Abstract
In several neurodegenerative diseases in which misfolded proteins accumulate there is impairment of the ubiquitin proteasome system (UPS). We tested if a similar disruption of proteostasis occurs in hereditary peripheral neuropathies. In sciatic nerves from mouse models of two human neuropathies, Myelin Protein Zero mutation (S63del) and increased copy number (P0 overexpression), polyubiquitinated proteins accumulated, and the overall rates of protein degradation were decreased. 26S proteasomes affinity-purified from sciatic nerves of S63del mice were defective in degradation of peptides and a ubiquitinated protein, unlike proteasomes from P0 overexpression, which appeared normal. Nevertheless, cellular levels of 26S proteasomes were increased in both, through the proteolytic-activation of the transcription factor Nrf1, as occurs in response to proteasome inhibitors. In S63del, increased amounts of the deubiquitinating enzymes USP14, UCH37, and USP5 were associated with proteasomes, the first time this has been reported in a human disease model. Inhibitors of USP14 increased the rate of protein degradation in S63del sciatic nerves and unexpectedly increased the phosphorylation of eIF2α by Perk. Thus, proteasome content, composition and activity are altered in these diseases and USP14 inhibitors have therapeutic potential in S63del neuropathy.
Keywords: Schwann cell, deubiquitinase, myelin, protein quality control
Introduction
Myelin protein zero (P0) is synthesized by Schwann cells and represents between 20–50% of total protein in peripheral nerves (Patzig et al. 2011). Over 180 different mutations of P0 cause hereditary peripheral neuropathies with widely varying phenotypes in humans (Sanmaneechai et al. 2015; Scherer and Wrabetz 2008), mostly through gain of toxic function mechanisms (Wrabetz et al. 2006). The deletion of serine 63 (P0S63del) causes Charcot Marie Tooth 1B disease (Kulkens et al. 1993), a demyelinating neuropathy characterized by myelin destruction and failed attempts at remyelination (Miller et al. 2012). P0S63del accumulates in the endoplasmic reticulum (ER), inducing ER stress and a canonical unfolded protein response (UPR) (Pennuto et al. 2008; Wrabetz et al. 2006). In the S63del transgenic mouse model (S63del L), genetic ablation of the UPR transcription factor CHOP, or attenuation of translation by increasing phosphorylated eIF2α (p-eIF2α) through genetic or pharmacological inhibition of GADD34 phosphatase improves the neuropathy (D’Antonio et al. 2013; Das et al. 2015; Pennuto et al. 2008).
Increasing Myelin Protein Zero (Mpz) copy number causes neuropathies in humans (Maeda et al. 2012; Speevak and Farrell 2013). A transgenic mouse model, which over expresses P0 due to an increase in Mpz copy number (P0 OE) has a dysmyelinating neuropathy (Wrabetz et al. 2000) caused by premature adhesion in myelin sheath formation (Yin et al. 2000), but unlike S63del L, this condition lacks a canonical UPR (Pennuto et al. 2008). Thus, S63del L and P0 OE model two human neuropathies with different pathogenetic mechanisms that lead to distinct morphological phenotypes.
In eukaryotic cells most intracellular proteins are degraded by the ubiquitin proteasome system (UPS). Proteins are first modified by the attachment of a ubiquitin chain via an enzymatic cascade and are then degraded by the 26S proteasome in an ATP-dependent manner (Hershko and Ciechanover 1992). The 26S proteasome is composed of a 20S core particle (CP) with a 19S regulatory particle (RP) bound to one or both ends. Ubiquitinated proteins are bound by the 19S RP, deubiquitinated, unfolded, and translocated into the hollow 20S core particle for proteolytic digestion (Finley 2009). In higher eukaryotes, the 26S proteasome has three deubiquitinases (DUBs) associated with the 19S RP: the subunit Rpn11; and the cysteine-protease DUBs, UCH37, and USP14/Ubp6 (Lee et al. 2011). By deubiquitinating substrates, UCH37 and USP14 allow some ubiquitinated proteins to escape degradation (Lee et al. 2016; Lee et al. 2010; Lee et al. 2011). However, these enzymes can also allosterically activate the peptidase and ATPase activities of the proteasome (Peth et al. 2009; Peth et al. 2013).
Mutant proteins cause impairment of proteasome function in neurodegenerative diseases caused by PrPSC (Deriziotis et al. 2011; Kristiansen et al. 2007) and mutant Tau (Myeku et al. 2016). This impairment leads to an accumulation of misfolded proteins in the neurons and can disrupt normal cellular functions, contributing to the disease state (Sherman and Goldberg 2001). It has been suggested that there is proteasome impairment in the sciatic nerves of mouse models of CMT1A hereditary peripheral neuropathies caused by alterations in PMP22 (Fortun et al. 2006; Fortun et al. 2005). However, these previous studies did not examine degradation rates in the tissue, the 26S proteasome or its capacity to degrade proteins. Therefore, it is not known if there is proteasome impairment in models of hereditary peripheral neuropathies. It is also not known if alterations in P0 that cause hereditary peripheral neuropathies in humans impair proteasome function. Such studies are important because improving 26S proteasome function through phosphorylation of 26S proteasome subunits (Lokireddy et al. 2015; Myeku et al. 2016) or pharmacological inhibition of the DUB USP14 (Lee et al. 2010), have therapeutic potential against proteotoxic diseases by accelerating the proteasome-mediated degradation of misfolded proteins.
Cellular responses to impairment of the proteasome have been primarily investigated by treating cells in culture with proteasome inhibitors. If the proteasome is inhibited, polyubiquitinated proteins accumulate, protein degradation slows, proteasome peptidase activity decreases, and degradation rate of ubiquitinated proteins by the proteasome diminishes. Additionally, if the proteasome is partially inhibited, the transcription factor Nrf1 is proteolytically-activated and mediates the compensatory production of new proteasomes (Radhakrishnan et al. 2010; Sha and Goldberg 2014). Pharmacological inhibition of proteasomes can also lead to alterations in proteasome composition, such as an increased association of the DUB USP14/Ubp6 with the proteasome (Besche et al. 2014; Borodovsky et al. 2001). It is currently unclear if proteasome levels and DUBs are also altered in human diseases.
Here we investigate if protein degradation and UPS function are impaired in the sciatic nerves of two distinct mouse models of human neuropathies: S63del L and P0 OE. To rigorously investigate the function of 26S proteasomes, we affinity-purified them from mouse sciatic nerves and found clear defects only in S63del L. We further investigated the cellular response to the deficits in protein degradation and surprisingly found that the cellular content of proteasomes was increased in both neuropathies. These findings suggest activation of a cellular response to increase proteasome levels to compensate for the decreased degradative capacity. Additionally, more DUBs associated with the proteasomes in S63del L and pharmacological inhibitors of USP14 increased the overall degradation rate of proteins in ex vivo sciatic nerves. This is the first demonstration of a DUB increase on the proteasome in a model of human disease in vivo. Thus, an increased USP14 level on the 26S proteasome is likely contributing to the reduced protein degradation in S63del L sciatic nerves and may represent a therapeutic target.
Materials and Methods
Animal Models
All experiments involving animals followed protocols approved by Institutional Animal Care and Use Committees. P0 OE, S63del L, S63del H, S63del L//P0+/−, S63del L//P0−/−, P0−/−, P0cre/Perkflox/flox, and S63del L//P0cre/Perkflox/flox transgenic mice have been described previously (Fratta et al. 2011; Sidoli et al. 2016; Wrabetz et al. 2006; Wrabetz et al. 2000). All mice were maintained on the congenic FVB background and both sexes were used equally. Genotyping of transgenic mice was performed by PCR on tail genomic DNA extracted with phenol and chloroform. Unless otherwise indicated, all experiments were performed on sciatic nerves dissected between post-natal day 28 (P28) and P32.
Lysis and Immunoblotting
Sciatic nerves were flash-frozen and pulverized with mortar and pestle in liquid nitrogen. The pulverized tissue was suspended in CHAPS lysis buffer (25mM HEPES-KOH, 150 mM NaCl, 1 mM EDTA, and 0.3% CHAPS) containing the following inhibitors: 10 mM N-Ethylmalmeimide, 1 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and 1 μM Bortezomib. The crude lysate was then rotated for 15 minutes at 4°C 4°C and centrifuged for 20 minutes at 14,000 × g at 4°C. Total protein was quantified with Bradford Assay (Life Technologies). Lysates were standardized based on total protein measurements and separated by Bis Tris SDS-PAGE. Proteins were transferred to either nitrocellulose (Protran; VWR) or PVDF (Immobilin FL; EMD Millipore) membranes and immunoblotting was performed using the following antibodies per manufacturer’s instructions: rabbit anti ubiquitin (VU-1) (LifeSensors); rabbit anti PSMA6, rabbit anti USP14, rabbit anti PSMD11 (Bethyl Laboratories); rabbit anti UCH37 (Abcam); rabbit anti USP5, rabbit anti PSMA7, rabbit anti PSMB5, rabbit anti RPT4, rabbit anti GADD34, mouse anti BAX (ProteinTech); goat anti USP14 (E-17), rabbit anti NRF1 (H-285), rabbit anti NRF2 (C-20), goat anti PSMC1 (C-16), rabbit anti ubiquitin (FL-76), rabbit anti PA200 (A-13), rabbit anti PA28beta (L-19) (Santa Cruz); rabbit anti ADRM1, mouse anti eIF2alpha, rabbit anti-phospho eIF2alpha, rabbit anti K48-ubiquitin, rabbit anti K63-ubiquitin (Cell Signaling); mouse anti α1234567 (MCP), mouse anti RPT5, mouse anti RPT1, rabbit anti RPN6 (Enzo); rabbit anti GRP78 (Novus); mouse anti PA28gamma, mouse anti VCP (BD Biosciences); mouse anti HA (Covance); chicken anti P0 (Aves). When detecting ubiquitin, the lysates were separated with Tricine SDS-PAGE and the nitrocellulose membranes were autoclaved after transfer. Visualization was performed with ECL, ECL Prime (GE Healthcare) or Odyssey CLx infrared imaging system (LiCor) and quantification was performed with ImageJ software (NIH).
Immunoprecipitation
For P0 immunoprecipitation, sciatic nerve lysates were prepared as described for immunoblotting. Equal amounts of total protein were pre-cleared with 40 uL Preciphen beads (Aves Labs). Chicken anti P0 (Aves Labs) was added to the supernatant at a final concentration of 200 ug/mL and samples rocked at 4°C for 3 hours. 80 uL of Preciphen beads were added and after 1 hour rocking at 4°C the beads were collected by centrifugation. The slurry was washed 3X in CHAPS lysis buffer and proteins were eluted by a 5 minute boil in 2X Sample buffer (3% SDS, 200 mM DTT, 10% glycerol, 120 mM Tris-HCl pH 6.8).
For immunoprecipitation of the 20S CP α1 subunit sciatic nerve lysates were prepared as described for Ubl proteasome purification. Equal amounts of total protein were pre-cleared with 15 uL Protein G beads (GE). Rabbit anti PSMA6 (Bethyl Laboratories) was added to the supernatant at a final concentration of 2 ug/mL and samples rocked at 4°C for 2 hours. 25 uL of Protein G beads were added and after 1 hour rocking at 4°C the beads were collected by centrifugation. The slurry was washed 3X in APB buffer and proteins were eluted by a 5 minute boil in 2X Sample buffer.
