Abstract
Several reports have demonstrated that mouse Cx3cr1 signaling promotes monocyte/macrophage survival. In agreement, we previously found that, in a mouse model of systemic candidiasis, genetic deficiency of Cx3cr1 resulted in increased mortality and impaired tissue fungal clearance associated with decreased macrophage survival. We translated this finding by showing that the dysfunctional CX3CR1 variant CX3CR1-M280 was associated with increased risk and worse outcome of human systemic candidiasis. However, the impact of this mutation on human monocyte/macrophage survival is poorly understood. Herein, we hypothesized that CX3CR1-M280 impairs human monocyte survival. We identified WT (CX3CR1-WT/WT), CX3CR1-WT/M280 heterozygous, and CX3CR1-M280/M280 homozygous healthy donors of European descent, and we show that CX3CL1 rescues serum starvation–induced cell death in CX3CR1-WT/WT and CX3CR1-WT/M280 but not in CX3CR1-M280/M280 monocytes. CX3CL1-induced survival of CX3CR1-WT/WT monocytes is mediated via AKT and ERK activation, which are both impaired in CX3CR1-M280/M280 monocytes, associated with decreased blood monocyte counts in CX3CR1-M280/M280 donors at steady state. Instead, CX3CR1-M280/M280 does not affect monocyte CX3CR1 surface expression or innate immune effector functions. Together, we show that homozygocity of the M280 polymorphism in CX3CR1 is a potentially novel population-based genetic factor that influences human monocyte signaling.
Keywords: Immunology, Infectious disease
Keywords: Chemokines, Monocytes
Homozygocity of the M280 polymorphism in CX3CR1 is a population-based genetic factor that influences human monocyte signaling.
Introduction
The mouse Cx3cl1/Cx3cr1 signaling axis has been mechanistically implicated in promoting the survival of mononuclear phagocytes in the blood, kidney, liver, and aorta (1–8). Consistent with this, genetic deficiency of mouse Cx3cr1 or its ligand Cx3cl1 (fractalkine) results in decreased blood monocyte counts and reduced numbers of resident macrophages in certain tissues (1–9). Of interest, the survival effects of Cx3cl1 in mice appear to be mediated via the full-length membrane–anchored, but not the shed soluble, component of the chemokine (10). In humans, in contrast to mice, both the recombinant soluble and the full-length CX3CL1 have been shown to protect primary human CD14+ monocytes from serum starvation–induced death (8, 11–13). The mechanism has been shown to be Gαi-dependent via amelioration of cellular oxidative stress (12).
We previously showed that in a mouse model of systemic candidiasis, a leading cause of nosocomial bloodstream infection in the US (14, 15), Cx3cr1-deficient mice had increased mortality and were unable to clear the fungal load from the kidney, associated with impaired renal macrophage survival and accumulation (6). We also showed that the dysfunctional CX3CR1 variant CX3CR1-M280, caused by a cytosine-to-thymidine substitution at nucleotide 839 that results in a threonine-to-methionine substitution at position 280 (16, 17), was associated with increased risk and worse outcome of human systemic candidiasis in 2 independent patient cohorts (6). In the present study, we aimed to test the hypothesis that carrying the CX3CR1-M280 allele impairs survival in primary human mononuclear phagocytes.
Results
CX3CR1-M280/M280 monocytes have decreased survival.
We screened 108 professional healthy donors of European descent from the NIH Blood Bank and stratified them according to their CX3CR1 genotype. We identified 67 (62%) WT CX3CR1-WT/WT, 37 (34.3%) CX3CR1-WT/M280 heterozygous, and 4 (3.7%) CX3CR1-M280/M280 homozygous donors, an allelic distribution comparable with that previously reported for individuals of European descent (6, 16–20). The CX3CR1-M280/M280 homozygous donors were available for repeated testing on different days for all experimental assays examined. Because it is not feasible to obtain freshly isolated CX3CR1+ macrophages from humans, we focused on studying CX3CR1+CD14+ monocytes by employing a negative immunomagnetic sorting strategy that resulted in harvesting highly pure and viable CD14+ monocytes from fresh whole blood of healthy donors (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.95417DS1).
