ABSTRACT
Contamination from anthropogenic activities has significantly impacted Earth’s biosphere. However, knowledge about how environmental contamination affects the biodiversity of groundwater microbiomes and ecosystem functioning remains very limited. Here, we used a comprehensive functional gene array to analyze groundwater microbiomes from 69 wells at the Oak Ridge Field Research Center (Oak Ridge, TN), representing a wide pH range and uranium, nitrate, and other contaminants. We hypothesized that the functional diversity of groundwater microbiomes would decrease as environmental contamination (e.g., uranium or nitrate) increased or at low or high pH, while some specific populations capable of utilizing or resistant to those contaminants would increase, and thus, such key microbial functional genes and/or populations could be used to predict groundwater contamination and ecosystem functioning. Our results indicated that functional richness/diversity decreased as uranium (but not nitrate) increased in groundwater. In addition, about 5.9% of specific key functional populations targeted by a comprehensive functional gene array (GeoChip 5) increased significantly (P < 0.05) as uranium or nitrate increased, and their changes could be used to successfully predict uranium and nitrate contamination and ecosystem functioning. This study indicates great potential for using microbial functional genes to predict environmental contamination and ecosystem functioning.
KEYWORDS: groundwater microbiome, random forest, ecosystem functioning, environmental contamination, metagenomics, microbial functional gene
IMPORTANCE
Disentangling the relationships between biodiversity and ecosystem functioning is an important but poorly understood topic in ecology. Predicting ecosystem functioning on the basis of biodiversity is even more difficult, particularly with microbial biomarkers. As an exploratory effort, this study used key microbial functional genes as biomarkers to provide predictive understanding of environmental contamination and ecosystem functioning. The results indicated that the overall functional gene richness/diversity decreased as uranium increased in groundwater, while specific key microbial guilds increased significantly as uranium or nitrate increased. These key microbial functional genes could be used to successfully predict environmental contamination and ecosystem functioning. This study represents a significant advance in using functional gene markers to predict the spatial distribution of environmental contaminants and ecosystem functioning toward predictive microbial ecology, which is an ultimate goal of microbial ecology.
INTRODUCTION
Anthropogenic activities have impacted Earth’s biosphere through climate change, contamination of air, water, and soil environments, introduction of invasive species, depletion of natural resources, and alterations of biogeochemical cycling (1, 2). These activities have reduced biodiversity, destabilized ecosystem functions such as carbon (C) and nitrogen (N) cycles, and threatened human health (3–7). A recent study showed that several distinct factors, such as concentrations of sulfate, iron, and dissolved CH4 and H2, might control the composition of groundwater microbiomes and that the microbial functional diversity (FD) could explain groundwater chemistry in a pristine aquifer (8). However, the ecological consequences and mechanisms of environmental contamination in the biodiversity of microbial communities and ecosystem functioning remain largely unclear. Even more challenging is to establish linkages between microbial biodiversity and ecosystem functioning.
It is generally believed that FD is better than taxonomic diversity (TD) and/or phylogenetic diversity (PD) for predicting ecosystem functioning (9–12). For example, a recent study across a gradient of sites from the subarctic to the tropics showed that a reduction of decomposer FD consistently decreased the rate of litter decomposition and carbon and nitrogen cycling (13). However, how to select molecular functional predictors (e.g., functional genes) remains a challenging question (11). Functional gene arrays (e.g., GeoChip) target key genes involved in geochemical cycles, bioremediation, stress responses, and other environmental processes and have been widely used to functionally profile microbial communities (14–19). Therefore, GeoChip is an ideal tool to examine the impacts of environmental contaminants on groundwater microbiomes.
The Oak Ridge Integrated Field Research Challenge (OR-IFRC) experimental site, located in Bear Creek Valley, Oak Ridge, TN, is a legacy site for the early development of enriched uranium (U) under the Manhattan Project. At this site, numerous studies have been conducted to examine the impact of contaminants on biological communities and ecosystem functioning (20–29). For example, a metagenome analysis of FW106, a highly contaminated well, showed that high relative levels of abundance of key genes encoding geochemical resistance functions were required for microbial survival in the presence of known environmental contaminants at the site (20). Also, key functional groups have been isolated and identified from the OR-IFRC site, including sulfate-reducing bacteria (SRB), nitrate-reducing bacteria (NRB), and metal-reducing bacteria (MRB) like Anaeromyxobacter, Clostridium, Desulfovibrio, Desulfitobacterium, Geobacter, Hyphomicrobium, Intrasporangium, Pseudomonas, and Rhodanobacter species (20, 23, 25–29). Recently, groundwater from 93 noncontaminated and contaminated wells along the Bear Creek Valley at the OR-IFRC site were sampled. Those wells had a wide range of environmental gradients and associated ecosystem data (22), thus making it possible to use microbial community data for predicting groundwater contamination. The results showed that 16S rRNA gene-sequencing analysis of groundwater microbiomes could accurately identify environmental contaminants (e.g., uranium or nitrate) at the OR-IFRC site (22). However, taxonomic information alone may not be enough to reflect the functional aspects of microbial communities or ecosystems, as not all members of a taxon may carry certain functional genes, making it difficult to predict geochemical properties, especially ecosystem functioning. The following important questions remain to be addressed. (i) How does the functional diversity of groundwater microbiomes change across a range of environmental gradients (e.g., pH, uranium, and nitrate)? (ii) What specific functional genes/populations are stimulated under high concentrations of uranium and nitrate? (iii) Is it possible to predict environmental contamination (e.g., uranium or nitrate) and ecosystem functioning using microbial functional genes?
In this study, we hypothesized the following: (i) FD would decrease with increased environmental contamination (e.g., uranium or nitrate) or a significant change in environmental conditions (e.g., pH); (ii) under conditions of uranium and nitrate contamination, the abundance of some key functional genes/populations (e.g., dsrA and cytochrome genes for uranium reduction or nirK and napA for nitrate reduction) would increase, while the rest would decrease or remain unchanged; and (iii) the relationship between FD and environmental contamination or ecosystem functioning would be predictable based on key microbial functional genes. To test those hypotheses, we used a new version of a functional-gene microarray (GeoChip 5.0) to analyze groundwater microbiomes from 69 wells at the OR-IFRC site. GeoChip is able to quantitatively detect known microbial functions but generally does not target unknown functions from known or unknown microbial groups. Our results indicate that the overall FD decreased as uranium (but not nitrate) concentrations increased or at low or high pH; however, some specific functional genes/populations were stimulated in response to uranium and nitrate contamination. Such microbial functional genes could be used to successfully predict uranium and nitrate contamination and ecosystem functioning. This study provides new insights for our understanding of the impacts of environmental contaminants on groundwater microbiomes and demonstrates the predictive power of microbial functional genes for environmental contamination and ecosystem functioning.
RESULTS
Geochemical properties and ecosystem function indicators.
A total of 38 environmental variables were measured, including pH, contaminant (e.g., uranium and nitrate) concentrations, dissolved gases (e.g., CO2, CH4, N2O, and H2S) as ecosystem function indicators, dissolved C and N, and direct cell counts, which were largely used in this study (see Table S1 in the supplemental material). The 69 wells had wide ranges of uranium, nitrate, and pH levels, with uranium at 0 to 55.3 mg/liter (average, 1.5 mg/liter), nitrate at 0 to 11,648 mg NO3−-N (nitrate as nitrogen)/liter (average, 641 mg NO3−-N/liter), and pHs of 3 to 10.5 (average, pH 6.9). Furthermore, wells with high concentrations of uranium (e.g., >3 mg/liter) also had high concentrations of nitrate (1,516 to 11,648 mg NO3−-N/liter) and low pHs (3.0 to 5.2). Also, dissolved gases varied greatly, with CO2 at 0 to 29,739 mg/liter (average, 476 mg/liter), N2O at 0 to 1.2 mg/liter (average, 0.1 mg/liter), CH4 at 0 to 0.6 mg/liter (average, close to 0 mg/liter), and H2S at 0 to 4.2 mg/liter (average, 0.1 mg/liter). The amounts of bacterial biomass in groundwater samples ranged from 3.5 × 102 to 1.8 × 106 cells/ml (average, 1.2 × 105 cells/ml), while the levels of dissolved organic carbon (DOC) were 0.2 to 128.2 mg/liter (average, 7.8 mg/liter) and the levels of dissolved inorganic carbon (DIC) were 9.4 to 179.2 mg/liter (average, 58.3 mg/liter). Such large ranges of environmental gradients provide an advantage in testing the relationships between functional gene diversity and environmental contamination, as well as ecosystem functioning.
Key geochemical and ecosystem data for the 69 wells selected. Download TABLE S1, DOCX file, 0.03 MB (32.7KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The relationships between functional richness/diversity/abundance, microbial biomass, and contaminant concentrations.