Immunohistochemistry on teased sciatic nerve fibers
Teased nerve fibers from sciatic nerve were prepared and immunostained as previously described (Pennuto et al. 2008). Briefly, sciatic nerves were dissected and fixed immediately by immersion in 4% paraformaldehyde for 30 minutes and stored in PBS at 4°C until time of teasing (less than seven days). Nerve fibers were gently separated with 26 gauge needles and transferred onto 3-aminopropyltriethoxy-sylane-treated slides. For immunostaining, fibers were permeabilized in methanol for 5 minutes at −20°C, blocked in 5% fish skin gelatin and 0.5% Triton X-100 in PBS and labeled with the following primary antibodies: Mouse anti-FK1 (Enzo) and Chicken anti-Neurofilament (Biolegend). After the addition of appropriate secondary antibodies: anti-mouse IgM-Cy3 and anti-chicken-488 (Jackson ImmunoResearch Laboratories, Inc); the fibers were washed, stained with DAPI (Sigma), dried, and mounted with Vectashield (Vector Laboratories). Nonspecific staining was determined by omission of the primary antibody.
Electron Microscopy on sciatic nerves
Sciatic nerves for electron microscopy were prepared as previously described (Quattrini et al. 1996). Images were acquired on a FEI Tecnai G2 Spirit bio twin electron microscope.
Ex vivo sciatic nerve protein degradation measurements
This method to measure protein degradation rates was adapted from a previously described assay (Zhao et al. 2007). Segments from the right and left sciatic nerves of equal length were dissected and placed on ice in DMEM containing 10% FBS and 100 ng/mL nerve growth factor (NGF). The nerve segments were desheathed (to remove blood nerve barrier) and lightly teased apart with a 26 gauge needle. To measure the rate of long-lived protein degradation, the nerve segments were incubated with 5 uCi/mL L-[3,53H]Tyrosine (Perkin Elmer) or [35S]cysteine and methionine (Express protein labeling mix, Easy Tag) for 16 hours at 37°C in 5% CO2. Segments were then washed 1X with DMEM containing 10% FBS, 100 ng/mL NGF and 2 mM tyrosine or 1 mM cysteine and 1mM methionine pre-warmed to 37°C (chase media) and incubated for 2 hours in chase media. Segments were washed 1X with chase media and then incubated in chase media for the length of the experiment. One nerve segment per animal was removed at this point, washed in cold PBS, and flash frozen on dry ice (input nerve). Aliquots of media were taken at indicated times and stored at 4°C until the completion of the experiment. To measure the rate of short-lived protein degradation, the nerve segments were incubated for 3 hours post dissection in DMEM containing 10% FBS and 100 ng/mL NGF at 37°C in 5% CO2 prior to pulse, the pulse time was reduced to 30 minutes, the washes were increased to 2, and the collection of media aliquots began immediately after the washes.
To determine the degradation rate of long-lived proteins by the ubiquitin proteasome system or the autophagy lysosome system, the sciatic nerves were prepared and pulsed as above. At the end of the pulse period the nerve segments were chased for one hour in chase media without inhibitors. The media was aspirated and new chase media containing either 5 μM Bortezomib or 150 μM chloroquine was added for an hour incubation. The inhibitor-containing media was aspirated and fresh media containing either Bortezomib or chloroquine was added. The nerve segments remained in this media for the remainder of the experiment. One nerve segments per animal was removed after the second hour of the chase period, washed in cold PBS, and flash frozen on dry ice (input nerve). Aliquots of media (5% of total media volume) were taken every hour for 4 hours and stored at 4°C until the completion of the experiment.
When long-lived protein degradation rates were measured in the presence of IU1 (SelleckChem) or IU1-47 (kindly provided by Dan Finley), IU1 or IU1-47 was added to the pulse media and all chase media at a final concentration of 200 μM or 2 μM.
Proteins in the media aliquots were precipitated with TCA for 1 hour at 4°C and pelleted by centrifugation for 15 minutes at 10,000 × g at 4°C. TCA-soluble radioactivity was then measured in glass vials in EcoScint scintillation fluid (National Diagnostics) on Tri-Carb 2800TR scintillation counter (PerkinElmer). The radiolabeled nerve segments were lysed in a buffer containing 25 mM HEPES-KOH, 150 mM NaCl, 2% SDS, 10 mM N-Ethylmalmeimide, 5 mM Iodoacetamide, 1 mM NaF, 1 mM Na3VO4, and 1 mM PMSF, precipitated with TCA for 1 hour at 4°C and pelleted by centrifugation for 15 minutes at 10,000 × g at 4°C. The pellets were washed 2x with acetone and resuspended in lysis buffer. Protein concentrations were determined by Bradford assay (Thermo Scientific) and then NaOH was added to a final concentration of 0.2 N. Radioactivity was measured as above. Proteolysis was expressed as amount of TCA soluble radioactivity released over time as percentage of total incorporated TCA insoluble radioactivity.
Cycloheximide protein degradation assay
Sciatic nerves were dissected, desheathed and lightly teased as previously described and then incubated at 37°C at 5% CO2 in DMEM plus 10% FBS and 100 ng/mL NGF for 2-3 hours. The nerves were transferred to media containing 2.5 mg/mL cycloheximide (Sigma), removed at indicated times, washed 1X in cold PBS and flash frozen on dry ice. Disappearance of proteins was observed with SDS PAGE and quantitative western blot.
Affinity purification of proteasomes using a ubiquitin-like domain
The gentle purification of proteasomes from sciatic nerves was performed as described previously described (Besche et al. 2009), with slight modifications. Briefly, the mouse sciatic nerves were flash frozen and pulverized in liquid nitrogen, suspended in Affinity Purify Buffer (APB) (25mM HEPES-KOH, 150 mM NaCl, 5 mM MgCl2, 1 mM ATP, 1 mM DTT and 10% glycerol) and lysed by sonication. The lysate was centrifuged for 20 minutes at 14,000 × g at 4°C. The supernatant was centrifuged for 60 minutes at 100,000 × g at 4°C. The supernatant was then incubated with purified GST-Ubl protein (Besche et al. 2009) and glutathione sepharose for 2 hours, rotating at 4°C. After two hour incubation the slurry containing 26S proteasomes bound to GST sepharose was washed 3× for 5 minutes at 4° with APB. Proteasomes were eluted by incubating the beads with purified His-UIM for 30 minutes rotating at 4°C. Excess His-UIM was removed from the eluate by incubating with Ni2+-NTA-Agarose for 15 minutes rotating at 4°C. Relative concentrations of proteasomes was determined using western blot with the MCP antibody (which detects 20S CP α subunits) or A280.
Measuring chymotrypsin-like peptidase activity of purified proteasomes
The chymotrypsin-like peptidase activity of purified proteasomes was measured with 100 μM Suc-LLVY-AMC (Enzo) in a buffer containing 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, and 0.05 mg/mL BSA. When indicated, 0.1 mM ATPᵧS (Sigma) or 1 μM ubiquitin aldehyde (Boston Biochem) was added to the reaction buffer. Fluorogenic hydrolysis was measured at excitation=380 nm and emission=460 nm at 37°C for 60 minutes in a BioTek Synergy plate reader. Rate of hydrolysis was calculated based on the slope of fluorescence increase over time during the linear phase of the reaction.
Proteasome Native Gel Electrophoresis
The native gel electrophoresis method was adapted from previously published protocols (Elsasser et al. 2005). In brief, sciatic nerves were pulverized on liquid nitrogen, suspended in APB without NaCl, lysed by sonication, centrifuged for 20 minutes at 14,000 × g at 4°C and normalized based on total protein concentration. 4% acrylamide resolving gels were prepared (90 mM Tris-HCl pH 7.5, 90 mM Boric Acid, 0.5 mM EDTA, 5 mM MgCl2, 1 mM DTT, 1 mM ATP, 10% glycerol, 0.1 % APS, 0.01% TEMED) and 2.5% acrylamide stacking gels (90 mM Tris-HCl pH 7.5, 90 mM Boric Acid, 0.5 mM EDTA, 5 mM MgCl2, 1 mM ATP, 0.1 % APS, 0.01% TEMED) were overlaid. Equal amounts of sciatic nerve lysate were added and gels were run in a TBE-based buffer (90 mM Tris-HCl pH 7.5, 90 mM Boric Acid, 0.5 mM EDTA, 5 mM MgCl2, 1 mM DTT, 1 mM ATP) at 145 V for 3-4 hours at 4°C. Activity was measured by incubating gel in 100 μM Suc-LLVY-AMC (50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM ATP and 1 mM DTT) for 25 minutes at 37°C. Imaging was performed on a VersaDoc (Biorad). For western analysis gels were soaked for 15 minutes at room temperature in a buffer containing 25 mM Tris,190 mM glycine and 1% SDS and then transferred to PVDF membranes in transfer buffer (25 mM Tris and 190 mM glycine) containing 1% MeOH overnight at 15V at 4°C.
Measuring DUB activity of purified proteasomes
The DUB activity of purified proteasomes was measured with 500 nM Ub-AMC (Fisher Scientific) in a buffer containing 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM ATP, 2.5 mM DTT, and 0.05 mg/mL BSA. For experiments with inhibitors, 1 mM TPEN (Sigma) and 2 μM IU1 (Selleckchem), inhibitors were added to the buffer prior to the addition of the buffer to the proteasomes and the beginning of the reaction. Fluorogenic hydrolysis was measured at excitation=380 nm and emission=460 nm at 37°C for 60 minutes in a BioTek Synergy plate reader. Rate of hydrolysis was calculated based on the slope of fluorescence increase over time during the linear phase of the reaction.
HA-Ub-VS (Boston Biochem) was added to purified proteasomes at a final concentration of 250 nM in a buffer containing 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM ATP, 2.5 mM DTT. The reaction took place at room temperature for 25 minutes and was stopped by the addition of 5X Laemmli Buffer (10% SDS, 600 mM DTT, 30% glycerol, 0.01% bromophenol blue, and 360 mM Tris-HCl pH 6.8). Samples were resolved by SDS-PAGE and transferred to PVDF membranes for western analysis.
Degradation of Ub5-DHFR by purified proteasomes
Ub5-DHFR (dihydrofolate reductase) was provided by Millennium Pharmaceuticals (Takeda Oncology Company) and radiolabeled using PKA and [gamma-32P]ATP as described previously (Peth et al. 2009). Degradation of Ub5-DHFR was measured in a buffer containing 50 mM Tris-HCl pH 7.5, 5mM MgCl2, 1 mM ATP, 1 mM DTT and 0.01 mg/mL BSA by the conversion of substrate to TCA-soluble radiolabeled peptides.
Measuring peptidase activity of the proteasomes in sciatic nerve lysate
Sciatic nerves were prepared in APB and the lysate was clarified by centrifugation for 20 minutes at 14,000 × g at 4°C. Protein concentration was determined with Bradford assay and 5 ug was used per reaction. The chymotrypsin-like peptidase activity was measured as described. Proteasome-specific cleavage of Suc-LLVY-AMC was calculated by subtracting the rate of hydrolysis by the rate of hydrolysis measured in the presence of 5 μM Epoxomicin (Enzo).
Measuring DUB-activity in total lysate
Sciatic nerves were prepared in APB as described above and the lysates were clarified by centrifugation for 20 minutes at 14,000 × g at 4°C. Protein concentration was determined with Bradford assay (Life Technologies) and 2.5 ug was used per reaction. The DUB activity in total lysate was measured with 1 μM Ub-AMC (Fisher Scientific) in a buffer containing 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM ATP, 2.5 mM DTT, and 0.05 mg/mL BSA as described above.
HA-Ub-VS (Boston Biochem) was added to the lysate at a final concentration of 1 μM in a buffer containing 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM ATP, 2.5 mM DTT. The reaction took place at room temperature for 25 minutes and was stopped by the addition of 5X Laemmli Buffer. Samples were resolved by SDS-PAGE and transferred to PVDF membranes for western analysis.
Statistics
Unless otherwise specified, error bars represent SEM and one-way ANOVA with Bonferroni post hoc analysis was applied. * = p<.05; **= p<.01; *** = p<.001.