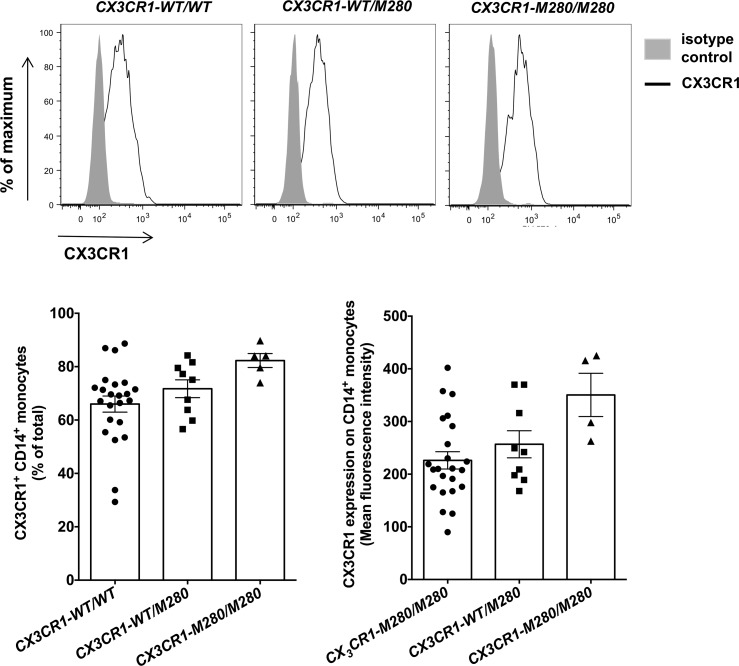
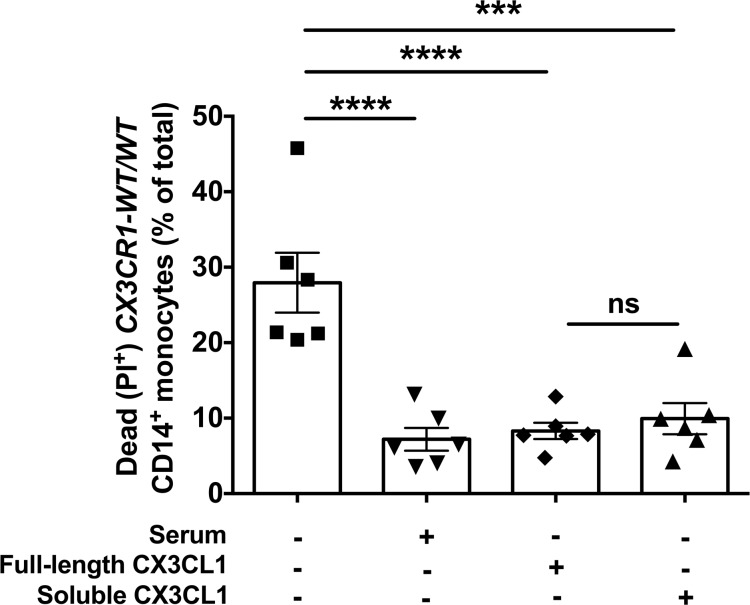
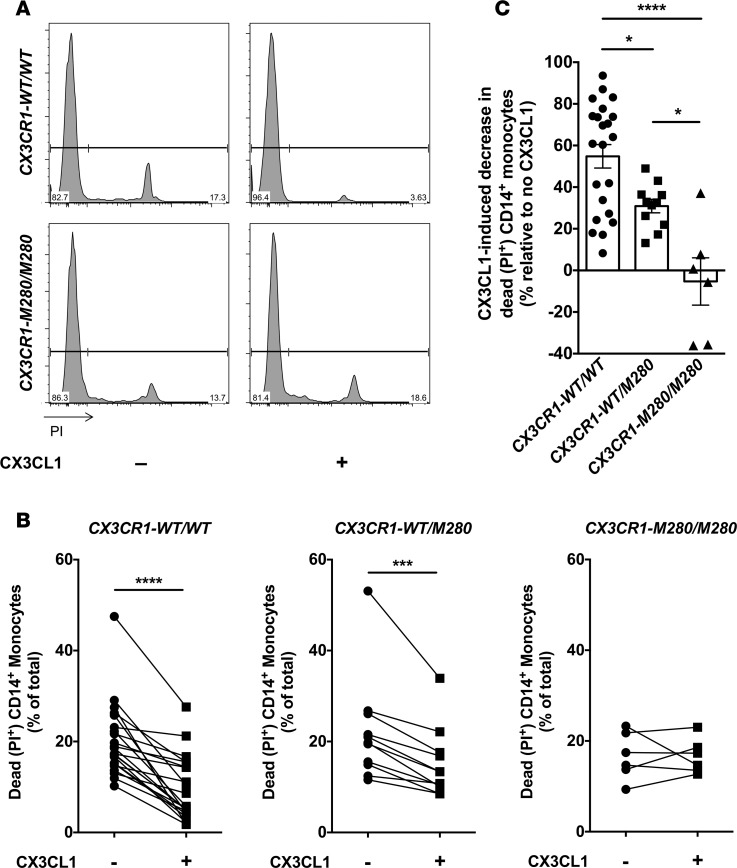
We first examined whether the CX3CR1-M280 mutation affects expression of CX3CR1 on the surface of CD14+ monocytes. We used flow cytometry and found that CX3CR1-M280/M280 and CX3CR1-WT/M280 monocytes did not have decreased CX3CR1 surface expression compared with CX3CR1-WT/WT cells (Figure 1). In agreement with previous reports, we found that both recombinant full-length and soluble CX3CL1 rescues serum starvation–induced cell death in human CX3CR1-WT/WT CD14+ monocytes (Figure 2) (8, 12). To test the hypothesis that CX3CR1-M280/M280 and CX3CR1-WT/M280 monocytes may have a survival defect relative to CX3CR1-WT/WT cells, we induced cell death by serum starvation and examined the ability of recombinant full-length CX3CL1 to rescue cell death in CX3CR1-WT/WT, CX3CR1-WT/M280, and CX3CR1-M280/M280 monocytes. In agreement with prior reports, we found that CX3CL1 promotes a ~55% reduction of cell death in CX3CR1-WT/WT monocytes following serum starvation (Figure 3, A–C) (8, 12). In addition, CX3CR1-WT/M280 monocytes were also rescued by CX3CL1 from serum starvation–induced cell death, although to a lesser extent (~30%; Figure 3, B and C) (12). However, CX3CL1 had no effect in rescuing CX3CR1-M280/M280 monocytes from serum starvation–induced cell death (Figure 3, A–C).
Figure 1. The CX3CR1-M280 mutation does not decrease CX3CR1 surface expression on CD14+ monocytes.
Representative histograms (upper panel) and summary data of percent (left lower panel) and mean fluorescent intensity (right lower panel) of CX3CR1 expression on the surface of CD14+ monocytes. n = 4–23. Statistical analysis was performed using Kruskal-Wallis with Dunn’s multiple comparisons test. Quantitative data represent the mean ± SEM.
Figure 2. Soluble and full-length CX3CL1 rescue CX3CR1-WT/WT CD14+ monocytes from serum starvation–induced death.
Percent of dead CD14+ monocytes elicited by serum starvation in the presence or absence of 100 nM of soluble or full-length CX3CL1 in CX3CR1-WT/WT monocytes. Monocytes cultured in serum-containing media were assayed as control. n = 6. ***P < 0.001; ****P < 0.0001. Statistical analysis was performed using 1-way ANOVA with Tukey’s multiple comparisons test. Data represent the mean ± SEM.
Figure 3. CX3CL1 rescues CX3CR1-WT/WT and CX3CR1-WT/M280, but not CX3CR1-M280/M280, CD14+ monocytes from serum starvation–induced death.
(A) Representative FACS histograms of propidium iodine (PI) staining in CX3CR1-WT/WT (upper panels) and CX3CR1-M280/M280 (lower panels) CD14+ monocytes following serum starvation in the presence or absence of 100 nM of CX3CL1. (B) Percent of dead CD14+ monocytes elicited by serum starvation in the presence or absence of 100 nM of CX3CL1 in CX3CR1-WT/WT (left panel), CX3CR1-WT/M280 (middle panel), and CX3CR1-M280/M280 (right panel) cells. Shown are paired experimental results with or without CX3CL1. (C) The percent decrease in cell death conferred by CX3CL1 exposure in serum-starved CD14+ monocytes is greater in CX3CR1-WT/WT compared with CX3CR1-WT/M280 cells, while no CX3CL1-induced decrease in cell death is seen in CX3CR1-M280/M280 cells. n = 22 CX3CR1-WT/WT, 11 CX3CR1-WT/M280 and 6 CX3CR1-M280/M280. *P < 0.05; ***P < 0.001; ****P < 0.0001. Statistical analysis was performed using paired 2-tailed t tests (B) or 1-way ANOVA with Tukey’s multiple comparisons test (C). Quantitative data represent the mean ± SEM.
CX3CL1-induced AKT and ERK activation promotes CX3CR1-WT/WT monocyte survival and is impaired in CX3CR1-M280/M280 cells.
We next examined the mechanism of impaired CX3CL1-mediated survival of CX3CR1-M280/M280 monocytes at the biochemical level. The CX3CL1/CX3CR1 axis has been reported to mediate survival of human vascular smooth muscle cells and rat microglial cells via AKT activation, while ERK activation was dispensable in the former context (11, 21, 22). In a mouse model of systemic candidiasis, where monocytes/macrophages are critical for host defense (23), we found that Cx3cr1 deficiency leads to impaired macrophage survival associated with decreased cellular AKT activation (6). However, the molecular basis of CX3CL1-dependent human monocyte survival is unknown.
