Abstract
Most previous studies of interferon-alpha/beta (IFN-α/β) response antagonism by alphaviruses have focused upon interruption of IFN-α/β induction and/or receptor signaling cascades. Infection of mice with Venezuelan equine encephalitis alphavirus (VEEV) or Sindbis virus (SINV) induces serum IFN-α/β, that elicits a systemic antiviral state in uninfected cells successfully controlling SINV but not VEEV replication. Furthermore, VEEV replication is more resistant than that of SINV to a pre-existing antiviral state in vitro. While host macromolecular shutoff is proposed as a major antagonist of IFN-α/β induction, the underlying mechanisms of alphavirus resistance to a pre-existing antiviral state are not fully defined, nor is the mechanism for the greater resistance of VEEV. Here, we have separated viral transcription and translation shutoff with multiple alphaviruses, identified the viral proteins that induce each activity, and demonstrated that VEEV nonstructural protein 2-induced translation shutoff is likely a critical factor in enhanced antiviral state resistance of this alphavirus.
Keywords: Alphavirus, Venezuelan equine encephalitis virus, eastern equine encephalitis virus, Sindbis virus, Chikungunya virus, nonstructural protein 2, transcription shutoff, translation shutoff, antiviral state resistance, Interferon
Introduction
The Alphavirus genus of the Togaviridae family of viruses consists of positive-sense single-stranded RNA viruses broadly classified into arthritogenic (e.g. Sindbis virus (SINV) and chikungunya virus (CHIKV)) and encephalitic (e.g. Venezuelan and eastern equine encephalitis viruses (VEEV, EEEV)) disease-causing groups. Members of this genus are responsible for millions of annual infections and ongoing epidemic outbreaks in several parts of the world, such as the current CHIKV epidemic in the Indian Ocean region (1) which has recently spread to the Caribbean, United States and Central and South America (2-4). Infection with arthritogenic alphaviruses causes a febrile illness, which can lead to arthralgia/arthritis lasing for months or years after infection (5). In contrast, encephalitic alphavirus infection results in prodromal disease of varying duration and severity which can progress to fatal encephalitis in a significant number of cases depending upon the virus (6).
Alphavirus replication and disease severity in mouse models is dependent on their resistance to or avoidance of the antiviral state generated following IFN-α/β induction, and it has been proposed that human disease severity is also associated with resistance to or avoidance of the antiviral effects of IFN (6-8). Infection of mice with VEEV elicits the highest levels of induced systemic IFN-α/β while significantly lower levels are observed following SINV infection (6), and little to no IFN is induced by EEEV infection (9). For CHIKV, robust IFN induction is observed in the serum of infected patients (10, 11), and infected non-human primates (12), whereas little IFN is detected in the serum of infected mice (6). However, non-hematopoietic cells are the primary source of IFN during CHIKV infection (13). Mice with functional IFN-α/β responses efficiently control SINV (14-16) and CHIKV infection (1, 5). In contrast infection with VEEV (17, 18) or EEEV (19, 20) is usually fatal. While the severity of EEEV infection is linked to its avoidance of replication in myeloid lineage cells and consequent suppression of IFN and other innate immune responses (21, 22), mortality and disease progression observed following VEEV infection is proposed to reflect greater resistance to the antiviral state induced by IFN (8).
IFN signaling upregulates hundreds of Interferon stimulated genes (ISG’s), many of which possess antiviral activities (23, 24), of which several have been shown to inhibit alphavirus replication (25-27). Notably, in conditions where replication of other alphaviruses is highly restricted by IFN-α/β priming, successful replication of VEEV can be observed (8, 28). The resistance of VEEV to many antiviral effectors which comprise the antiviral state in IFN-primed cells suggests a global mechanism that overcomes their inhibitory activities, rather than resistance to the activity of each ISG individually. To suppress the induction of cell stress responses, alphaviruses have been shown to block host cell transcription (29, 30) and translation (8, 31), and it is possible that the induction of one or more such processes during infection of IFN-primed cells by VEEV is able to suppress the pre-existing antiviral state. The Old world alphaviruses mediate host transcription and translation shutoff through an activity of the nonstructural protein nsP2 (8, 31, 32), while the capsid protein of New world alphaviruses shuts off host cell transcription (30, 33). The viral protein involved in host translation arrest during New World alphavirus infection has not been determined conclusively.
Most previous studies exploring the mechanisms of alphavirus mediated IFN-α/β antagonism were performed in unprimed cells, cells treated with IFN-α/β post infection, or cells over-expressing individual ISGs such as Interferon-inducible protein with tetratricopeptide repeat 1 (IFIT1) (26, 34-36). However, rapid induction of serum IFN-α/β in mice after VEEV and SINV infection upregulates an antiviral state in most cells at sites where the infection has not progressed, causing the of majority cells infected by these viruses in vivo to be primed to resist infection. Thus, previous in vitro work in unprimed cells primarily represents the few cells initially infected after inoculation of mice. The interaction of VEEV and SINV with a pre-established antiviral state was explored in recent studies (6, 8), which demonstrated that VEEV was far more resistant to a pre-existing antiviral state than SINV.
Previous studies have also focused on the effect of a generalized shutoff, or when specific, virus-induced transcription shutoff on induction of IFN-α/β responses (32, 34), while the role of translation shutoff in antiviral state antagonism has not been emphasized. For SINV, both transcription and translation shutoff are induced by the same protein (31), and the relative contribution of these functions in resisting the antiviral state is difficult to explore. Similarly, most previous work with VEEV or EEEV has implicated capsid induced transcription shutoff to play a major role in suppression of IFN-α/β induction, despite the temporally deferred synthesis of this viral protein during infection (35, 37). Induction of host translation shutoff by VEEV has been localized to the nonstructural protein region of the genome, which is translated before the capsid region during infection (8), suggesting a role for this activity in the antiviral state resistance of VEEV.
Here we have examined possible mechanisms underlying resistance of VEEV to an IFN-α/β induced, pre-established antiviral state and identified/confirmed the proteins that mediate host transcription and translation shutoff with CHIKV, SINV, VEEV and EEEV through individual protein expression. In in vitro testing, VEEV was more resistant than SINV, CHIKV and EEEV to the global antiviral state in mouse and human cells, and this resistance became evident at a point after initial translation of the incoming virus genome. Furthermore, a panel of mutant viruses deficient in host macromolecular synthesis shutoff demonstrated that sensitivity to the antiviral state was correlated with slower rates of this activity. Using a plasmid expression system to study host macromolecular synthesis shutoff independent of virus replication rates, we found that expression of VEEV, CHIKV or SINV nsP2, or VEEV or EEEV capsid expression, but not control nsPs or GFP was sufficient to block host translation, with VEEV and EEEV capsid translation blockade likely secondary to transcription shutoff. VEEV and EEEV nsP2 did not inhibit transcription. VEEV or EEEV capsid and CHIKV or SINV nsP2 expression directly inhibited host transcription. EEEV nsP2 also failed to block host translation revealing a stark difference between VEEV and EEEV. Importantly, in the absence of transcription shutoff, host translation in IFN-primed cells was inhibited more efficiently by VEEV nsP2 than that of SINV nsP2. Furthermore, when VEEV nsP2 was expressed in IFN-primed cells, levels of ISG’s were lower, and replication of an unrelated IFN-sensitive virus (yellow fever virus 17-D) was enhanced over IFN-primed control cells. Overall, we conclude that VEEV nsP2-induced host translation shutoff early after infection downregulates the antiviral state by decreasing levels of ISG’s and creating an environment more permissive for viral replication.
Materials and Methods
Cell culture
Neuro 2a, Vero and Huh7 cells (acquired from American Type Culture Collection (ATCC)) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 200mM L-glutamine (L-glut; Sigma, 10,000 units/mL penicillin (Sigma), and 10 mg/mL streptomycin (Sigma). Tetracycline-inducible murine embryonic fibroblasts (MEF; Clontech) were maintained in the above medium supplemented additionally with 50 mg/mL G418. The generation of, and target gene induction from, tetracycline-inducible murine embryonic fibroblasts overexpressing GFP, IFIT1 and Interferon-stimulated gene 20 (ISG20) has been previously described (38). BHK-21 cells (ATCC) were maintained in RPMI supplemented with 10% donor bovine serum (DBS), 10% tryptose phosphate broth (TPB), and supplements as above. All cells were grown at 37°C with 5% CO2.
Viruses and replicons
Construction of cDNA clones for VEEV ZPC738 (18), EEEV FL93-939 (39), SINV TR339 (40) and CHIK-LR (41) has been previously described. Mutant VEEV viruses P713G, P713S, P713K and Q739L were generated by site-directed mutagenesis using appropriate overlapping primers and Quikchange kit and according to manufacturer’s guidelines (Agilent). VEEV mutants CD and CD/nsP2 739L were generated similarly by deleting amino acids 64-68 from capsid using site-directed mutagenesis. Yellow fever virus (YFV) vaccine strain 17-D was used to construct a reporter virus expressing Nano-luciferase (nLuc) (42) by inserting the nLuc gene followed by the Thosea asigna virus (Tav) 2A-like protease in frame between Capsid and prM, as previously described for alphaviruses (43). Viruses were generated by electroporation of capped in vitro-transcribed RNA (mMessage mMachine; Ambion) into BHK-21 cells (20). Titer was determined using a BHK-21 plaque assay. Construction and packaging of VEEV, EEEV and SINV replicons expressing the firefly luciferase gene (fLuc) has previously been described (9). A CHIKV replicon expressing fLuc, was constructed using methods similar to those previously described. Nano luciferase activity on lysates from 17-D nLuc infected cells was measured using the Nano-Glo Luciferase Assay kit (Promega). Protein levels were quantified using a BCA assay (Pierce) and data expressed as relative light units (RLU)/μg of cell protein.
Protein expression plasmids
Individual virus proteins were cloned into the pCAGGS vector containing a hemagglutinin tag at the N-terminus proximal to the insertion site (gift from Chris Basler) (44). The nsP and capsid proteolytic cleavage sites were chosen as the boundaries of the protein genes and a stop codon was added at the end of each gene. Viral genes were PCR-amplified from full-length cDNA clones using suitable primers with Not1 and Nhe1/Xma1 restriction sites added at the 5’ and 3’ ends respectively. Additionally, GFP was PCR-amplified from a previously constructed plasmid using suitable primers with Not1 and Nhe1 restriction sites added at the 5’ and 3’ ends respectively. PCR products and pCAGGS vector were digested and ligated, and individual clones were selected and confirmed by restriction digestion and sequencing. Huh7 and MEF cells were transiently transfected with plasmids (10 μg per plasmid) using the Nucleofector II machine and manufacturer’s protocols (Amaxa). Huh7 cells (1×106 per reaction) were nucleofected using Kit V and protocol T-022. MEF cells (1×106 per reaction) were nucleofected using Kit V and protocol T-020. Each transfection reaction was divided into two or three wells. A GFP-expressing plasmid was used as transfection control and reactions were used for experiments if GFP positive cells numbered >90%.
Translation reporters
Reporters were constructed as described (27, 45, 46). Briefly, the firefly luciferase (fLuc) gene was fused in frame with 5’ and 3’ NTR and poly (A) tail from SINV, CHIKV, VEEV or EEEV, such that reporter RNA would initiate translation of fLuc from authentic virus translation start sites. RNA was synthesized using in vitro transcription kits (mMessage, mMachine; Ambion). Cells were electroporated with virus derived reporter RNA’s (5μg per reaction) using a BioRad Gene Pulser II machine (for each reaction, two pulses at 220 Volts and 1 mFarad). Each electroporation reaction was divided into three wells. Lysates were collected in passive lysis buffer (Promega). Firefly or Renilla luciferase activity was measured using the Dual Luciferase Reporter Assay kit (Promega). Protein levels were quantified by bicinchoninic acid assay (BCA; Pierce) and data expressed as RLU/μg.