Results
Increase in ubiquitinated proteins in neuropathic sciatic nerves
To initially investigate whether the UPS was impaired in P0S63del or P0 OE neuropathies, we performed immunoblot analysis for ubiquitin on lysates of sciatic nerves. The expression of P0S63del caused an increase in polyubiquitinated proteins (Figure 1A). The S63del Low (S63del L) mouse expresses P0S63del mRNA at approximately 60% the level of wild type P0 mRNA, and therefore expresses 160% total P0 mRNA (100% WT P0; 60% P0S63del) (Wrabetz et al. 2006). Increasing the level of P0S63del mRNA to 210% in the S63del High (S63del H) mouse, which has additional copies of the P0S63del transgene, further increased the amount of polyubiquitinated proteins (Figure 1A). Additionally, the level of polyubiquitinated proteins was increased in sciatic nerve lysates from P0 OE transgenic mice (Figure 1A). The Schwan cells in the nerves of P0 OE mice express wild type P0 mRNA at 80% the level of endogenous wild type P0 mRNA and therefore expresses 180% total P0 mRNA (Wrabetz et al. 2000). Immunohistochemical analysis of sciatic nerve fibers showed that this increase in polyubiquitinated proteins in S63del L and P0 OE was localized to Schwann cells and not to the ensheathed axons (Supplemental Figure 1A).
Figure 1. Ubiquitinated proteins are increased in neuropathic sciatic nerves.
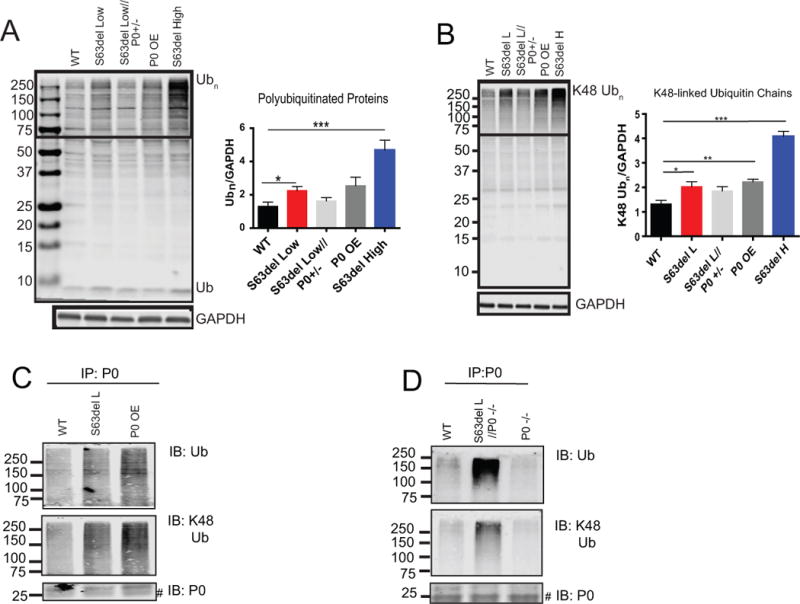
(A) Low and high expression of P0S63del and overexpression of only WT P0 (P0 OE) increased amounts of polyubiquitinated proteins. The box indicates the area used for densitometric analysis of polyubiquitin (Ubn) signal. n=4 mice.
(B) Ubiquitin chains that target proteins for proteasomal degradation (K48-linked) were increased by low and high expression of P0S63del and the overexpression of WT P0. Western blot analysis with a linkage-specific ubiquitin antibody was performed as an (A). n=4 mice.
(C) Polyubiquitinated P0 proteins accumulated in S63del L and P0 OE sciatic nerves. P0 was immunoprecipitated from sciatic nerve lysates and western analysis was performed for ubiquitin, K48-linked ubiquitin and P0. # indicates nonspecific bands which were present in the P0−/− immunoprecipitation control. The experiment was repeated at least 3 times with similar results.
(D) Polyubiquitinated P0S63del protein accumulated in sciatic nerves. P0 was immunoprecipitated from sciatic nerves that contained P0S63del but lacked WT P0 (S63del L//P0−/−). The immunoprecipitation was also performed from sciatic nerves lacking WT P0 (P0−/−) as a control. Western analysis was performed as in (C).
Ubiquitin contains seven lysines, and the lysine used in the formation of the polyubiquitin chain determines the fate of the conjugated protein. Using linkage-specific antibodies, we tested which types of ubiquitin chains accumulate in neuropathic sciatic nerves and found increases in K48-linked polyubiquitin chains (Figure 1B), which target proteins to proteasomal degradation (Komander and Rape 2012). No change in K63-linked polyubiquitin chains was observed in S63del L nerves, but these chains were increased in P0 OE (Supplemental Figure 1B). K63-linked ubiquitin chains do not increase in response to pharmacological inhibition of the proteasome (Kim et al. 2011), but do serve other roles such as intracellular signaling or DNA repair, and can promote degradation by the endosomal-lysosomal system or autophagy (Komander and Rape 2012). The reason for the accumulation of K63-linked chains in P0 OE is unclear, but it does suggest that the ubiquitin system is altered differently by overexpression of P0 than by expression of P0S63del.
To learn if P0 accumulates in a polyubiquitinated form in WT, S63del L, and P0 OE nerves, we immunoprecipitated P0 from nerve lysates. There was an increase in polyubiquitinated and K48-linked polyubiquitinated P0 in S63del L and P0 OE (Figure 1C). Since the S63del L mouse expresses both wild type P0 and P0S63del, both P0 proteins may be immunoprecipitated in the assay. To determine if the P0S63del mutant protein is ubiquitinated, we crossed S63del L onto the P0−/− background (S63del L//P0−/−), generating a mouse with sciatic nerves that that contain only P0S63del protein (Wrabetz et al. 2006). Immunoprecipitation of P0 from these sciatic nerves showed that the P0S63del protein is also polyubiquitinated and attached to K48-linked chains (Figure 1D).
Decreased rates of protein degradation in neuropathic sciatic nerves
The accumulation of polyubiquitinated proteins in the sciatic nerves led us to test if protein degradation was impaired. S63del L and P0 OE conditions were chosen for further investigation because they model two human neuropathies with different disease mechanisms, a UPR in S63del L and premature myelin adhesion in P0 OE (Wrabetz et al. 2006; Wrabetz et al. 2000) that lead to distinct nerve morphologies (Supplemental Figure 2). In order to measure overall rates of protein degradation, we modified a well-validated pulse chase approach using [3H]tyrosine (Zhao et al. 2007) to label proteins in the sciatic nerve ex vivo. The application of these techniques to explanted sciatic nerve primarily measures protein turnover in Schwann cells. Schwann cells represent approximately 90% of cells in peripheral nerves (Rutkowski et al. 1992) and perform the majority of protein synthesis ex vivo because the sciatic nerve lacks neuronal cell bodies.
The degradation of long-lived proteins, which represent the bulk of cellular proteins, was measured after labeling proteins synthesized in the explanted sciatic nerve for 16 hours and chasing for 2 hours with a large excess of nonradioactive tyrosine to prevent the reincorporation of [3H]tyrosine released by the degradation of labeled proteins (Figure 2A). The degradation of these long-lived proteins was decreased by approximately 60% in S63del L and 40% in P0 OE (Figure 2B). Inclusion of either bortezomib, a proteasome inhibitor, or chloroquine, an inhibitor of lysosomal acidification, in the chase media showed that the proteasome-mediated, but not the lysosome-mediated degradation of long-lived proteins, was decreased in S63del L sciatic nerves (Figure 2C).
Figure 2. The rate of protein degradation is decreased in S63del L and P0 OE sciatic nerves.
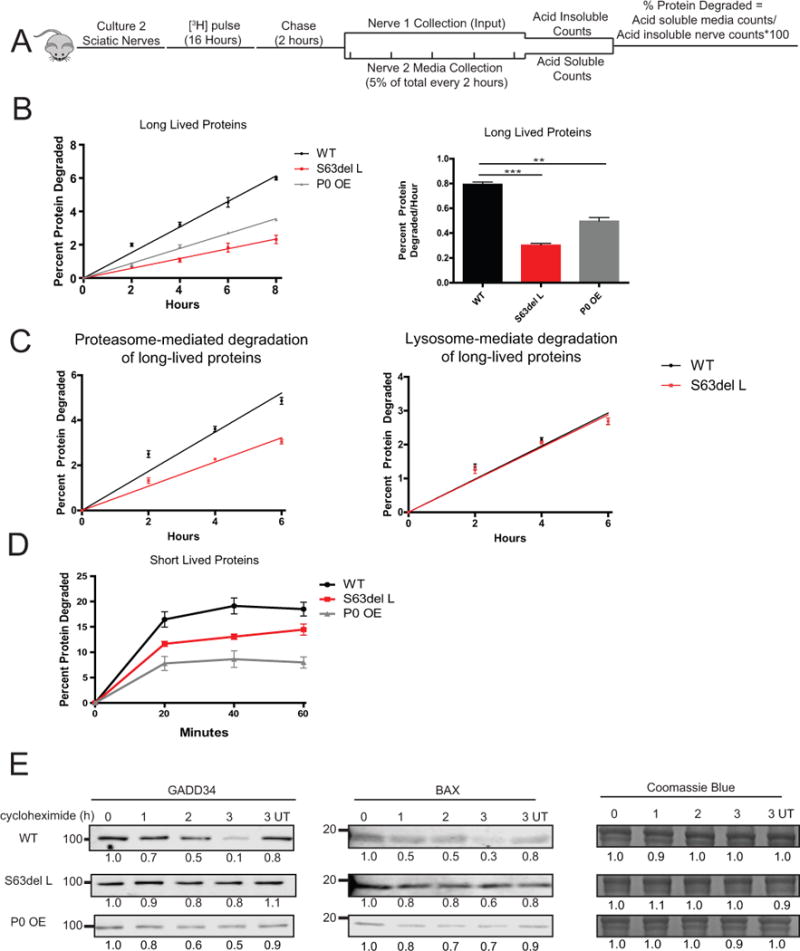
(A) To measure the rates of degradation of long-lived proteins, two sciatic nerve segments per mouse were pulse labeled ex vivo for 16 hours with 5 μM [3H]tyrosine, chased for 2 hours in media containing 2 mM nonradioactive tyrosine, and then acid-soluble counts per minute (CPM) in the media were determined. One nerve segment per mouse was lysed after the chase period and the [3H]tyrosine incorporation into protein was determined (input). Fresh chase media was added to the second nerve and 5% of the total media volume was removed every two hours, and the TCA soluble [3H]tyrosine in the media was determined. The percent protein degraded was calculated by dividing the TCA soluble radioactivity (degraded proteins) in the media at each time point by the TCA insoluble radioactivity in the nerve (radiolabeled proteins) at the end of the initial 2 hour chase period.
(B) S63del L and P0 OE sciatic nerves showed a reduction in the rate of degradation of long-lived proteins. The rate of proteolysis was calculated from the linear slopes. The mean rate of proteolysis for three individual mice per genotype was analyzed.
(C) The rate of proteasome-mediated degradation of long-lived proteins was decreased in S63del L sciatic nerves. The TCA soluble radioactivity in the media was plotted as a percentage of the total radioactivity incorporated into protein in the nerve. Proteasome-mediated degradation was determined in the presence of chloroquine and lysosome-mediated degradation was determined in the presence of bortezomib. N=3 mice per genotype, per condition.
(D) The degradation rate of short-lived proteins was reduced in S63del L and P0 OE. n=3 mice per genotype.