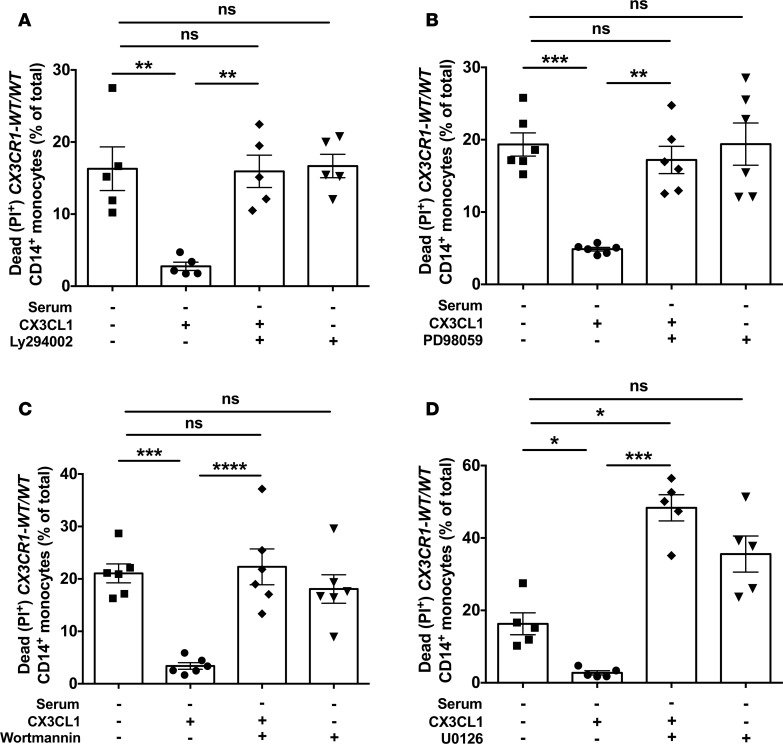
We thus examined CX3CL1-mediated human monocyte survival following serum starvation in the presence of AKT or ERK pharmacological inhibition with the PI3K inhibitor LY294002 or the MEK inhibitor PD98059, respectively. We found that inhibition of either AKT or ERK fully abrogated the CX3CL1-mediated survival rescue in CX3CR1-WT/WT monocytes (Figure 4, A and B). We then attempted to inhibit AKT or ERK via siRNA in human monocytes to independently verify the role of AKT- and ERK-dependent signaling on CX3CL1-mediated human monocyte survival following serum starvation. However, consistent with previous reports (24), we found significant downregulation of CX3CR1 on the surface of CD14+ monocytes as early as within 1 day of culture after monocyte harvesting, with almost complete abolishment of receptor surface expression at days 2 and 3 of monocyte culture (Supplemental Figure 2A). Importantly, this CX3CR1 downregulation was accompanied by abrogation of CX3CL1-mediated rescue of monocyte survival at days 1, 2, and 3 after incubation (Supplemental Figure 2B). Therefore, because siRNA-induced downregulation of AKT or ERK at the protein level is only feasible at day 3 after transfection (data not shown) when CX3CR1 is not expressed on the monocyte surface and when CX3CL1 no longer rescues monocytes from cell death, we were unable to use siRNA-mediated knockdown of AKT or ERK to confirm the role of these molecules in CX3CL1-mediated survival in monocytes. Instead, we tested an independent set of PI3K (wortmannin) and MEK (U0126) inhibitors and found that, similar to the use of LY294002 or PD98059 (Figure 4, A and B), wortmannin or U0126 also abrogated the CX3CL1-mediated survival rescue of CX3CR1-WT/WT monocytes (Figure 4, C and D).
Figure 4. CX3CL1-induced survival rescue after serum starvation in CX3CR1-WT/WT monocytes is mediated by activation of AKT and ERK.
Shown are percent of dead monocytes elicited by serum starvation in the presence or absence of 100 nM of CX3CL1 after exposure to 50 μM of the PI3K inhibitor LY294002 (A, n = 5) or 20 μM of the MEK inhibitor PD98059 (B, n = 6), or 100 nM of the PI3K inhibitor wortmannin (C, n = 6) or 20 μM of the MEK inhibitor U0126 (D, n = 5). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Statistical analysis was performed using 1-way ANOVA with Tukey’s multiple comparisons test. Data represent the mean ± SEM.
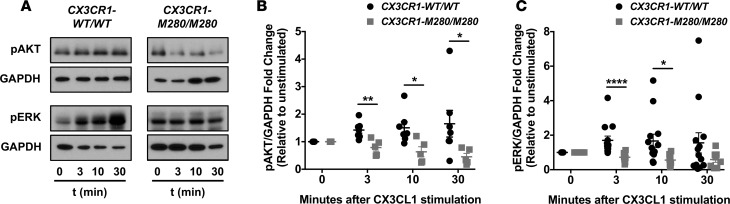
We then reasoned that AKT and/or ERK activation might be impaired in CX3CR1-M280/M280 monocytes upon CX3CL1 ligation. Indeed, while AKT and ERK phosphorylation were induced after stimulation of CX3CR1-WT/WT monocytes with CX3CL1, no induction of pAKT or pERK was seen in CX3CR1-M280/M280 monocytes using Western blot at 3, 10, or 30 minutes after CX3CL1 stimulation (Figure 5). These data collectively demonstrate that CX3CR1-M280/M280 monocytes have impaired survival associated with defective AKT and ERK activation after CX3CL1 stimulation.
Figure 5. CX3CR1-M280/M280 monocytes exhibit impaired AKT and ERK activation following CX3CL1 stimulation.
(A) Representative Western blots. (B and C) Quantitation of Western blot data. n = 5–7 (B), n = 8–13 (C). *P < 0.05; **P < 0.01; ****P < 0.0001. Statistical analysis was performed using an unpaired 2-tailed t test (with or without Welch’s correction) with the exception of the comparison of pERK/GADPH fold change at 3 minutes between CX3CR1-WT/WT and CX3CR1-M280/M280 which was performed using Mann-Whitney test. Quantitative data represent the mean ± SEM.
CX3CR1-M280/M280 donors have decreased monocyte counts.
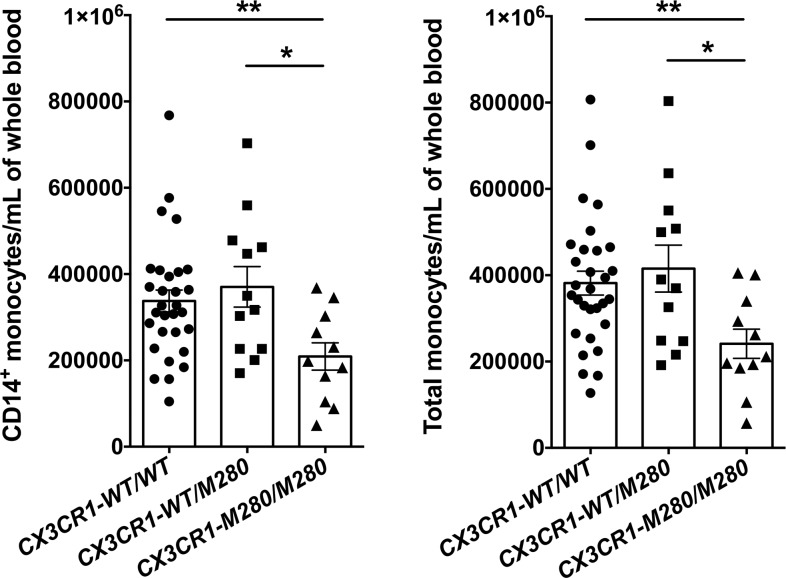
Because CX3CR1-M280/M280 monocytes have impaired survival, and Cx3cr1–/– and Cx3cl1–/– mice have decreased blood monocyte counts at steady state (8), we asked whether healthy donors carrying the CX3CR1-M280 allele might be monocytopenic at steady state. We used flow cytometry to quantify the numbers of CD14+ monocytes, total monocytes, and neutrophils in whole blood of CX3CR1-WT/WT, CX3CR1-WT/M280, and CX3CR1-M280/M280 healthy donors. Neutrophils were studied as a CX3CR1-negative control myeloid cell population in blood. Importantly, CX3CR1-M280/M280 donors exhibited a significant ~40% reduction in numbers of both CD14+ monocytes and total monocytes compared with CX3CR1-WT/WT donors (Figure 6), while their neutrophil counts were similar (Supplemental Figure 3). Instead, no decrease was observed in the numbers of CD14+ monocytes, total monocytes, or neutrophils of CX3CR1-WT/M280 relative to CX3CR1-WT/WT donors (Figure 6 and Supplemental Figure 3). Together, these data show that genetic variation at CX3CR1 influences human monocyte counts at steady state, along with the IRF8, GATA2, and WHIM-CXCR4 mutations previously reported by others (25–27).
