Abstract
Type I IFN (IFN-I) dysregulation contributes to type 1 diabetes (T1D) development, and although increased IFN-I signals are pathogenic at the initiation of autoimmune diabetes, IFN-I dysregulation at later pathogenic stages more relevant for therapeutic intervention is not well understood. We discovered that 5 key antigen-presenting cell subsets from adult prediabetic NOD mice have reduced responsiveness to IFN-I that is dominated by a decrease in the tonic-sensitive subset of IFN-I response genes. Blockade of IFNAR1 in prediabetic NOD mice accelerated diabetes and increased Th1 responses. Therefore, IFN-I responses shift from pathogenic to protective as autoimmunity progresses, consistent with chronic IFN-I exposure. In contrast, IL-1–associated inflammatory pathways were elevated in prediabetic mice. These changes correlated with human T1D onset-associated gene expression. Prostaglandin E2 (PGE2) and prostaglandin receptor 4 (PTGER4), a receptor for PGE2 that mediates both inflammatory and regulatory eicosanoid signaling, were higher in NOD mice and drive innate immune dysregulation. Treating prediabetic NOD mice with a PTGER4 antagonist restored IFNAR signaling, decreased IL-1 signaling, and decreased infiltration of leukocytes into the islets. Therefore, innate cytokine alterations contribute to both T1D-associated inflammation and autoimmune pathogenesis. Modulating innate immune balance via signals such as PTGER4 may contribute to treatments for autoimmunity.
Keywords: Autoimmunity, Inflammation
Keywords: Autoimmune diseases, Dendritic cells, Innate immunity
Innate cytokine alterations contribute to both T1D-associated inflammation and autoimmune pathogenesis, suggesting potential treatments for autoimmunity.
Introduction
Innate immune responses are an essential first-line defense against infection and other perturbations, but regulation of these responses is needed to avoid direct tissue damage. Therefore, increased inflammatory signals, especially in chronic settings, can trigger multiple layers of immune regulation that help in preventing self-tissue injury due to hyperinflammation. Type 1 diabetes (T1D) is attributed in part to the exacerbated innate immune activity that has been characterized by early elevation of proinflammatory cytokines, which contribute to subsequent activation of self-reactive islet-specific T cells (1–4). But the nature of the chronic innate signals and altered regulation of this response at later stages likely also contributes to autoimmune pathogenesis and is currently not well defined. The two main categories of innate immunity, IL-1 and type I IFN (IFN-I), elicit different responses and can counterregulate each other.
IFN-I has been shown to be an early pathogenic signal in T1D that contributes to breaking self-tolerance (5). However, less is understood about the role of IFN-I at the later prediabetic and new-onset phases of disease that may be more relevant for therapeutic intervention. Because peripheral antigen-presenting cells (APCs) are more likely to be targeted with antigen for tolerogenic treatment, it is important to understand the systemic changes in APC innate responsiveness. We have recently shown that DCs from prediabetic 8- to 10-week-old NOD mice display muted IFN-I gene expression (6), despite increased levels of IFN-I. Increased IFN-I–inducible gene expression is associated with the earliest stages of T1D pathogenesis, defined in humans by the initial appearance of islet autoantibodies in the serum and in mice by initiation of insulitis (7). But surprisingly, this effect is transient, as it is not found in patients with established T1D or older 8- to 10-week-old NOD mice, despite their higher IFN-I levels (6, 8). Determining how this chronically elevated IFN-α/β and reduced cellular IFNAR response contribute to the loss of regulation that drives pathogenesis will help define how to modulate innate signals for disease intervention at the prediabetic and new-onset phases. At later stages of chronic autoimmunity, the role IFN-I responses is less clear and could be important for maintaining self-tolerance, as shown for multiple sclerosis (9). In this regard, to prevent T1D development or to delay the pathogenesis a better understanding of the mechanisms underlying the altered innate responses is needed. Specifically, measuring innate mediators without exogenous stimulation (baseline) to focus on chronic signals and determining defective counterregulation during later stages of chronic innate activation can inform how modulation of these signals could restore a normal cytokine balance to limit autoimmune pathology.