(E) The degradation of GADD34 and BAX, two well-described UPS substrate proteins, is decreased in S63del L and P0 OE sciatic nerves. Sciatic nerves were treated ex vivo with cycloheximide to block protein synthesis and collected at 1, 2 and 3 hours post cycloheximide addition. The levels of GADD34 and BAX remaining at each time point were measured by quantitative western analysis. The numbers below the blots correspond to the relative signal normalized to time 0. Coomassie blue staining (CBB) was used to estimate the total protein per lane.
With a shorter pulse followed by immediate measuring of the release of [3H]tyrosine from labeled proteins (Zhao et al. 2016), it is possible to primarily monitor the degradation of short-lived proteins, which include regulatory and misfolded proteins. All proteins synthesized in the sciatic nerve were labeled with a 30 minute [3H]tyrosine pulse and then the rates of degradation were measured by the release of radiolabeled peptides into the chase medium. Breakdown of short-lived proteins was decreased in S63del L and markedly decreased in P0 OE (Figure 2D).
In addition, the degradation rates of two UPS substrates, GADD34 (Brush and Shenolikar 2008) and BAX (Li and Dou 2000), were measured in explanted sciatic nerves in the presence of cycloheximide by quantitative western blot. The degradation of both proteins was slower in S63del L and P0 OE than in WT (Figure 2E). Combined, these data demonstrate a general reduction in the degradation of proteins in both neuropathies.
Increase in proteasome content and active Nrf1
The accumulation of polyubiquitinated proteins and the decreased rates of protein degradation together suggested an impairment in proteasome function in the S63del L and P0 OE nerves. We therefore measured the proteasomal chymotrypsin-like peptidase activity in lysates of sciatic nerves using the fluorogenic peptide substrate, Suc-LLVY-AMC. Surprisingly, the activity per μg of protein was similar in WT and S63del L lysates, and approximately 2-fold greater in P0 OE lysates (Figure 3A).
Figure 3. Increase in proteasome expression and content in S63del L and P0 OE sciatic nerves.
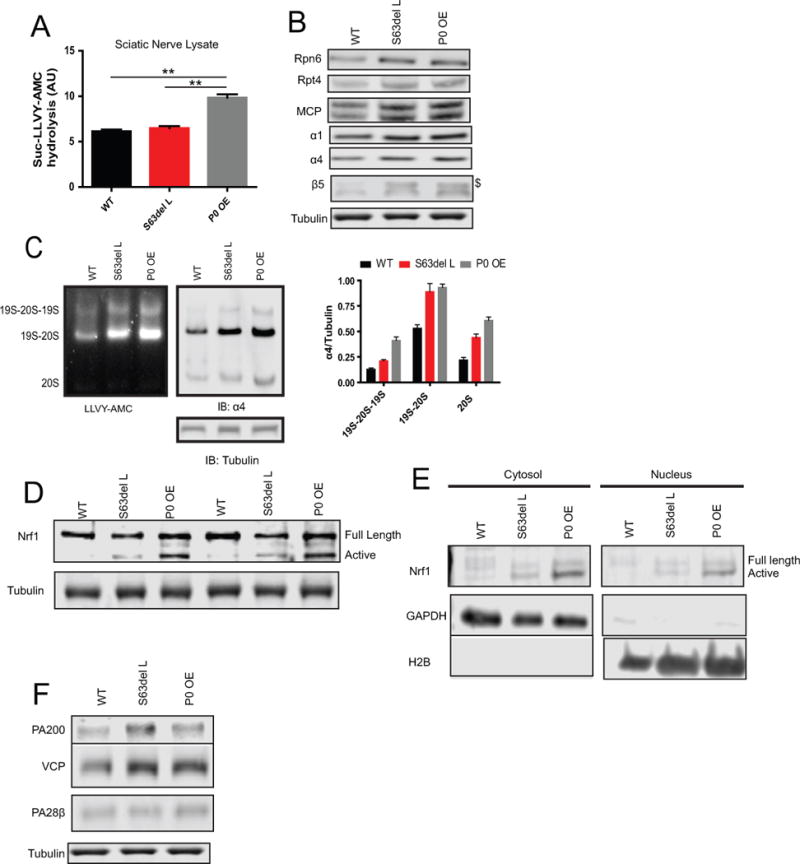
(A) Proteasome chymotrypsin-like peptidase activity was unchanged in the lysates of S63del L sciatic nerves and increased in P0 OE sciatic nerves. The hydrolysis of Suc-LLVY-AMC was measured in sciatic nerve lysates. Non-proteasomal hydrolysis of Suc-LLVY-AMC was measured in the presence of 5 μM epoxomicin and subtracted from the total activity. This value was slightly greater in the neuropathic lysates than in the WT.
(B) The levels of proteasome subunits were increased in S63del L and P0 OE sciatic nerve lysates by western blot. The MCP antibody recognizes all 20S CP ɑ subunits. $ indicates the precursor form of β5. See Supplemental Figure 3B for quantification.
(C) Total 26S proteasome levels are increased in S63del L and P0 OE. The levels of assembled proteasomes in sciatic nerve lysate were analyzed by native electrophoresis and western blot. The hydrolysis of Suc-LLVY-AMC and western analysis for the 20S α4 subunit were used to detect the proteasome populations. Proteasomes were identified by relative migration and normalized by analyzing an equal volume of each lysate by SDS PAGE and western blot for tubulin.
D) The levels of active Nrf1 transcription factor are increased in S63del L and even more so in P0 OE. Full length and active (proteolytically-processed) Nrf1 were analyzed by western blot. The experiment was repeated at least 3 times with at least 2 mice per genotype.
(E) The active form of Nrf1 is only found in the nuclear fraction in S63del L and P0 OE sciatic nerves. Lysates were separated into cytoplasmic and nuclear fractions and the levels of Nrf1 were analyzed by western blot. GAPDH and H2B were used to identify the cytoplasmic and nuclear fractions, respectively. The experiment was repeated at least 3 times with at least two animals per genotype each time.
(F) Increased protein levels of the alternative proteasome activator PA200 and the VCP/p97 complex, which are under Nrf1 transcriptional control, in S63del L and P0 OE. Protein levels of the alternative activator PA28β, which is not under Nrf1 transcriptional control, were not increased. See Supplemental Figure 3B for quantification
Since peptidase activity was not decreased as expected, we hypothesized that there were more proteasomes in S63del L and P0 OE sciatic nerves. In response to pharmacological inhibition of the proteasome, cells adapt by increasing degradative capacity through the generation of more proteasomes (Radhakrishnan et al. 2010; Sha and Goldberg 2014). Transcriptomic analysis performed on sciatic nerves (D’Antonio et al. 2013) revealed that six proteasome mRNA transcripts were increased by at least 1.5 fold in S63del L over WT levels and seven in P0 OE (Supplemental Figure 3A). Accordingly, western analysis in both S63del L and P0 OE nerve lysates showed an increase in 26S proteasome subunits above WT levels (Figure 3B). The larger precursor form of the catalytic subunit β5 (Figure 3B, see $) was detected only in S63del L and P0 OE, indicating that proteasome assembly was occurring in both. To verify that this increase in subunits was resulting in an increase in intact proteasomes, we performed native gel electrophoresis on sciatic nerve lysate. Both S63del L and P0 OE had more doubly-capped and singly-capped 26S proteasomes, and free 20S proteasomes, than WT (Figure 4C). Collectively, these data show that production of proteasomes is increased in both neuropathic conditions, presumably as a response to the decreased protein degradation or reduced proteasome activity. However, the increase does not seem to compensate, as polyubiquitinated and K48-linked polyubiquitinated proteins still accumulated in both neuropathies.
Figure 4. Proteasome function is decreased in S63del L, but not in P0 OE.
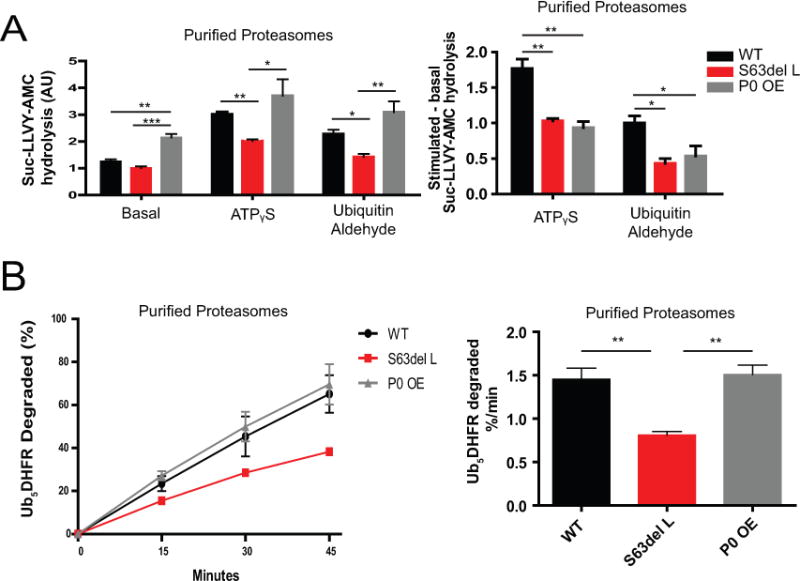
(A) Proteasomes purified from S63del L sciatic nerves show reduced basal activity and reduced activity while stimulated, whereas proteasomes from P0 OE show higher basal activity and WT-levels of activity while stimulated. 26S proteasomes were purified from sciatic nerves by the Ubl-method (Besche et al, 2009) and the chymotrypsin-like peptidase activity was measured with Suc-LLVY-AMC. To learn about the regulation of proteasome function in these neuropathies, proteasome activities were assayed with or without the non-hydrolyzable ATP analog, ATPγS, or the DUB inhibitor, ubiquitin aldehyde, which mimic activation by nucleotides or ubiquitin conjugates, respectively. The rates of peptide hydrolysis were normalized to the amounts of 20S ɑ subunits as determined by western blot analysis with an MCP antibody. The difference between basal and activated Suc-LLVY-AMC hydrolysis by purified 26S proteasomes is shown.
(B) Proteasomes purified from S63del L, but not P0 OE, have a reduced capacity to degrade ubiquitinated proteins. Proteasomes were purified as in (A) and their capacity to degrade the ubiquitinated protein Ub5DHFR was measured.
Error bars represent SEM among independent proteasome purifications. n=3 purifications. The sciatic nerves of 4 mice were pooled for one proteasome purification.
These observations suggest an increase of new proteasomes in S63del L and P0 OE sciatic nerves in response to an impairment of proteasome activity, as occurs in all cells in response to pharmacological proteasome inhibition. Partial impairment of proteasome activity by proteasome inhibitors leads to the proteolytic processing and activation of the transcription factor Nrf1, and the Nrf1-mediated transcription of proteasome subunits, PA200 and the VCP/p97 complex (Radhakrishnan et al. 2010; Sha and Goldberg 2014). The processed active form of Nrf1 was detected in S63del L and P0 OE lysates (Figure 3D) and nuclear fractions (Figure 3E), but not in the WT. Additionally, protein levels of the alternative proteasome activator PA200 and the VCP/p97 complex were increased in S63del L and P0 OE, but not the alternative activator PA28β, whose expression is IFNγ-inducible (Kloetzel 2001) and not upregulated via Nrf1 after proteasome inhibition (Sha and Goldberg 2014)(Figure 4F). Thus, Nrf1 is likely responsible for the increase in 26S proteasomes, PA200, and VCP/p97.
Decreased proteasome function in neuropathic sciatic nerves
Since proteasome levels are increased in S63del L and P0 OE sciatic nerves the peptidase activity measurements performed in lysate do not reflect proteasome activity per proteasome. In order to directly evaluate the chymotrypsin-like activity per proteasome, we affinity-purified 26S proteasomes from sciatic nerves by the Ubl method (Besche et al. 2009), determined their content by western analysis with an antibody that recognizes the 20S CP α subunits, and normalized activities to this value. The proteasomes purified from S63del L sciatic nerves showed a small, but reproducible decrease in specific peptidase activity compared to WT, whereas proteasomes purified from P0 OE surpisingly showed approximately 2-fold greater specific activity than WT (Figure 4A, left panel).
