of mapatumumab (fully human monoclonal antibody to TRAIL-R1) in patients with advanced solid malignancies. Clin Cancer Res. 2008;14:3450–5. doi: 10.1158/1078-0432.CCR-07-1416. [DOI] [PubMed] [Google Scholar]
- 12.Merchant MS, Geller JI, Baird K, Chou AJ, Galli S, Charles A. et al. Phase I trial and pharmacokinetic study of lexatumumab in pediatric patients with solid tumor. J Clin Oncol. 2012;30:4141–7. doi: 10.1200/JCO.2012.44.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trivedi R, Mishra DP. Trailing TRAIL Resistance: Novel Targets for TRAIL Sensitization in Cancer Cells. Front Oncol. 2015;5:69. doi: 10.3389/fonc.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim SC, Jeon HJ, Kee KH, Lee MJ, Hong R, Han SI. Involvement of DR4/JNK pathway-mediated autophagy in acquired TRAIL resistance in HepG2 cells. Int J Oncol. 2016;49:1983–90. doi: 10.3892/ijo.2016.3699. [DOI] [PubMed] [Google Scholar]
- 15.Thorburn A, Behbakht K, Ford H. TRAIL receptor-targeted therapeutics: resistance mechanisms and strategies to avoid them. Drug Resist Updat. 2008;11:17–24. doi: 10.1016/j.drup.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sha O, Niu J, Ng TB, Cho EY, Fu X, Jiang W. Anti-tumor action of trichosanthin, a type 1 ribosome-inactivating protein, employed in traditional Chinese medicine: a mini review. Cancer Chemother Pharmacol. 2013;71:1387–93. doi: 10.1007/s00280-013-2096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins EJ, Robertus JD, Mary LoPrestig, Kathy L. Stoneg, Williams KR, Paul Wu, et al. Primary amino acid sequence of alpha-trichosanthin and molecular models for abrin A-chain and alpha-trichosanthin. J Biol Chem. 1990;265:8665–9. [PubMed] [Google Scholar]
- 18.Pan K, Lin Yj, Zhou K, Fu Z, Chen Mh, Huang D. et al. The crystal and molecular structure of trichosanthin at 2.6 A resolution. Sci China B. 1993;36:1069–81. [PubMed] [Google Scholar]
- 19.Byers VS, Levin AS, Malvino A, Waites L, Robins RA, Baldwin RW. A phase II study of effect of addition of trichosanthin to zidovudine in patients with HIV disease and failing antiretroviral agents. AIDS Res Hum Retrovir. 1994;10:413–20. doi: 10.1089/aid.1994.10.413. [DOI] [PubMed] [Google Scholar]
- 20.Sha O, Yew DT, Ng TB, Yuan L, Kwong WH. Different in vitro toxicities of structurally similar type I ribosome-inactivating proteins (RIPs) Toxicol In Vitro. 2010;24:1176–82. doi: 10.1016/j.tiv.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Ng TB, Wong JH, Wang H. Recent progress in research on ribosome inactivating proteins. Curr Protein Pept Sci. 2010;11:37–53. doi: 10.2174/138920310790274662. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Li X, Li JC. Possible mechanisms of trichosanthin-induced apoptosis of tumor cells. Anat Rec (Hoboken) 2010;293:986–92. doi: 10.1002/ar.21142. [DOI] [PubMed] [Google Scholar]
- 23.Nishant Bhatia, McDonald KA, Alan P. Jackman, Dandekar AM. A Simplified Procedure for the Purification of Trichosanthin (A Type 1 Ribosome Inactivating Protein) from Trichosanthes kirilowii Root Tubers. Protein Expr Purif. 1996;7:143–6. doi: 10.1006/prep.1996.0020. [DOI] [PubMed] [Google Scholar]
- 24.Sharma N, Park SW, Vepachedu R, Barbieri L, Ciani M, Stirpe F. et al. Isolation and characterization of an RIP (ribosome-inactivating protein)-like protein from tobacco with dual enzymatic activity. Plant Physiol. 2004;134:171–81. doi: 10.1104/pp.103.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou T-C. Frequently asked questions in drug combinations and the mass-action law-based answers. Synergy. 2014;1:3–21. [Google Scholar]
- 26.Stegehuis JH, de Wilt LH, de Vries EG, Groen HJ, de Jong S, Kruyt FA. TRAIL receptor targeting therapies for non-small cell lung cancer: current status and perspectives. Drug Resist Updat. 2010;13:2–15. doi: 10.1016/j.drup.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA. et al. Safty and antitumor activity of recombinant soluble Apo 2 ligand. J Cli Invest. 1999;104:155–62. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao C, Beguinot F, Condorelli G, Trencia A, Van Meir EG, Yong VW. et al. Induction and intrancellular regulaton of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) mediated apotosis in human malignant glioma cells. Cancer Res. 2001;61:1162–70. [PubMed] [Google Scholar]