In parallel with this impaired IFNAR response, endogenous ligands for innate receptors, such as TLRs, RIG-I–like helicases (immunoreceptors for viral RNA), and nucleotide-binding oligomerization domain–like receptors (NLRs), can enhance inflammatory responses and pathogenesis in T1D (10–12). Elevated inflammatory responses have been demonstrated by the transcriptome analysis of whole blood and immune cells as well as through increased circulating proinflammatory cytokines, such as TNF-α, IL-6, IL-1β, in NOD mice and patients at onset of T1D (13–16). IL-1 is an important cytokine that enhances adaptive immune responses by inducing Th1 and Th17 differentiation and proliferation, and the IL-1 gene has been implicated in risk for T1D development (17). Circulating monocytes from T1D patients spontaneously produce more IL-6 and IL-1β and induce more Th17/Th1 cells (18). Although antagonism of IL-1 showed improved glycemic control and β cell function in T2D (19), in T1D, blockade of IL-1 failed to significantly alter clinical symptoms of T1D (20, 21) but did quench the IL-1 gene expression signature associated with new-onset T1D. However, NOD mice deficient in NLRP3, an inflammasome-associated NLR family member that regulates innate inflammation by activating both IL-1 and IL-18 (22), have limited trafficking of immune cells to the islets and are protected from T1D development (12, 23). Therefore, blocking IL-1 along with related inflammatory cytokines can block diabetes pathogenesis.
IL-1 is a key signal for induction of prostaglandin E2 (PGE2) synthesis, a critical signal for protection against bacterial infections such as Mycobacterium tuberculosis (24). PGE2 interacts with 4 G protein–coupled PGE2 receptors (EP1–4). PTGS2 (encoding prostaglandin synthase 2/COX2) or PGE2-PTGER4 interactions have been specifically shown to be directly linked with inflammatory responses and diabetes pathogenesis (25–27). Both human and mouse studies showed enhanced plasma PGE2 and monocyte-derived COX2 levels before and after the onset of T1D (28–30). However, PGE2 can also inhibit NLRP3 activation and reduce IL-1 production in certain conditions of homeostasis, and signals mediated by receptors for PGE2 can be antiinflammatory in some contexts (31, 32). Therefore, a better understanding of the cross-talk between these pathways in chronic autoimmunity is needed.
In the present study, we separated the IFN-I gene signature from other inflammatory gene signatures at the basal level (tonic signals) for different APC subsets to better understand the regulation (activation/inhibition) that occurs during the chronic inflammatory prediabetic stage in NOD mice. We found that multiple inflammatory molecules are elevated just prior to the onset of T1D, including PGE2, EP4 (also known as prostaglandin receptor 4 [PTGER4]), and IL-1β, but the IFN-I response is decreased, suggesting a failure in the balance of innate signal. In particular, it is the previously defined tonic-sensitive subset of IFN response genes that were most significantly decreased in NOD APCs. Importantly, treating prediabetic NOD mice with a PTGER4 antagonist restored IFNAR signaling; decreased IL-1, NLRP3, and IFN-γ; and, in fact, caused a significant (P < 0.01) reduction in infiltration of leukocytes into the islets. Therefore, we have identified a signal that can rebalance innate signals in chronic autoimmunity, leading to inhibition of autoreactive T cell differentiation and infiltration.
Results
DCs from adult prediabetic NOD mice have decreased expression of IFN-I–dependent tonic-sensitive genes that utilize canonical IFNAR signaling.
We previously observed decreased expression of IFN-I response genes after CpG-A stimulation in 8- to 10-week-old prediabetic NOD mice compared with age-matched diabetes-resistant B6.g7 mice (6). These B6.g7 mice share the NOD MHC haplotype but not the other disease-susceptible genetic regions. This unexpected finding led us to investigate whether the basal IFN-I–dependent responses are also lower in prediabetic (8-week-old) NOD mice. Consistent with the in vivo CpG-stimulated response, 8-week-old NOD mice had a significantly lower basal IFN-I response. Earlier studies had described higher IFN-I–signature gene (ISGs) in very young NOD mice (33), and similarly, we measured ISGs in 3-week-old NOD mice that were either similar to or higher than those of 3-week-old B6.g7 mice, showing a shift in the response as diabetes progresses in NOD mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97843DS1). These results are consistent with muted IFNAR signaling in the adult prediabetic NOD mice that could be the results of chronic inflammation associated with altered immune cell activation.