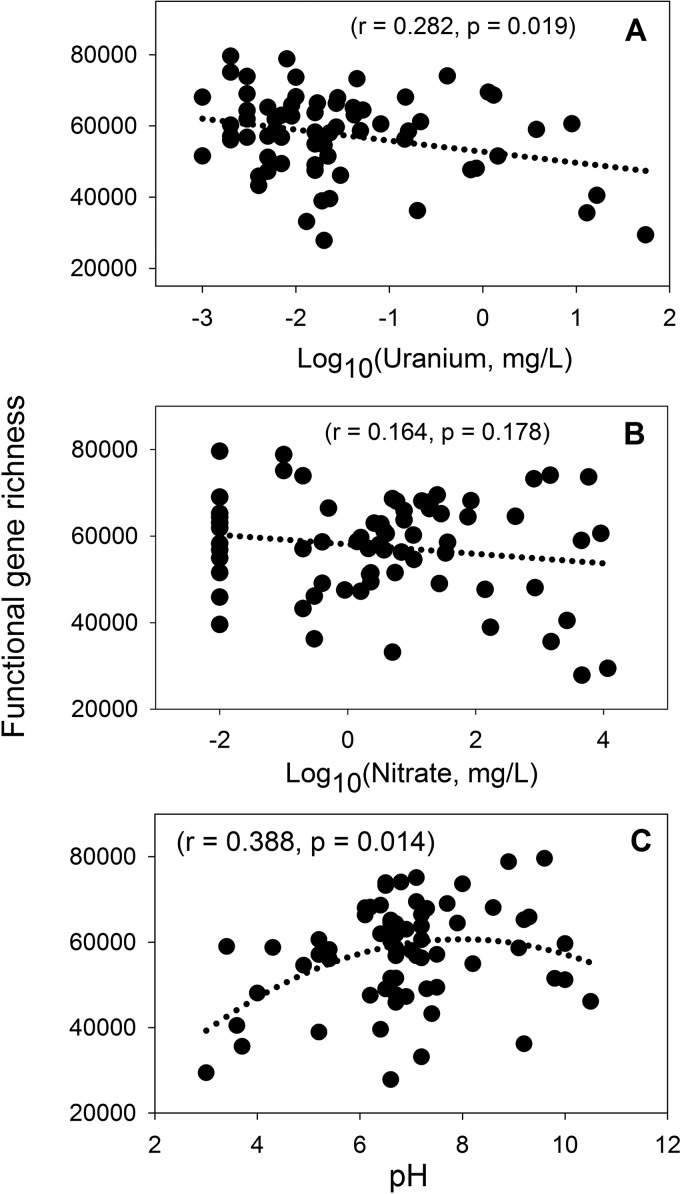
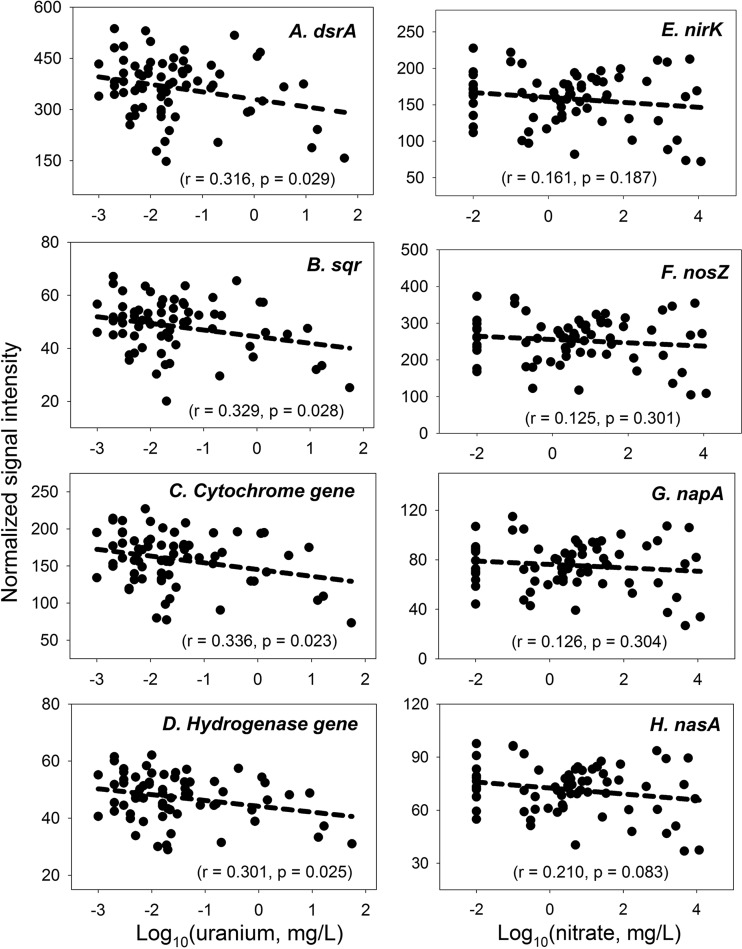
For this study, we defined functional richness as the number of functional genes detected by GeoChip 5.0 and functional diversity as the Shannon diversity index. The overall levels of functional gene richness and diversity decreased significantly (P < 0.05) as uranium concentrations increased. The functional diversity was highest at a neutral pH, not under low- or high-pH conditions, but it was not significantly (P < 0.05) impacted by nitrate concentrations in groundwater (Fig. 1; Fig. S1). We further examined the relationships between the levels of abundance of key gene families along the environmental gradients. For example, the abundances of sulfur (S) cycling genes (e.g., dsrA and sqr) and cytochrome and hydrogenase genes decreased significantly (P < 0.05) with increasing uranium concentrations (Fig. 2A to D). However, the abundances of denitrification (e.g., nirK and nosZ), dissimilatory N reduction (e.g., napA), and assimilatory N reduction (e.g., nasA) genes did not decrease significantly (r = 0.125 to 0.210, P > 0.05) with increasing nitrate concentrations (Fig. 2E to H). Further analysis of other key N cycling genes showed significantly (P < 0.05) decreased abundances with increased uranium or at low or high pHs, but no significant (P > 0.05) correlations were observed between N cycling gene abundances and nitrate concentrations (Table S2). In addition, the effects of uranium and pH on microbial biomass (measured by direct cell count) were not significant (P > 0.05), nor was there a significant correlation (P > 0.05) between biomass and functional richness, but it appeared that microbial biomass increased significantly (P = 0.001) with increased nitrate concentrations, suggesting that nitrate consumers (e.g., nitrate reducers) may be dominant in the environment (Fig. S2). Further analysis showed that the abundance of ~95% of genes detected by GeoChip 5.0 decreased, while only about 5% of them increased, indicating that most of the functional genes were inhibited or remained unchanged as uranium and nitrate concentrations increased.
FIG 1 .
Relationships between the overall functional richness and concentrations of uranium (A) and nitrate (B), as well as pH (C), in groundwater. Uranium and nitrate concentrations were first log transformed, and then linear regressions were performed for functional richness and uranium or nitrate concentrations. Nonlinear regression was used for functional richness and pH.
FIG 2 .
Linear relationships between the levels of abundance of specific functional gene families and log-transformed Uranium (A to D) or nitrate (E to H) concentrations in groundwater, including data for dsrA, encoding the alpha subunit of sulfite reductase for dissimilatory sulfite reduction (A), sqr, encoding sulfide-quinone reductase (B), cytochrome genes from well-known organisms, e.g., Geobacter, Anaeromyxobacter, Dechloromonas, Desulfovibrio, Shewanella, Desulfurobacterium, Desulfobacterium, Rhodobacter, Pseudomonas, Enterobacter, and Ochrobactrum (C), hydrogenase genes from well-known organisms, e.g., Geobacter, Desulfovibrio, Desulfurobacterium, Desulfobacterium, and Rhodobacter (D), nirK, encoding nitrite reductase for denitrification (E), nosZ, encoding nitrous oxide reductase for denitrification (F), napA, encoding nitrate reductase for dissimilatory nitrate reduction (G), and nasA, encoding nitrate reductase for assimilatory nitrate reduction (H).
Relationships between the overall functional diversity (Shannon diversity index) and concentrations (log transformed) of uranium (A) and nitrate (B), as well as pH (C), in groundwater. Linear regression was used for the Shannon index and uranium or nitrate concentrations, and nonlinear regression for the Shannon index and pHs. Download FIG S1, TIF file, 0.7 MB (697.2KB, tif) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relationships between log-transformed uranium (A) or nitrate (B) concentrations, pH (C), or functional richness (D) and groundwater microbial biomass determined by the acridine orange direct count (AODC) method. Linear regression was used for such relationships. Download FIG S2, TIF file, 0.5 MB (521KB, tif) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Regressions of key nitrogen cycling gene abundance with uranium, nitrate, or pH. Uranium and nitrate concentrations were first log transformed and then used for linear regressions, while nonlinear regression without log transformation was used for pHs. P values of <0.05 are in boldface. Download TABLE S2, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Key functional populations stimulated in response to a uranium gradient.