To address whether 26S proteasomes show similar regulatory properties in the neuropathies, we measured proteasome peptidase activity during activation by either the non-hydrolyzable ATP analog, ATPᵧS or the DUB inhibitor, ubiquitin aldehyde. In the presence of ATPᵧS, the 19S ATPase subunits relocate above the 20S (Sledz et al. 2013), and there is increased peptide entry and hydrolysis (Peth et al. 2009). ATPᵧS stimulated peptidase activity was approximately 3-fold in the WT and 2-fold in the neuropathies. Consequently, the activity of S63del L proteasomes was less than WT in the presence of ATPᵧS while the activity of P0 OE proteasomes was similar to WT in the presence of ATPᵧS (Figure 4A, middle panel).
The interaction of ubiquitinated proteins with proteasomal DUBs, USP14 or UCH37, opens the 20S gate and causes rearrangement of the 19S RP (Aufderheide et al. 2015), leading to greater peptide hydrolysis (Peth et al. 2009; Peth et al. 2013). Ubiquitin aldehyde, a DUB inhibitor, induces a conformational change in USP14 (Hu et al. 2005) and stimulates USP14-mediated gate opening in a similar manner as ubiquitinated proteins (Peth et al. 2009). The 26S peptidase activity was lower in the presence of ubiquitin aldehyde in S63del L, but similar to WT in P0 OE (Figure 4A, right panel). Thus, the activity of activated proteasomes is impaired in S63del L. In P0 OE 26S proteasomes, the stimulation by ATPᵧS or ubiquitin aldehyde seemed to be lower than WT, but because these particles had greater activity in the basal state, there was no reduction in activity when stimulated. Proteasomes purified from P0 OE sciatic nerves may have been already in a partially activated state and thus exhibited high basal activity that could not be as stimulated as WT.
Ubiquitinated proteins are the primary substrates of 26S proteasomes in the cell. Therefore, a physiologically relevant measure of proteasomal function is to determine their capacity to degrade a ubiquitinated protein. Using the well-characterized substrate protein ubiquitinated-dihydrofolate reductase (Ub5-DHFR), we found that S63del L proteasomes showed a slower degradation rate than WT, whereas P0 OE proteasomes showed a similar degradation rate as WT (Figure 4B). Thus, these findings with ubiquitin conjugates and model peptides indicate a clear defect in proteasome function only in S63del L.
Increase in DUBs at the proteasome in neuropathic nerves
Since proteasome function was clearly impaired in S63del L, we further investigated the 26S proteasomes to determine if the proteasomes were altered in a way that might affect protein degradation. During protein degradation, the ubiquitin chains on proteins are removed by proteasome-associated DUBs Rpn11, UCH37 and USP14. To evaluate the activities of UCH37 and USP14, we used HA-Ub-VS, which reacts with the active site cysteines in the DUBs that co-purified with 26S proteasomes from sciatic nerves. Western analysis showed five HA-positive bands, four of which were increased in S63del L (Figure 5A). Based on molecular weights and reactions with specific antibodies, three of these bands corresponded to HA-Ub-VS-bound USP5, HA-Ub-VS-bound USP14, and HA-Ub-VS-bound UCH37 (Figure 5A, bands 2, 3, and 4), suggesting increased activity or greater amounts of USP5, USP14 and UCH37 on S63del L proteasomes. The amounts of USP5, USP14, and UCH37 co-purifying with the proteasomes were also increased in S63del L (Figure 5A, right panel). To our knowledge, this is the first time where increased amounts of these enzymes on the proteasome have been seen in human disease models.
Figure 5. Increase in DUBs on 26S proteasomes in S63del L sciatic nerves.
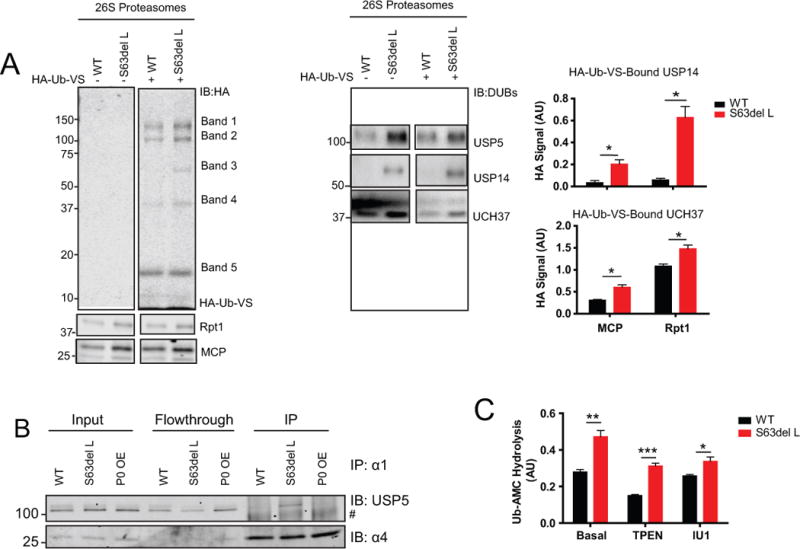
(A) The DUBs USP5, USP14 and UCH37 are increased on the 26S proteasome in S63del L. Proteasomes were incubated with HA-Ub-VS to modify the active site cysteines of the associated DUBs that copurified with them. Levels of modification, and thus activity of the DUB, were analyzed by western blot for HA and normalized to both the 20S ɑ subunits and the 19S subunit Rpt1. 5 HA-positive bands were detected and re-probing the blots with DUB-specific antibodies enabled the identification of Band 2 as USP5, Band 3 as USP14, and Band 4 as UCH37. The specific antibodies poorly recognized the HA-Ub-VS-bound DUBs, so primarily unmodified forms were detected. HA-positive bands 1 and 5 could not be identified with specific antibodies. The relative molecular weight of Band 1 is approximately ~140 kDa and antibodies against USP7 were tried. Band 5 is approximately 18 kDA and is potentially an HA-Ub-VS dimer.
(B) USP5 is only increased at the 26S proteasome in S63del L. 26S and 20S proteasomes were immunoprecipitated using an antibody against the 20S CP subunit, α1. The lysates used to perform the immunoprecipitation were prepared as described for the Ubl-method of 26S proteasome purification. The protein levels of USP5 in the input, flow through, and immunoprecipitate were evaluated by western blot. # indicates a nonspecific band. The sciatic nerves of 2 mice were pooled for one immunoprecipitation and the experiment was repeated at least 3 times with similar results.
(C) Proteasomal-DUB activity, as measured by hydrolysis of Ub-AMC, is increased in S63del L and UCH37 is primarily responsible for the increase. The zinc chelator TPEN (1 mM) was used to inhibit Rpn11 and IU1 (2 μM) was used to inhibit USP14. The proteasome concentrations were determined by both western analysis for the 20S CP ɑ subunits (MCP) and by A280. The method used for normalization did not change the calculated rates of Ub-AMC hydrolysis; the values obtained by normalizing with A280 are shown. Error bars represent SEM among independent proteasome purifications. n=3 purifications. The sciatic nerves of 4 mice were pooled for one proteasome purification.
The finding that USP5 co-purified with 26S proteasomes from WT and S63del L sciatic nerves nerves (Figure 5A, band 2) was verified by another technique because USP5 may bind the GST-Ubl resin that was used to purify the proteasomes for this assay (Besche et al. 2009). 26S and 20S proteasomes were immunoprecipitated using an antibody against the 20S CP subunit α1 and the levels of co-immunoprecipitating USP5 were analyzed by western blot. USP5 co-immunoprecipitated with and was enriched on proteasomes from S63del L sciatic nerves, but little or no USP5 co-immunoprecipitated with proteasomes from WT or P0 OE sciatic nerves (Figure 5B). Thus, USP5 appears to associate with the proteasome only in S63del L nerves. The significance of this interesting finding is currently unclear, as the functions of USP5 at the proteasome are not well elucidated. There were two other HA-positive bands, labeled 1 and 5, but we were unable to identify them using specific antibodies (Figure 5A).
Western analysis on the sciatic nerve lysates showed that the total levels of USP5, USP14 and UCH37 were not increased in S63del L (Supplemental Figure 4A), and evaluation of active DUBs in the lysates with HA-Ub-VS showed similar banding patterns and intensities in WT and S63del L (Supplemental Figure 4B). Therefore, the observed increases in DUBs on the 26S proteasomes were due to a greater binding to the proteasome in S63del L and not an increase in their cellular content.
We also measured proteasomal-DUB activity using Ub-AMC as the substrate. The proteasomes purified from S63del L sciatic nerves showed greater Ub-AMC hydrolysis than WT (Figure 5C), as expected from the higher levels of DUBs and greater reactivity with HA-Ub-VS (Figure 5A). By contrast, Ub-AMC hydrolysis was not increased in the lysates of S63del L sciatic nerves (Supplemental Figure 4C). Thus, this increased activity was specific to the proteasome.
Rpn11 is a zinc metalloprotease and an intrinsic proteasome DUB subunit (Lee et al. 2011). The zinc chelator TPEN blocks Rpn11 activity, but did not abolish the increased Ub-AMC hydrolysis in S63del L (Figure 5C, second panel). Thus, Rpn11 is not responsible for the greater Ub-AMC hydrolysis.
Selective inhibition of USP14 with IU1 did not decrease Ub-AMC hydrolysis by proteasomes from WT sciatic nerves (Figure 5C), in accord with our previous observation that very little HA-Ub-VS bound USP14 could be detected on WT proteasomes (Figure 5A). Because inhibition of USP14 did not eliminate the difference in Ub-AMC hydrolysis between the WT and the S63del proteasomes, the increase in UCH37 is most likely the major contributor to the increased Ub-AMC hydrolysis, as was also suggested by our finding of increased HA-Ub-VS-bound UCH37 in S63del L proteasomes (Figure 5A).
Pharmacological inhibitors of USP14 increase protein degradation in S63del L
USP14, and its yeast ortholog Ubp6, regulate proteasomal degradation through catalytic and noncatalytic mechanisms (Hanna et al. 2006; Lee et al. 2010; Peth et al. 2009; Peth et al. 2013). Pharmacological inhibition of USP14 with IU1 enhances degradation of some disease-associated proteins in cultured cells (Lee et al. 2010), including the prion protein PrPSC (Homma et al. 2015), but possible effects on overall protein degradation rates have not been studied. Since more USP14 was associated with the 26S proteasomes in S63del L (Figure 5A), we hypothesized that this enzyme may be contributing to the decreased degradation of proteins in these neuropathic nerves. To test this hypothesis, we initially treated sciatic nerves ex vivo with 200 μM IU1, a similar concentration to what was used in previous publications (Homma et al. 2015; Lee et al. 2010) and measured the degradation rate of long-lived proteins. Although this treatment did not change the rate of degradation of long-lived proteins in WT or P0 OE, it did clearly increase the rate of proteolysis in the S63del L nerves (Figure 6A). These findings suggest that the impact of IU1 treatment on protein degradation depends on the population of misfolded or mutant proteins in the cell, since both neuropathies had an accumulation of polyubiquitinated proteins. A similar increase in overall protein degradation in S63del L sciatic nerves was also seen with 2 μM IU1 and 2 μM IU1-47 (a more potent derivative of IU1) (Figure 6B).
Figure 6. USP14 inhibitors IU1 and IU1-47 increase protein degradation in S63del L nerves.