Figure 6. CX3CR1-M280/M280 healthy donors have decreased blood monocyte counts at steady state.
CX3CR1-M280/M280 healthy donors have decreased CD14+ (left panel) and total (right panel) monocyte counts at steady state compared with CX3CR1-WT/WT and CX3CR1-WT/M280 healthy donors. n = 11–30. *P < 0.05; **P < 0.01. Statistical analysis was performed using Kruskal-Wallis with Dunn’s multiple comparisons test (left panel) or 1-way ANOVA with Tukey’s multiple comparisons test (right panel). Data represent the mean ± SEM.
The CX3CR1-M280 allele does not impair the capacity of monocytes for fungal killing or cytokine production upon fungal stimulation.
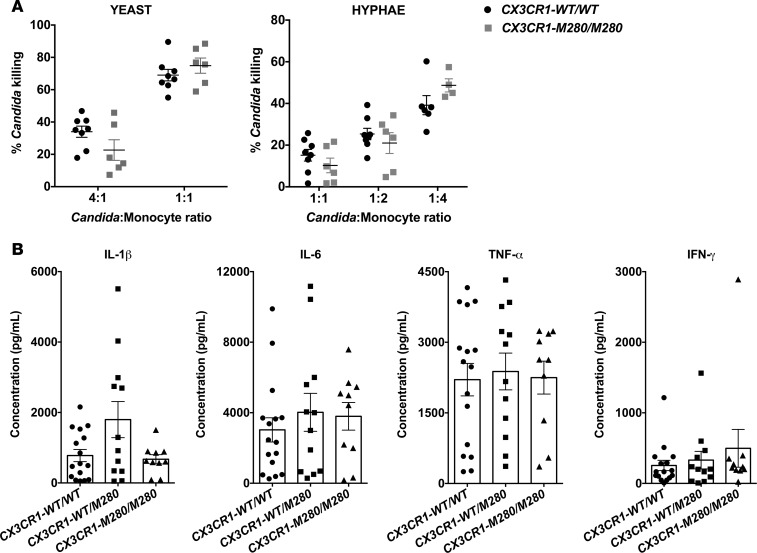
We next investigated the impact of carrying the CX3CR1-M280 allele on monocyte innate immune effector function. CD14+ monocytes exert direct anti–Candida albicans killing (28), and we previously reported that Cx3cr1-deficient mouse resident kidney macrophages exhibit a modest, yet significant, impairment in fungal killing (6). We therefore compared the ability of CD14+ monocytes from CX3CR1-WT/WT and CX3CR1-M280/M280 donors to inactivate opsonized yeast and hyphal forms of Candida albicans ex vivo and found it to be similar (Figure 7A). Besides direct killing, monocytes and macrophages are a major cellular source of proinflammatory mediators during infection (28, 29). Thus, we stimulated CD14+ monocytes from CX3CR1-WT/WT, CX3CR1-WT/M280, and CX3CR1-M280/M280 donors with Candida albicans ex vivo and determined the production of 31 cytokines and chemokines via Luminex. We found no significant differences in the production of IL-1β, IL-6, TNF-α, and IFN-γ (Figure 7B) or any other cytokine or chemokine tested (data not shown) among monocytes of the 3 groups of healthy donors. Collectively, the CX3CR1-M280 mutation does not adversely affect monocyte fungal killing or production of proinflammatory mediators.
Figure 7. The CX3CR1-M280/M280 mutation does not impair innate immune effector functions of CD14+ monocytes.
(A) Monocytes from CX3CR1-WT/WT and CX3CR1-M280/M280 donors exhibit similar ability to inactivate opsonized yeast (left panel) and hyphal forms (right panel) of Candida albicans ex vivo, assessed using the alamarBlue cell viability assay. n = 6–8. (B) Monocytes from CX3CR1-WT/WT, CX3CR1-WT/M280, and CX3CR1-M280/M280 donors exhibit a similar ability to produce IL-1β, IL-6, TNF-α, and IFN-γ following Candida albicans stimulation ex vivo, assessed by a Luminex-array assay. Statistical analysis was performed using Mann-Whitney test or unpaired t test, where appropriate (A), or Kruskal-Wallis with Dunn’s multiple comparisons test or 1-way ANOVA with Tukey’s multiple comparisons test, where appropriate (B). n = 10–16. Data represent the mean ± SEM.
Discussion
In the present study, we demonstrate that primary human CD14+ monocytes from CX3CR1-M280/M280 healthy donors exhibit impaired CX3CL1-induced activation of AKT and ERK, defective survival, and decreased blood monocyte counts at steady state, while monocyte innate immune effector functions appear preserved. Instead, heterozygocity of the M280 polymorphism in CX3CR1 results in no defect in cell survival or decrease in blood monocyte counts. Taken together, we show that homozygosity of the M280 polymorphism in the human CX3CR1 gene is a phenocopy of mouse Cx3cr1 genetic deficiency at the level of monocyte survival signaling and congenital monocytopenia.
Of note, we found that impaired CX3CL1-induced signaling and survival in CX3CR1-M280/M280 monocytes is not due to decreased cell surface expression of CX3CR1. This finding is consistent with a potential model of decreased number of CX3CR1 binding sites per cell and delayed kinetics of CX3CL1 binding on CX3CR1-M280/M280 monocytes, as shown before in human PBMCs and in CX3CR1-M280–transfected HEK293 cells (17, 19).
Our data demonstrate that CX3CR1-M280/M280 monocytes are not rescued by CX3CL1 from serum starvation–induced cell death, in contrast to the CX3CL1-dependent cell survival effects observed in CX3CR1-WT/WT and CX3CR1-WT/M280 monocytes. We examined the biochemical basis of CX3CL1-mediated protection from serum starvation in CX3CR1-WT/WT monocytes and found that both the AKT and ERK pathways are involved. The PI3K/AKT pathway is a well-recognized signal transduction pathway that promotes cell survival in response to chemokines and other extracellular signals. This includes the AKT-mediated survival in the setting of CX3CL1-CX3CR1 signaling in rat microglia, mouse renal macrophages, and human vascular smooth muscle cells (6, 11, 21, 22, 30). The ERK pathway also modulates cell survival depending on the examined cell and experimental condition. For example, CCL19/CCL21 engagement of the chemokine receptor CCR7 was recently shown to promote ERK-mediated survival in human mature DCs (31). However, the role of ERK in promoting cell survival upon CX3CL1 stimulation appears cell specific. Indeed, whereas we show dependence on ERK for CX3CL1-mediated survival in human monocytes, White and colleagues showed that the ERK pathway is dispensable for CX3CL1-mediated survival in human coronary artery vascular smooth muscle cells (22). Therefore, additional research in different human and other mammalian CX3CR1–expressing cell subsets will be required to determine whether CX3CL1/CX3CR1 signaling promotes survival in other cell types and, if so, by which cell-specific biochemical mechanisms.