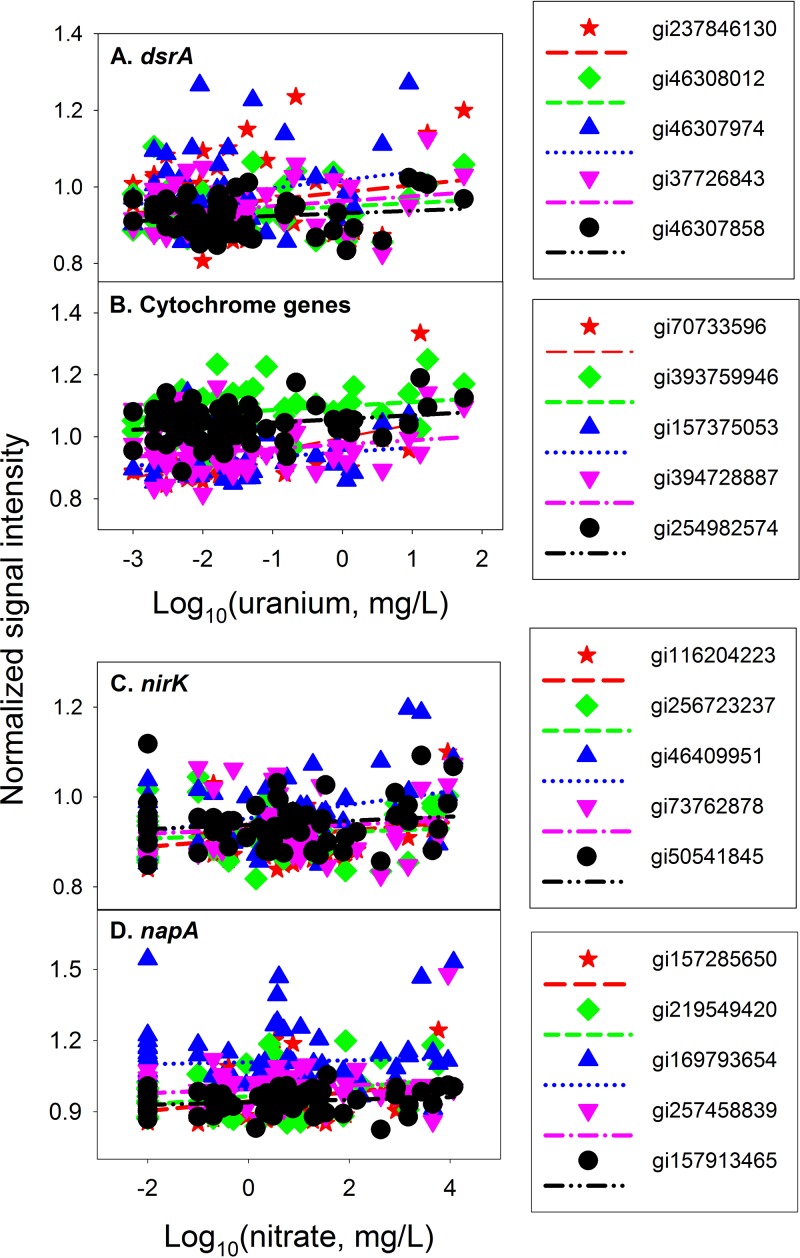
Although the richness and diversity of functional genes generally decreased as uranium concentrations increased in groundwater, some specific populations of certain functional gliders did increase significantly (P < 0.05) (Fig. 3A and B; Table S3). For example, the abundance of 43 dsrA-bearing populations (~5.8% of total dsrA detected by GeoChip 5), mostly uncultured SRB with a few sequenced species (e.g., Halorhodospira halophila, Desulfobulbus propionicus, Pelodictyon luteolum, and Vibrio rotiferianus), increased significantly (P < 0.05) (Table S3). In particular, five abundant dsrA probes/gene variants (gi237846130, gi46308012, gi46307974, gi37726843, and gi46307858) derived from uncultured SRB were identified as being significantly (P < 0.05) increased as uranium increased (Fig. 3A). Increased levels of abundance of 21 cytochrome (~4.6%) and 6 hydrogenase (~7.3%) gene variants were also observed, specifically from well-known microorganisms like Geobacter, Dechloromonas, Enterobacter, Pseudomonas, Alcaligenes, Desulfovibrio, Desulfitobacterium, Rhodobacter, Ochrobactrum, and Anaeromyxobacter (Table S3). Also, five abundant cytochrome genes (gi70733596, gi393759946, gi157375053, gi394728887, and gi254982574) were significantly (P < 0.05) increased as uranium concentrations increased in groundwater (Fig. 3B). These stimulated populations could play important roles in uranium bioremediation at this site.
FIG 3 .
Significantly (P < 0.05) positive correlations between the levels of abundance of stimulated populations and log-transformed uranium (A and B) or nitrate (C and D) concentrations, including data for dsrA gene variants gi237846130, gi46308012, gi46307974, gi37726843, and gi46307858, derived from uncultured sulfate-reducing bacteria (A), cytochrome genes gi70733596 from Pseudomonas fluorescens, gi393759946 from Alcaligenes faecalis, gi157375053 from Shewanella sediminis, gi394728887 from Enterobacter sp., and gi254982574 from Geobacter sp. (B), nirK gene variants gi116204223 from Chaetomium globosum, gi256723237 from Nectria haematococca, and gi46409951, gi73762878, and gi50541845 from uncultured denitrifying bacteria (C), and napA gene variants gi219549420 from Vibrio parahaemolyticus, gi257458839 from Campylobacter gracilis, gi157913465 from Dinoroseobacter shibae, and gi157285650 and gi169793654 from uncultured nitrate-reducing bacteria (D).
Relationships between the abundances of significantly increased or decreased populations (bearing key genes) and uranium concentrations by linear regression. Significantly increased slopes are in boldface, and the relative abundances are presented as mean ratios. Download TABLE S3, DOCX file, 0.03 MB (33.3KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Key functional populations stimulated in response to a nitrate gradient.
We also found that the abundance of many specific functional genes/populations involved in N cycling increased significantly (P < 0.05) as nitrate increased (Fig. 3C and D; Table S4). For example, the abundance of 13 nirK-bearing (4.9%) populations increased significantly (P < 0.05), with most being uncultured bacteria and a few sequenced microbes (e.g., Chaetomium, Arthroderma, Nectria, and Pseudomonas); the abundance of 9 napA (6.0%) gene variants for dissimilatory N reduction, derived from Beggiatoa, Vibrio, Campylobacter, and Dinoroseobacter species, as well as uncultured NRB, also increased significantly (P < 0.05) as nitrate increased (Table S4). Five abundant nirK gene variants (gi116204223, gi256723237, gi46409951, gi73762878, and gi50541845) (Fig. 3C) and five abundant napA gene variants (gi157285650, gi219549420, gi169793654, gi257458839, and gi157913465) increased significantly (P < 0.05) as nitrate increased (Fig. 3D). In addition, populations stimulated by high concentrations of nitrate were observed for other N cycling genes, such as amoA, nifH, narG, nirS, norB, nasA, nosZ, and nrfA (Table S4). These stimulated populations are expected to play important roles in bioremediation of this nitrate-contaminated site.
Relationships between the abundances of significantly increased or decreased populations (bearing key genes) and nitrate concentrations by linear regression. Significantly increased slopes are in boldface, and the relative abundances are presented as mean ratios. Download TABLE S4, DOCX file, 0.1 MB (54.1KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prediction of uranium contamination in groundwater using microbial functional genes.
As significant relationships were observed between functional richness, diversity, and/or populations and uranium concentrations in groundwater, we attempted to predict groundwater contamination by the presence of microbial functional genes using random forest, a machine learning method (30). First, we selected a total of 2,361 of the functional genes detected that could predict uranium contamination on the basis of being involved in S cycling and electron transfer (e.g., dsrA, dsrB, sir, cytochrome, hydrogenase, and cytochrome P-450 genes). Cross-validation by out-of-bagging (OOB) estimation of errors for classification of uranium contamination was 28.99%. Second, we selected a subset of 1,521 specific functional genes from the first set of 2,361 genes for predicting uranium contamination, including 892 dsrA, 536 cytochrome, and 93 hydrogenase genes. OOB estimation of errors was 24.64% for all three functional gene families and 24.64%, 26.09%, and 28.99% for dsrA, cytochrome, and hydrogenase genes, respectively, indicating that the best predictor for uranium contamination was dsrA or a combination of all three gene families, each with an error rate of 24.64%. Third, we used the significantly changed populations bearing the best predictor, dsrA (Table S3), and the same results were observed for uranium contamination prediction (Table 1). To further improve our prediction, we used the area under the receiver operating characteristic curve as the predictive accuracy for random forest (AUC-RF) (31) to automatically select 50 predictors (Table S5) from the initial 2,361 functional probes related to uranium reduction, which dramatically decreased the OOB estimate of error rate, from 28.99% to 11.59% (Table 1). These results indicated that microbial functional genes were able to successfully predict groundwater uranium contamination.