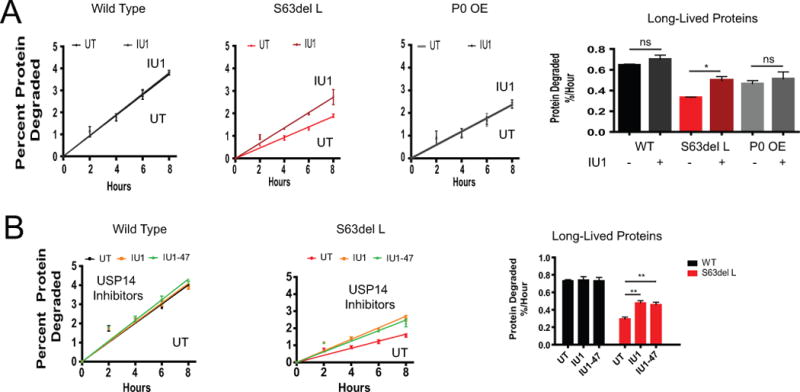
(A) The USP14 inhibitor IU1 increased the degradation rate of long-lived proteins in S63del L nerves at a similar concentration (200 μM) used in previous publications (Homma et al. 2015; Lee et al. 2010). The rate of degradation was calculated as in Figure 2. The mean rate of proteolysis for three individual mice was graphed.
(B) IU1 and IU1-47 increased the degradation rate of long-lived proteins in S63del L nerves at a concentration of 2 μM. The rate of long-lived protein degradation was measured as in Figure 2. Error bars represent SEM between mice. A Student’s t-test was performed between IU1 treated and untreated conditions of the same genotype.
Pharmacological inhibitors of USP14 lead to phosphorylation of eif2α by Perk
Since S63del L manifests a UPR, we measured the levels of UPR mediator proteins by western blot to evaluate if IU1 treatment reduced ER stress. Surprisingly, the treatment of the sciatic nerves ex vivo with 200 μM IU1 increased the levels of the translational attenuator, p-eIF2α, approximately 2-fold in WT and S63del L (Figure 7A). Accordingly, the level of CHOP, a downstream effector of p-eIF2α, also increased (Figure 7B). However, the levels of the UPR proteins BiP and GADD34 were unchanged (Figure 7C), suggesting that the increase in p-eIF2α was not simply due to the IU1 treatment increasing ER stress.
Figure 7. IU1 treatment increased levels of p-eIF2α and CHOP in a Perk-dependent manner.
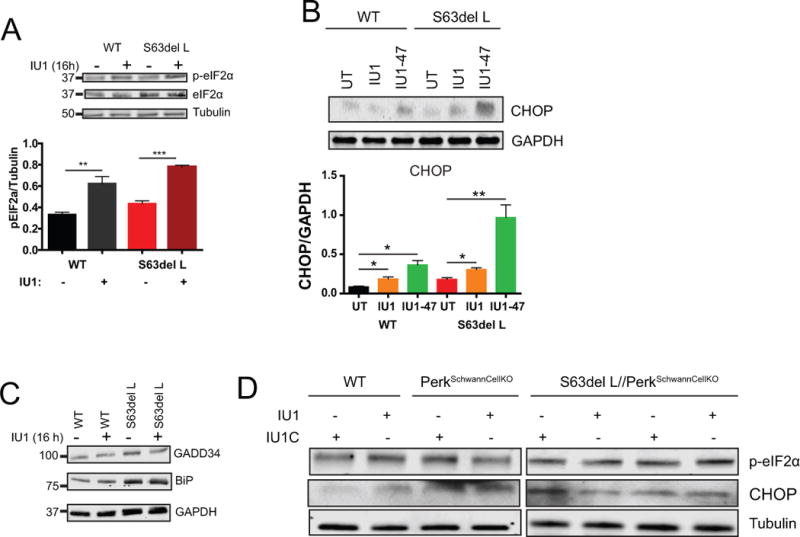
(A) IU1 treatment increased levels of p-eIF2α in both WT and S63del L. Sciatic nerves were treated ex vivo with 200 μM IU1 for 16 hours and the levels of p-eIF2α and eIF2α were analyzed by western blot. A Student’s t-test was performed between treated and untreated conditions of the same genotype.
(B) Levels of CHOP, a protein downstream of p-eIF2α, were increased by IU1 and IU1-47 treatment. Sciatic nerves were treated ex vivo with 2 μM IU1 or 2 μM IU1-47. n=3 sciatic nerves per genotype, per condition. A Student’s t-test was performed between treated and untreated conditions of the same genotype.
(C) IU1 treatment did not increase the levels of UPR proteins BiP or GADD34. Sciatic nerves were treated ex vivo with 200 μM IU1 for 16 hours and the levels of BiP and GADD34 were analyzed by western blot. The experiment was repeated at least 3 times with similar results.
(D) The genetic ablation of Perk in Schwann cells abrogates the IU1-induced increase in p-eIF2α and CHOP. Sciatic nerves were treated ex vivo with 2 μM IU1 or 2 μM IU1-C (an inactive form of IU1) for 16 hours.
Four kinases have been documented to phosphorylate eIF2α in response to different types of cellular stress (Donnelly et al. 2013). To investigate whether Perk was responsible for the increased phosphorylation of eIF2α caused by USP14 inhibitors, we treated sciatic nerves ex vivo from WT, Perk Schwann cell-specific knockout (P0cre/Perkflox/flox), and S63del//Perk Schwann cell-specific KO mice (Sidoli et al. 2016) with IU1 or the inactive analog IU1-C. An IU1-mediated increase in p-eIF2α or CHOP was not seen in the absence of Perk (Figure 7D). This finding that the treatment with IU1 led to the phosphorylation of eIF2α by Perk is intriguing, but the connection between USP14 inhibition, proteasome function, and control of protein synthesis by Perk-eIF2α remains to be elucidated.
Discussion
Proteasome function in two distinct neuropathy models
Our results clearly show that proteasome function and protein degradation are impaired in Schwann cells in the sciatic nerves of S63del L mice (Figure 8). Levels of polyubiquitinated proteins were increased, and the rates of degradation of short- and long-lived cell proteins were decreased in S63del sciatic nerves. Importantly, the proteasome-mediated degradation, and not the lysosome-mediated degradation, of long-lived proteins was diminished in S63del L nerves. 26S proteasomes purified from these nerves exhibited decreased degradation of peptides and importantly, a ubiquitinated protein. Our in-depth findings in this study confirm preliminary observations that modulation of UPS or proteasome function in glial cells is associated with both physiological and pathological effects in glia (Goldbaum et al. 2008; Karim et al. 2010; Stohwasser et al. 2000).
Figure 8. Defects in protein degradation caused by P0S63del and P0 overexpression.
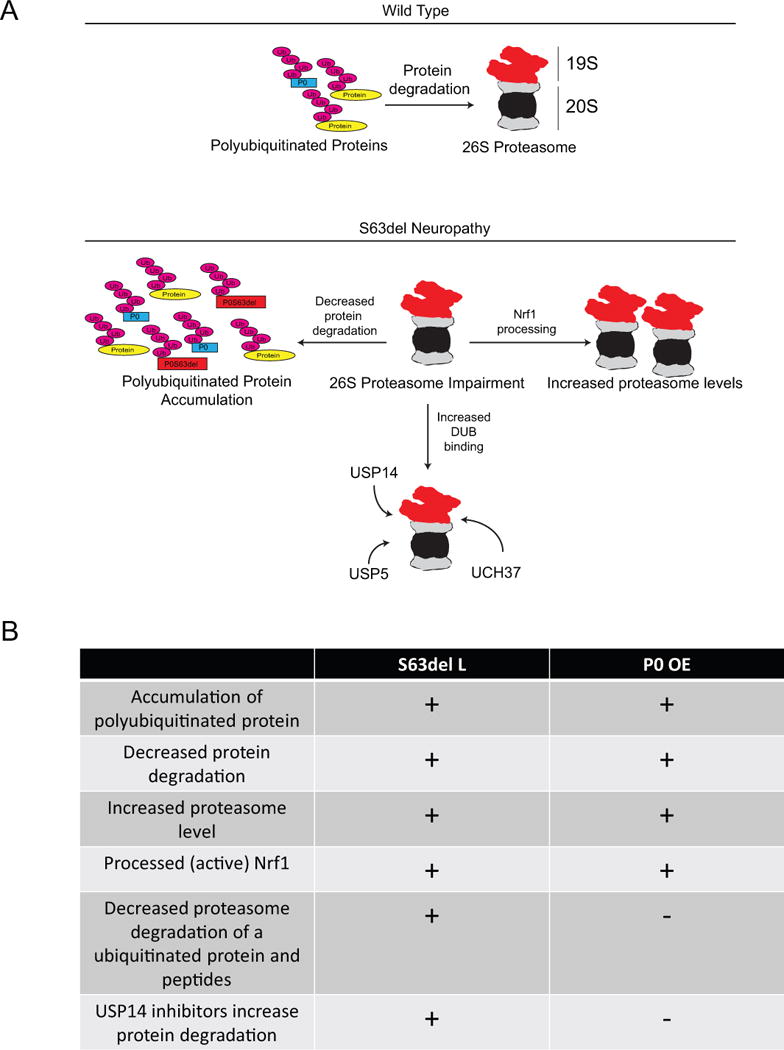
(A) A schematic diagram of the effects of P0S63del expression on the ubiquitin proteasome system.
(B) A comparison of protein degradation defects in S63del L and P0 OE sciatic nerves.
Similar defects in proteasomal substrate degradation were reported in the brain of a mouse model that expresses an aggregation-prone mutant tau that causes frontotemporal dementia (Myeku et al. 2016). Direct inhibition of proteasome function has also been seen with aggregated forms of PrPSC (Deriziotis et al. 2011; Kristiansen et al. 2007). Thus, expression of different mutant proteins can impair proteasome function, potentially through related mechanisms. The question remains as to how the mutant protein disrupts proteasome function. A tempting hypothesis is that a mutant protein, like P0S63del, binds the proteasome after ubiquitination, fails to be degraded, and its continued presence interferes with proteasome function and the degradation of other substrates. However, because of its localization in the myelin sheath, its immunoglobulin-like domain and its hydrophobic nature, purification of P0 is challenging (Shapiro et al. 1996) and it remains unclear if P0 proteins are bound to the proteasome.
S63del L sciatic nerves also exhibited increased levels of proteasomes and an increase in the active, proteolytically-processed form of the Nrf1 transcription factor. This response has been reported with pharmacological inhibitors of the proteasome and is a compensatory response to increase cellular degradative capacity when proteins are not being efficiently degraded (Radhakrishnan et al. 2010; Sha and Goldberg 2014). However, increased proteasome level is not seen with other mutant proteins that impair proteasome function, like tau (Myeku et al. 2016), or in sciatic nerves from other mouse models of hereditary peripheral neuropathies (Fortun et al. 2005). It is not clear how the expression of P0S63del causes the proteolytic processing of Nrf1. It is most likely that the proteasome impairment caused by P0S63del promotes the Nrf1 processing, as demonstrated by several previous studies with proteasome inhibitors (Radhakrishnan et al. 2010; Sha and Goldberg 2014). However, the processed form of Nrf1, which enters the nucleus and promotes transcription of proteasome genes, was also detected in P0 OE, actually to a greater extent than in S63del L, and P0 OE proteasomes did not show any deficiencies in the proteasome functions we measured. Additionally, in P0 OE, but not S63del L, the increase in proteasome subunits actually preceded the accumulation of polyubiquitinated proteins (Supplemental Figure 5). These findings suggest that in disease settings there may be more factors than proteasome impairment that result in Nrf1 proteolytic processing, and consequently, the Nrf1-mediated upregulation of proteasomes.