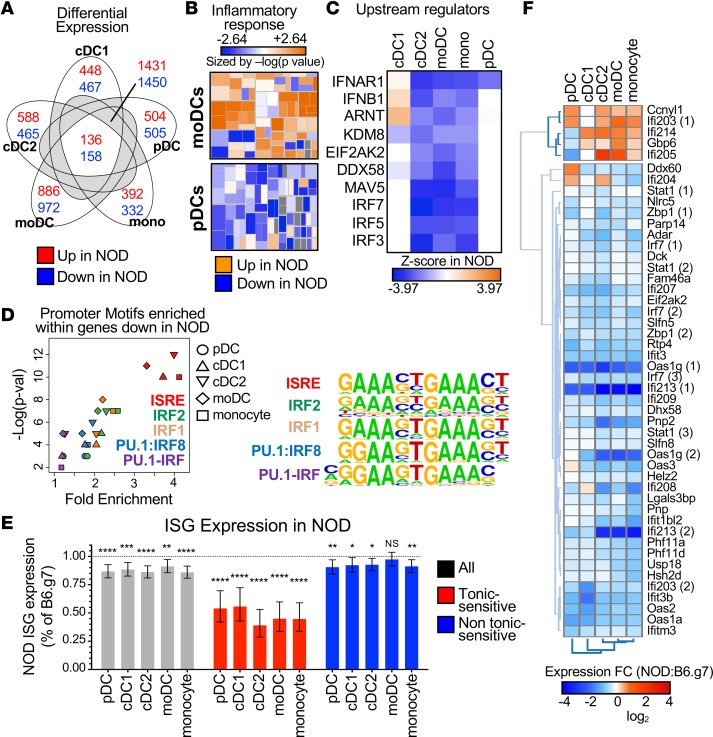
To better define innate alterations present in adult prediabetic NOD mice, we sorted a set of 5 key previously defined spleen APC populations from prediabetic NOD mice (8 weeks old) and age-matched diabetic-resistant B6.g7 mice (34). These 5 populations represent the 3 major bona fide DC subsets (one subset of conventional DCs [cDC1], a second subset of cDCs [cDC2], and plasmacytoid DC [pDC]) along with monocytes and MHCII monocyte-derived cells (moDCs) (35). The B6.g7 strain was used as the reference to define genes differentially expressed (DE) either up or down in prediabetic NOD mice for individual APC populations, using >1.5-fold induction and FDR <0.1 as filters (Supplemental Table 1). As shown in Figure 1A, the number of DE genes was higher in moDCs than other DC subsets, suggesting these cells may be a central contributor to the autoimmune process. Indeed, NOD mice have more moDCs that produce more TNF-α without exogenous stimulation (34), and Ingenuity pathway analysis (IPA) downstream effects analysis predicted an increase in inflammatory response functions for moDCs in prediabetic NOD mice compared with those in B6.g7 mice, which in contrast had decreased pDCs (Figure 1B). This upregulation of inflammatory pathways was also observed in other key APC subsets.
Figure 1. Tonic-sensitive IFN-I response genes are lower in 5 key APC subsets in adult prediabetic NOD mice.
(A) Venn diagram of upregulated (red) and downregulated (blue) DE genes in APC subsets. Numbers are shown for DE genes unique to one subset, shared by all subsets, or shared by at least 2 subsets but not all. (B) IPA heat map for inflammatory gene pathways for moDCs and pDCs in NOD mice compared with B6.g7 mice. Each box was sized by the log(P value) and colored by the Z score. (C) Hierarchical clustering of significant IPA upstream regulators with lower expression in NOD mice, as ranked by the Z score. Z score indicates the predicted activity of the upstream regulator in NOD mice compared with B6.g7 mice. (D) Fold enrichment and –log(P value) are plotted for promoters motifs enriched in transcripts expressed lower in NOD mice compared with B6.g7 mice. Motifs shown are enriched (P < 0.1) in at least 4 of the DC subsets. Motifs are color coded, and DC subsets are represented by shape. Aligned logos represent the enriched motifs. (E) NOD ISG expression as a percentage of B6.g7 ISG expression for all ISGs (gray), tonic-sensitive ISGs (red), and ISGs that are not tonic-sensitive (blue). (F) Heat map of average fold change (NOD-B6.g7) for tonic-sensitive ISGs that are significant in at least 1 subset. Experiments were performed twice with 3 mice/group for each time. All replicates were averaged for each gene to obtain the average log2 expression value. Paired t test was performed for each DC subset on the ISG groups by comparing B6.g7 with NOD mice. Data are plotted as mean with 95% confidence intervals. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
IPA upstream regulator analysis, which can be used to identify potential key upstream signals that may be driving the observed gene expression changes, identified lower signals mediated by IFNAR and related transcription factors in almost all NOD APC populations (Figure 1C). This analysis does not indicate that IFNAR itself has lower expression, and our prior data indicate that IFNAR protein expression levels in some of these APCs are not altered in NOD mice (6). cDC2, moDCs, and monocytes were equally affected, with lower IFNAR-mediated signaling, but the effect was less evident in pDCs and cDC1. Motif enrichment analysis also identified that the genes expressed at lower levels in NOD DCs were significantly enriched with binding motifs of interferon-regulatory factors (Figure 1D). Several signaling branches downstream of IFNAR have been described (33, 36), including the IFN-γ–activated sequence (GAS) elements that bind STAT1 homodimers and are shared with IFN-γ signaling and canonical IFN-I signaling that uses interferon-stimulated response elements (ISREs). ISRE are specific binding sites for the IFN-stimulated gene factor 3 (ISGF3) complex that controls both expression of IFN-I and ISGs. The ISRE motif was the most significantly enriched within the promoter regions of the downregulated DE genes in 4 APC subsets except pDCs, with highest P values and fold induction. Therefore, the key APC subsets in NOD mice expressed lower levels of ISG, implicating lower IFNAR signaling in 8- to 10-week-old NOD APCs.