After determining that AKT and ERK are involved in CX3CL1-mediated survival rescue from serum starvation–induced cell death in CX3CR1-WT/WT monocytes, we examined whether the inability of CX3CL1 to rescue CX3CR1-M280/M280 monocytes from serum starvation–induced cell death is the result of impaired CX3CL1-induced AKT and ERK activation. Indeed, AKT and ERK were not induced upon CX3CL1 stimulation in CX3CR1-M280/M280 monocytes, in contrast to CX3CR1-WT/WT monocytes. The impaired CX3CL1-induced AKT activation in CX3CR1-M280/M280 monocytes is in contrast to the previously reported unaffected AKT activation upon CX3CL1 stimulation in CX3CR1-M280–transfected Chinese hamster ovary cells (32). This discrepancy may be due to the different experimental conditions and differential cell-specific effects of the CX3CR1-M280 mutation in inducing the AKT signal transduction pathway. Alternatively, the defect in CX3CL1-mediated AKT and ERK induction that we observed in human primary CX3CR1-M280/M280 monocytes could be caused by an independent molecular defect resulting from linkage with another yet-unknown gene polymorphism leading to the loss of the CX3CL1-mediated cell survival.
In the present study, we used the soluble chemokine domain and full-length forms of CX3CL1, but not membrane-bound CX3CL1. The membrane-bound form of CX3CL1 has been shown to exert different actions compared with the soluble CX3CL1 forms. For example, membrane-bound CX3CL1 specifically stimulates production of IL-6, CCL2, and MMP-9 by human monocytes and production of IFN-γ by human NK cells (33, 34). Therefore, because our work does not address potential effects of membrane-bound CX3CL1 on monocyte survival, future studies using coated CX3CL1 or cells expressing surface CX3CL1 will be required to examine the role of membrane-bound CX3CL1 in mediating AKT and ERK activation and promoting survival of CX3CR1-WT/WT and CX3CR1-M280/M280 monocytes.
The decreased CX3CL1-mediated survival rescue seen in CX3CR1-M280/M280 monocytes led us to hypothesize that the observed survival defect may render CX3CR1-M280/M280 individuals monocytopenic. We examined peripheral blood myeloid cell numbers at steady state and found decreased numbers of total and CD14+ monocytes in CX3CR1-M280/M280 individuals, whereas, as control, the numbers of CX3CR1-negative neutrophils were not decreased in these healthy donors. Although the identified survival defect upon CX3CL1 stimulation may contribute to the decreased monocyte counts in the peripheral blood of CX3CR1-M280/M280 donors, other factors may play a role, such as the previously described stronger adhesion of CX3CR1-M280/M280 monocytes to membrane-bound CX3CL1 (35) or other factors that remain unknown.
Beyond a Cx3cr1-dependent survival effect on mouse renal macrophages, our prior work in the mouse model of invasive candidiasis had also revealed a modest, yet statistically significant, defect in fungal killing in Cx3cr1–/– kidney macrophages (6). Herein, we found that CX3CR1-M280/M280 monocytes did not exhibit a killing defect against Candida yeast or hyphal morphotypes, nor did they exhibit an impairment in proinflammatory cytokine production upon Candida stimulation. The differences in fungal killing between Cx3cr1–/– kidney macrophages and CX3CR1-M280/M280 monocytes may reflect a differential role of CX3CR1 signaling in modulating antifungal effector function in human monocytes versus mouse macrophages, or it could be accounted for by the degree of CX3CR1 impairment of Cx3cr1–/– macrophages (i.e., complete deficiency) versus CX3CR1-M280/M280 monocytes (i.e., incomplete deficiency) (6, 19).
Our study provides potentially novel insight into the biology of CX3CR1-M280 and further underscores the observation that this mutation exerts cell-specific effects on various cellular functions, which may be either loss-of-function or gain-of-function, depending on the experimental context, as previously demonstrated for monocyte adhesion, for NK cell, peripheral blood mononuclear cell (PBMC), and CX3CR1-M280–transfected HEK293 cell chemotaxis, and for PBMC and CX3CR1-M280-transfected HEK293 cell calcium release (19, 35). In addition, our study further extends the previously reported observations that heterozygosity of the M280 polymorphism in CX3CR1 may not result in similarly pronounced alterations in cellular function when compared with CX3CR1-M280 homozygosity (12, 35). Indeed, although we found that CX3CR1-WT/M280 monocytes display a modest, yet statistically significant, reduction in CX3CL1-mediated rescue from serum starvation–induced cell death relative to the degree of survival rescue seen in CX3CR1-WT/WT monocytes, the monocyte numbers in the peripheral blood of CX3CR1-WT/M280 donors are not decreased at the steady state.
At the population genetic level, the CX3CR1-M280 mutation has thus far been shown to modulate the risk for development of cardiovascular disease, the leading cause of death in the US (17, 19, 20, 36); age-related macular degeneration, a leading cause of blindness in adults > 50 years old in the US (18); systemic candidiasis, the leading cause of invasive fungal infection in the US (6, 14, 15); and progression of HIV to AIDS (16). Given the pleiotropic effects that the CX3CR1-M280 mutation may exert on various tissues and immune cell subsets, additional studies will be required to further delineate the cell-specific consequences and molecular and biochemical mechanisms by which CX3CR1-M280 heterozygosity and homozygosity may modulate the functions of different CX3CR1-expressing human hematopoietic and nonhematopoietic cells beyond CD14+ monocytes, such as CD16+ monocytes, DCs, macrophages, NK cells, CD4+ and CD8+ T lymphocytes, endothelial cells, and epithelial cells (37). Moreover, future studies in a larger number of healthy donors that would also include different ethnic groups, as well as in patients with various infectious or inflammatory conditions, will be required to verify our findings and to further elucidate the role of the CX3CR1-M280 variant in human health and various disease states.
Methods
Fungal strain.
The Candida albicans strain SC5314 (gift from John Bennett, NIAID) was grown in yeast extract, peptone, and dextrose medium containing penicillin and streptomycin (Mediatech Inc.) in a shaking incubator at 30°C (38). Cells were centrifuged at 427 g for 5 minutes (Sorvall Legend XTR centrifuge), washed in PBS, counted with a hemocytometer, and used for fungal killing and stimulation assays by human monocytes.
Isolation of DNA from peripheral whole blood and CX3CR1-M280 genotyping.