It is also interesting that this pathway to increase proteasome content is activated in vivo in the sciatic nerves of these neuropathy models. With proteasome inhibitors, this response is compensatory, but it is currently not clear if the increase in proteasome content is beneficial in models of chronic diseases like S63del L and P0 OE. Even with increased proteasome number, degradation was still decreased and polyubiquitinated proteins accumulated. To better understand this response, it will be important to investigate whether the Nrf1-mediated pathway to increase proteasome content is active in other diseases.
Sciatic nerves from P0 OE mice, like S63del L, had increased polyubiquitinated proteins and decreased rates of protein degradation, as well as an increase in proteasome content. These findings suggested decreased proteasome function in both neuropathies. However, proteasome function in P0 OE, as measured by degradation of ubiquitin conjugates and peptides, was comparable to WT, indicating a clear defect only in S63del L. Thus, the proteasome impairment in S63del L was specifically caused by the mutation in P0, and not overexpression of P0, which was similar in both models (Wrabetz et al. 2006; Wrabetz et al. 2000). The proteasome impairment only in S63del L also highlights the differences in the disease mechanisms causing these two neuropathies and may contribute to the distinct nerve morphologies (Figure 8).
It will be important to determine why P0 OE sciatic nerves exhibited signs commonly associated with proteasome impairment, like accumulated polyubiquitinated proteins, without demonstrable deficits in proteasome function. The most likely explanation is that proteasome function is impaired in P0 OE, but in a manner we do not yet know how to measure (e.g. degradation of specific types of ubiquitin conjugates). We investigated two activation mechanisms of the proteasome to try and understand if they were dysregulated, to our knowledge we are the first to explore this possibility in disease, but activation with either ATPᵧS or ubiquitin aldehyde did not yield activities different than WT. The increased levels of polyubiquitinated proteins may not be indicative of decreased proteasomal degradation, but of increased ubiquitination, as seen in muscle atrophy or with mTOR inhibition (Zhao et al. 2015). Although possible, this does not seem likely in P0 OE because in the aforementioned contexts, increased ubiquitination was associated with increased protein degradation. There could also be impairment of processes preceding proteasomal function, such as retrotranslocation of misfolded proteins from the ER (Christianson and Ye 2014).
Increase in DUBs associated with the 26S proteasome in S63del neuropathy
Examination of 26S proteasomes purified from the sciatic nerves of S63del L mice revealed an intriguing increase in the amount of associated DUBs. This novel finding demonstrates that increased levels of DUBs accumulate on the proteasome in diseased tissues, perhaps due to the higher levels of ubiquitin conjugates. These observations highlight the fact that the proteasome composition can be altered in diseases and raise the possibility of selective pharmacological treatment. The mechanisms through which these DUBs accumulate on the proteasome in S63del is currently unclear, but treatment of cells with proteasome inhibitors also increased association of DUBs with the proteasome (Besche et al. 2014; Borodovsky et al. 2001), suggesting that DUBs accumulate on the proteasome when proteasome activity is reduced. We found increased USP14, UCH37, and USP5 in S63del L. How, or if, increased amounts of UCH37 or USP5 effects proteasome function and the disease progression remains an intriguing question for future study.
Pharmacological inhibitors of USP14 increased the degradation rate of cellular proteins specifically in S63del L sciatic nerves. USP14 inhibitors have been shown to increase proteasomal degradation of certain misfolded proteins associated with neurodegenerative disease (Homma et al. 2015; Lee et al. 2010), but this is the first time an effect on global degradation of cell proteins has been investigated and observed. Presumably the increased amounts of USP14 on the proteasome in S63del L contributes to the decrease in protein degradation. However, increased DUB levels cannot be the only cause of the decreased degradation in S63del L sciatic nerves because defects in proteasome activity that cannot be caused by USP14 binding, such as decreased peptidase activity, were also observed. USP14 prevents the proteasomal degradation of substrate proteins bearing more than one ubiquitin chain by removing ubiquitin chains before the protein is committed to degradation (Lee et al. 2016). Inhibiting the catalytic activity of USP14 with IU1 or IU1-47 may be preventing this ‘rescue’ of ubiquitinated proteins in S63del L sciatic nerves, and as a result, increasing their degradation. These findings, along with the possible stimulation of proteasome activity by phosphorylation (Lokireddy et al. 2015; Myeku et al. 2016), present new opportunities to therapeutically stimulate proteasomal protein degradation and enhance the clearance of the misfolded proteins that accumulate in proteotoxic diseases.
USP14 inhibitors increased p-eIF2ɑ
We unexpectedly found that USP14 inhibitors increased eIF2ɑ phosphorylation in WT and S63del L sciatic nerves (Figure 7). eIF2ɑ phosphorylation attenuates protein synthesis by inhibiting translational initiation but also promotes translation of mRNAs containing upstream open reading frames (uORFs) in their 5′ untranslated regions (5′ UTRs) (Walter and Ron 2011). Since increasing the levels of p-eIF2α reduces the unfolded protein response and improves myelination and motor function in the S63del L mouse model (D’Antonio et al. 2009; Das et al. 2015), the mechanism behind this unexpected observation, and its connection to USP14 inhibition, deserves further investigation. Since IU1 treatment both increased protein degradation and the phosphorylation of eif2α in the nerves, it has clear potential to be a doubly beneficial treatment for S63del neuropathy.
Supplementary Material
Supplemental Figure 1. Increase in polyubiquitinated proteins in neuropathic sciatic nerves
(A) The ubiquitin signal is increased in the Schwann cells and not in the axons. Sciatic nerves were teased into individual nerve fibers and immunohistochemical analysis was performed. Nuclear staining (DAPI) is in blue, polyubiquitin (FK1) is in red and a marker of axons, neurofilament (NF) is in green. The scale bar represents 5 μm.
(B) K63-linked ubiquitin chains that do not target proteins for proteasomal degradation are increased in P0 OE and S63del H, but not S63del L. The box indicates area used for densitometric analysis of K63-linked polyubiquitin (K63 Ubn) signal. The area of the box is decreased compared to ubiquitin and K48-linked ubiquitin blots in order to avoid including the intense, possibly non-specific band located around 75 kDa in the analysis.
Supplemental Figure 2. S63del L and P0 OE sciatic nerves have distinct morphologies
(A) Electron microscopic images of ultrathin sections of sciatic nerves. In the images, # indicates an amyelinated fiber, which contains an axon greater than 1 μm in diameter that should be myelinated. Arrows indicate hypomyelinated fibers, nerve fibers that have thinner myelin than predicted, based on the diameter of the axon. Note that the S63del L nerves manifest primarily hypomyelinated fibers with only occasional amyelinated fibers, consistent with demyelination followed by remyelination, whereas P0 OE nerves contain more amyelinated fibers, consistent with developmental amyelination. At 6 months of age (not shown), S63del L nerves contain many onion bulbs, the hallmark of demyelination, whereas P0 OE nerves do not (Wrabetz et al. 2006; Wrabetz et al. 2000). Collectively, these observations indicate that S63del L mice manifest a demyelinating neuropathy, whereas P0 OE mice manifest a developmental, hypomyelinating neuropathy. The scale bar represents 2 μm.
Supplemental Figure 3. Proteasome expression and content are increased in S63del L and P0 OE sciatic nerves.
(A) mRNAs for multiple proteasome subunits are increased in S63del L and P0 OE sciatic nerves. A list of the proteasome subunit and activator mRNAs in S63del L and P0 OE that were increased by at least 1.5 fold over WT in transcriptomic analysis performed on an Affymetrix Mouse GeneChip-MOE430A 2.0 (D’Antonio et al. 2013). Only one proteasome subunit or activator mRNA transcript was decreased by at least 1.5 fold compared to WT in the neuropathic conditions, PSMB1, in S63del L.
(B) Proteasome content is increased in both S63del L and P0 OE. Shown are the normalized levels of all examined proteasome activators, proteasome subunits, and the VCP/p97 complex analyzed in Figures 3B and 3F.
Supplemental Figure 4. No increase in DUB level in S63del sciatic nerve lysates.
(A) USP5, USP14 and UCH37 are not increased in the sciatic nerve lysates of S63del L. The experiment was repeated at least 3 times with at least 2 mice per genotype each time.
(B) The amount of active DUBs is similar in sciatic nerve lysates of WT and S63del L. Sciatic nerve lysates were treated with HA-Ub-VS to modify the cysteines of active DUBs. Levels of modification, indicating DUB activity, were analyzed by western blot for HA. The levels of USP14 and UCH37 were also evaluated with specific antibodies. The USP14 antibodies did not recognize HA-Ub-VS-bound USP14 so only unmodified USP14 is detected with the USP14-specific antibody in the HA-Ub-VS lanes. The UCH37 antibody did recognize HA-Ub-VS-bound UCH37, as indicated by the approximately 10 kDa-shift in the HA-Ub-VS lanes compared to the control lanes when the UCH37-specific antibody was used. The experiment was repeated at least 3 times with 2 mice per genotype each time.
(C) The DUB activity is similar in the sciatic nerve lysates from WT and S63del L. The hydrolysis of Ub-AMC was measured in sciatic nerve lysate and was normalized to total protein. n=3.
Supplemental Figure 5: In P0 OE, but not S63del L, the increase in levels of proteasome subunits precedes the accumulation of polyubiquitinated proteins.
(A) The levels of K48-linked polyubiquitinated proteins and proteasome subunits were analyzed at post natal day 10 (p10) or 20 (p20) in S63del L or P0 OE, and compared to WT littermates.
Main points.
Proteasome function and protein degradation are impaired in Schwann cells of sciatic nerves from S63del neuropathy mice. Increased levels of three deubiquitinases are found on the proteasome and inhibition of one, USP14, improves protein degradation in Schwann cells of S63del sciatic nerves.
Acknowledgments
The authors would like to thank Dr. Daniel Finley (Harvard Medical School) for the generous gift of IU1-C and IU1-47, and Edward Hurley, Joshua Lewis and Courtney Williamson for excellent technical assistance. This study was supported by grants from the National Institutes of Health (1R01NS100464 and 2R01NS045630 to M.L.F; 2R01GM051923 to A.L.G.; R01NS055256 and R56NS096104 to L.W.), as well as grants from the Muscular Dystrophy Association and Target ALS to A.L.G.
Footnotes
Author Contributions
J.V., A.L.G., and L.W. designed the research; J.V. and S.R. performed the research; J.V., S.R., A.L.G., and L.W. analyzed the data; and J.V., M.L.F., A.L.G., and L.W. wrote the paper.
Conflict of Interest
The authors have no conflict of interest to declare.