The endogenous or tonic IFN-I signal that occurs in the absence of a specific pathogen–associated signal can increase the sensitivity of cell response to acute IFN-I stimulation. Chronic high tonic IFN-I, however, can lead to dysregulated IFN-I responses (37). To further elucidate the effect of altered IFNAR signaling in NOD APCs, we analyzed the subset of ISGs referred to as tonic sensitive, as defined by Mostafavi et al., that are characterized as the ISGs most sensitive to the baseline activity of the IFN-α receptor (defined as the subset of IFN-α–stimulated genes that are most affected by the absence of IFNAR); additionally, they appear to be highly dependent on the canonical ISGF3-mediated IFNAR signaling. We applied this tonic-sensitive gene list to identify differences in gene expression related to chronically elevated IFN-I exposure among key APC subsets in NOD mice. As shown in Figure 1, E and F, tonic-sensitive genes were significantly decreased (≈50%) in all NOD APC subsets compared with nondiabetic mice. In contrast, however, NOD APCs had similar or mildly decreased expression of nontonic-sensitive genes (Figure 1E), suggesting that diminution of this tonic-sensitive response is responsible for most of the impaired NOD ISG expression. Similar results were obtained when tonic-sensitive gene expression was analyzed from NOD Rag1–/– mice (Supplemental Figure 2, A and B). This shows that alteration in the IFNAR signaling is independent of the adaptive immune responses associated with diabetes and suggests that changes in innate signaling may drive the T cell–mediated pathogenesis. Together, these data define the decreased IFN-I response in NOD APCs as focused on the tonic-sensitive ISG subset and dependent on canonical ISRE signals.
Blockade of IFNAR signaling in adult prediabetic NOD mice increases IFN-γ–producing CD4+ T cells and accelerates diabetes development.
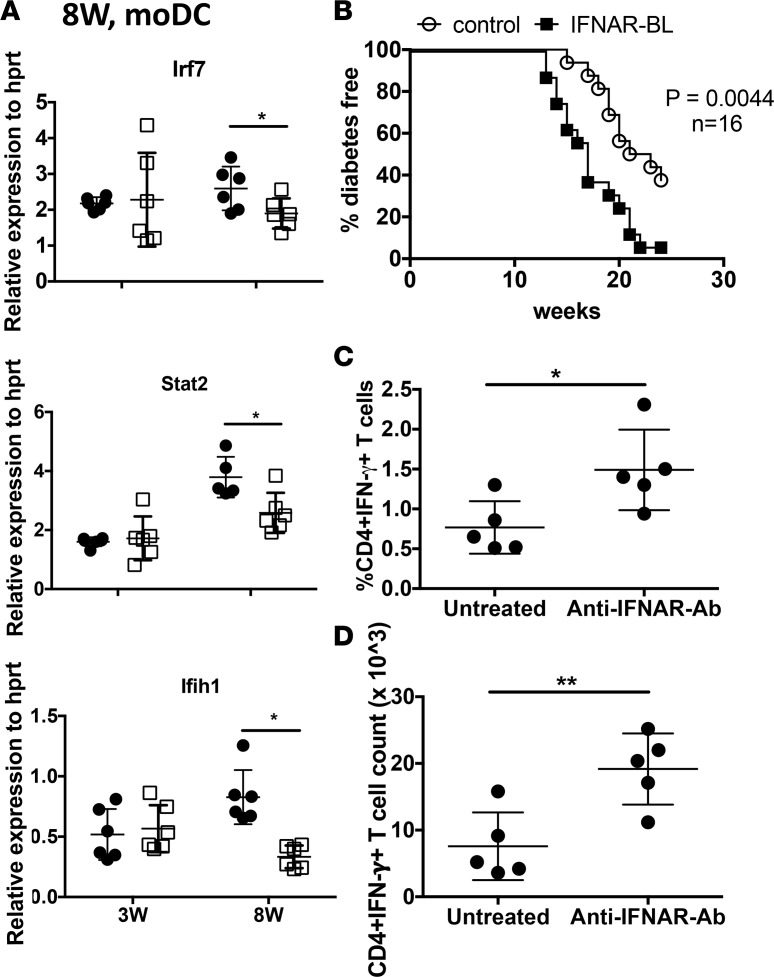
The observed diminution of basal IFN-I response in prediabetic NOD APCs was confirmed by sorting spleen moDCs from NOD and B6.g7 mice and measuring expression of ISGs. This decrease in Irf7, Stat2, and Ifih1 was observed in moDCs at 8-week-old but not 3-week-old mice (Figure 2A), consistent with our data from whole spleen cells (Supplemental Figure 