TABLE 1 .
Performance of the random forest model for predicting environmental contamination by uranium or nitrate in 69 wells at the OR-IFRC site using microbial functional genes as predictors
| Contaminant | Predictora | OOB error rate (%) |
No. of wells predicted/no. of wells defined |
|
|---|---|---|---|---|
| Background wellsb | Contaminated wellsc | |||
| Uranium | All S cycling and metal-related genes | 28.99 | 47/47 | 2/22 |
| All dsrA, cytochrome, and hydrogenase genes | 24.64 | 47/47 | 5/22 | |
| All dsrA genes | 24.64 | 47/47 | 5/22 | |
| All cytochrome genes | 26.09 | 46/47 | 5/22 | |
| All hydrogenase genes | 28.99 | 41/47 | 8/22 | |
| Key dsrA, cytochrome, and hydrogenase genes | 27.54 | 45/47 | 5/22 | |
| Key dsrA genes | 24.64 | 45/47 | 7/22 | |
| Key cytochrome genes | 39.13 | 38/47 | 4/22 | |
| Key hydrogenase genes | 42.03 | 33/47 | 7/22 | |
| AUC-RF selection | 11.59 | 47/47 | 14/22 | |
| Nitrate | All N cycling genes | 36.23 | 39/44 | 5/25 |
| All nifH, amoA, narG, nasA, and napA genes | 34.78 | 40/44 | 5/25 | |
| All nifH genes | 33.33 | 41/44 | 5/25 | |
| All amoA genes | 27.54 | 41/44 | 9/25 | |
| All narG genes | 36.23 | 40/44 | 4/25 | |
| All nasA genes | 36.23 | 37/44 | 7/25 | |
| All napA genes | 34.78 | 41/44 | 4/25 | |
| Key nifH, amoA, narG, nasA, and napA genes | 30.43 | 40/44 | 8/25 | |
| Key nifH genes | 27.54 | 41/44 | 9/25 | |
| Key amoA genes | 28.99 | 39/44 | 10/25 | |
| Key narG genes | 37.68 | 37/44 | 6/25 | |
| Key nasA genes | 40.58 | 32/44 | 9/25 | |
| Key napA genes | 40.58 | 32/44 | 9/25 | |
| AUC-RF selection | 15.94 | 42/44 | 16/25 | |
Key functional genes detected from each family are listed in Tables S3 and S4 in the supplemental material.
In background wells, the concentrations of uranium or nitrate were 30 µg/liter or below or 10 mg/liter or below, respectively.
In contaminated wells, the concentrations of uranium or nitrate were higher than 30 µg/liter or 10 mg/liter, respectively.
Fifty predictors from 2,361 detected functional genes related to uranium reduction automatically selected by AUS-RF for predicting uranium contamination in groundwater. Items in boldface were also identified as belonging to populations significantly increased/decreased with increasing uranium concentrations in groundwater (see Table S3). Download TABLE S5, DOCX file, 0.02 MB (19.2KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prediction of nitrate contamination in groundwater using microbial functional genes.
Similarly, we predicted nitrate contamination in groundwater. First, we selected a total of 5,273 functional genes involved in N cycling and showed that the error rate for nitrate contamination prediction was 36.23%. Second, we selected a subset of 2,239 specific functional genes from that first set that were involved in N fixation (1,044 nifH genes), nitrification (173 amoA genes), denitrification (705 narG genes), and assimilatory (134 nasA genes) and dissimilatory (183 napA genes) N reduction, and the error rates were 34.79% for all the gene families selected and 33.33%, 27.54%, 36.23%, 36.23%, and 34.78%, respectively, for individual functional gene families, indicating that the best predictor for nitrate contamination was amoA, with an error rate of 27.54%. Third, we used the best predictor, amoA, and the significantly changed populations bearing it for the same prediction, and the error rate for nitrate contamination prediction was 28.99% (Table 1), which was not an improvement from the previous test. To reduce the collinearity, we again used AUC-RF (31) to automatically select 54 predictors (Table S6) from the original 5,273 N cycling genes. This substantially improved our prediction, decreasing the OOB estimate of error rate to 15.94% (Table 1). These results indicated that microbial functional genes were able to accurately predict nitrate contamination in groundwater.
Fifty-four predictors from 5,273 detected N cycling genes automatically selected by AUC-RF for predicting nitrate contamination in groundwater. Items in boldface were also identified as belonging to populations significantly increased/decreased with increasing nitrate concentrations in groundwater (see Table S4). Download TABLE S6, DOCX file, 0.02 MB (19.9KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prediction of ecosystem functioning using microbial functional genes.
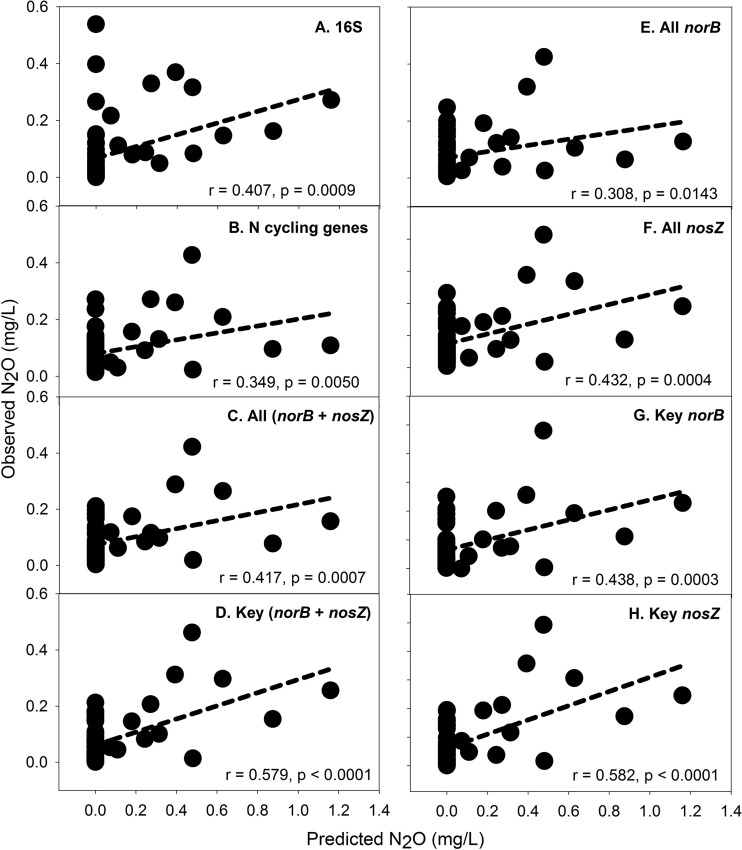
We also attempted to select specific microbial functional genes, as well as 16S rRNA genes (for a comparison), to predict ecosystem functions that may be occurring based on the concentrations of dissolved gases (e.g., CO2, CH4, and N2O) in the groundwater (Table S1). No significant correlations were observed either between the predicted CH4 concentration and the observed CH4 concentration or between the predicted CO2 concentration and the observed CO2 concentration (data not shown). However, when 16S rRNA genes, N cycling genes, all norB or nosZ genes, key norB or nosZ genes, all norB plus nosZ genes, or key norB plus nosZ genes were used to predict N2O concentrations in groundwater, significant correlations between the predicted N2O concentration and the observed N2O concentration were evident, and among those sets of genes or combinations of genes, key norB plus nosZ genes or key nosZ genes were the best predictors for N2O concentrations in groundwater based on the r and P values of linear regressions (Fig. 4). The results suggest that microbial functional genes are potentially useful and better than 16S rRNA genes for predicting ecosystem functions (e.g., N2O concentrations in groundwater).
FIG 4 .
Random forest predictions of N2O concentrations in groundwater using different sets of genes, including 16S rRNA genes (A); all N cycling genes (B); all norB and nosZ genes (C); key (significantly increased/decreased) norB and nosZ genes (D); all norB genes (E); all nosZ genes (F); key norB genes (G); and key nosZ genes (H). All norB and nosZ key genes are listed in Table S4 in the supplemental material.