Virus infections
Cells were seeded in plates overnight before infections. Viruses were diluted to the multiplicity of infection (MOI) indicated in figure legends in virus diluent (PBS with Calcium and Magnesium, supplemented with 1% DBS). Cells were infected for one hour at 37°C, following which medium was added to cells. Supernatants were collected at indicated times and viral titers were determined as described above. For semi-quantitative PCR measuring viral replication, lysates were collected using Trizol reagent and total cellular RNA was extracted using manufacturer’s protocols (Ambion). 100ng total RNA was reverse-transcribed into cDNA using a virus specific primer for VEEV (5’-GCGTAATACGACTCACTATACTGGTACTAGATTTATGCGC -3’), EEEV (5’-GCGTAATACGACTCACTATATGACAACCAACGAGTGTGGG-3’), SINV (5’-GCGTAATACGACTCACTATACGTGAGGAAGATTGCGGTTC-3’) and CHIKV (5’-GCGTAATACGACTCACTATAGCTGTCTAGATCCACCCCATACATG-3’). For the semi-quantitative step, a primer for T7 (5’-GCGTAATACGACTCACTATA-3’) and virus specific primers for VEEV (5’-TCCGTCAGCTCTCCCGCAGG-3’), EEEV (5’-AGAGTGGCTGACGTTCGCAC-3’), SINV (5’-CTGGGAAGGGCACACAACTT-3’) and CHIKV (5’-GGCAGTGGTCTCAGATAATTCAAG-3’) were used. Additionally, 100ng RNA was reverse-transcribed using random hexamers, and 18S primers were used as loading control (sense, 5’-CGCCGCTAGAGGTGAATTTCT-3’; antisense, 5’-CGAACCTCCGACTTTCGTTCT-3’). Values obtained were normalized to 18S levels using the ddCT method (47). DNA contamination was ruled out by performing reactions excluding reverse transcriptase.
Interferon (IFN) bioassay and ELISA
The concentration of biologically active mouse IFN was measured using a bioassay as previously described (48). Briefly, 3×104 L929 cells were seeded per well in a 96-well plate. Samples (200 μL) were acidified to pH 2.0 using 1M HCl for 24h at 4°C. Samples were then neutralized to pH 7.0 using 2M NaOH and 100 μL was added to the first well in duplicate. Samples were diluted 2-fold across the plate and incubated at 37°C for 24h. EMCV (4×103 PFU/well) was added to each well and incubated 24h at 37°C and then stained with crystal violet. The IFN concentration in samples (cell supernatants or mouse sera) was set as the dilution of sample required for 50% protection from cytopathic effect (CPE), compared with protection conferred by an IFN standard. The IFN-α ELISA was performed on mouse sera as per manufacturer’s instructions.
Western blotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1 mM EDTA, 1 mM EGTA) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1μg/ml leupeptin, and 1 μg/ml pepstatin), and a phosphatase inhibitor cocktail (Sigma). Protein concentrations were determined as above. Equal amounts of protein (25 μg) from each lysate was resolved on an 8% SDS-polyacrylamide gel (SDS-PAGE) and transferred to a PVDF membrane (BioRad). Membranes were blocked for 1h in 5% milk in Tris-buffered saline (TBS) with 0.1% Tween 20 (TBST) and incubated overnight with primary antibody at 4°C. Primary antibodies were diluted in 3% BSA-TBST. Membranes were washed in TBST four times (15min each) and incubated for 1h in appropriate horseradish peroxidase-conjugated secondary antibody (Thermo Fisher) diluted in 2% milk-TBST. Membranes were washed in TBST four times and probed with an ECL chemiluminescence kit (Pierce). Densitometry was performed using Image J software. Membranes were probed with following antibodies: rabbit polyclonal against IFIT1 (1:1000, Sigma), goat polyclonal against TGTP (1:250, Santa Cruz) and mouse monoclonal against actin (1:5000, Millipore) and HA-tag (1:1000, Thermo Scientific).
Metabolic labeling
Cells were infected with viruses or transfected with plasmids for the times indicated in figure legends. Thirty minutes before labeling, growth medium was replaced with starvation medium (Cys/Met free DMEM (Cellgro) supplemented with 1% FBS, L-glut, PS). Next, cells were incubated in starvation medium complemented with 100μCi/mL [35S] Cys/Met (MP Biomedicals) for 2h at 37°C. Cells were washed with phosphate-buffered saline (PBS) and lysed in RIPA buffer. An equal volume of sample was resolved on an 8% SDS-PAGE gel. Gels were fixed and dried and exposed to photographic film (GE Healthcare) for 7 days at −80°C for visualizing labeled proteins. Densitometry was performed as described above. As a loading control, equal volumes of lysate were resolved and stained for levels of actin using western blot.
Immunofluorescence
MEF cells were pre-treated with 150IU/mL mouse IFN for 16h and transfected with expression plasmids as described above. Cells were fixed at 24h post-transfection in 4% paraformaldehyde (PFA) overnight at 4°C and permeabilized with ice-cold methanol or 0.1% Triton X-100 at room temperature for 15 min. Cells were blocked with blocking buffer (BB, PBS supplemented with 1% bovine serum albumin and 0.3% Triton X-100) mixed with 10% donkey serum for 1h at room temperature. Primary antibody was diluted in BB and incubated overnight at 4°C. Next, cells were washed in PBS and incubated in fluorophore-conjugated secondary antibody in the dark for 1h at room temp. DAPI (10μL per well) was added before cells were observed with a confocal fluorescence microscope. The following primary antibody was used: mouse monoclonal anti-HA tag (1:200, Thermo Fisher). Secondary antibody used was Alexa 488 donkey anti-mouse (1:1000, Jackson Immunological). Images were acquired using Olympus Fluoview 1000 microscope at a magnification of 60× using Fluoview software version 3.1.
Transcription shutoff analysis
Lysates were collected from transfected cells using Trizol reagent and total cellular RNA was extracted using manufacturer’s protocols (Ambion). 100 ng total RNA was reverse-transcribed into cDNA using random hexamers. Primers were designed to detect levels of human gamma actin intron #3 (sense, 5’-TTCTTTCGCTGTTCCAGGCT-3’; antisense, 5’-AGGCTTCAGGGAGGAAATGC-3’) or mouse gamma actin intron #3 (sense, 5’-ACAGAACGCAAGCAGAAACG-3’; antisense, 5’-TGGCATTTCCTCCCTGAAGC-3’). 18S primers were used as loading control (sense, 5’-CGCCGCTAGAGGTGAATTTCT-3’; antisense, 5’-CGAACCTCCGACTTTCGTTCT-3’). Actinomycin D treatment (15 μg/mL treatment for 6h) was used as a positive control for transcription shutoff.
In vivo experiments
Inocula (10μL) containing 100 or 1000 PFU virus or PBS was administered subcutaneously to the hind leg footpad of CD-1 mice using a 27 gauge needle and a gastight Hamilton syringe. Mice were observed and scored for degree of sickness at 24h intervals and average survival times (AST) and percent mortality were calculated.
Statistics
Statistical tests used were Student’s t-test, one-way ANOVA, two-way ANOVA, as indicated in figure legends. Graph Pad Prism software was used for all statistical analyses.
Ethics statement
All animal research was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal procedures were performed according to a protocol (Protocol no. 101195) approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. For viral infections, mice were anesthetized with Isoflurane. Mice reaching euthanasia criteria were euthanized by Isoflurane overdose and cervical dislocation. For serum collection, mice were overdosed with Isoflurane and exsanguinated by cardiac puncture.
Results
VEEV is more resistant to the IFN-induced antiviral state than other alphaviruses
Previously, we compared the relative resistance of SINV and VEEV to an IFN-induced anti-viral state in primary mouse neurons (8). Treatment of primary neuron cultures with 1000 IU IFN α/β post-infection had limited effect on viral growth; however, pre-treatment with 1000 IU IFN α/β for 24h prior to infection substantially inhibited growth of SINV but not VEEV (8). Here we determined the relative resistance of multiple Old world (SINV and CHIKV) and New world (VEEV and EEEV) alphaviruses to a pre-existing antiviral state in both mouse and primate cells.
Treatment of Vero cells with 5000 IU human leukocyte IFN for 24h prior to infection significantly (P<0.0005) reduced the growth of all tested alphaviruses at 6h post-infection (p.i.) (Fig 1A). By 24h p.i., this early blockade on virus growth was subsequently overcome by VEEV and replication in IFN-pretreated cells was comparable to untreated cells. In contrast, the antiviral effects of IFN significantly (P<0.0005) virus growth of all other tested alphaviruses. Similar results were obtained in mouse embryonic fibroblast (MEF) cells (data not shown), indicating that the resistance phenotype of VEEV is not mouse- or primate-specific. These results indicate that VEEV is resistant to a global IFN-induced anti-viral state.
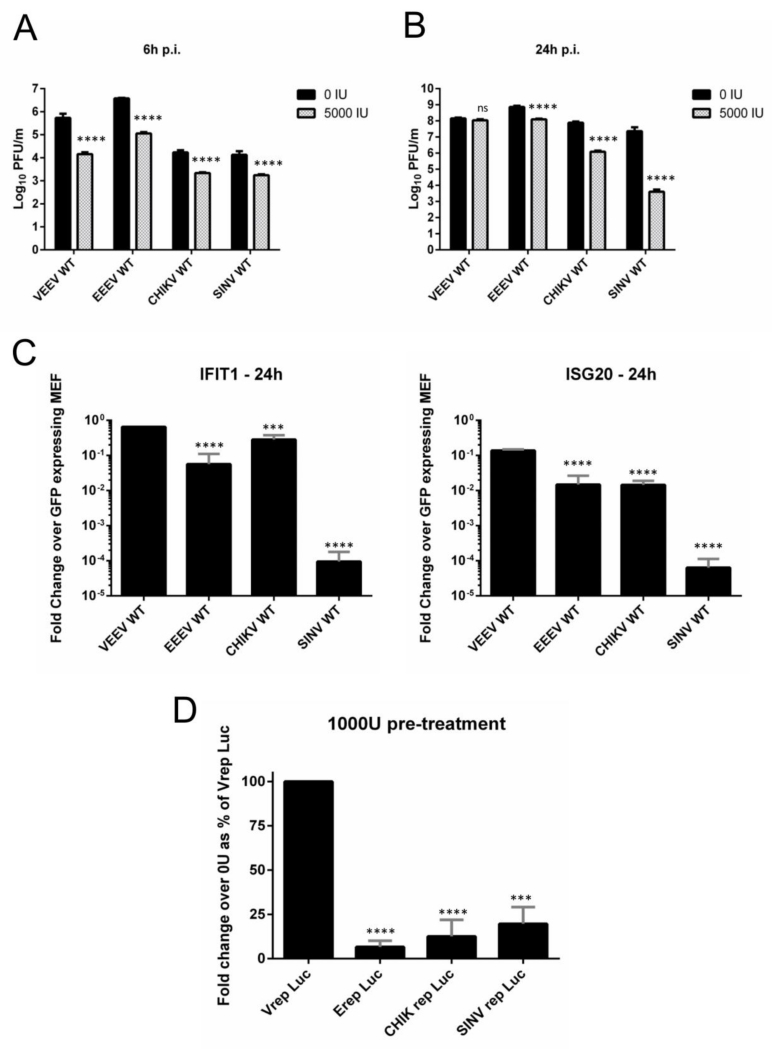
Fig 1. VEEV is resistant to a pre-established anti-viral state and to individual overexpressed IFN effectors.