1) and with other studies showing no decrease in ISGs at early time points (38). To understand how IFN-I signaling affects pathogenesis in an established autoimmune environment, we treated adult prediabetic NOD mice with anti-IFNAR1 antibody from 6–8 weeks of age. Age-matched control mice received isotype-matched control monoclonal antibody (GIR-208). Diabetes was significantly accelerated and the incidence was higher in NOD mice treated with anti-IFNAR1 (Figure 2B). Similarly, anti-IFNAR1 antibody–treated NOD mice showed a significantly higher percentage and number of CD4+IFN-γ+ T cells in the pancreatic lymph nodes (pLNs) after PMA and ionomycin stimulation (Figure 2, C and D, and Supplemental Figure 3A). Prior studies showed that IFNAR blocking during the initial break in tolerance (2–4 weeks of age) inhibits diabetes and shifts cytokine production toward IL-4 and IL-10 (5). Therefore, the effect of blocking IFNAR signaling in NOD mice is age dependent and any further decrease of IFNAR signaling in adult prediabetic NOD mice could be detrimental, as this could increase pathogenic T cell responses.
Figure 2. Lower IFN response genes correlate with diabetes pathogenesis, and blocking IFNAR in prediabetic NOD mice accelerates diabetes and increases Th1 responses.
(A) Type I IFN gene expression in sorted splenic moDCs from age-matched NOD and B6.g7 mice. (B) NOD mice were treated i.p. with anti-IFNAR1 antibody or control monoclonal antibody 2 times/week from 6–8 weeks of age. The percentage of diabetes-free mice was recorded and compared between the anti–IFNAR antibody–treated group and untreated control group. Statistical analysis was performed with log-rank test. Summation of 2 experiments with n = 16 mice (C) Intracellular IFN-γ staining in lymph node–derived CD4+ T cells, after PMA plus ionomycin (Ion) stimulation (total 6 hours), where brefeldin A was added for the last 4 hours. (D) Counts of intracellular CD4+IFN-γ+ T cells in lymph node after PMA plus Ion stimulation. Data were pooled from 2 independent experiments, with 5–6 mice per group (A, C, and D). Values show individual mice, with mean ± SD. P values in A, C and D were calculated using t test. *P < 0.05, **P < 0.01.
Inflammatory pathways associated with IL-1 are activated in prediabetic NOD APCs.