DISCUSSION
Understanding the impacts of contaminants on biological communities and predicting the effects of those communities on ecosystem functioning are important topics in ecology and environmental management. In this study, we surveyed the functional diversity and composition of groundwater microbial communities and their linkages with environmental contamination or ecosystem functioning at the OR-IFRC experimental site. Our results showed that the overall functional diversity/richness of groundwater microbiomes decreased as uranium (but not nitrate) concentrations increased or at low or high pHs. However, some specific functional genes/populations involved in uranium and/or nitrate reduction and denitrification were stimulated, and these functional genes could be used to predict environmental contamination (e.g., uranium or nitrate) and ecosystem functioning. In addition, unlike previous studies, which only had a limited number of samples/wells, this study analyzed 69 microbial communities from a large range of environmental gradients (e.g., uranium, nitrate, and pH), providing a more robust picture of the impact of human activities on biodiversity. The experimental results from this study generally support our hypotheses (with the exception of the relationship between nitrate and functional diversity).
Our first hypothesis was that the overall functional diversity/richness of groundwater microbiomes would decrease with an increase in environmental contamination (e.g., uranium or nitrate) or under extreme pH conditions. A previous clone library analysis of nirS and nirK genes from the same site found that novel nirK and nirS sequences were present in the contaminated groundwater and that the diversity of both gene families changed with contaminant (e.g., uranium or nitrate) concentrations (32). Also, a comparison of metagenomes from FW106 (a highly contaminated well) and FW301 (a background well) revealed that long-term exposure to low pHs and high concentrations of uranium, nitrate, and organic solvents resulted in decreased species diversity and loss of functional diversity (20, 24). Additionally, GeoChip analysis of a landfill leachate-contaminated aquifer showed that leachate from an unlined landfill impacted the diversity, composition, structure, and functional potential of groundwater microbiomes as a function of groundwater pH, DOC, and concentrations of sulfate and ammonia (33). In this study, we found that the overall functional diversity of groundwater microbial communities decreased under uranium contamination or extreme pH conditions, which is consistent with previous observations in groundwater (20, 32–36), as well as in the soil environment (37–40). Several possible mechanisms might be responsible for such a reduction in the functional diversity/richness. First, most microorganisms may not have developed efficient strategies for surviving/growing in such stressed environments, so their abundances would decrease to below detection level or even to extinction (20, 24). Second, if there are no appropriate mechanisms to deal with high uranium concentrations in the environment, uranium may accumulate in or be deposited on the cell surface, which could directly or indirectly inhibit specific key functional genes/enzymes, as well as associated pathways (41), resulting in a decrease in functional richness/diversity. Third, low pHs might reduce intracellular pH and disrupt the chemiosmotic gradient (42), impairing cellular metabolism. Fourth, high concentrations of uranium and nitrate and low pHs coexist in some wells (e.g., FW-021, FW-106, FW-126, and FW-410), which may cause additive impacts, further reducing the overall functional diversity/richness. These possibilities may lead to a decreased functional richness/diversity of groundwater microbial communities. However, the functional richness/diversity of certain specific gene families did not decrease significantly as nitrate concentrations increased. One possible explanation is that most microbes (e.g., nitrate reducers) might use nitrate or related N compounds (e.g., NO2−, NO, N2O, or NH4+) as electron donors/acceptors and sources of energy and assimilatory N, so that they were able to cope with such high nitrate concentrations. Indeed, a previous study indicated that elevated nitrate could stimulate microorganisms, especially those with diverse metabolic capabilities (43). Therefore, our results generally support the hypothesis that the overall functional richness/diversity of groundwater microbial communities decreases as uranium concentrations increase or under extreme pH conditions in groundwater.
Although the overall functional diversity/richness decreased as uranium concentrations increased or remained unchanged as nitrate concentrations increased, some key functional genes/populations involved in uranium or nitrate reduction/resistance would be expected to increase under high concentrations of uranium and nitrate. The dsrA gene, encoding the alpha subunit of dissimilatory sulfite reductase, an SRB biomarker indicating the ability to reduce sulfate and heavy metals (e.g., uranium) (44–47), and cytochrome genes (48, 49) were enriched. Previous studies also indicated that some of these functional genes/populations were stimulated under conditions of high concentrations of heavy metals (e.g., uranium and chromate) in this OR-IFRC site (50–53), the Uranium Mill Tailings Remedial Action site in Rifle, CO (54), and the chromate-contaminated Hanford site (55), suggesting the important role of these functions in metal (e.g., uranium and chromate) reduction. As nitrate is an important nutrient and electron acceptor for microorganisms, adequately high concentrations of nitrate in groundwater are expected to stimulate N cycling genes and associated processes. For example, a recent study indicated that elevated nitrate could enrich functional genes involved in C, N, S, and phosphorus (P) cycling, thus leading to the potential in situ bioremediation of polybrominated diphenyl ether (PBDE)- and polycyclic aromatic hydrocarbon (PAH)-contaminated sites (43). In the current study, we found that the abundances of about 5 to 6% dsrA, cytochrome, and N cycling genes were positively correlated with the uranium or nitrate concentrations. These genes were largely derived from SRB, NRB, and MRB, particularly those microorganisms with versatile metabolic capabilities (e.g., Rhodanobacter, Geobacter, Pseudomonas, Alcaligenes, Desulfovibrio, Desulfitobacterium, Rhodobacter, and Anaeromyxobacter). Some of these key microorganisms have been isolated from the OR-IFRC site (23, 25–29), and several key genes have been identified by shotgun metagenome sequencing (20, 24). The results generally support our second hypothesis, that key functional genes/populations involved in uranium reduction, nitrate reduction, and denitrification could be stimulated under high concentrations of uranium and nitrate. These significantly increased or decreased functional genes or populations were used to predict uranium and nitrate contamination and ecosystem functioning in this study, as they are expected to play important roles in this groundwater system.
Two recent studies compared different machine learning methods, one aimed at finding predictors of bacterial vaginosis (56) and the other at identifying environmental sensors in groundwater contamination (22), and both showed that random forest was a suitable approach for predictive analysis of microbial communities. Another study showed that 16S rRNA gene sequencing data of human fecal communities were good predictors of a city’s obesity level using random forest algorithms (57). Also, 16S rRNA gene sequencing of fecal samples was used to distinguish pediatric patients with inflammatory bowel disease (IBD) from patients with similar symptoms (58). At the OR-IFRC site, a recent study found that 16S rRNA gene sequencing data could be used to successfully predict most (26 out of 38) of the groundwater geochemical properties, such as uranium and nitrate concentrations and pHs (22). Although all these studies used 16S rRNA genes as predictors, it is believed that functional genes may be better predictors of ecosystem functions. Currently, some challenges remain in the use of functional genes as predictors. One challenge is to determine which functional genes or sets of functional genes are appropriate choices for given functions, phenotypes (e.g., disease), or processes (e.g., CO2 production), and another challenge is to accurately identify or measure a specific phenotype or functional process.
In this study, our results indicated that uranium and nitrate contamination were accurately predicted, specifically with AUC-RF (31), and we also successfully predicted dissolved N2O in groundwater. However, several challenges still remain in predicting other ecosystem functions, such as CO2 and CH4 concentrations in groundwater. First, only a few wells had relatively high concentrations of CH4 or CO2, while most wells had undetectable concentrations of these gases in the groundwater. Such a skewed distribution of data may affect our prediction accuracy. Second, the high diversity of functional genes/populations may present multiple instances of collinearity in the community, thus compromising our predictions. Indeed, when we used AUC-RF to reduce collinearity, the prediction error rates decreased dramatically, from approximately 29% to 12% for uranium contamination and from 36% to 16% for nitrate contamination. Third, it is hard to identify the specific functional genes responsible for some general functional processes. For example, groundwater CO2 could be generated from many C decomposition pathways and other physical or chemical pathways or consumed by autotrophy and chemical reactions, making it difficult to select specific genes for predicting this functional process and, thus, limiting the predictive power. Fourth, the relationship between dissolved gases and functional gene abundance may be subtle. The concentrations of gases in groundwater may not accurately reflect ecosystem functioning, or functional gene abundance may not reflect actual activity. Perhaps due to these challenges, a recent study also showed that adding functional information did not improve classification accuracy (59). Therefore, to accurately predict ecosystem functioning, more studies need to be conducted to optimize methods, select appropriate functional predictors, reduce skewed sample distribution, decrease multiple incidences of collinearity, and/or increase the reliability of ecosystem functional process data.
Conclusions.
Our results indicated that the overall functional richness/diversity decreased with increased uranium (but not nitrate) concentrations or at low or high pHs. Some specific functional genes/populations were stimulated under high concentrations of uranium or nitrate and could be used to successfully predict uranium and nitrate contamination and, potentially, ecosystem functioning. This study provides new insights for our understanding of the impacts of environmental contaminants on the functional richness/diversity of groundwater microbiomes and demonstrates the predictive power of microbial functional genes to identify environmental contamination and ecosystem functioning.
MATERIALS AND METHODS
More detailed descriptions of the site, sampling methods, physical, geochemical and microbiological measurements, groundwater biomass collection, DNA extraction, and random forest analysis was provided previously (22).
Site description and sampling.