(A and B) Vero cells mock-treated or pre-treated with 5000 IU human leukocyte IFN for 24h were infected in triplicate with indicated viruses (M.O.I. = 2.5). Supernatants were collected at 6h (A) and 24h (B) p.i. and virus replication was quantified using plaque assays. ****, P<0.0005 using t-test. Data is representative of two independent experiments. (C) Tet-inducible mouse embryonic fibroblasts (MEFs) stably expressing IFIT1, ISG20 and GFP (control) were infected in triplicate with indicated viruses (M.O.I. = 1). Cell lysates were collected at 24h p.i and viral RNA levels were measured using RT PCR as described in Materials and Methods. Data is viral RNA levels from IFIT1 or ISG20 expressing cells as a fold change of viral RNA levels from GFP expressing cells. ****, P<0.0001; ***, P<0.001 using One-way ANOVA. All error bars are standard deviations. (D) MEF cells were mock-treated or pre-treated with 1000 IU mouse IFN for 24h and infected with indicated replicons (equal dilution). Lysates were collected in passive lysis buffer at 16h p.i. and luciferase activity was measured. Data is RLUs per μg total protein expressed as a fold change over no IFN for each replicon. Fold change for Vrep was set as 100% and all replicons were compared to Vrep. Infection was performed in triplicate. ****, P<0.0001; ***, P<0.0003 using t-test.
We next tested the ability of alphaviruses to replicate in the presence of individually overexpressed IFN effectors that had been previously shown to possess anti-alphaviral activity (38). Tet-inducible MEF cells over-expressing IFIT1 and ISG20 were infected with VEEV, EEEV, SINV and CHIKV and viral replication was measured using qRT-PCR at 24h p.i. Similar to pre-treatment experiments, VEEV replicated successfully; in contrast, EEEV, CHIKV and SINV were significantly inhibited (P<0.001, Fig 1C). Taken together, these results indicate that VEEV is resistant to both a global anti-viral state and individual IFN induced effector proteins.
During alphavirus infection, nonstructural proteins are produced first, and are required for production of structural proteins (49). To identify the role of nonstructural proteins in the resistance phenotype of VEEV, we used VEEV, SINV, CHIKV and EEEV replicons lacking structural proteins and expressing fLuc. Luciferase activity in MEF cells pre-treated with IFN was reduced during infection of all replicons when compared to untreated cells. However, luciferase activity in IFN primed cells infected with Vrep Luc was significantly higher (P<0.0003) than those infected with other replicons (Fig 1D). We concluded that the resistance phenotype of VEEV is localized, at least in part, to the nonstructural protein region of the genome.
Translation of incoming genomes of all tested alphaviruses is similarly affected by IFN
The previous experiments demonstrated the ability of VEEV to more efficiently initiate replication in the presence of an antiviral state compared to other tested alphaviruses and suggested a role for noncoding regions or nonstructural proteins in the antiviral state resistance of VEEV. Subsequently, we sought to determine the step(s) in the replication cycle where VEEV escaped suppressing effects of the antiviral state. Possible points where upregulated antiviral effectors could potentially block alphavirus replication initiation are virus entry, virion envelope fusion with the endosomal membrane, sequestration/degradation of incoming viral RNA and suppression of initial translation of the incoming viral genome. We previously observed that translation of mRNA messages entering an IFN-primed cell across the cytoplasm was substantially reduced, while that of nuclear originating mRNA was not (46). Additionally, this block was at the step of initial translation of the mRNA. Furthermore, recent work using chimeric alphaviruses has shown that, in general, attachment, entry and nucleocapsid dissociation are not affected by IFN (36). Therefore, we speculated that the IFN induced antiviral state might block virus replication at the point of initial genome translation. We used capped and poly-adenylated reporter RNA molecules in which the fLuc gene was flanked by authentic 5’ and 3’ non-translated regions (NTR) and fused to truncated nsP1 of VEEV, EEEV, CHIKV and SINV (Fig 2A) to measure the effect of antiviral activity on translation of incoming virus genomes as previously described (46). Translation of reporter RNA mimics initial translation of an incoming viral genome as the reporter is incapable of replication. In addition, electroporation of reporter RNA’s bypasses the entry and fusion steps of the viral infection cycle by delivering RNA directly into the cytoplasm.
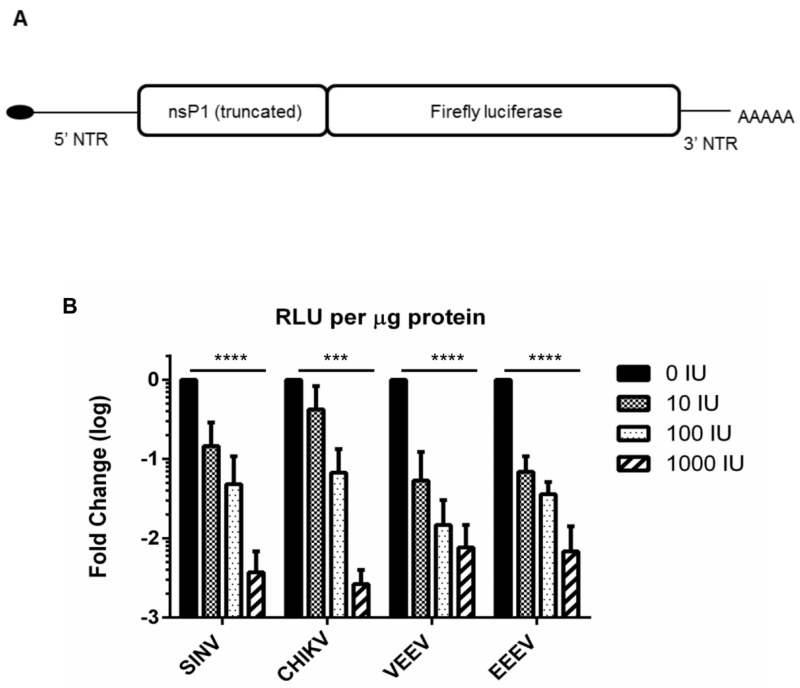
Fig 2. Translation of incoming genomes is attenuated in IFN pre-treated cells.
(A) Schematic of translation reporter RNA with wild-type virus 5’ and 3’ NTRs. (B). MEF cells were untreated or pre-treated with 0, 10, 100 or 1000 IU mouse IFN for 16h and electroporated with reporter RNAs of indicated viruses as described in Materials and Methods. Cells were lysed 2h post-electroporation and luciferase activity was measured. Data is RLUs per μg total protein expressed as a fold change over no IFN; average of three experiments for per treatment. ***, P<0.0015; ****, P<0.0001 using One way ANOVA. All error bars are standard deviations.
The activity of fLuc in electroporated MEF cells pre-treated with IFN revealed a dose dependent decrease in translation of incoming RNA independent of the origin of the reporter (Fig 2B), similar to published observations using chimeric viruses (36). Activity was significantly lower (reduced >100 fold; P<0.0015) in cells pre-treated with the highest dose of IFN (1000 IU) compared to untreated cells for all viral reporter RNA’s. We conclude that replication of alphaviruses is significantly diminished in IFN-primed cells after entry and at the point of initial translation of the genome and production of nsPs. Notably, translation of all reporter RNA’s, while being heavily suppressed was not completely ablated even in cells receiving the highest dose of IFN. While, recently, IFIT-1 activity was demonstrated to inhibit translation differentially between alphaviruses (36), VEEV does not appear more resistant to the overall effect of IFN priming in this assay. This suggests that low-level translation of viral nonstructural proteins occurs following infection of IFN-primed cells. Based upon this result, and consistent with replicon data (Fig 1D), we hypothesized that an activity or activities of one or more VEEV nsPs may contribute to the antiviral state resistance of VEEV.
Phenotypes of mutant VEE viruses suggest a role for host macromolecular synthesis shutoff in antiviral state antagonism
Alphaviruses can inhibit host macromolecular synthesis by causing transcription and translation shutoff in many replication-permissive cells (8, 33, 39, 50). We hypothesized that VEEV induces host macromolecular synthesis shutoff in IFN-primed cells to inhibit the antiviral state during infection. To test this hypothesis, we created a panel of VEEV viruses incorporating published mutations in the nsP2 or capsid of VEEV and other alphaviruses known to reduce host macromolecular synthesis shutoff (Table 1). The wild-type nsP2 sequences of most alphaviruses contain a conserved proline residue at amino acid position 713 in VEEV, 726 in SINV and 718 in CHIKV, with the notable exception of EEEV which possesses a lysine at the analogous position. Published data with mutations at or near this position (SINV P726G, CHIKV P718S, VEEV Q739L) have been reported to decrease host macromolecular synthesis shutoff or, with Q739L, cytopathic activity, which may reflect shutoff efficiency (32, 51-53). In addition, a five amino acid deletion in VEEV capsid greatly reduces its ability to shut off host cell transcription (33, 54).
Table 1. Summary of published and generated mutants.
| Virus | Wild Type | Phenotype |
|---|---|---|
| VEEV WT | HLNPGGTCVSIGYGYADRASESIIGAIARQFKF | Wild type |
| SINV WT | CLNPGGTLVVKSYGYADRNSEDVVTALARKFVR | Wild type |
| CHIK WT | LLKPGGSLLIRAYGYADRTSERVICVLGRKFRS | Wild type |
| Published mutations in nsP2 and Capsid | ||
| SINV P726G | CLNGGGTLVVKSYGYADRNSEDVVTALARKFVR | Non-cytopathic in cells (51, 52) |
| CHIKV P718S |
LLKSGGSLLIRAYGYADRTSERVICVLGRKFRS | Does not antagonize macromolecular synthesis shutoff (32) |
| VEEV Q739L | HLNPGGTCVSIGYGYADRASESIIGAIARLFKF | Non-cytopathic in BHK cells (53) |
| VEEV CD | Deletion of Amino acids 64-68 in Capsid | No transcription shutoff (54) |
| Generated mutations in nsP2 and Capsid | ||
| VEEV CD | Deletion of Amino acids 64-68 in Capsid | No transcription shutoff (54) |
| VEEV CD/739L |
Deletion in capsid + nsP2 739L | Unknown |
| VEEV P713G | HLNGGGTCVSIGYGYADRASESIIGAIARQFKF | Unknown |
| VEEV P713S | HLNSGGTCVSIGYGYADRASESIIGAIARQFKF | Unknown |
| VEEV Q739L | HLNPGGTCVSIGYGYADRASESIIGAIARLFKF | Non-cytopathic in BHK cells (53) |
Bold red letters indicate mutated amino acid residues.
We measured the following phenotypes of the panel of mutant viruses: 1.) replication in unprimed and IFN-primed cells; 2.) the ability to shutoff host macromolecular synthesis by measuring translation shutoff and IFN induction; and 3.) replication efficiency in IFN-primed cells compared to WT VEEV. We reasoned that a correlation of one or more mutant phenotypes with the ability of mutants to replicate in the presence of an antiviral state would implicate those activities in the antiviral state resistance of WT VEEV.