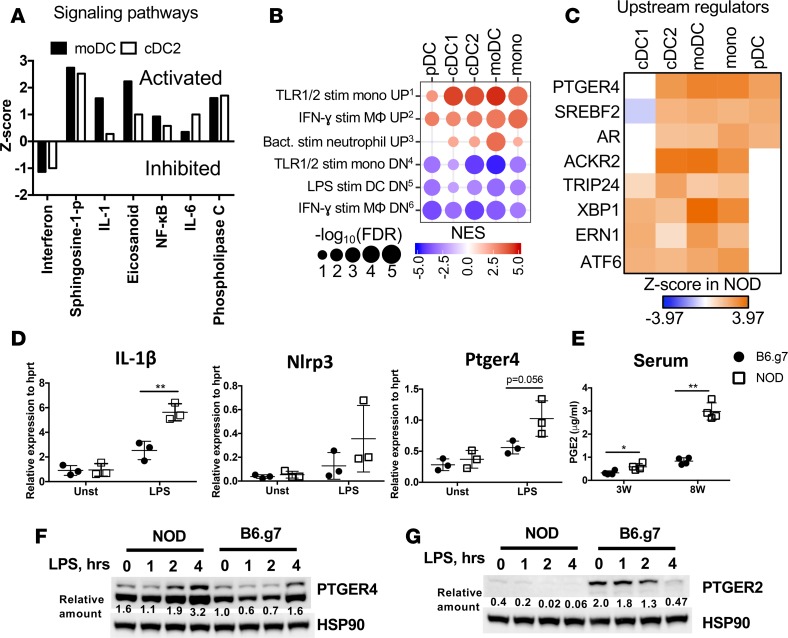
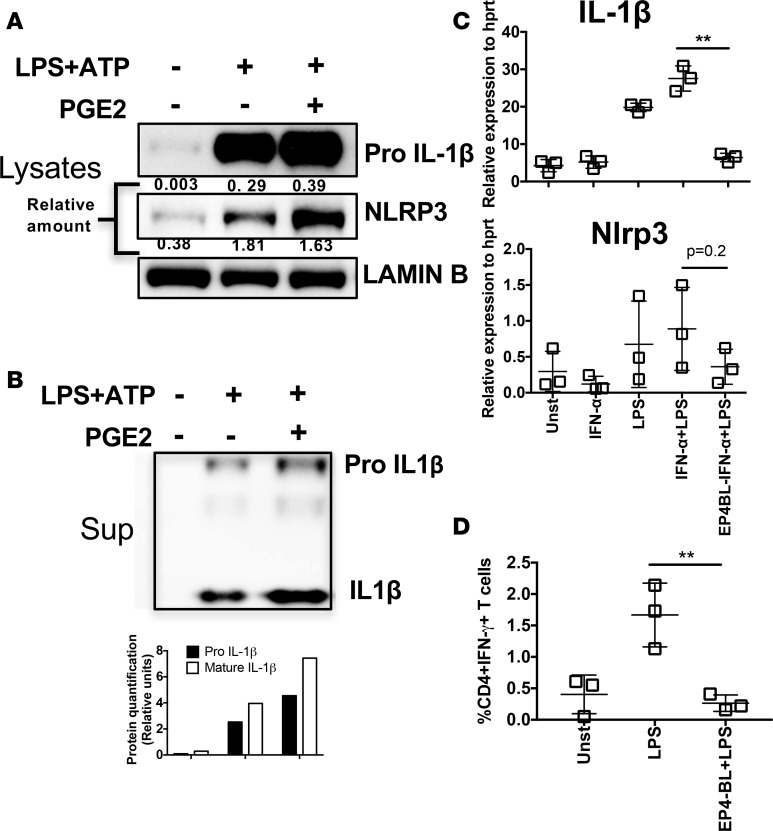
Next, genes with increased expression in NOD APCs were analyzed to identify activated pathways that might also have relevance to diabetes pathogenesis. Several inflammatory pathways were identified as highly activated in NOD cDCs and moDCs, including sphingosine, IL-1, IL-6, NF-κB, eicosanoids, and phospholipase C signaling (Figure 3A). Gene set enrichment analysis (GSEA) analysis for all 5 subsets of APCs showed significant enrichment of gene expression changes associated with inflammatory stimuli in NOD mice (Figure 3B). For example, genes upregulated in macrophages after IFN-γ stimulation were also higher in NOD DCs, and genes downregulated in DCs after LPS stimulation were lower in NOD DCs. PTGER4 was identified as the upstream regulator most likely to drive genes upregulated in NOD mice across all APCs populations (Figure 3C). LPS-stimulated expression of IL-1β, Nlrp3, and Ptger4 genes was measured and found to be higher in NOD spleen cells (Figure 3D). In fact, serum PGE2 levels showed a dramatic increase between week 3 and week 8, especially in NOD mice, which was consistent with cells secreting PGE2 (Figure 3E and Supplemental Figure 3B). Transcripts of Ptger4 were higher in 3-week-old NOD mice compared with B6.g7 mice (in sorted moDCs) but showed comparable levels at 8 week (Supplemental Figure 3, C and E). No difference in the level of Ptegr2 transcripts was observed between the strains (Supplemental Figure 3D). To determine protein expression levels of PTGER4, Western blot was performed on bone marrow–derived GM-CSF DCs, an in vitro model of moDCs (39, 40), which showed higher PTGER4 expression in NOD mice compared with B6.g7 mice (Figure 3F). Interestingly, EP2 protein, which also can bind PGE2, was much lower in DCs from NOD mice (Figure 3G). Similarly, spleen cells from NOD mice showed higher PTGER4 protein expression (Supplemental Figure 4A). These data suggest that increased PGE2-PTGER4 interactions in NOD may begin in early life and progress into chronic stimulation in adulthood, contributing to altered cytokine responses in NOD APCs that may contribute to pathologic inflammation.