The U.S. Department of Energy’s (DOE) Oak Ridge Integrated Field Research Challenge (OR-IFRC) site has a 243-acre contaminated area and a 402-acre uncontaminated background area located within the Bear Creek Valley watershed in Oak Ridge, TN. This site has been contaminated with radionuclides (e.g., uranium and technetium), nitrate, sulfide, and volatile organic compounds. The major source of contamination is the former S-3 waste disposal ponds within the Y-12 national security complex, which has been continuously monitored and documented over the past several decades (25, 60). Further information regarding the plume and sources of contamination can be found at https://public.ornl.gov/orifc/orfrc1_fieldchallenge.cfm.
Physical, geochemical, and microbiological measurements.
In this study, 93 groundwater wells were carefully selected to cover the maximum geochemical diversity of this site without exhaustively sampling all available wells. However, we were only able to obtain enough DNA from 69 wells for GeoChip analysis (see Table S1 in the supplemental material). Groundwater samples were collected from the OR-IFRC experimental site between November 2012 and February 2013. A variety of physical, geochemical, and microbiological properties were measured on site or in the laboratory as previously described (22); a brief summary follows. (i) Bulk water parameters, including temperature, pH, dissolved oxygen (DO), conductivity, and redox, were measured at the wellhead using an In-Situ Troll 9500 sensor (In-Situ, Inc., Fort Collins, CO). (ii) Dissolved gases, including He, H2, N2, O2, CO, CO2, CH4, and N2O, were measured on an SRI 8610C gas chromatograph with argon carrier gas using a method derived from EPA RSK-175 and USGS Reston Chlorofluorocarbon Laboratory procedures. (iii) Dissolved organic carbon (DOC) and inorganic carbon (DIC) concentrations were determined with a Shimadzu TOC-V CSH analyzer (Tokyo, Japan). (iv) Anions, including bromide, chloride, nitrate, phosphate, and sulfate, were determined using a Dionex 2100 with an AS9 column and carbonate eluent. (v) Concentrations of metals (and trace elements) in the groundwater were determined on an inductively coupled plasma-mass spectrometry (ICP-MS) instrument (Elan 6100) (61). Finally, (vi) the amounts of bacterial biomass in groundwater samples were determined using the acridine orange direct count (AODC) method (62).
Groundwater biomass collection, DNA extraction, and template preparation.
Microbial biomass was collected and DNA extracted as described previously (11). Briefly, 4.0 liters of groundwater was filtered through 0.2-µm filters to collect biomass. Filters containing biomass were placed into 50-ml Falcon tubes, immediately stored on dry ice, transferred to the laboratory, and stored at −80°C until DNA extraction. DNA was extracted and purified using a modification of the Miller method (62).
GeoChip hybridization and data preprocessing.
The GeoChip 5.0 microarray chip contains 167,044 distinct functional gene probes, covering 395,894 coding sequences (CDS) from ~1,600 functional gene families involved in microbial carbon (e.g., degradation, methane metabolism, and fixation) and nitrogen (e.g., nitrification, denitrification, reduction, and fixation) cycling, electron transfer, organic remediation, secondary metabolism, stress responses, and virulence. To obtain sufficient DNA for microarray analysis, 10 ng of template DNA from each sample was amplified using whole-community genome amplification (WCGA) (63). After amplification, 2.5 μg of DNA was labeled, resuspended in hybridization buffer, and hybridized on a GeoChip 5.0 microarray chip with 10% formamide at 67°C for 24 h in an Agilent microarray hybridization oven (Agilent Technologies, Santa Clara, CA). The array was then washed, dried, and scanned at 100% laser power at wavelengths of 532 nm and 635 nm. Intensity data were collected using the Agilent Feature Extraction program. Raw intensity data were uploaded to the Functional Gene Microarray analysis pipeline (http://ieg2.ou.edu/Agilent) for preprocessing, including normalization and log transformation.
GeoChip data analysis.
The preprocessed GeoChip data and environmental variables were used for further statistical analyses, including (i) α diversity and evenness indexes of microbial communities as previously described (16), (ii) linear and nonlinear regressions between measures of functional gene diversity/abundances of selected genes and geochemical properties by SigmaPlot (Systat Software, Inc., San Jose, CA), and (iii) linear regressions between each probe (normalized signal intensity profile across all samples) and environmental variables and calculations of slopes and R2 and P values using R (64).
Random forest for predicting environmental contamination and ecosystem functioning.
Random forest was used for classification and regression as it does not require extensive tuning and recent studies have demonstrated that it is a suitable tool in microbial community analysis (22, 58, 65). This method included three major steps: feature selection, modeling (classification or regression), and error rate estimation by out-of-bag (OOB) data.
(i) Feature selection.
Different sets of functional genes were selected as features for predicting environmental (uranium and nitrate) contamination and ecosystem functioning (e.g., N2O), including related functional gene categories (e.g., all N cycling genes), specific functional gene families (e.g., norB or nosZ), and key functional genes that were significantly increased or decreased as contamination increased. For the classification of environmental (uranium and nitrate) contamination, we also used the receiver operating characteristic curve and the area under the curve (AUC) as the predictive accuracy for random forest (RF) and then selected the set of features with the highest AUC values, termed AUC-RF (31), thus reducing the multiple collinearity among features. An AUC of around 0.5 indicates that the classification is only as good as a random guess, while the classification is perfect if the AUC is 1.0. This was performed by using the R package AUCRF.
(ii) Modeling.
The random forest models were constructed using the R package “randomForest” as described by Leo Breiman (66). The algorithm is briefly summarized below. First, bootstrap samples were drawn from the original data n times. Second, for each set of bootstrap samples, an unpruned classification or regression tree was grown, and at each node, rather than choosing the best split among all features, we randomly sampled the mtry (number of features randomly sampled as candidates at each split) of the features and chose the best split among those features. By default, mtry equals one-third the number of all features. Third, new data were predicted by aggregating the predictions of n trees (i.e., majority votes for classification and averages for regression).
(iii) Error rate estimation.
The estimate of the error rate was obtained without independent test data sets. At each bootstrap iteration, the data not included in the bootstrap samples, also known as out-of-bag (OOB) data, were used for prediction with the tree constructed from the bootstrap samples. Then, the error rate was calculated by aggregating the OOB predictions to obtain the OOB estimate of error rate.
ACKNOWLEDGMENTS
This material by ENIGMA (Ecosystems and Networks Integrated with Genes and Molecular Assemblies [http://enigma.lbl.gov]), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory, is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under contract number DE-AC02-05CH11231 and by funding from the Thousand Talents Program (grant number 38000-18821105) to Zhili He through Sun Yat-Sen University, China.
Footnotes
Citation He Z, Zhang P, Wu L, Rocha AM, Tu Q, Shi Z, Wu B, Qin Y, Wang J, Yan Q, Curtis D, Ning D, Van Nostrand JD, Wu L, Yang Y, Elias DA, Watson DB, Adams MWW, Fields MW, Alm EJ, Hazen TC, Adams PD, Arkin AP, Zhou J. 2018. Microbial functional gene diversity predicts groundwater contamination and ecosystem functioning. mBio 9:e02435-17. https://doi.org/10.1128/mBio.02435-17.