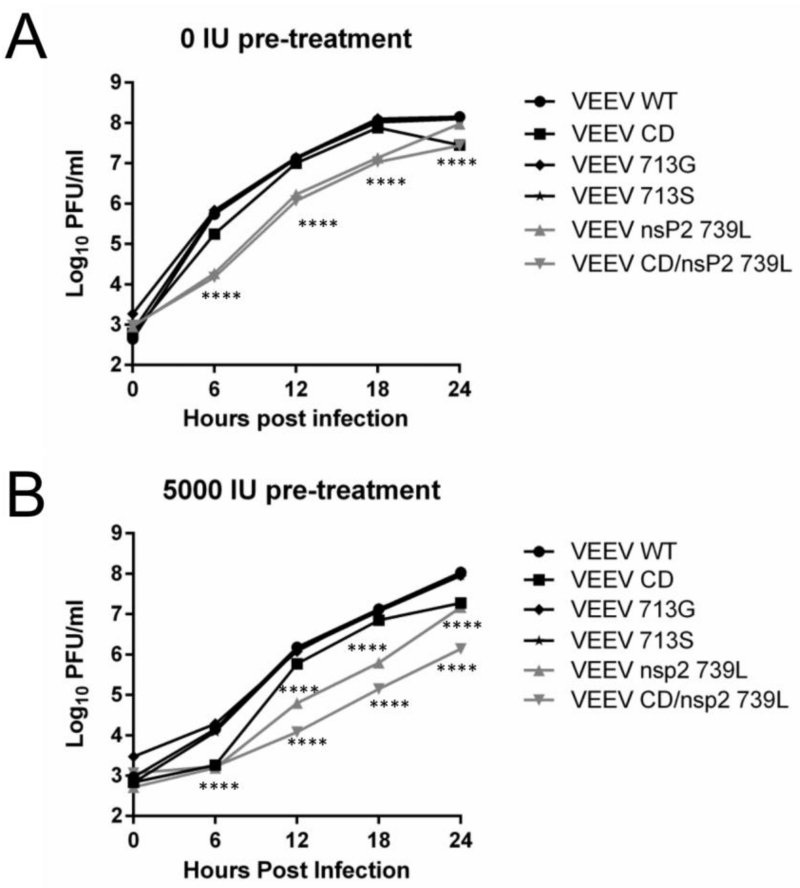
We introduced these mutations into an enzootic wild-type VEEV virus (Table 1) and tested their effects on viral replication by performing a one-step growth curve in untreated and IFN pre-treated Vero cells (Fig 3A and B). The rate and efficiency of infection was similar between WT and mutant viruses. In untreated cells the mutants divided into two groups (Fig 3A); slower growing mutants (Q739L, CD/739L) which significantly (P<0.0001) lagged WT VEEV replication at early times p.i., and mutants which grew to levels and at rates not significantly different from WT VEEV (CD, P713G, P713S). Most mutants achieved levels of replication similar to WT VEEV by 24h p.i. Substitution of the conserved proline at position 713 in VEEV nsP2 with glycine had no impact on replication kinetics, in stark contrast to the substantial effect of the analogous mutation in SINV (52). Additionally, the effect of the capsid deletion mutation on viral replication was observed only in conjunction with Q739L. The replication of mutants in IFN-primed cells (Fig 3B) was similar to that observed in unprimed cells. 713G and 713S replication was not significantly different from VEEV WT. The mutant CD was slower significantly only at 6h and 24h p.i. (P<0.0001) compared to VEEV WT. In contrast, growth of 739L and CD/739L was significantly (P<0.0001) inhibited compared to VEEV WT at all times p.i., and similar to the effect observed in unprimed cells, the combined effect of two mutations significantly inhibited viral replication. Finally, we titered all viruses on multiple cell types (BHK, Vero and Huh7, data not shown). We observed different titers between cell types, but crucially, the difference between WT and mutant titers was similar in each cell type, suggesting that the viral infectivity was similar between WT and mutant viruses.
Fig 3. Growth characteristics of VEEV mutants.
Vero cells untreated (A) or pre-treated with 5000 IU IFN (B) were infected with indicated viruses (M.O.I. = 2.5) and supernatants were collected at 6, 12, 18 and 24h p.i. Virus replication was quantified using plaque assays. ****, P<0.0001 using Two-way ANOVA. All error bars are standard deviations. Data is representative of two independent experiments.
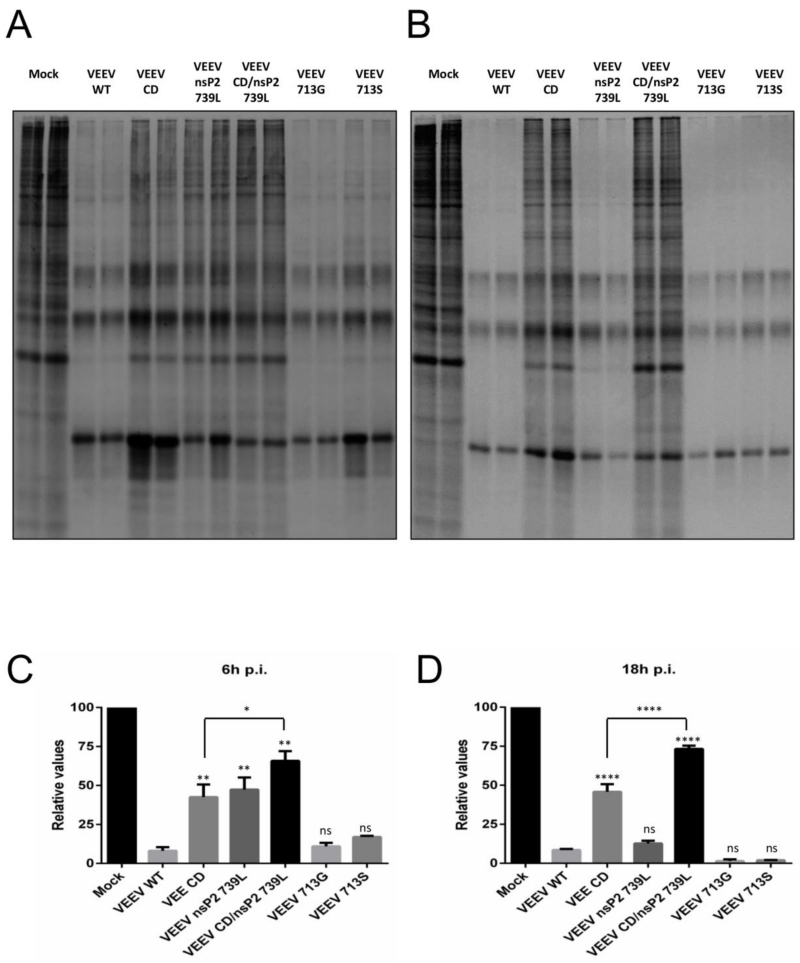
We next tested the ability of mutant viruses to shut off host macromolecular synthesis, measured in this case as translation shutoff which represents cumulative inhibition of transcription and translation activities. Neuro 2a cells were used as a substitute for primary neurons, which were previously used to study the resistance of VEEV to the antiviral state (8). Infected Neuro 2a cells were labeled with [35S] Cys/Met for 2h at 6h and 18h p.i and lysates were resolved on polyacrylamide gels to measure total protein synthesis (Fig 4A and B). As previously reported, WT VEEV efficiently shut off host translation by 6h. Shutoff induced by mutants 713G and 713S was not significantly different from WT VEEV early or late after infection. The mutants 739L, CD and CD/739L were significantly (P<0.01) impaired at inducing shutoff early after infection, and while the extent of shutoff achieved by 739L was not significantly different from WT VEEV by 18h, CD and CD/739L were unable to induce a complete shutoff even by 18h p.i. (P<0.0001). Shutoff induced by CD/739L was significantly lower than CD alone both early (P<0.05) and late (P<0.0001) p.i., reflecting the combined effects of the two mutations present in the virus. Similar results were obtained in BHK and Huh7 cells (data not shown).
Fig 4. Host macromolecular synthesis shutoff by VEEV mutants.
(A-D) Neuro 2a cells were infected with indicated viruses (M.O.I. = 2.5) and labeled with 100μCi/ml of [35S] Cys/Met for 2h at 6h (A) and 18h (B) p.i. Lysates were collected and resolved on SDS-PAGE gels and visualized as described in Materials and Methods. (C and D) Densitometry performed on gels from (A) and (B) respectively. ****, P<0.0001; **, P<0.01; *, P<0.05 using One-way ANOVA, compared to VEEV WT. Ns = not significant. Data is representative of two independent experiments. All error bars are standard deviations.
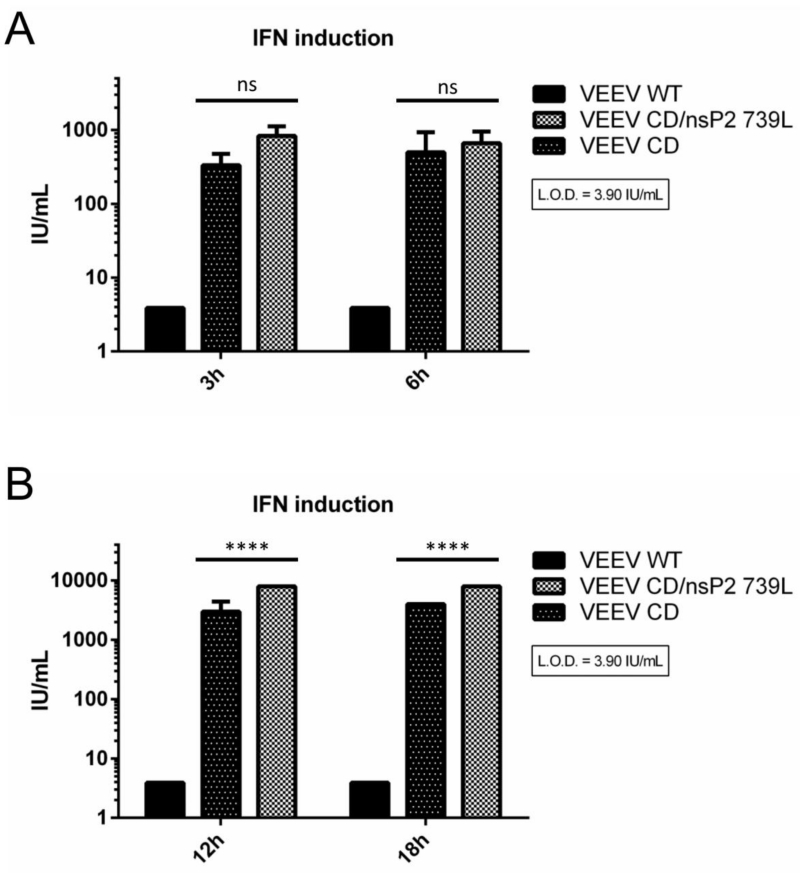
IFN induction following infection can result in upregulation of an antiviral state via autocrine or paracrine signaling (55). The ability of VEEV to shutoff host macromolecular synthesis would also prevent IFN production and subsequent establishment and sustenance of an antiviral state. We explored the role of host macromolecular synthesis shutoff in antagonism of IFN induction by measuring secreted IFN levels following infection of MEF cells with WT and mutant VEEV viruses as this might affect rates of virus replication in cell types competent for IFN production. MEF cells were chosen as they are capable of secreting IFN following treatment with appropriate stimuli. VEEV WT infection did not result in IFN secretion at early or late times p.i. (Fig. 5A and B). IFN was only detected in CD and CD/739L supernatants at both early and late times p.i. (Fig 5A and B). CD/739L induced slightly, but not significantly, greater amounts of IFN than CD early (3h; 830IU vs 330IU), and significantly (P<0.0001) greater amounts late during infection (24h; 8000IU vs 4000IU). Notably, while 739L infected cells did not secrete IFN, CD/739L infected cells secreted the greatest amount of IFN at all measured times p.i, suggesting that different components of the host macromolecular synthesis shutoff mechanism have a combined effect on antiviral state antagonism. Taken together, these results suggest, consistent with previous reports with SINV (34), that host macromolecular synthesis shutoff plays a role in suppression of IFN induction in fibroblast type cells by VEEV. These results also demonstrate that both VEEV nsP2 and capsid contribute to this suppression during infection
Fig 5. IFN induction by VEEV mutants.
MEF cells were infected with indicated viruses (M.O.I = 5) and supernatants were collected at early (A) or late (B) times p.i. IFN bioassays were performed as described in Materials and Methods to determine secreted IFN levels. ****, P<0.0001 using Two way ANOVA. Ns = not significant. Data is the representative of at least two independent experiments for each virus. All error bars are standard deviations.