Figure 3. Inflammatory signature identified in NOD APCs correlates with increased PTGER4 and PGE2.
(A) Z scores for IPA signaling pathways significantly altered in moDCs and cDC2 from NOD versus B6.g7 mice. (B) Bubble plots of key inflammatory gene sets from GSEA of NOD APCs compared with B6.g7 APCs using the immunologic signatures gene sets from MSigDB. Gene sets enriched with genes with lower expression in NOD mice compared with B6.g7 mice are depicted as blue circles, while gene sets with higher expression in NOD mice are depicted as red circles. Circle size indicates FDR-adjusted P values, and color intensity indicates normalized enrichment scores. (C) Hierarchical clustering of significant IPA upstream regulators with increased expression in NOD mice as ranked by Z score, which indicates the predicted enrichment in NOD mice compared with B6.g7 mice. (D) Gene expression measured by qPCR for IL-1β, Nlrp3, and Ptger4 after 4 hours of LPS stimulation of spleen cells from 8-week-old prediabetic NOD and B6.g7 mice. Plots are representative of 2 independent experiments with 3 mice per group. (E) PGE2 levels in sera from NOD and B6.g7 mice (representative of 2 experiments, 3 mice per group) at weeks 3 and 8. Comparison was made between NOD and B6.g7 mice; P values were calculated using t test. Data show mean ± SD. (F and G) Protein expression level by Western blot of PTGER4 and PTGER2 in bone marrow–derived DCs from NOD and B6.g7 mice, from 0–4 hours after LPS (500 ng/ml) stimulation. Relative quantification of bands normalization to HSP90 is indicated underneath. *P < 0.05, **P < 0.01.
NOD APC expression changes are enriched in human T1D IL-1–associated gene signatures.
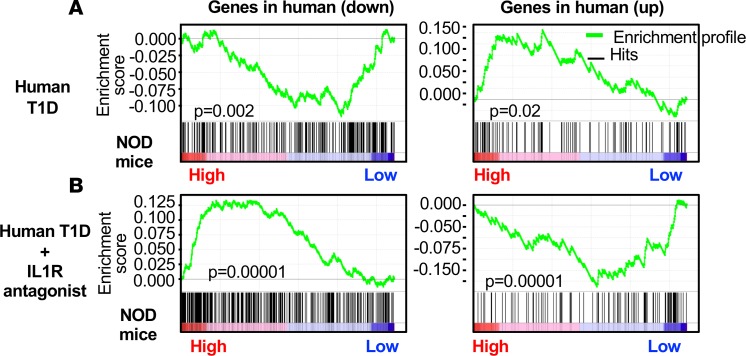
The gene and protein expression data in NOD APCs suggest that, despite early dominance of IFN-I responses, at later stages, IFNAR-dependent responses diminish and IL-1–associated inflammatory responses increase, which parallel observations in human T1D. To determine if these differentially expressed genes from prediabetic NOD APCs related to human T1D, we used GSEA to test if our DE gene list was also enriched in gene sets associated with T1D onset. We therefore applied GSEA to published microarray data sets from two studies (human trials) performed by Chen et al. (41) and Cabrera et al. (21) independently. The gene set published by Chen et al. was derived from responder PBMCs stimulated with sera from T1D patients or healthy controls (41). As shown in Figure 4A, GSEA showed that genes upregulated in human T1D at the time of onset were similarly upregulated in NOD moDCs. A second analysis was performed comparing a gene set derived from responder PBMCs cultured with sera from diabetic patients who were treated with IL-1R antagonist (21). Interestingly, DE genes from NOD moDCs showed a highly significant negative correlation with the genes that were upregulated or downregulated after IL-1R antagonism in humans (Figure 4B). Unlike IFN-I response genes, which have been well defined, the set of gene expression changes resulting from IL-1R signal are not as well characterized. Therefore, these data also add to our analysis by showing that IL-1R–dependent gene expression is higher in prediabetic NOD APCs, parallel to the analysis in Figure 1E, showing decreased IFNAR-dependent gene expression. This suggests that the inflammatory signature observed in NOD APCs is IL-1–dependent and matches human T1D pathogenesis. Interestingly, these sets of genes contained IFN-I–related genes, suggesting that the inhibitory effect of IL-1 on gene expression downstream of IFNAR is consistent between humans with T1D and prediabetic NOD mice.