References
- Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W, Forster F. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A. 2015;112:8626–31. doi: 10.1073/pnas.1510449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of Mammalian 26S Proteasomes and p97/VCP Complexes Using the Ubiquitin-like Domain from HHR23B Reveals Novel Proteasome-Associated Proteins†. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, Gygi S, Gutierrez JA, Liao H, Dick L. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. The EMBO journal. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site‐directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. The EMBO journal. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Shenolikar S. Control of cellular GADD34 levels by the 26S proteasome. Molecular and cellular biology. 2008;28:6989–7000. doi: 10.1128/MCB.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature structural & molecular biology. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antonio M, Feltri ML, Wrabetz L. Myelin under stress. Journal of neuroscience research. 2009;87:3241–3249. doi: 10.1002/jnr.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antonio M, Musner N, Scapin C, Ungaro D, Del Carro U, Ron D, Feltri ML, Wrabetz L. Resetting translational homeostasis restores myelination in Charcot-Marie-Tooth disease type 1B mice. The Journal of experimental medicine. 2013;210:821–838. doi: 10.1084/jem.20122005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I, Krzyzosiak A, Schneider K, Wrabetz L, D’Antonio M, Barry N, Sigurdardottir A, Bertolotti A. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science. 2015;348:239–242. doi: 10.1126/science.aaa4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriziotis P, Andre R, Smith DM, Goold R, Kinghorn KJ, Kristiansen M, Nathan JA, Rosenzweig R, Krutauz D, Glickman MH, et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. Embo j. 2011;30:3065–77. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci. 2013;70:3493–511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005;398:353–63. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry. 2009;78:477. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortun J, Go JC, Li J, Amici SA, Dunn WA, Notterpek L. Alterations in degradative pathways and protein aggregation in a neuropathy model based on PMP22 overexpression. Neurobiology of disease. 2006;22:153–164. doi: 10.1016/j.nbd.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Fortun J, Li J, Go J, Fenstermaker A, Fletcher BS, Notterpek L. Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. Journal of neurochemistry. 2005;92:1531–1541. doi: 10.1111/j.1471-4159.2004.02987.x. [DOI] [PubMed] [Google Scholar]
- Fratta P, Saveri P, Zambroni D, Ferri C, Tinelli E, Messing A, D’Antonio M, Feltri ML, Wrabetz L. P0S63del impedes the arrival of wild-type P0 glycoprotein to myelin in CMT1B mice. Human molecular genetics. 2011;20:2081–2090. doi: 10.1093/hmg/ddr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbaum O, Jensen PH, Richter-Landsberg C. The expression of tubulin polymerization promoting protein TPPP/p25alpha is developmentally regulated in cultured rat brain oligodendrocytes and affected by proteolytic stress. Glia. 2008;56:1736–46. doi: 10.1002/glia.20720. [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annual review of biochemistry. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Homma T, Ishibashi D, Nakagaki T, Fuse T, Mori T, Satoh K, Atarashi R, Nishida N. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Scientific reports. 2015:5. doi: 10.1038/srep11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chernova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome‐associated deubiquitinating enzyme USP14. The EMBO journal. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim SA, Barrie JA, McCulloch MC, Montague P, Edgar JM, Iden DL, Anderson TJ, Nave KA, Griffiths IR, McLaughlin M. PLP/DM20 expression and turnover in a transgenic mouse model of Pelizaeus-Merzbacher disease. Glia. 2010;58:1727–38. doi: 10.1002/glia.21043. [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2:179–87. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, Clarke AR, van Leeuwen FW, Menendez-Benito V, Dantuma NP, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–88. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Kulkens T, Bolhuis PA, Wolterman RA, Kemp S, te Nijenhuis S, Valentijn LJ, Hensels GW, Jennekens FG, de Visser M, Hoogendijk JE. Deletion of the serine 34 codon from the major peripheral myelin protein P0 gene in Charcot–Marie–Tooth disease type 1B. Nature genetics. 1993;5:35–39. doi: 10.1038/ng0993-35. [DOI] [PubMed] [Google Scholar]
- Lee B-H, Lu Y, Prado MA, Shi Y, Tian G, Sun S, Elsasser S, Gygi SP, King RW, Finley D. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature. 2016;532:398–401. doi: 10.1038/nature17433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10:R110.003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proceedings of the National Academy of Sciences. 2000;97:3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A. 2015;112:E7176–85. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda MH, Mitsui J, Soong BW, Takahashi Y, Ishiura H, Hayashi S, Shirota Y, Ichikawa Y, Matsumoto H, Arai M. Increased gene dosage of myelin protein zero causes Charcot‐Marie‐Tooth disease. Annals of neurology. 2012;71:84–92. doi: 10.1002/ana.22658. [DOI] [PubMed] [Google Scholar]
- Miller LJ, Patzko A, Lewis RA, Shy ME. Phenotypic presentation of the Ser63Del MPZ mutation. Journal of the Peripheral Nervous System. 2012;17:197–200. doi: 10.1111/j.1529-8027.2012.00398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, Duff KE. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22:46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzig J, Jahn O, Tenzer S, Wichert SP, de Monasterio-Schrader P, Rosfa S, Kuharev J, Yan K, Bormuth I, Bremer J. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. The Journal of neuroscience. 2011;31:16369–16386. doi: 10.1523/JNEUROSCI.4016-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennuto M, Tinelli E, Malaguti M, Del Carro U, D’Antonio M, Ron D, Quattrini A, Feltri ML, Wrabetz L. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot-Marie-Tooth 1B mice. Neuron. 2008;57:393–405. doi: 10.1016/j.neuron.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Molecular cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Kukushkin N, Bosse M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem. 2013;288:7781–90. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrini A, Previtali S, Feltri ML, Canal N, Nemni R, Wrabetz L. Beta 4 integrin and other Schwann cell markers in axonal neuropathy. Glia. 1996;17:294–306. doi: 10.1002/(SICI)1098-1136(199608)17:4<294::AID-GLIA4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski JL, Tennekoon GI, McGillicuddy JE. Selective culture of mitotically active human Schwann cells from adult sural nerves. Annals of neurology. 1992;31:580–586. doi: 10.1002/ana.410310603. [DOI] [PubMed] [Google Scholar]
- Sanmaneechai O, Feely S, Scherer SS, Herrmann DN, Burns J, Muntoni F, Li J, Siskind CE, Day JW, Laura M. Genotype–phenotype characteristics and baseline natural history of heritable neuropathies caused by mutations in the MPZ gene. Brain. 2015;138:3180–3192. doi: 10.1093/brain/awv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SS, Wrabetz L. Molecular mechanisms of inherited demyelinating neuropathies. Glia. 2008;56:1578–1589. doi: 10.1002/glia.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol. 2014;24:1573–83. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17:435–49. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- Sidoli M, Musner N, Silvestri N, Ungaro D, D’Antonio M, Cavener DR, Feltri ML, Wrabetz L. Ablation of Perk in Schwann Cells Improves Myelination in the S63del Charcot-Marie-Tooth 1B Mouse. J Neurosci. 2016;36:11350–11361. doi: 10.1523/JNEUROSCI.1637-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Forster F, Baumeister W. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A. 2013;110:7264–9. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speevak MD, Farrell SA. Charcot-Marie-Tooth 1B caused by expansion of a familial myelin protein zero (MPZ) gene duplication. Eur J Med Genet. 2013;56:566–9. doi: 10.1016/j.ejmg.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Stohwasser R, Giesebrecht J, Kraft R, Muller EC, Hausler KG, Kettenmann H, Hanisch UK, Kloetzel PM. Biochemical analysis of proteasomes from mouse microglia: induction of immunoproteasomes by interferon-gamma and lipopolysaccharide. Glia. 2000;29:355–65. [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wrabetz L, D’Antonio M, Pennuto M, Dati G, Tinelli E, Fratta P, Previtali S, Imperiale D, Zielasek J, Toyka K. Different intracellular pathomechanisms produce diverse Myelin Protein Zero neuropathies in transgenic mice. The Journal of neuroscience. 2006;26:2358–2368. doi: 10.1523/JNEUROSCI.3819-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrabetz L, Feltri ML, Quattrini A, Imperiale D, Previtali S, D’Antonio M, Martini R, Yin X, Trapp BD, Zhou L. P0 glycoprotein overexpression causes congenital hypomyelination of peripheral nerves. The Journal of cell biology. 2000;148:1021–1034. doi: 10.1083/jcb.148.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Kidd G, Wrabetz L, Feltri M, Messing A, Trapp B. Schwann cell myelination requires timely and precise targeting of P0 protein. The Journal of cell biology. 2000;148:1009–1020. doi: 10.1083/jcb.148.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell metabolism. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Zhao J, Garcia GA, Goldberg AL. Control of proteasomal proteolysis by mTOR. Nature. 2016;529:E1–2. doi: 10.1038/nature16472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A. 2015;112:15790–7. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Increase in polyubiquitinated proteins in neuropathic sciatic nerves
(A) The ubiquitin signal is increased in the Schwann cells and not in the axons. Sciatic nerves were teased into individual nerve fibers and immunohistochemical analysis was performed. Nuclear staining (DAPI) is in blue, polyubiquitin (FK1) is in red and a marker of axons, neurofilament (NF) is in green. The scale bar represents 5 μm.
(B) K63-linked ubiquitin chains that do not target proteins for proteasomal degradation are increased in P0 OE and S63del H, but not S63del L. The box indicates area used for densitometric analysis of K63-linked polyubiquitin (K63 Ubn) signal. The area of the box is decreased compared to ubiquitin and K48-linked ubiquitin blots in order to avoid including the intense, possibly non-specific band located around 75 kDa in the analysis.
Supplemental Figure 2. S63del L and P0 OE sciatic nerves have distinct morphologies
(A) Electron microscopic images of ultrathin sections of sciatic nerves. In the images, # indicates an amyelinated fiber, which contains an axon greater than 1 μm in diameter that should be myelinated. Arrows indicate hypomyelinated fibers, nerve fibers that have thinner myelin than predicted, based on the diameter of the axon. Note that the S63del L nerves manifest primarily hypomyelinated fibers with only occasional amyelinated fibers, consistent with demyelination followed by remyelination, whereas P0 OE nerves contain more amyelinated fibers, consistent with developmental amyelination. At 6 months of age (not shown), S63del L nerves contain many onion bulbs, the hallmark of demyelination, whereas P0 OE nerves do not (Wrabetz et al. 2006; Wrabetz et al. 2000). Collectively, these observations indicate that S63del L mice manifest a demyelinating neuropathy, whereas P0 OE mice manifest a developmental, hypomyelinating neuropathy. The scale bar represents 2 μm.
Supplemental Figure 3. Proteasome expression and content are increased in S63del L and P0 OE sciatic nerves.
(A) mRNAs for multiple proteasome subunits are increased in S63del L and P0 OE sciatic nerves. A list of the proteasome subunit and activator mRNAs in S63del L and P0 OE that were increased by at least 1.5 fold over WT in transcriptomic analysis performed on an Affymetrix Mouse GeneChip-MOE430A 2.0 (D’Antonio et al. 2013). Only one proteasome subunit or activator mRNA transcript was decreased by at least 1.5 fold compared to WT in the neuropathic conditions, PSMB1, in S63del L.
(B) Proteasome content is increased in both S63del L and P0 OE. Shown are the normalized levels of all examined proteasome activators, proteasome subunits, and the VCP/p97 complex analyzed in Figures 3B and 3F.
Supplemental Figure 4. No increase in DUB level in S63del sciatic nerve lysates.
(A) USP5, USP14 and UCH37 are not increased in the sciatic nerve lysates of S63del L. The experiment was repeated at least 3 times with at least 2 mice per genotype each time.
(B) The amount of active DUBs is similar in sciatic nerve lysates of WT and S63del L. Sciatic nerve lysates were treated with HA-Ub-VS to modify the cysteines of active DUBs. Levels of modification, indicating DUB activity, were analyzed by western blot for HA. The levels of USP14 and UCH37 were also evaluated with specific antibodies. The USP14 antibodies did not recognize HA-Ub-VS-bound USP14 so only unmodified USP14 is detected with the USP14-specific antibody in the HA-Ub-VS lanes. The UCH37 antibody did recognize HA-Ub-VS-bound UCH37, as indicated by the approximately 10 kDa-shift in the HA-Ub-VS lanes compared to the control lanes when the UCH37-specific antibody was used. The experiment was repeated at least 3 times with 2 mice per genotype each time.
(C) The DUB activity is similar in the sciatic nerve lysates from WT and S63del L. The hydrolysis of Ub-AMC was measured in sciatic nerve lysate and was normalized to total protein. n=3.
Supplemental Figure 5: In P0 OE, but not S63del L, the increase in levels of proteasome subunits precedes the accumulation of polyubiquitinated proteins.
(A) The levels of K48-linked polyubiquitinated proteins and proteasome subunits were analyzed at post natal day 10 (p10) or 20 (p20) in S63del L or P0 OE, and compared to WT littermates.