DNA was isolated from whole blood using the Gentra Puregene Blood Kit (Qiagen) following the manufacturer’s instructions. To determine whether healthy donors carried the mutant CX3CR1-M280 allele, a 25-μl PCR reaction was performed using the following conditions as previously described (6, 20): 100 ng of genomic DNA, 1× buffer, 2.0 mM MgCl2, 175 μM each of dNTP, 0.75 U Taq polymerase, and 15 pmoles of the following primers: 5′-AGAATCATCCAGACGCTGTTTTCC-3′ and 5′-CACAGGACAGCCAGGCATTTCC-3′. Amplification conditions consisted of a single cycle of 95°C for 3 minutes, followed by 35 cycles of 94°C, 69°C, and 72°C for 30 seconds each, and then by a single cycle of 72°C for 10 minutes. This amplification protocol produced a single amplicon of 311 bp. The alleles at codon 280 were genotyped via overnight digestion at 37°C by incubating the resulting PCR reaction product with 1.5 U of Bst4CI (New England BioLabs, catalog R0618S) in a 20-μl reaction as per the manufacturer’s instructions. This resulted in 107- and 204-bp fragments when a C was present at nucleotide 839 of the open reading frame, whereas the amplicon remained uncut at 311-bp if a T was present. The genotype was determined via electrophoresis on a 2% agarose gel stained with Gelstar (FMC Bioproducts) according to the manufacturer’s instructions.
Isolation of CD14+monocytes from peripheral whole blood. PBMCs were harvested from sodium heparin–treated whole blood via gradient centrifugation using Lymphocyte Separation Media, according to the manufacturer’s protocol (Lonza). PBMCs were washed in PBS, and CD14+ monocytes were isolated by a negative immunomagnetic selection approach using the Monocyte Isolation Kit II (Miltenyi Biotec) according to the manufacturer’s instructions. Purity and viability of CD14+ monocytes were 84%–94% and >90%, respectively (Supplemental Figure 1).
CD14+ monocyte stimulation and cytokine/chemokine determination.
A 31-plex Luminex-array assay was employed to determine proinflammatory cytokine and chemokine production by monocytes after Candida albicans stimulation ex vivo. In brief, 1 × 106 CD14+ monocytes from either CX3CR1-WT/WT, heterozygous CX3CR1-WT/M280, or homozygous CX3CR1-M280/M280 donors were plated in a round-bottom 96-well plate (Corning Inc.) at 37°C in a 5% CO2 incubator in RPMI 1640 containing 10% FBS (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin (unstimulated), or RPMI 1640 with 10% FBS/antibiotics containing heat-killed C. albicans SC5314 yeasts (1 × 106/ml), as previously described (39). After 24 hours of stimulation, the supernatant was collected and stored at –80°C until analysis. Luminex was performed via a multiplex bead array assay with antibodies and cytokine standards to generate known concentration curves (R&D Systems, Peprotech). According to the manufacturer’s protocols, individual Luminex bead sets were coupled with specific biotinylated capture polyclonal antibodies, which were used at twice the recommended concentrations for a standard ELISA. The assay was run in a 50-μl volume with 1,500 beads per set of cytokines. The plates were read on a Luminex MAGPIX platform, with greater than 50 beads collected per bead region. The median fluorescence intensity of the beads were measured for each individual bead and analyzed with the Millipex software using a 5-parameter regression algorithm. The following cytokines and chemokines were determined: IL-1α, IL-1β, IL-2, IL-4, IL-7, IL-12p70, IL-16, IL-17A, IL-17F, IL-22, CCL2, CCL3, CCL4, CCL5, CCL7, CCL8, CCL11, CCL13, CCL20, CXCL1, CXCL2, CXCL5, CXCL8 (IL-8), CXCL9, CXCL10, CXCL12, CXCL16, IFN-γ, TNF-α, TGF-β1, and GM-CSF.
Monocyte CX3CR1 surface expression and quantification of monocyte and neutrophil counts in whole blood using flow cytometry.
Whole blood was obtained from CX3CR1-WT/WT, heterozygous CX3CR1-WT/M280, and homozygous CX3CR1-M280/M280 donors, and RBCs were lysed with 1× BD PharmLyse in sterile water per the manufacturer’s protocol. Leukocytes were washed in PBS and stained with Live/Dead fluorescent dye (Invitrogen) for 10 minutes in PBS at 4°C. Fc Blocking Reagent (Miltenyi Biotec) was added for 10 minutes in FACS buffer at 4°C. Surface antigen staining was then performed by incubating fluorochrome-conjugated (eFluor 605NC, Biotin, PE, FITC, PerCP-Cy5.5) antibodies against human CD45 (HI30, eBioscience), CX3CR1 (2A9-1, eBioscience), rat IgG2b K isotype control (eB149/10H5, eBioscience), CD16 (3G8, BD Biosciences), CD14 (M5E2, BD Biosciences), and HLA-DR (G46-6, BD Biosciences) for 30 minutes at 4°C. Cells were washed with FACS buffer, and cells were incubated with BV570-conjugated Streptavidin (BioLegend) for 20 minutes at 4°C. Cells were then washed with FACS buffer and analyzed on a 5-laser LSRFortessa with FACS Diva software (BD Biosciences). Data was analyzed with FlowJo software (Tree Star Inc.). Only live single cells were analyzed, and cell numbers were quantified using PE-conjugated fluorescent counting beads (Spherotech) as previously described (38).
In some experiments, we compared the expression of CX3CR1 on the surface of CD14+ monocytes in freshly harvested cells and in monocytes after 1, 2, or 3 days of culture. Specifically, CD14+ monocytes were isolated by negative immunomagnetic selection as mentioned above. For freshly isolated monocytes, (day 0), 1 × 106 cells were blocked with Fc Blocking Reagent (Miltenyi Biotec) in PBS for 10 minutes, and FACS staining was performed using APC-conjugated CD14 (M5E2, BD Biosciences) and PE-conjugated CX3CR1 (2A9-1, eBioscience) or rat IgG2b K isotype control (eB149/10H5, eBioscience), for 30 minutes at 4°C. Cells were then washed with FACS buffer and analyzed on a 5-laser LSRFortessa with FACS Diva software. For assessment of monocyte CX3CR1 expression at days 1, 2 ,and 3 after harvesting, CD14+ monocytes from the same donors were cultured in a 6-well culture plate in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin in a 37oC incubator with 5% CO2. Cells were trypsinized, collected at the indicated day, washed with RPMI serum free medium, and stained for CX3CR1 or isotype control antibodies as described above. Trypsinization alone did not alter monocyte CX3CR1 surface expression (data not shown).
Candida albicans killing by CD14+ monocytes.
CD14+ monocytes (5 × 104) from CX3CR1-WT/WT and homozygous CX3CR1-M280/M280 donors were incubated in a flat-bottom 96-well plate (Corning) with opsonized Candida albicans yeast cells at a monocyte/Candida ratio of 1:4 and 1:1 or opsonized Candida albicans hyphae at a monocyte/Candida albicans ratio of 4:1, 2:1, and 1:1 in duplicate for 2.5 hours at 37°C. Human serum (10%) obtained from the same healthy donor pool was used for Candida albicans opsonization in all experiments. Wells were then treated with 0.02% Triton X-100 in water for 7 minutes and subsequently washed twice with PBS. AlamarBlue (1×, Invitrogen) diluted in PBS was then added to each well and incubated for 18 hours at 37°C as previously described (6). Fluorescence in each well was measured using a PolarStar Optima Plate Reader (BMG Labtech). Fungal inactivation by monocytes was calculated by comparing the fluorescence of Candida albicans incubated with monocytes to that of Candida albicans incubated without monocytes.
Monocyte survival analysis using flow cytometry.