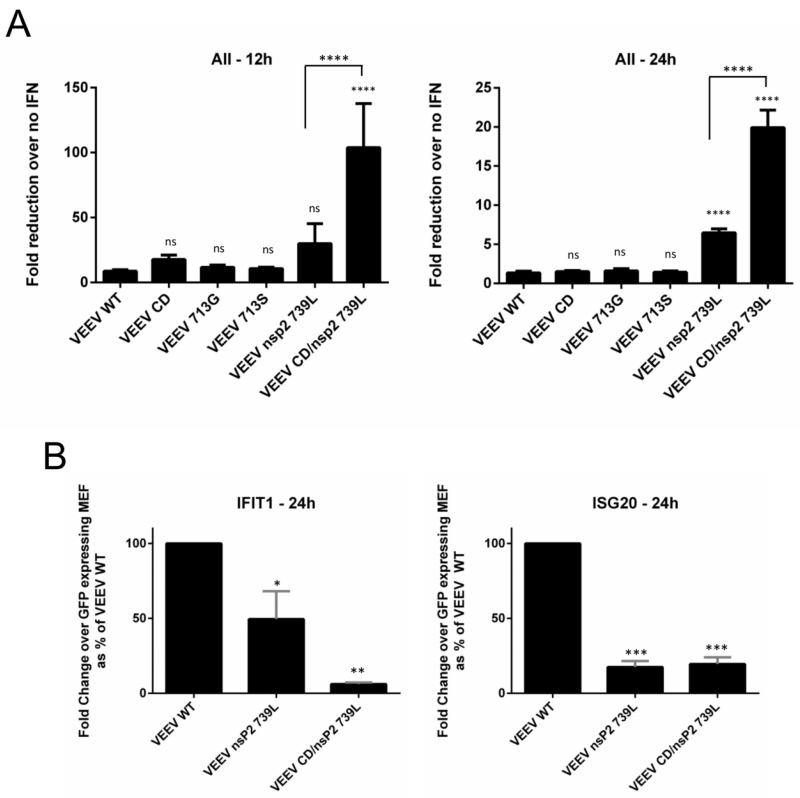
We tested the ability of VEEV mutant viruses to replicate in the presence of a pre-established antiviral state in order to identify whether macromolecular synthesis shutoff played a role in antiviral state resistance. Vero cells were selected as they are incapable of producing IFN upon stimulus (56, 57); thus the antiviral state generated by IFN pre-treatment would be equivalent for all VEEV mutants and would not be affected by virus induced IFN. Vero cells were pre-treated with 5000 IU human leukocyte IFN for 24h and infected with VEEV WT and mutants at equal M.O.I. The fold reduction in replication in IFN-treated cells versus replication in untreated cells was used to determine the sensitivity of VEEV mutants to IFN-priming (Fig 6A). The mutants CD, 713G and 713S were not significantly different from WT VEEV in resisting the effects of the anti-viral state, whereas 739L and CD/739L were significantly (P<0.0001) more sensitive than VEEV WT at late times p.i. Additionally, CD/739L was significantly (P<0.0001) more sensitive to the antiviral state than 739L, reflecting the presence of mutations affecting both components of macromolecular synthesis shutoff. Growth of 739L and CD/739L was also significantly inhibited (P<0.05) in cells over-expressing single antiviral effector proteins IFIT1 or ISG20 (Fig 6B) when compared to VEEV WT.
Fig 6. Sensitivity of VEEV mutants to a pre-established anti-viral state.
(A) Vero cells were mock-treated or treated with 5000 IU human leukocyte IFN for 24h before infection in triplicate with indicated viruses (M.O.I. = 2.5). Supernatants were collected at 12h and 24h p.i. and virus replication was quantified using plaque assays. For each virus, data represents fold change in titer over no IFN. ****, P<0.0001 using One-way ANOVA. Ns = not significant. Data is representative of two independent experiments. All error bars are standard deviations. (B) Tet-inducible mouse embryonic fibroblasts (MEFs) stably expressing IFIT1, ISG20 and GFP (control) were infected in triplicate with indicated viruses (M.O.I. = 1). Cell lysates were collected at 24h p.i and viral RNA levels were measured using RT PCR as described in Materials and Methods. Data represent viral RNA levels from IFIT1 or ISG20 expressing cells as a fold change of viral RNA levels from GFP expressing cells. ***, P<0.0002; **, P<0.006; *, P<0.05 using One-way ANOVA. All error bars are standard deviations.
Summarizing the results of in vitro experiments with the VEEV mutants (Table 2), we found that the efficiency of replication early during infection was the viral phenotype most closely associated with resistance to the antiviral state during late stages of infection, and was positively associated with host shutoff. Lesser shutoff of host macromolecular synthesis likely contributed to the reduced growth of 739L and CD/739L observed in IFN-primed cells. Viruses containing mutations negatively affecting host macromolecular synthesis shutoff (739L and CD/739L) were thus more sensitive to the antiviral state, while mutations that had a negligible to slightly positive, albeit non-significant, effect on these viral activities (713G, 713S) were similarly resistant as VEEV WT to the antiviral state.
Table 2. Summary of VEEV mutant phenotypes at early and late times p.i.
| Early times p.i. (6h) | Late times p.i. (18-24h) | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| No IFN | Priming | No IFN | Priming | |||||
|
|
||||||||
| Virus | Replication efficiency |
Macromolecular synthesis shutoff |
IFN induction |
Resistance to anti- viral state |
Replication efficiency |
Macromolecular synthesis shutoff |
IFN induction |
Resistance to anti- viral state |
| WT | +++ | +++ | None | + | +++ | +++ | None | +++ |
|
nsP2
713G |
+++ | +++ | None | + | +++ | +++ | None | +++ |
|
nsP2
713S |
+++ | +++ | None | + | +++ | +++ | None | +++ |
|
nsP2
739L |
+ | + | None | + | +++ | +++ | None | ++ |
| CD | +++ | + | High | + | +++ | + | High | +++ |
| CD/739L | + | + | High | + | ++ | + | High | ++ |
VEEV nsP2 promotes host translation shutoff
Due to the potential for pleiotropic effects of nsP2 and/or capsid mutations on multiple viral activities confounding interpretation of virus infection experiments, we developed a plasmid expression system similar to that previously described (58) to assess the role individual VEEV proteins in host macromolecular synthesis shutoff and their involvement in antiviral state resistance. To ensure the observed effects of mutations in viral proteins were not due to differential expression levels in this system, we measured expression levels of WT and mutant nsP2 and capsid and VEEV nsP1 and nsP3 following transfection of plasmids (Fig 7A). There were no apparent differences observed between WT and mutant VEEV, SINV and CHIKV or WT EEEV nsP2. The expression level of EEEV capsid appeared lower than that of VEEV capsid, while VEEV nsP1 and nsP3 expression levels appeared similar to each other.
Fig 7. Individually expressed viral proteins block transcription and translation.
(A) Huh7 cells were transfected with indicated plasmids and lysates were collected at 18h post transfection. Western blots for HA-tag were performed as described in Materials and Methods. (B and C) Huh7 cells were transfected with plasmids coding for indicated viral proteins and labeled with 100μCi/ml of [35S] Cys/Met for 2h at 8-24h post transfection. Lysates were collected and resolved on SDS-PAGE gels and visualized as described in Materials and Methods. (B) Representative image of nsP and capsid induced shutoff compared to GFP control. (C) Densitometry was performed to quantify the extent of shutoff following transfection of indicated plasmids. ****, P<0.0001; **, P<0.01 using t-test and One-way ANOVA. Ns = not significant. Data is the average of four independent experiments. All error bars are standard deviations. (D) Huh7 cells were transfected with plasmids coding for indicated viral proteins and lysates were collected in triplicate at 12h post transfection. RT PCR for human gamma actin intron #3 was performed as described in Materials and Methods. ****, P<0.0001; ***, P<0.01; **, P<0.02 using t-test. Ns = not significant. Data is representative of four independent experiments. All error bars are standard deviations.
We had previously observed shutoff of host translation, but not transcription, in cells infected with a VEEV replicon expressing only nsPs (8), and hypothesized that one or more VEEV nsPs induced this activity during infection. We transfected Huh7 cells with plasmids encoding capsid and WT/mutant nsP2 proteins from different alphaviruses, and measured their ability to repress host translation using [35S] pulse-labeling to detect steady-state translation. Expression of VEEV/EEEV capsid and VEEV/SINV/CHIKV WT nsP2 was sufficient to significantly (P<0.0001) diminish host translation (by >50%) when compared to GFP expressing control cells (Fig 7B and C). The extent of translation shutoff is lower than that observed during replicating virus infections (Fig 4), likely due to lower efficiency of the transfection process and/or additional effects of infection on host cell viability. Additionally, the viral proteins expressed from plasmids likely arrest their own transcription/translation. VEEV nsP2 739L delayed induction of host translation shutoff during viral infection, but exhibited only a consistent, but non-significant decrease in translation shutoff compared to VEEV nsP2 WT when expressed from plasmid. SINV nsP2 726G did not shutoff host cell translation. VEEV nsP1 and VEEV nsP3 did not affect host translation (Fig 7B). Notably, expression of WT EEEV nsP2 was also unable to shutoff host translation, in contrast to WT VEEV nsP2.
We also observed ablation of host translation in cells expressing VEEV capsid. In order to determine if translation shutoff by viral proteins was an independent activity, or potentially caused by shutoff of host transcription, we tested the ability of individually expressed viral proteins to induce host transcription shutoff. We used a quantitative RT-PCR (qRT-PCR) assay to measure levels of an intron from a highly expressed, constitutively active gene whose half-life in cells is thought to be in the order of minutes. This allows use of intron levels as a measure of pol II transcription levels in a cell (59) and this type of assay has previously been used to analyze transcription of the human insulin gene (59, 60) and to measure transcription shutoff during La Crosse virus infection (61, 62). We transfected Huh7 cells with plasmids expressing SINV, CHIKV, EEEV and VEEV nsP2 and VEEV and EEEV capsid proteins and used qRT-PCR to measure levels of gamma actin intron #3 (61) at 12h post-transfection (Fig 7D). Levels of this intron were reduced 10-fold (P<0.0001) with 6h of Actinomycin D treatment as a control for pol II transcription inhibition. SINV and CHIKV nsP2 and EEEV and VEEV capsid significantly inhibited transcription (by >50%; P<0.02) when compared to a GFP control. In contrast, EEEV and VEEV nsP2 did not inhibit transcription in transfected cells. These observations with individually expressed proteins confirm and extend published observations using viruses (29, 31, 32, 39, 63). From our results we concluded that VEEV nsP2 was the viral nsP responsible for host translation, but not transcription shutoff.
Induction of host translation shutoff by VEEV nsP2 in IFN pre-treated cells contributes to its resistance to the antiviral state
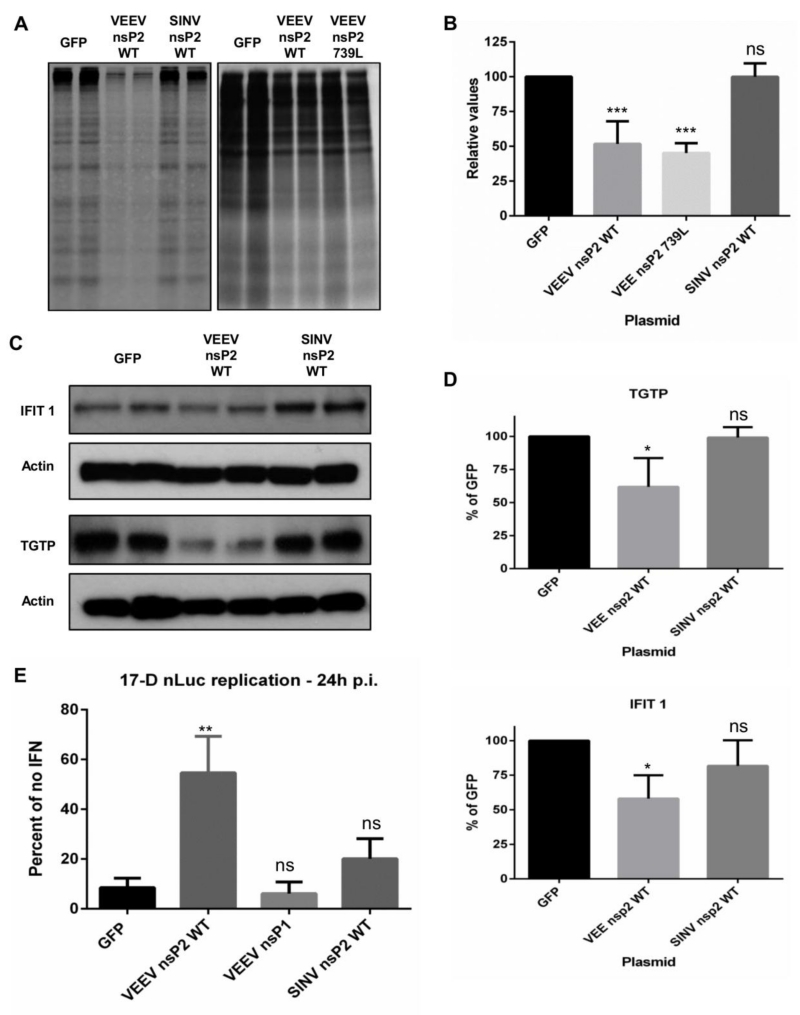
Our previous experiments using propagation-competent viruses indicated a role for both components of host macromolecular synthesis shutoff in the antiviral state resistance of VEEV, mediated by nsP2 (translation shutoff) and capsid (transcription shutoff). As the nonstructural proteins are produced earlier than structural proteins during viral infection (49), we addressed the role of translation shutoff in the antiviral state resistance of VEEV in a context independent of replication rates and absent other viral proteins using the nsP overexpression system. MEF cells were used in these experiments as they establish a potent antiviral state in response to IFN treatment in contrast to Huh7 cells. MEF cells were pre-treated with mouse IFN for 16h and transfected with plasmids encoding GFP (control) or VEEV/SINV WT nsP2. Cells were labeled with [35S] Cys/Met at 24h post transfection. Similar to data in untreated Huh7 cells, host translation in untreated MEF cells was significantly inhibited by the expression of either VEEV or SINV nsP2 WT when compared to GFP expressing control cells (data not shown). However, a significant reduction in host translation in IFN-primed cells (>50%; P<0.0004) was only observed in VEEV nsP2 WT transfected cells (Fig 8A and B), indicating that VEEV was capable of inhibiting translation despite the presence of an antiviral state, whereas SINV nsP2 was not. This is consistent with the published observation that VEEV successfully induced global translation shutoff during infection of IFN-primed cells (8). This data, in conjunction with the observation that VEEV successfully replicates in the presence of an antiviral state (Fig 1), and our mutant virus studies (Table 2) supports the conclusion that the ability of VEEV nsP2 to induce translation shutoff in IFN-primed cells contributes to a host cell environment permissive to virus replication. We also tested the ability of VEEV nsP2 739L to shutoff translation in IFN-primed cells. Similar to observations made in untreated cells, VEEV nsP2 739L expressed from plasmid was successful at inducing translation shutoff (Fig 8A and B).