REFERENCES
- 1.Vitousek PM, Mooney HA, Lubchenco J, Melillo JM. 1997. Human domination of Earth’s ecosystems. Science 277:494–499. doi: 10.1126/science.277.5325.494. [DOI] [Google Scholar]
- 2.Halpern BS, Walbridge S, Selkoe KA, Kappel CV, Micheli F, D’Agrosa C, Bruno JF, Casey KS, Ebert C, Fox HE, Fujita R, Heinemann D, Lenihan HS, Madin EMP, Perry MT, Selig ER, Spalding M, Steneck R, Watson R. 2008. A global map of human impact on marine ecosystems. Science 319:948–952. doi: 10.1126/science.1149345. [DOI] [PubMed] [Google Scholar]
- 3.Sahney S, Benton MJ, Ferry PA. 2010. Links between global taxonomic diversity, ecological diversity and the expansion of vertebrates on land. Biol Lett 6:544–547. doi: 10.1098/rsbl.2009.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.May RM. 1988. How many species are there on Earth? Science 241:1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- 5.Worm B, Barbier EB, Beaumont N, Duffy JE, Folke C, Halpern BS, Jackson JBC, Lotze HK, Micheli F, Palumbi SR, Sala E, Selkoe KA, Stachowicz JJ, Watson R. 2006. Impacts of biodiversity loss on ocean ecosystem services. Science 314:787–790. doi: 10.1126/science.1132294. [DOI] [PubMed] [Google Scholar]
- 6.Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, Schindler DW, Schlesinger WH, Tilman DG. 1997. Human alteration of the global nitrogen cycle: sources and consequences. Ecol Appl 7:737–750. doi: 10.1890/1051-0761(1997)007[0737:HAOTGN]2.0.CO;2. [DOI] [Google Scholar]
- 7.Canadell JG, Ciais P, Dhakal S, Dolman H, Friedlingstein P, Gurney KR, Held A, Jackson RB, Le Quéré C, Malone EL, Ojima DS, Patwardhan A, Peters GP, Raupach MR. 2010. Interactions of the carbon cycle, human activity, and the climate system: a research portfolio. Curr Opin Environ Sustain 2:301–311. doi: 10.1016/j.cosust.2010.08.003. [DOI] [Google Scholar]
- 8.Flynn TM, Sanford RA, Ryu H, Bethke CM, Levine AD, Ashbolt NJ, Santo Domingo JW. 2013. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol 13:146. doi: 10.1186/1471-2180-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn DFB, Mirotchnick N, Jain M, Palmer MI, Naeem S. 2011. Functional and phylogenetic diversity as predictors of biodiversity—ecosystem-function relationships. Ecology 92:1573–1581. doi: 10.1890/10-1245.1. [DOI] [PubMed] [Google Scholar]
- 10.Petchey OL, Gaston KJ. 2006. Functional diversity: back to basics and looking forward. Ecol Lett 9:741–758. doi: 10.1111/j.1461-0248.2006.00924.x. [DOI] [PubMed] [Google Scholar]
- 11.Krause S, Le Roux X, Niklaus PA, Van Bodegom PM, Lennon JT, Bertilsson S, Grossart H-P, Philippot L, Bodelier PLE. 2014. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol 5:251. doi: 10.3389/fmicb.2014.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardinale BJ, Matulich KL, Hooper DU, Byrnes JE, Duffy E, Gamfeldt L, Balvanera P, O’Connor MI, Gonzalez A. 2011. The functional role of producer diversity in ecosystems. Am J Bot 98:572–592. doi: 10.3732/ajb.1000364. [DOI] [PubMed] [Google Scholar]
- 13.Handa IT, Aerts R, Berendse F, Berg MP, Bruder A, Butenschoen O, Chauvet E, Gessner MO, Jabiol J, Makkonen M, McKie BG, Malmqvist B, Peeters ETHM, Scheu S, Schmid B, van Ruijven J, Vos VCA, Hättenschwiler S. 2014. Consequences of biodiversity loss for litter decomposition across biomes. Nature 509:218–221. doi: 10.1038/nature13247. [DOI] [PubMed] [Google Scholar]
- 14.He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Huang Z, Wu W, Gu B, Jardine P, Criddle C, Zhou J. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J 1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, Li X, Wu L, Gentry TJ, Yin Y, Liebich J, Hazen TC, Zhou J. 2010. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J 4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- 16.He Z, Xu MY, Deng Y, Kang SH, Kellogg L, Wu LY, Van Nostrand JD, Hobbie SE, Reich PB, Zhou JZ. 2010. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett 13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 17.Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, Zhou A, Voordeckers J, Lee YJ, Qin Y, Hemme CL, Shi Z, Xue K, Yuan T, Wang A, Zhou J. 2014. GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour 14:914–928. doi: 10.1111/1755-0998.12239. [DOI] [PubMed] [Google Scholar]
- 18.He Z, Deng Y, Zhou J. 2012. Development of functional gene microarrays for microbial community analysis. Curr Opin Biotechnol 23:49–55. doi: 10.1016/j.copbio.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 19.He Z, Van Nostrand JD, Zhou J. 2012. Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotechnol 23:460–466. doi: 10.1016/j.copbio.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 20.Hemme CL, Deng Y, Gentry TJ, Fields MW, Wu L, Barua S, Barry K, Tringe SG, Watson DB, He Z, Hazen TC, Tiedje JM, Rubin EM, Zhou J. 2010. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J 4:660–672. doi: 10.1038/ismej.2009.154. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkin AP. 2014. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci U S A 111:E836–E845. doi: 10.1073/pnas.1324044111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith MB, Rocha AM, Smillie CS, Olesen SW, Paradis C, Wu L, Campbell JH, Fortney JL, Mehlhorn TL, Lowe KA, Earles JE, Phillips J, Techtmann SM, Joyner DC, Elias DA, Bailey KL, Hurt RA, Preheim SP, Sanders MC, Yang J, Mueller MA, Brooks S, Watson DB, Zhang P, He Z, Dubinsky EA, Adams PD, Arkin AP, Fields MW, Zhou J, Alm EJ, Hazen TC. 2015. Natural bacterial communities serve as quantitative geochemical biosensors. mBio 6:e00326-15. doi: 10.1128/mBio.00326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akob DM, Mills HJ, Gihring TM, Kerkhof L, Stucki JW, Anastácio AS, Chin KJ, Küsel K, Palumbo AV, Watson DB, Kostka JE. 2008. Functional diversity and electron donor dependence of microbial populations capable of U(VI) reduction in radionuclide-contaminated subsurface sediments. Appl Environ Microbiol 74:3159–3170. doi: 10.1128/AEM.02881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemme CL, Tu Q, Shi Z, Qin Y, Gao W, Deng Y, Van Nostrand JD, Wu L, He Z, Chain PSG, Tringe SG, Fields MW, Rubin EM, Tiedje JM, Hazen TC, Arkin AP, Zhou J. 2015. Comparative metagenomics reveals impact of contaminants on groundwater microbiomes. Front Microbiol 6:1205. doi: 10.3389/fmicb.2015.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green SJ, Prakash O, Jasrotia P, Overholt WA, Cardenas E, Hubbard D, Tiedje JM, Watson DB, Schadt CW, Brooks SC, Kostka JE. 2012. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl Environ Microbiol 78:1039–1047. doi: 10.1128/AEM.06435-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostka JE, Green SJ, Rishishwar L, Prakash O, Katz LS, Mariño-Ramírez L, Jordan IK, Munk C, Ivanova N, Mikhailova N, Watson DB, Brown SD, Palumbo AV, Brooks SC. 2012. Genome sequences for six Rhodanobacter strains, isolated from soils and the terrestrial subsurface, with variable denitrification capabilities. J Bacteriol 194:4461–4462. doi: 10.1128/JB.00871-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bollmann A, Palumbo AV, Lewis K, Epstein SS. 2010. Isolation and physiology of bacteria from contaminated subsurface sediments. Appl Environ Microbiol 76:7413–7419. doi: 10.1128/AEM.00376-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields MW, Yan T, Rhee SK, Carroll SL, Jardine PM, Watson DB, Criddle CS, Zhou J. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol Ecol 53:417–428. doi: 10.1016/j.femsec.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Cardenas E, Wu WM, Leigh MB, Carley J, Carroll S, Gentry T, Luo J, Watson D, Gu B, Ginder-Vogel M, Kitanidis PK, Jardine PM, Zhou J, Criddle CS, Marsh TL, Tiedje JM. 2010. Significant association between sulfate-reducing bacteria and uranium-reducing microbial communities as revealed by a combined massively parallel sequencing-indicator species approach. Appl Environ Microbiol 76:6778–6786. doi: 10.1128/AEM.01097-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 31.Calle ML, Urrea V, Boulesteix AL, Malats N. 2011. AUC-RF: a new strategy for genomic profiling with random forest. Hum Hered 72:121–132. doi: 10.1159/000330778. [DOI] [PubMed] [Google Scholar]
- 32.Yan T, Fields MW, Wu L, Zu Y, Tiedje JM, Zhou J. 2003. Molecular diversity and characterization of nitrite reductase gene fragments (nirK and nirS) from nitrate- and uranium-contaminated groundwater. Environ Microbiol 5:13–24. doi: 10.1046/j.1462-2920.2003.00393.x. [DOI] [PubMed] [Google Scholar]
- 33.Lu Z, He Z, Parisi VA, Kang S, Deng Y, Van Nostrand JD, Masoner JR, Cozzarelli IM, Suflita JM, Zhou J. 2012. GeoChip-based analysis of microbial functional gene diversity in a landfill leachate-contaminated aquifer. Environ Sci Technol 46:5824–5833. doi: 10.1021/es300478j. [DOI] [PubMed] [Google Scholar]
- 34.Tiago I, Veríssimo A. 2013. Microbial and functional diversity of a subterrestrial high pH groundwater associated to serpentinization. Environ Microbiol 15:1687–1706. doi: 10.1111/1462-2920.12034. [DOI] [PubMed] [Google Scholar]
- 35.Roadcap GS, Sanford RA, Jin Q, Pardinas JR, Bethke CM. 2006. Extremely alkaline (pH > 12) ground water hosts diverse microbial community. Ground Water 44:511–517. doi: 10.1111/j.1745-6584.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 36.Méndez-García C, Peláez AI, Mesa V, Sánchez J, Golyshina OV, Ferrer M. 2015. Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:475. doi: 10.3389/fmicb.2015.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhalnina K, Dias R, de Quadros PD, Davis-Richardson A, Camargo FA, Clark IM, McGrath SP, Hirsch PR, Triplett EW. 2015. Soil pH determines microbial diversity and composition in the Park Grass experiment. Microb Ecol 69:395–406. doi: 10.1007/s00248-014-0530-2. [DOI] [PubMed] [Google Scholar]