CD14+ monocytes (1 × 105) from CX3CR1-WT/WT, heterozygous CX3CR1-WT/M280, and homozygous CX3CR1-M280/M280 donors were serum starved in 5 ml polystyrene round-bottom tubes (BD Falcon) by resuspension in either RPMI 1640 containing 100 U/ml of penicillin and 100 μg/ml of streptomycin or RPMI 1640 containing antibiotics and 100 nM final concentration of recombinant human full-length CX3CL1 (R&D Systems, catalog 365-FR) or 100 nM final concentration of recombinant soluble CX3CL1 (R&D Systems, catalog 362-CX-025) at 37°C for 4 hours. Monocytes were then resuspended in annexin V binding buffer (BD Biosciences) and incubated at room temperature with FITC-conjugated annexin V, propidium iodide, and APC-conjugated mouse anti–human CD14 (M5E2, BD Biosciences). The percentage of propidium iodide–positive cells was determined by flow cytometry. Cells were analyzed on a 5-laser LSRFortessa with FACS Diva software (BD Biosciences), and data were analyzed using FlowJo. In some experiments, we assessed the ability of full-length CX3CL1 to rescue serum starvation–induced cell death of CD14+ monocytes after 1, 2 ,or 3 days of monocyte culture.
AKT and ERK inhibition in monocytes.
Monocytes (1 x 105) were pretreated for 20 minutes at 37oC with either the PI3K inhibitor LY294002 (50 μM; Tocris Bioscience, catalog 1130) or the PI3K inhibitor wortmannin (100 nM; Tocris Bioscience, catalog 1232), or the MEK inhibitor PD98059 (20 μM; Tocris Bioscience, catalog 1213) or the MEK inhibitor U0126 (20 μM; Tocris Bioscience, catalog 1144) in serum free medium. Recombinant human full-length CX3CL1 (100 nM; R&D Systems, catalog 365-FR) was then added, and the samples were further incubated for 4 hours at a 37oC incubator with 5% CO2. Monocytes in serum free media with the above-mentioned inhibitors but without CX3CL1, and monocytes in serum free media without the above-mentioned inhibitors with or without addition of CX3CL1 served as controls. After 4 hours, the cells were resuspended in annexin V binding buffer and stained with FITC-labeled annexin V and propidium iodide at room temperature, and the percentage of propidium iodide–positive cells was determined by flow cytometry.
Western blot analysis of CD14+ monocytes.
To define impairments in ERK signaling downstream of CX3CL1-CX3CR1 ligation in CD14+ monocytes from homozygous CX3CR1-M280/M280 donors relative to CX3CR1-WT/WT donors, 1.25 × 106 cells were incubated in an ultra low attachment flat-bottomed 96-well plate (Costar) in RPMI 1640 containing 100 U/ml of penicillin and 100 μg/ml of streptomycin for 3 hours at 37°C. Recombinant human soluble CX3CL1 (R&D Systems, catalog 362-CX) was then added at a final concentration of 100 nM for 3, 10, or 30 minutes. Cells not exposed to soluble CX3CL1 were used as a control. After corresponding exposures to CX3CL1 or control buffer, the monocytes were washed with PBS, and the cells were resuspended in RIPA buffer (MilliporeSigma) containing Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). After 20 minutes of incubation at 4°C, cell lysates were clarified by centrifugation at 15,000 g for 10 minutes at 4°C. Supernatants were transferred to a new Eppendorf tube and stored at –20°C until further analysis. Western blot lysates were resolved on 10% SDS-PAGE gels, transferred onto PVDF membranes (MilliporeSigma, catalog IPVH304F0), probed with the indicated antibodies and developed with ECL (MilliporeSigma, catalog WBKLS0500) and film (Agfa X-Ray Film). Antibodies against p-ERK1/2 (Thr202/Tyr204; catalog 9101), p-AKT (Ser473; catalog 4060), and GAPDH (catalog 5174) were all obtained from Cell Signaling Technology. Goat anti-rabbit secondary Ab was from Southern Biotech (catalog 4010-05). Western blots were quantified by densitometry analysis using ImageJ analysis software (NIH) and Scion Image analysis software (Scion Corporation).
Study approval.
The healthy donors were enrolled in protocols approved by the NIAID and National Cancer Institute (NCI) IRB committees, and provided written informed consent for participation in the study. This study was conducted in accordance with the Declaration of Helsinki .
Statistics.
Statistical analyses were performed using GraphPad Prism 6.0 software. Data was analyzed by paired or unpaired 2-tailed or paired 2-tailed t test, Mann Whitney U test, 1-way ANOVA test, or Kruskal-Wallis test, as appropriate. P < 0.05 was considered significant.
Author contributions
MSL conceived the project and contributed to the design and supervision of the experiments. ALC and MSL wrote the manuscript. ALC, MS, MOH, MSS, KWH, SPS, JKL, CMM, and MSL conducted experiments and generated manuscript figures. ALC, MS, MOH, SPS, AM, MDJ, JMF, CMM, JSG, and MSL contributed to the interpretation of the data. EMNF contributed to healthy donor enrollment.
Supplementary Material
Acknowledgments
We thank the healthy donors for their participation in our study. This work was supported by the Division of Intramural Research, NIAID, and NIDCR, NIH.
Version 1. 02/08/2018
Electronic publication
Funding Statement
This work was supported by the Division of intramural Research, NIAID and NIDCR, NIH
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(3):e95417. https://doi.org/10.1172/jci.insight.95417.
Contributor Information
Muthulekha Swamydas, Email: muthulekha.swamydas@nih.gov.
Md Sanaullah Sajib, Email: s.sajib@ttuhsc.edu.
Ahmad Mourad, Email: ahmad.mourad@duke.edu.
J. Silvio Gutkind, Email: sgutkind@ucsd.edu.
Michail S. Lionakis, Email: michail.lionakis@nih.gov.