Fig 8. VEEV nsP2 can shut off global host macromolecular synthesis and downregulate the antiviral state.
(A and B) MEF cells were treated with 100-150 IU/mL (10mL total) mouse IFN in a 60mm dish or 400 IU/mL (10mL total) mouse IFN in a 100 mm dish for 16h and transfected with indicated plasmids. Cells were labeled with 100μCi/ml of [35S] Cys/Met for 2h at 24h post transfection. Lysates were collected and resolved on SDS-PAGE gels and visualized as described in Materials and Methods. (A) Representative images of nsP2 induced shutoff in IFN primed cells compared to Mock or GFP control. (B) Densitometry was performed to quantify the extent of shutoff. ***, P<0.0004 using t-test. Ns = not significant. Data is the average of four independent experiments. (C and D) MEF cells were treated with 125-150 IU/mL mouse IFN in a 60mm dish for 16h and transfected with indicated plasmids. Lysates were collected at 24h post transfection and western blots for ISGs were performed as described in Materials and Methods. (C) Representative blot of IFIT1 and TGTP levels in VEEV and SINV nsP2 transfected cells. (D) Densitometry was performed to normalize IFIT1 and TGTP levels to actin and compared to GFP transfected control. *, P<0.04 using t-test. (E) Huh7 cells were treated with 2000 IU/mL for 16h and transfected with plasmids expressing indicated proteins. Cells were infected with 17-D nLuc at 8h following transfection (MOI = 1.5) and lysates were collected for luciferase assay at 24h p.i. Data is expressed as RLU/μg in IFN-primed cells as a percentage of untreated cells for each plasmid. Results are average of three independent experiments. **, P<0.002 using t-test. Ns = not significant. All error bars are standard deviations.
We speculated that if this model were correct, expression of VEEV nsP2 in IFN-primed cells should result in modulation of the cellular environment to favor viral replication by decreasing protein levels of antiviral ISG’s. Several ISG’s have been shown to have short half-lives (64-67) between 6-18h, which makes the antiviral state potentially vulnerable to a sustained translation shutoff. In order to test this hypothesis, we quantified protein levels of two ISG’s in IFN-primed MEF cells transfected with plasmids expressing VEEV nsP2 and SINV nsP2 (Fig 8C and D). IFIT1 levels in VEEV nsP2 expressing MEF’s were significantly (by 40-50%; P<0.05) lower than levels in GFP expressing control cells, while levels in SINV nsP2 expressing cells did not significantly differ from those of GFP expressing cells. Similarly, levels of TGTP were significantly lower (by 25%; P<0.001) in VEEV nsP2 expressing MEF’s when compared to GFP and SINV nsP2 expressing cells (Fig 8C and D). This data taken together with the resistance of VEEV to individual overexpressed antiviral proteins (Fig 1C) suggests the resistance phenotype of VEEV is at least partially mediated through a global effect such as host macromolecular synthesis shutoff rather than the presence of multiple separate viral resistance factors.
To demonstrate the functional significance of host translation shutoff in diminishing the antiviral state, we measured the replication of an IFN sensitive virus, yellow fever virus (YFV) 17-D strain, engineered to express nano-Luciferase (nLuc) as a cleavable fusion with the YFV capsid protein (described in Watson et.al., manuscript in preparation) in IFN-primed Huh7 cells transfected with plasmids expressing GFP (control) or virus nsP2 proteins. Replication of 17-D virus was inhibited 50% in Huh7 cells primed with only 5IU IFN when compared to replication in untreated cells (data not shown). Huh7 cells were treated with 2000 IU IFN for 16h and infected with 17-D nLuc 8h post transfection. The replication level of 17-D in VEEV nsP2 expressing IFN-primed cells was significantly (P<0.002) higher than that observed in GFP or VEEV nsP1 expressing control cells (Fig 8E). In contrast, 17-D replication in SINV nsP2 expressing cells was not significantly different from replication in controls. Similar results were observed in Vero cells (data not shown). We conclude that VEEV nsP2 induced translation shutoff in primed cells reduces the efficacy of the antiviral state, supporting the replication of an IFN-sensitive virus.
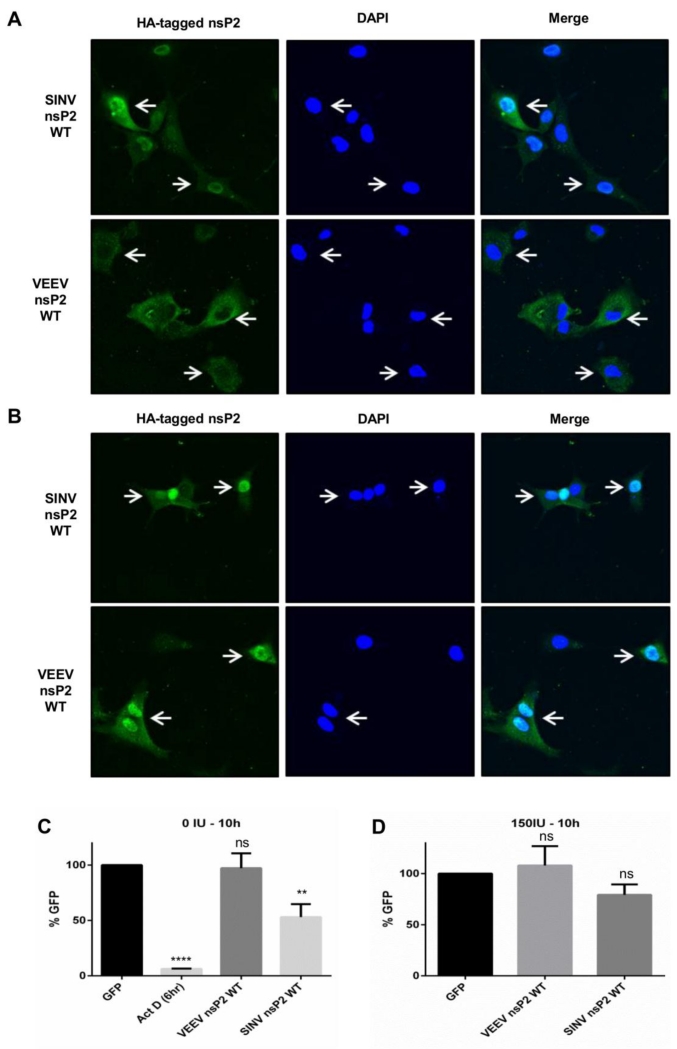
To determine if the difference in functionality between these proteins was due to altered localization or activity in IFN pre-treated cells, we investigated whether a change in localization of VEEV/SINV nsP2 occurred in response to IFN pre-treatment (Fig 9A and B). In unprimed cells, SINV nsP2 was present in the nucleus and cytoplasm, which is consistent with published observations (68). Similarly, VEEV nsP2 was present mainly in the cytoplasm in most transfected cells (69) (Fig 9A). Pre-treatment with IFN did not noticeably alter the distribution of SINV nsP2 (Fig 9B). Surprisingly, VEEV nsP2 localized to the nucleus in nearly all primed and transfected cells (Fig 9B). This raised the possibility that nucleus-localized VEEV nsP2 induced transcription shutoff in IFN-primed cells which indirectly ablated host translation (Fig 8A and B). However, expression of SINV nsP2 significantly (P<0.003) inhibited host transcription compared to GFP expressing control in unprimed cells, while VEEV nsP2 did not (Fig 9C). However, no inhibition of host transcription was observed upon expression of VEEV or SINV nsP2 in primed cells (Fig 9D). Therefore, the movement of VEEV nsP2 into the nucleus in IFN-primed cells does not induce host transcription shutoff.
Fig 9. Localization of VEEV nsP2 is altered in IFN pre-treated MEF cells.
(A and B) MEF cells were untreated (A) or pre-treated (B) with 150 IU/mL mouse IFN in a 60mm dish for 16h and transfected with plasmids expressing indicated viral proteins. Cells were fixed at 24h post transfection and stained for HA tag as described in Materials and Methods. (C and D) MEF cells were untreated (C) or pre-treated (D) with 150 IU/mL mouse IFN in a 60mm dish for 16h and transfected with plasmids expressing indicated proteins. Lysates were collected at indicated times post transfection. RT PCR for human gamma actin intron #3 was performed as described in Materials and Methods. Results are average of three independent experiments. ****, P<0.0001; **, P<0.003 using t-test. Ns = not significant. All error bars are standard deviations.
VEEV nsP2 or capsid mutants are attenuated in mice but do not increase systemic IFN induction
To determine if alterations in translation shutoff were attenuating in vivo, we infected mice with VEEV WT, CD, 739L and CD/739L viruses. Morbidity and mortality profiles and disease phenotypes demonstrated that attenuation co-varied with replication rates and IFN sensitivity measured in vitro (Fig 1 and 6; Fig 10A and B). IFN-α4 induction in the serum of infected mice at 12h p.i. (peak IFN is typically 12-18h p.i. (6)) as measured by ELISA was similar between viruses, while IFN-α/β measured by bioassay showed the mutant viruses to induce generally lower levels than the WT (Fig 10C), likely reflecting more limited replication. Therefore, systemic IFN induction appears to be independent of differential macromolecular synthesis shutoff characteristics in vivo and unlikely to determine virulence between viruses.
Fig 10. VEEV mutants are attenuated in vivo.
(A) CD-1 mice were infected subcutaneously in the hind leg footpad with 100 PFU of indicated viruses and scored for degree of sickness at 24h intervals. (B) CD-1 mice were infected subcutaneously in the hind leg footpad with 1000 PFU of indicated viruses and average survival time was determined. 4 mice per group. (C and D) Serum from mice infected with indicated viruses was collected at 12h p.i. and used to measure IFN-α using ELISA (C) or used to measure IFN-α/β using a bioassay (D) as described in Materials and Methods. **, P<0.006 using One-way ANOVA. Ns = not significant. All error bars are standard deviations.