- 38.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Y, Zhao H, Zhang X, Zhou J, Li G. 2014. Contrasting microbial functional genes in two distinct saline-alkali and slightly acidic oil-contaminated sites. Sci Total Environ 487:272–278. doi: 10.1016/j.scitotenv.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 41.Antunes SC, Pereira R, Marques SM, Castro BB, Gonçalves F. 2011. Impaired microbial activity caused by metal pollution: a field study in a deactivated uranium mining area. Sci Total Environ 410–411:87–95. doi: 10.1016/j.scitotenv.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Bearson S, Bearson B, Foster JW. 1997. Acid stress responses in enterobacteria. FEMS Microbiol Lett 147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 43.Xu M, Zhang Q, Xia C, Zhong Y, Sun G, Guo J, Yuan T, Zhou J, He Z. 2014. Elevated nitrate enriches microbial functional genes for potential bioremediation of complexly contaminated sediments. ISME J 8:1932–1944. doi: 10.1038/ismej.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovley DR, Phillips EJP. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl Environ Microbiol 58:850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovley DR, Phillips EJP. 1994. Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome. Appl Environ Microbiol 60:726–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tebo BM, Obraztsova AY. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol Lett 162:193–198. doi: 10.1111/j.1574-6968.1998.tb12998.x. [DOI] [Google Scholar]
- 47.Suzuki Y, Kelly SD, Kemner KM, Banfield JF. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl Environ Microbiol 69:1337–1346. doi: 10.1128/AEM.69.3.1337-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Payne RB, Gentry DM, Rapp-Giles BJ, Casalot L, Wall JD. 2002. Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl Environ Microbiol 68:3129–3132. doi: 10.1128/AEM.68.6.3129-3132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lovley DR, Widman PK, Woodward JC, Phillips EJP. 1993. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl Environ Microbiol 59:3572–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu M, Wu WM, Wu L, He Z, Van Nostrand JD, Deng Y, Luo J, Carley J, Ginder-Vogel M, Gentry TJ, Gu B, Watson D, Jardine PM, Marsh TL, Tiedje JM, Hazen T, Criddle CS, Zhou J. 2010. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J 4:1060–1070. doi: 10.1038/ismej.2010.31. [DOI] [PubMed] [Google Scholar]
- 51.Zhang P, Wu W-M, Van Nostrand JD, Deng Y, He Z, Gihring T, Zhang G, Schadt CW, Watson D, Jardine P, Criddle CS, Brooks S, Marsh TL, Tiedje JM, Arkin AP, Zhou J. 2015. Dynamic succession of groundwater functional microbial communities in response to emulsified vegetable oil amendment during sustained in situ U(VI) reduction. Appl Environ Microbiol 81:4164–4172. doi: 10.1128/AEM.00043-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Nostrand JD, Wu L, Wu W-M, Huang Z, Gentry TJ, Deng Y, Carley J, Carroll S, He Z, Gu B, Luo J, Criddle CS, Watson DB, Jardine PM, Marsh TL, Tiedje JM, Hazen TC, Zhou J. 2011. Dynamics of microbial community composition and function during in situ bioremediation of a uranium-contaminated aquifer. Appl Environ Microbiol 77:3860–3869. doi: 10.1128/AEM.01981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Nostrand JD, Wu WM, Wu L, Deng Y, Carley J, Carroll S, He Z, Gu B, Luo J, Criddle CS, Watson DB, Jardine PM, Marsh TL, Tiedje JM, Hazen TC, Zhou J. 2009. GeoChip-based analysis of functional microbial communities during the reoxidation of a bioreduced uranium-contaminated aquifer. Environ Microbiol 11:2611–2626. doi: 10.1111/j.1462-2920.2009.01986.x. [DOI] [PubMed] [Google Scholar]
- 54.Liang Y, Van Nostrand JD, N’Guessan LA, Peacock AD, Deng Y, Long PE, Resch CT, Wu LY, He ZL, Li GH, Hazen TC, Lovley DR, Zhou JZ. 2012. Microbial functional gene diversity with a shift of subsurface redox conditions during in situ uranium reduction. Appl Environ Microbiol 78:2966–2972. doi: 10.1128/AEM.06528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang P, Van Nostrand JD, He Z, Chakraborty R, Deng Y, Curtis D, Fields MW, Hazen TC, Arkin AP, Zhou J. 2015. A slow-release substrate stimulates groundwater microbial communities for long-term in situ Cr(VI) reduction. Environ Sci Technol 49:12922–12931. doi: 10.1021/acs.est.5b00024. [DOI] [PubMed] [Google Scholar]
- 56.Beck D, Foster JA. 2014. Machine learning techniques accurately classify microbial communities by bacterial vaginosis characteristics. PLoS One 9:e87830. doi: 10.1371/journal.pone.0087830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML. 2015. Sewage reflects the microbiomes of human populations. mBio 6:e02574-14. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, Schauer DB, Ward DV, Korzenik JR, Xavier RJ, Bousvaros A, Alm EJ. 2012. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 7:e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Z, Malmer D, Langille MGI, Way SF, Knight R. 2014. Which is more important for classifying microbial communities: who is there or what they can do? ISME J 8:2357–2359. doi: 10.1038/ismej.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Green SJ, Prakash O, Gihring TM, Akob DM, Jasrotia P, Jardine PM, Watson DB, Brown SD, Palumbo AV, Kostka JE. 2010. Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl Environ Microbiol 76:3244–3254. doi: 10.1128/AEM.03069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorgersen MP, Lancaster WA, Vaccaro BJ, Poole FL, Rocha AM, Mehlhorn T, Pettenato A, Ray J, Waters RJ, Melnyk RA, Chakraborty R, Hazen TC, Deutschbauer AM, Arkin AP, Adams MWW. 2015. Molybdenum availability is key to nitrate removal in contaminated groundwater environments. Appl Environ Microbiol 81:4976–4983. doi: 10.1128/AEM.00917-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D’haeseleer P, Holman HY, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 63.Wu L, Liu X, Schadt CW, Zhou J. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol 72:4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R Core Team 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 65.Metcalf JL, Wegener Parfrey L, Gonzalez A, Lauber CL, Knights D, Ackermann G, Humphrey GC, Gebert MJ, Van Treuren W, Berg-Lyons D, Keepers K, Guo Y, Bullard J, Fierer N, Carter DO, Knight R. 2013. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. eLife 2:e01104. doi: 10.7554/eLife.01104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Key geochemical and ecosystem data for the 69 wells selected. Download TABLE S1, DOCX file, 0.03 MB (32.7KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relationships between the overall functional diversity (Shannon diversity index) and concentrations (log transformed) of uranium (A) and nitrate (B), as well as pH (C), in groundwater. Linear regression was used for the Shannon index and uranium or nitrate concentrations, and nonlinear regression for the Shannon index and pHs. Download FIG S1, TIF file, 0.7 MB (697.2KB, tif) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relationships between log-transformed uranium (A) or nitrate (B) concentrations, pH (C), or functional richness (D) and groundwater microbial biomass determined by the acridine orange direct count (AODC) method. Linear regression was used for such relationships. Download FIG S2, TIF file, 0.5 MB (521KB, tif) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Regressions of key nitrogen cycling gene abundance with uranium, nitrate, or pH. Uranium and nitrate concentrations were first log transformed and then used for linear regressions, while nonlinear regression without log transformation was used for pHs. P values of <0.05 are in boldface. Download TABLE S2, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relationships between the abundances of significantly increased or decreased populations (bearing key genes) and uranium concentrations by linear regression. Significantly increased slopes are in boldface, and the relative abundances are presented as mean ratios. Download TABLE S3, DOCX file, 0.03 MB (33.3KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relationships between the abundances of significantly increased or decreased populations (bearing key genes) and nitrate concentrations by linear regression. Significantly increased slopes are in boldface, and the relative abundances are presented as mean ratios. Download TABLE S4, DOCX file, 0.1 MB (54.1KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fifty predictors from 2,361 detected functional genes related to uranium reduction automatically selected by AUS-RF for predicting uranium contamination in groundwater. Items in boldface were also identified as belonging to populations significantly increased/decreased with increasing uranium concentrations in groundwater (see Table S3). Download TABLE S5, DOCX file, 0.02 MB (19.2KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fifty-four predictors from 5,273 detected N cycling genes automatically selected by AUC-RF for predicting nitrate contamination in groundwater. Items in boldface were also identified as belonging to populations significantly increased/decreased with increasing nitrate concentrations in groundwater (see Table S4). Download TABLE S6, DOCX file, 0.02 MB (19.9KB, docx) .
Copyright © 2018 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.