References
- 1.Ensan S, et al. Self-renewing resident arterial macrophages arise from embryonic CX3CR1(+) precursors and circulating monocytes immediately after birth. Nat Immunol. 2016;17(2):159–168. doi: 10.1038/ni.3343. [DOI] [PubMed] [Google Scholar]
- 2.Peng X, Zhang J, Xiao Z, Dong Y, Du J. CX3CL1-CX3CR1 Interaction Increases the Population of Ly6C(-)CX3CR1(hi) Macrophages Contributing to Unilateral Ureteral Obstruction-Induced Fibrosis. J Immunol. 2015;195(6):2797–2805. doi: 10.4049/jimmunol.1403209. [DOI] [PubMed] [Google Scholar]
- 3.Poupel L, et al. Pharmacological inhibition of the chemokine receptor, CX3CR1, reduces atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 2013;33(10):2297–2305. doi: 10.1161/ATVBAHA.112.300930. [DOI] [PubMed] [Google Scholar]
- 4.Zheng J, Yang M, Shao J, Miao Y, Han J, Du J. Chemokine receptor CX3CR1 contributes to macrophage survival in tumor metastasis. Mol Cancer. 2013;12(1):141. doi: 10.1186/1476-4598-12-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlmark KR, et al. The fractalkine receptor CX3CR1 protects against liver fibrosis by controlling differentiation and survival of infiltrating hepatic monocytes. Hepatology. 2010;52(5):1769–1782. doi: 10.1002/hep.23894. [DOI] [PubMed] [Google Scholar]
- 6.Lionakis MS, et al. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest. 2013;123(12):5035–5051. doi: 10.1172/JCI71307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Łyszkiewicz M, Witzlau K, Pommerencke J, Krueger A. Chemokine receptor CX3CR1 promotes dendritic cell development under steady-state conditions. Eur J Immunol. 2011;41(5):1256–1265. doi: 10.1002/eji.201040977. [DOI] [PubMed] [Google Scholar]
- 8.Landsman L, et al. CX3CR1 is required for monocyte homeostasis and atherogenesis by promoting cell survival. Blood. 2009;113(4):963–972. doi: 10.1182/blood-2008-07-170787. [DOI] [PubMed] [Google Scholar]
- 9.Medina-Contreras O, et al. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest. 2011;121(12):4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KW, et al. In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. Blood. 2011;118(22):e156–e167. doi: 10.1182/blood-2011-04-348946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White GE, Greaves DR. Fractalkine: a survivor’s guide: chemokines as antiapoptotic mediators. Arterioscler Thromb Vasc Biol. 2012;32(3):589–594. doi: 10.1161/ATVBAHA.111.237412. [DOI] [PubMed] [Google Scholar]
- 12.White GE, McNeill E, Channon KM, Greaves DR. Fractalkine promotes human monocyte survival via a reduction in oxidative stress. Arterioscler Thromb Vasc Biol. 2014;34(12):2554–2562. doi: 10.1161/ATVBAHA.114.304717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panek CA, et al. Differential expression of the fractalkine chemokine receptor (CX3CR1) in human monocytes during differentiation. Cell Mol Immunol. 2015;12(6):669–680. doi: 10.1038/cmi.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lionakis MS. New insights into innate immune control of systemic candidiasis. Med Mycol. 2014;52(6):555–564. doi: 10.1093/mmy/myu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magill SS, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faure S, et al. Rapid progression to AIDS in HIV+ individuals with a structural variant of the chemokine receptor CX3CR1. Science. 2000;287(5461):2274–2277. doi: 10.1126/science.287.5461.2274. [DOI] [PubMed] [Google Scholar]
- 17.Moatti D, et al. Polymorphism in the fractalkine receptor CX3CR1 as a genetic risk factor for coronary artery disease. Blood. 2001;97(7):1925–1928. doi: 10.1182/blood.V97.7.1925. [DOI] [PubMed] [Google Scholar]
- 18.Combadière C, et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007;117(10):2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDermott DH, et al. Chemokine receptor mutant CX3CR1-M280 has impaired adhesive function and correlates with protection from cardiovascular disease in humans. J Clin Invest. 2003;111(8):1241–1250. doi: 10.1172/JCI16790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott DH, et al. Association between polymorphism in the chemokine receptor CX3CR1 and coronary vascular endothelial dysfunction and atherosclerosis. Circ Res. 2001;89(5):401–407. doi: 10.1161/hh1701.095642. [DOI] [PubMed] [Google Scholar]
- 21.Boehme SA, Lio FM, Maciejewski-Lenoir D, Bacon KB, Conlon PJ. The chemokine fractalkine inhibits Fas-mediated cell death of brain microglia. J Immunol. 2000;165(1):397–403. doi: 10.4049/jimmunol.165.1.397. [DOI] [PubMed] [Google Scholar]
- 22.White GE, Tan TC, John AE, Whatling C, McPheat WL, Greaves DR. Fractalkine has anti-apoptotic and proliferative effects on human vascular smooth muscle cells via epidermal growth factor receptor signalling. Cardiovasc Res. 2010;85(4):825–835. doi: 10.1093/cvr/cvp341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lionakis MS, Iliev ID, Hohl TM. Immunity against fungi. JCI Insight. 2017;2(11):93156. doi: 10.1172/jci.insight.93156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barlic J, Zhang Y, Foley JF, Murphy PM. Oxidized lipid-driven chemokine receptor switch, CCR2 to CX3CR1, mediates adhesion of human macrophages to coronary artery smooth muscle cells through a peroxisome proliferator-activated receptor gamma-dependent pathway. Circulation. 2006;114(8):807–819. doi: 10.1161/CIRCULATIONAHA.105.602359. [DOI] [PubMed] [Google Scholar]
- 25.Hsu AP, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDermott DH, et al. The CXCR4 antagonist plerixafor corrects panleukopenia in patients with WHIM syndrome. Blood. 2011;118(18):4957–4962. doi: 10.1182/blood-2011-07-368084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smeekens SP, et al. The classical CD14++ CD16- monocytes, but not the patrolling CD14+ CD16+ monocytes, promote Th17 responses to Candida albicans. Eur J Immunol. 2011;41(10):2915–2924. doi: 10.1002/eji.201141418. [DOI] [PubMed] [Google Scholar]
- 29.Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, van de Veerdonk FL. Immune defence against Candida fungal infections. Nat Rev Immunol. 2015;15(10):630–642. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- 30.Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003;22(56):8983–8998. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 31.López-Cotarelo P, et al. A novel MEK-ERK-AMPK signaling axis controls chemokine receptor CCR7-dependent survival in human mature dendritic cells. J Biol Chem. 2015;290(2):827–840. doi: 10.1074/jbc.M114.596551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis CN, Harrison JK. Proline 326 in the C terminus of murine CX3CR1 prevents G-protein and phosphatidylinositol 3-kinase-dependent stimulation of Akt and extracellular signal-regulated kinase in Chinese hamster ovary cells. J Pharmacol Exp Ther. 2006;316(1):356–363. doi: 10.1124/jpet.105.093039. [DOI] [PubMed] [Google Scholar]
- 33.Ancuta P, Wang J, Gabuzda D. CD16+ monocytes produce IL-6, CCL2, and matrix metalloproteinase-9 upon interaction with CX3CL1-expressing endothelial cells. J Leukoc Biol. 2006;80(5):1156–1164. doi: 10.1189/jlb.0206125. [DOI] [PubMed] [Google Scholar]
- 34.Yoneda O, et al. Membrane-bound form of fractalkine induces IFN-gamma production by NK cells. Eur J Immunol. 2003;33(1):53–58. doi: 10.1002/immu.200390007. [DOI] [PubMed] [Google Scholar]
- 35.Daoudi M, et al. Enhanced adhesive capacities of the naturally occurring Ile249-Met280 variant of the chemokine receptor CX3CR1. J Biol Chem. 2004;279(19):19649–19657. doi: 10.1074/jbc.M313457200. [DOI] [PubMed] [Google Scholar]
- 36.Lavergne E, et al. Adverse associations between CX3CR1 polymorphisms and risk of cardiovascular or cerebrovascular disease. Arterioscler Thromb Vasc Biol. 2005;25(4):847–853. doi: 10.1161/01.ATV.0000157150.23641.36. [DOI] [PubMed] [Google Scholar]
- 37.Bachelerie F, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2014;66(1):1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lionakis MS, Lim JK, Lee CC, Murphy PM. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun. 2011;3(2):180–199. doi: 10.1159/000321157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drummond RA, et al. CARD9-Dependent Neutrophil Recruitment Protects against Fungal Invasion of the Central Nervous System. PLoS Pathog. 2015;11(12):e1005293. doi: 10.1371/journal.ppat.1005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.