Discussion
Interaction of VEEV with the IFN-induced antiviral state
During infection of mice, and presumably humans, most alphaviruses initially interact with myeloid cells which are likely the major source of systemic serum IFN (9, 15, 70). Notably, VEEV infection induces the highest levels of serum IFN of all alphaviruses tested, with lower levels observed during SINV infection (6), while EEEV or CHIKV infection results in little to no IFN production (6, 9). IFN induction during CHIKV infection occurs from non-hematopoietic cells (13). This induction of large amounts of IFN during VEEV infection, and lesser amounts during SINV infection, occurs within 12h of inoculation in vivo (6), upregulates ISG’s and establishes an antiviral state at uninfected sites distal to the initial infection site, including the central nervous system (71, 72) an important site of replication during VEEV infection. Thus, it is likely that most, if not all, cells infected by VEEV (and possibly SINV) following the initial round of infection in vivo have already been exposed to IFN and are primed to resist infection.
The ability of the IFN induced antiviral state to limit virus infection is believed to be closely associated with severity of alphavirus induced disease and mortality. Alphaviruses overcome the antiviral state by either preventing its activation (EEEV; (9, 21)) or by resisting its inhibitory effects on viral replication (VEEV, SINV; (6)). Previous reports have suggested that the balance between virus replication and IFN secretion/response early during infection determines the severity of disease (34). However, those studies were performed using an in vitro model of SINV infection and did not take into account the induction of large amounts of IFN from myeloid cells following in vivo inoculation, which rapidly primes uninfected cells in vivo. Alphaviruses resistant to this priming successfully replicate, infect the brain and cause disease (VEEV), while the replication of sensitive alphaviruses is controlled (e.g., SINV). It is notable that SINV infection in normal adult mice is completely controlled with no visible signs of disease observed during the course of infection (15), while VEEV infection leads to essentially uniform mortality (17).
Our data show that VEEV replication is most resistant to the effects of IFN priming among all alphaviruses tested, and that this resistance localizes at least in part to the nonstructural protein/non-translated regions of the genome. VEEV replication in IFN-primed cells approached levels observed in untreated cells, while other alphaviruses were 10-4000 times more sensitive to the antiviral state. VEEV replication in IFN-primed cells lags by ~6h compared to replication in unprimed cells, which suggests VEEV modulates the cellular environment to favor viral replication during this phase. Using reporter RNA’s that mimicked initial translation of the incoming viral genome in an infected cell, we found that the antiviral state suppressed VEEV, EEEV, SINV and CHIKV replication at the point of initial translation, in agreement with and extending published observations (36, 45, 46). Nevertheless, low-level translation of reporters in primed cells indicates that the nsPs of alphaviruses that infect IFN-primed cells are indeed produced. This suggests that an activity or activities of one or more nsPs of VEEV would be present to successfully antagonize, in whole or in part, the antiviral state, while the activities of other tested alphavirus nsPs do so to a lesser degree.
NsP2 is a suppressor of multiple host responses but activity varies between alphaviruses
Previous studies, primarily with SINV, have demonstrated that mutations in the C-terminal region of nsP2 can impact shutoff of transcription and translation (31, 33, 52). With VEEV and EEEV New World viruses, the capsid protein affects host transcription (30, 54), which we confirm but our data suggest that this activity is delayed versus the translation inhibiting activity of nsP2. Additionally, data from CHIKV and SINV (31, 73) indicates that translation shutoff occurs before shutoff of transcription during infection, and the activities are mediated through distinct mechanisms (i.e. transcription shutoff does not directly mediate translation shutoff).
With SINV, a single mutation (726G) can render the virus defective in abrogation of each of these activities (34, 52). A widely conserved “PGG” domain in which the 726G (SINV) or 718S (CHIKV) mutation substitutes the proline has been suggested to be the critical site for these activities of nsP2 (32, 34, 52). We confirm that the “P-G” mutation at aa 726 affects each activity of the SINV TR339 consensus strain using both viruses and individually expressed nsP2 (nsP2 726G transcription shutoff data not shown). For CHIKV, in contrast to published data with P718S which suggest this mutation abrogates nsP2 mediated host macromolecular synthesis shutoff (32), our data indicates P718S has no effect on this activity of CHIKV nsP2 (data not shown). The substitution of “G” or “S” for “P” in the VEEV “PGG” domain (713G), analogous to the SINV/CHIKV mutations had no effect on these activities. Indeed it had a slight enhancing effect on translation shutoff (current studies and data not shown) as well as virulence in mice (not shown). EEEV does not have the canonical “PGG” domain; rather its sequence is “KGG.” It is tempting to speculate that the sequence differences result in the failure of EEEV nsP2 to inhibit translation. However, substitution of the “K” residue for “P” in the “PPG” site of VEEV had no effect on transcription or translation shutoff (data not shown). This along with the 713 “G” and “S” mutant data, suggest that this is not a critical residue with New World viruses and, perhaps, Old World viruses other than SINV. The 739L mutation of VEEV, selected for limited cytopathogenicity in BHK cells (53), lessened translation shutoff in the context of virus infection but the individually expressed protein affected translation shutoff in a reproducible but ultimately non-significant manner. Studies with WT and 739L VEEV replicons were inconclusive due to low infectivity of Vrep 739L (data not shown) for multiple cell types. It is possible that the effects of 739L on translation shutoff only manifest in the context of virus infection, and not when VEEV nsP2 is expressed alone. While VEEV nsP2 739L shut off host translation when expressed from a plasmid, the delayed induction of translation shutoff in VEEV 739L virus infected cells compared to VEEV WT implies that this mutation may be at least partially deficient at this activity independent of its effect on viral replication. While VEEV 739L virus replicated more slowly than WT, replication in 739L infected cells suggests that nsP2 levels were presumably sufficient to induce translation shutoff during infection. We conclude that the C-terminal region of New world alphavirus nsP2 but not the “PGG” domain is directly involved in translational shutoff, in contrast to the critical requirement of the “P” residue for efficient function of SINV nsP2. Further, the importance of the “PGG” domain or “P” residue appears to be limited to SINV nsP2, suggesting that activities attributed to nsP2 may be localized to unique regions of the protein in different alphaviruses.
Effects of VEEV induced translation shutoff on the antiviral state
Our virus mutagenesis data suggest that replication efficiency of VEEV is most closely associated with antiviral state resistance in vitro, which itself positively associates with macromolecular synthesis shutoff. The sensitivity of mutants to IFN priming correlates with their growth rates in vitro, suggesting that an inability to efficiently shutoff host macromolecular synthesis reduces the growth of mutant viruses and renders mutants more sensitive to cell stress responses. The viruses grow more slowly in cells that do not make IFN but other stress responses could be active as well as constitutive ISG induction. Our data show host macromolecular synthesis shutoff through its ability to prevent synthesis of new ISG’s, control IFN induction and enhance viral growth in infected cells plays a role in resistance to the anti-viral state, as defective mutants (739L and CD/739L) are more sensitive to the antiviral state when compared to mutants (713G, 713S) having no effect on this process.
During alphavirus infection, the nonstructural proteins including nsP2 are produced first, and are required for production of structural proteins including capsid from the subgenomic promoter (49). Thus it is likely that virus induced translation shutoff in IFN-primed cells is important early during resistance of VEEV to the antiviral state, and the effect of capsid mediated transcription shutoff on the antiviral state is additive to, and very likely delayed versus, the activity of nsP2 due to temporally deferred synthesis of capsid during infection. We demonstrate that VEEV nsP2 induces translation shutoff in IFN-primed cells while SINV nsP2 is unable to shutoff host translation or transcription. As expected, we observed lower levels of measured ISG’s in IFN-primed cells expressing VEEV nsP2 when compared to SINV nsP2. Translation shutoff also supports the replication of Yellow Fever vaccine strain 17-D, a virus highly susceptible to IFN priming, as replication of 17-D virus in IFN-primed cells expressing VEEV nsP2 approaches replication levels in unprimed cells. Our data suggest VEEV nsP2 induced translation shutoff in IFN-primed cells early after infection is an important factor in the resistance phenotype of VEEV by decreasing levels of pre-existing ISG’s and reducing production of new ISG molecules, which engenders a cellular environment permissive to viral replication. In contrast, the inability of SINV nsP2 to shutoff transcription or translation in IFN pre-treated cells likely explains in part its sensitivity to IFN priming in vitro, and lack of disease in immunocompetent adult mouse models where SINV induces significant serum IFN. It is unclear whether one or both these activities contributes to antiviral state resistance, and whether or not one predominates, as both are localized to a single domain of nsP2 in SINV. Nonetheless, the inhibition of nsP2 activity by IFN likely results in the sensitivity of SINV to the antiviral state. Similarly, the sensitivity of other alphaviruses to the antiviral state may in part result from their inefficient induction of translation shutoff early after infection. Interestingly, although EEEV nsP2 is unable to induce host translation shutoff, EEEV demonstrates mortality similar to VEEV in mice (9, 22), and is far more virulent in humans than VEEV (74). While EEEV capsid can shutoff transcription in cells (30), EEEV mediated suppression of systemic innate immune responses in vivo involves avoidance of lymphoid tissue targeting through heparan sulfate binding (22) as well as suppression of replication in myeloid cells via virus genome binding to a hematopoietic cell-specific microRNA, miR142-3p (21). Thus, very closely related alphaviruses utilize very different strategies to overcome innate immunity. Furthermore, our current and previous results suggest the efficacy of innate immunity suppression as major factors in the relative virulence of arthritogenic alphaviruses (rarely fatal), VEEV (occasionally fatal) and EEEV (frequently fatal).
Highlights.
VEEV is the alphavirus most resistant to the IFN-induced antiviral state.
Transcription and translation shutoff are independently mediated viral activities.
VEEV nsP2 shuts off host translation but not transcription.
VEEV nsP2, but not SINV nsP2, shuts of host translation in IFN-primed cells.
NsP2-mediated translation shutoff is critical for the resistance phenotype of VEEV.