- 29.Teng Y, Gao M, Wang J, Kong Q, Hua H, Luo T. et al. Inhibition of eIF2alpha dephosphorylation enhances TRAIL-induced apoptosis in hepatoma cells. Cell Death Dis. 2014;5:e1060. doi: 10.1038/cddis.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan S, Li Y, Yue P, Khuri FR, Sun S-Y. The eIF4E/eIF4G Interaction Inhibitor 4EGI-1 Augments TRAIL-Mediated Apoptosis through c-FLIP Down-regulation and DR5 Induction Independent of Inhibition of Cap-Dependent Protein Translation. Neoplasia. 2010;12:346–56. doi: 10.1593/neo.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mert U, Sanlioglu AD. Intracellular localization of DR5 and related regulatory pathways as a mechanism of resistance to TRAIL in cancer. Cell Mol Life Sci. 2017;74:245–55. doi: 10.1007/s00018-016-2321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Z, McDonald ER, Dicker DT, El-Deiry WS. Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem. 2004;279:35829–39. doi: 10.1074/jbc.M405538200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Gong Y, Ma H, An C, Chen D. Reactive oxygen species involved in trichosanthin-induced apoptosis of human choriocarcinoma cells. Biochem J. 2001;355:653–61. doi: 10.1042/bj3550653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He D, Jin J, Zheng Y, Bruce IC, Tam S, Ma X. Anti-angiogenesis effect of trichosanthin and the underlying mechanism. Biochem Biophys Res Commun. 2013;430:735–40. doi: 10.1016/j.bbrc.2012.11.080. [DOI] [PubMed] [Google Scholar]
- 35.Fang EF, Zhang CZ, Zhang L, Wong JH, Chan YS, Pan WL. et al. Trichosanthin inhibits breast cancer cell proliferation in both cell lines and nude mice by promotion of apoptosis. PLoS One. 2012;7:e41592. doi: 10.1371/journal.pone.0041592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Wang B, Wang Z, Yu S. Trichosanthin down-regulates Notch signaling and inhibits proliferation of the nasopharyngeal carcinoma cell line CNE2 in vitro. Fitoterapia. 2012;83:838–42. doi: 10.1016/j.fitote.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y, Sun Y, Cai Y, Sha O, Jiang W. Trichosanthin reduces the viability of SUDHL2 cells via the activation of the extrinsic and intrinsic apoptotic pathways. Mol Med Rep. 2016;13:403–11. doi: 10.3892/mmr.2015.4531. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Song H, Hu H, Cui L, You C, Huang L. Trichosanthin inhibits DNA methyltransferase and restores methylation-silenced gene expression in human cervical cancer cells. Mol Med Rep. 2012;6:872–8. doi: 10.3892/mmr.2012.994. [DOI] [PubMed] [Google Scholar]
- 39.Li CT, Lin CH, Kao TY, Wu MF, Yeh CS, Yeh KT. et al. The mechanisms of action of Tianhua on antitumor activity in lung cancer cells. Pharm Biol. 2010;48:1302–9. doi: 10.3109/13880201003789432. [DOI] [PubMed] [Google Scholar]
- 40.Cai Y, Xiong S, Zheng Y, Luo F, Jiang P, Chu Y. Trichosanthin enhances anti-tumor immune response in a murine Lewis lung cancer model by boosting the interaction between TSLC1 and CRTAM. Cell Mol Immunol. 2011;8:359–67. doi: 10.1038/cmi.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, El-Deiry WS. Silencing of KILLER/DR5 promotes bioluminescent colon tumor xenograft growth and confers resistance to 5-FU. Cancer Res. 2004;64:6666–72. doi: 10.1158/0008-5472.CAN-04-1734. [DOI] [PubMed] [Google Scholar]
- 42.Yan S, Qu X, Xu L, Che X, Ma Y, Zhang L. et al. Bufalin enhances TRAIL-induced apoptosis by redistributing death receptors in lipid rafts in breast cancer cells. Anticancer Drugs. 2014;25:683–9. doi: 10.1097/CAD.0000000000000095. [DOI] [PubMed] [Google Scholar]
- 43.Delmas D, Rebe C, Micheau O, Athias A, Gambert P, Grazide S. et al. Redistribution of CD95, DR4 and DR5 in rafts accounts for the synergistic toxicity of resveratrol and death receptor ligands in colon carcinoma cells. Oncogene. 2004;23:8979–86. doi: 10.1038/sj.onc.1208086. [DOI] [PubMed] [Google Scholar]
- 44.Bellail AC, Tse MC, Song JH, Phuphanich S, Olson JJ, Sun SY. et al. DR5-mediated DISC controls caspase-8 cleavage and initiation of apoptosis in human glioblastomas. J Cell Mol Med. 2010;14:1303–17. doi: 10.1111/j.1582-4934.2009.00777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu L, Qu X, Zhang Y, Hu X, Yang X, Hou K. et al. Oxaliplatin enhances TRAIL-induced apoptosis in gastric cancer cells by CBL-regulated death receptor redistribution in lipid rafts. FEBS Lett. 2009;583:943–8. doi: 10.1016/j.febslet.2009.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.