Acknowledgements
We thank Matthew Dunn, Jenna Girardi and Nicolas Garcia for excellent technical assistance. We thank Carolyn Coyne for assistance with confocal microscopy. We thank Scott Weaver, Michal Parker, Pamela Glass and Robert Johnston for providing reagents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010;8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 2.Van Bortel W, Dorleans F, Rosine J, Blateau A, Rousset D, Matheus S, Leparc-Goffart I, Flusin O, Prat C, Cesaire R, Najioullah F, Ardillon V, Balleydier E, Carvalho L, Lemaitre A, Noel H, Servas V, Six C, Zurbaran M, Leon L, Guinard A, van den Kerkhof J, Henry M, Fanoy E, Braks M, Reimerink J, Swaan C, Georges R, Brooks L, Freedman J, Sudre B, Zeller H. Chikungunya outbreak in the Caribbean region, December 2013 to March 2014, and the significance for Europe. Euro Surveill. 2014;19 doi: 10.2807/1560-7917.es2014.19.13.20759. [DOI] [PubMed] [Google Scholar]
- 3.Vega-Rua A, Zouache K, Girod R, Failloux AB, Lourenco-de-Oliveira R. High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of Chikungunya virus. J Virol. 2014;88:6294–6306. doi: 10.1128/JVI.00370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis. 2014;8:e2921. doi: 10.1371/journal.pntd.0002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner CL, Burke CW, Higgs ST, Klimstra WB, Ryman KD. Interferon-alpha/beta deficiency greatly exacerbates arthritogenic disease in mice infected with wild-type chikungunya virus but not with the cell culture-adapted live-attenuated 181/25 vaccine candidate. Virology. 2012;425:103–112. doi: 10.1016/j.virol.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryman KD, Klimstra WB. Host responses to alphavirus infection. Immunol Rev. 2008;225:27–45. doi: 10.1111/j.1600-065X.2008.00670.x. [DOI] [PubMed] [Google Scholar]
- 7.Suhrbier A, Jaffar-Bandjee MC, Gasque P. Arthritogenic alphaviruses--an overview. Nat Rev Rheumatol. 2012;8:420–429. doi: 10.1038/nrrheum.2012.64. [DOI] [PubMed] [Google Scholar]
- 8.Yin J, Gardner CL, Burke CW, Ryman KD, Klimstra WB. Similarities and differences in antagonism of neuron alpha/beta interferon responses by Venezuelan equine encephalitis and Sindbis alphaviruses. J Virol. 2009;83:10036–10047. doi: 10.1128/JVI.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner CL, Burke CW, Tesfay MZ, Glass PJ, Klimstra WB, Ryman KD. Eastern and Venezuelan equine encephalitis viruses differ in their ability to infect dendritic cells and macrophages: impact of altered cell tropism on pathogenesis. J Virol. 2008;82:10634–10646. doi: 10.1128/JVI.01323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wauquier N, Becquart P, Nkoghe D, Padilla C, Ndjoyi-Mbiguino A, Leroy EM. The acute phase of Chikungunya virus infection in humans is associated with strong innate immunity and T CD8 cell activation. J Infect Dis. 2011;204:115–123. doi: 10.1093/infdis/jiq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow A, Her Z, Ong EK, Chen JM, Dimatatac F, Kwek DJ, Barkham T, Yang H, Renia L, Leo YS, Ng LF. Persistent arthralgia induced by Chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J Infect Dis. 2011;203:149–157. doi: 10.1093/infdis/jiq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messaoudi I, Vomaske J, Totonchy T, Kreklywich CN, Haberthur K, Springgay L, Brien JD, Diamond MS, Defilippis VR, Streblow DN. Chikungunya virus infection results in higher and persistent viral replication in aged rhesus macaques due to defects in anti-viral immunity. PLoS Negl Trop Dis. 2013;7:e2343. doi: 10.1371/journal.pntd.0002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schilte C, Couderc T, Chretien F, Sourisseau M, Gangneux N, Guivel-Benhassine F, Kraxner A, Tschopp J, Higgs S, Michault A, Arenzana-Seisdedos F, Colonna M, Peduto L, Schwartz O, Lecuit M, Albert ML. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J Exp Med. 2010;207:429–442. doi: 10.1084/jem.20090851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrnes AP, Durbin JE, Griffin DE. Control of Sindbis virus infection by antibody in interferon-deficient mice. J Virol. 2000;74:3905–3908. doi: 10.1128/jvi.74.8.3905-3908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryman KD, Klimstra WB, Nguyen KB, Biron CA, Johnston RE. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J Virol. 2000;74:3366–3378. doi: 10.1128/jvi.74.7.3366-3378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryman KD, White LJ, Johnston RE, Klimstra WB. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 2002;15:53–76. doi: 10.1089/088282402317340233. [DOI] [PubMed] [Google Scholar]
- 17.Davis NL, Grieder FB, Smith JF, Greenwald GF, Valenski ML, Sellon DC, Charles PC, Johnston RE. A molecular genetic approach to the study of Venezuelan equine encephalitis virus pathogenesis. Arch Virol Suppl. 1994;9:99–109. doi: 10.1007/978-3-7091-9326-6_11. [DOI] [PubMed] [Google Scholar]
- 18.Anishchenko M, Paessler S, Greene IP, Aguilar PV, Carrara AS, Weaver SC. Generation and characterization of closely related epizootic and enzootic infectious cDNA clones for studying interferon sensitivity and emergence mechanisms of Venezuelan equine encephalitis virus. J Virol. 2004;78:1–8. doi: 10.1128/JVI.78.1.1-8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguilar PV, Paessler S, Carrara AS, Baron S, Poast J, Wang E, Moncayo AC, Anishchenko M, Watts D, Tesh RB, Weaver SC. Variation in interferon sensitivity and induction among strains of eastern equine encephalitis virus. J Virol. 2005;79:11300–11310. doi: 10.1128/JVI.79.17.11300-11310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis NL, Willis LV, Smith JF, Johnston RE. In vitro synthesis of infectious venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology. 1989;171:189–204. doi: 10.1016/0042-6822(89)90526-6. [DOI] [PubMed] [Google Scholar]
- 21.Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, Mildner A, Weaver SC, Ryman KD, Klimstra WB. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2014;506:245–248. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner CL, Ebel GD, Ryman KD, Klimstra WB. Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc Natl Acad Sci U S A. 2011;108:16026–16031. doi: 10.1073/pnas.1110617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 24.Sarkar SN, Sen GC. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther. 2004;103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O’Guin AK, Schmidt RE, Levine B, Virgin HWt. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, Huang C, Tonelli M, Paessler S, Takeda K, Klimstra WB, Amarasinghe GK, Diamond MS. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014;343:783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perri S, Greer CE, Thudium K, Doe B, Legg H, Liu H, Romero RE, Tang Z, Bin Q, Dubensky TW, Jr., Vajdy M, Otten GR, Polo JM. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J Virol. 2003;77:10394–10403. doi: 10.1128/JVI.77.19.10394-10403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frolova EI, Fayzulin RZ, Cook SH, Griffin DE, Rice CM, Frolov I. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J Virol. 2002;76:11254–11264. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguilar PV, Weaver SC, Basler CF. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J Virol. 2007;81:3866–3876. doi: 10.1128/JVI.02075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79:9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fros JJ, van der Maten E, Vlak JM, Pijlman GP. The C-terminal domain of chikungunya virus nsP2 independently governs viral RNA replication, cytopathicity, and inhibition of interferon signaling. J Virol. 2013;87:10394–10400. doi: 10.1128/JVI.00884-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol. 2007;81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frolov I, Akhrymuk M, Akhrymuk I, Atasheva S, Frolova EI. Early events in alphavirus replication determine the outcome of infection. J Virol. 2012;86:5055–5066. doi: 10.1128/JVI.07223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atasheva S, Krendelchtchikova V, Liopo A, Frolova E, Frolov I. Interplay of acute and persistent infections caused by Venezuelan equine encephalitis virus encoding mutated capsid protein. J Virol. 2010;84:10004–10015. doi: 10.1128/JVI.01151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynaud JM, Kim DY, Atasheva S, Rasalouskaya A, White JP, Diamond MS, Weaver SC, Frolova EI, Frolov I. IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon. PLoS Pathog. 2015;11:e1004863. doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atasheva S, Kim DY, Frolova EI, Frolov I. Venezuelan equine encephalitis virus variants lacking transcription inhibitory functions demonstrate highly attenuated phenotype. J Virol. 2015;89:71–82. doi: 10.1128/JVI.02252-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. 2007;81:11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguilar PV, Adams AP, Wang E, Kang W, Carrara AS, Anishchenko M, Frolov I, Weaver SC. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J Virol. 2008;82:4920–4930. doi: 10.1128/JVI.02514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klimstra WB, Ryman KD, Johnston RE. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsetsarkin K, Higgs S, McGee CE, De Lamballerie X, Charrel RN, Vanlandingham DL. Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis. 2006;6:325–337. doi: 10.1089/vbz.2006.6.325. [DOI] [PubMed] [Google Scholar]
- 42.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas JM, Klimstra WB, Ryman KD, Heidner HW. Sindbis virus vectors designed to express a foreign protein as a cleavable component of the viral structural polyprotein. J Virol. 2003;77:5598–5606. doi: 10.1128/JVI.77.10.5598-5606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 45.Ryman KD, Meier KC, Nangle EM, Ragsdale SL, Korneeva NL, Rhoads RE, MacDonald MR, Klimstra WB. Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells. J Virol. 2005;79:1487–1499. doi: 10.1128/JVI.79.3.1487-1499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tesfay MZ, Yin J, Gardner CL, Khoretonenko MV, Korneeva NL, Rhoads RE, Ryman KD, Klimstra WB. Alpha/beta interferon inhibits cap-dependent translation of viral but not cellular mRNA by a PKR-independent mechanism. J Virol. 2008;82:2620–2630. doi: 10.1128/JVI.01784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Trgovcich J, Aronson JF, Johnston RE. Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis. Virology. 1996;224:73–83. doi: 10.1006/viro.1996.0508. [DOI] [PubMed] [Google Scholar]
- 49.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorchakov R, Frolova E, Williams BR, Rice CM, Frolov I. PKR-dependent and - independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol. 2004;78:8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frolov I, Agapov E, Hoffman TA, Jr., Pragai BM, Lippa M, Schlesinger S, Rice CM. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73:3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burke CW, Gardner CL, Steffan JJ, Ryman KD, Klimstra WB. Characteristics of alpha/beta interferon induction after infection of murine fibroblasts with wild-type and mutant alphaviruses. Virology. 2009;395:121–132. doi: 10.1016/j.virol.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrakova O, Volkova E, Gorchakov R, Paessler S, Kinney RM, Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garmashova N, Atasheva S, Kang W, Weaver SC, Frolova E, Frolov I. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J Virol. 2007;81:13552–13565. doi: 10.1128/JVI.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desmyter J, Melnick JL, Rawls WE. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc Natl Acad Sci U S A. 1988;85:5259–5263. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang A, Masante C, Buchholz UJ, Dutch RE. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J Virol. 2012;86:3230–3243. doi: 10.1128/JVI.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clement JQ, Qian L, Kaplinsky N, Wilkinson MF. The stability and fate of a spliced intron from vertebrate cells. RNA. 1999;5:206–220. doi: 10.1017/s1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans-Molina C, Garmey JC, Ketchum R, Brayman KL, Deng S, Mirmira RG. Glucose regulation of insulin gene transcription and pre-mRNA processing in human islets. Diabetes. 2007;56:827–835. doi: 10.2337/db06-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verbruggen P, Ruf M, Blakqori G, Overby AK, Heidemann M, Eick D, Weber F. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J Biol Chem. 2011;286:3681–3692. doi: 10.1074/jbc.M110.154799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng C, Sharp PA. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol Cell Biol. 2003;23:1961–1967. doi: 10.1128/MCB.23.6.1961-1967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garmashova N, Gorchakov R, Frolova E, Frolov I. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J Virol. 2006;80:5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo J, Peters KL, Sen GC. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 65.Bogunovic D, Boisson-Dupuis S, Casanova JL. ISG15: leading a double life as a secreted molecule. Exp Mol Med. 2013;45:e18. doi: 10.1038/emm.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor JL, D’Cunha J, Tom P, O’Brien WJ, Borden EC. Production of ISG-15, an interferon-inducible protein, in human corneal cells. J Interferon Cytokine Res. 1996;16:937–940. doi: 10.1089/jir.1996.16.937. [DOI] [PubMed] [Google Scholar]
- 68.Akhrymuk I, Kulemzin SV, Frolova EI. Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol. 2012;86:7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Atasheva S, Garmashova N, Frolov I, Frolova E. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in Mammalian but not in mosquito cells. J Virol. 2008;82:4028–4041. doi: 10.1128/JVI.02330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacDonald GH, Johnston RE. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J Virol. 2000;74:914–922. doi: 10.1128/jvi.74.2.914-922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan W, Banks WA, Kastin AJ. Permeability of the blood-brain and blood-spinal cord barriers to interferons. J Neuroimmunol. 1997;76:105–111. doi: 10.1016/s0165-5728(97)00034-9. [DOI] [PubMed] [Google Scholar]
- 72.Habif DV, Lipton R, Cantell K. Interferon crosses blood-cerebrospinal fluid barrier in monkeys. Proc Soc Exp Biol Med. 1975;149:287–289. doi: 10.3181/00379727-149-38790. [DOI] [PubMed] [Google Scholar]
- 73.White LK, Sali T, Alvarado D, Gatti E, Pierre P, Streblow D, Defilippis VR. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J Virol. 2011;85:606–620. doi: 10.1128/JVI.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deresiewicz RL, Thaler SJ, Hsu L, Zamani AA. Clinical and neuroradiographic manifestations of eastern equine encephalitis. N Engl J Med. 1997;336:1867–1874. doi: 10.1056/NEJM199706263362604. [DOI] [PubMed] [Google Scholar]