Figure 4. Genes differentially expressed in NOD APCs correlate with IL-1–dependent gene signatures from new-onset human T1D.
GSEA for NOD DE genes with 2 human data sets. (A) Enrichment results from Chen et al. (41) showing genes DE lower (n = 282) or higher (n = 175) in patients with recent-onset type 1 diabetes positively correlate with DE genes from NOD APCs. (B) Enrichment results from Cabrera et al. (21) showing that genes expressed at lower (n = 368) or higher levels (n = 160) with IL-1R antagonist treatment of individuals with new-onset T1D negatively correlate with DE genes from NOD APCs. NES, normalized enrichment statistic. Nominal estimated P values determined using GSEA software.
PGE2-PTGER4–driven signals are inflammatory in NOD mice.
Identification of PTGER4 as an upstream regulator of NOD differential gene expression, and increased PTGER4 expression in NOD DCs, led us to investigate the effect of PTGER4-mediated signaling on NLRP3 and IL-1 production. PGE2 binds PTGER4 and can modulate LPS-stimulated NLRP3 or IL-1 levels (31). To understand the role of this signaling axis in NOD mice, we treated splenocytes with LPS for 4 hours, followed by stimulation with ATP to activate inflammasome (1 hour) and PGE2 (30 minutes), and measured the expression of IL-1 and NLRP3 protein by Western blotting. As shown in Figure 5, A and B, adding PGE2 to LPS and ATP enhanced IL-1 and NLRP3 levels in NOD mice. PGE2 decreased IL-1 and NLRP3 in B6.g7 mice, matching a published study using human macrophages from healthy donors (31) (Supplemental Figure 4B). Ex vivo treatment of cells with the PTGER4 antagonist, grapiprant, reduced IL-1 and Nlrp3 gene expression (enriched CD11C+ cells from spleen) (Figure 5C) and the percentage of IFN-γ+CD4+ T cells (spleen cells) (Figure 5D). Grapiprant can reduce inflammation and is an approved therapy for arthritis in dogs (42). Together, these results suggest that PGE2-PTGER4 signaling in NOD mice contributes to an elevated level of multiple inflammatory molecules. Next, we asked whether the PGE2 signaling could alter the IFNAR signaling pathway and change IFN-I gene expression. Cells pretreated with PGE2 and stimulated later with IFN-α showed no effect on IFN-I–induced STAT1/STAT2 levels in NOD mice (Supplemental Figure 4C). Interestingly, pretreatment with PGE2 decreased STAT1 but not STAT2 in B6.g7 mice. Therefore, under chronic autoimmune conditions, in vitro PGE2 treatment does not alter IFN-I–induced STAT1 or STAT2 molecules.
Figure 5. PGE2-PTGER4–driven signals are inflammatory in NOD mice.
(A and B) Spleen cells from 8-week-old NOD mice were stimulated with LPS-ATP. After 4 hours of LPS stimulation, where indicated PGE2 was added to the culture for 30 minutes, followed by ATP stimulation for 1 hour. Cells lysates (A) or supernatant (B) were analyzed by Western blotting for IL-1β and NLRP3. Protein bands were quantified relative to LAMIN-B (A) or total cell number (B). (C) CD11c+ cells were enriched and stimulated in vitro with IFN-α or LPS or both and PTGER4 (EP4) antagonist (EP4BL) as indicated. Transcripts of IL-1β and Nlrp3 were measured 2 hours after stimulation. (D) Total spleen cells were stimulated with or without LPS with or without PTGER4 antagonist. Intracellular IFN-γ was measured 6 hours after stimulation in CD4+ T cells. Data are representative of at least 2 (A and B) or 3 (C and D) independent experiments performed with pooled mice (n = 3), mean ± SD. Statistical analysis was performed with 1-way ANOVA with Bonferroni post-test, and only the relevant comparison was depicted (C and D). **P < 0.01.
In vivo blocking of PTGER4 increases IFNAR signaling and reduces Nlrp3, IL-1β, and IFN-γ levels.
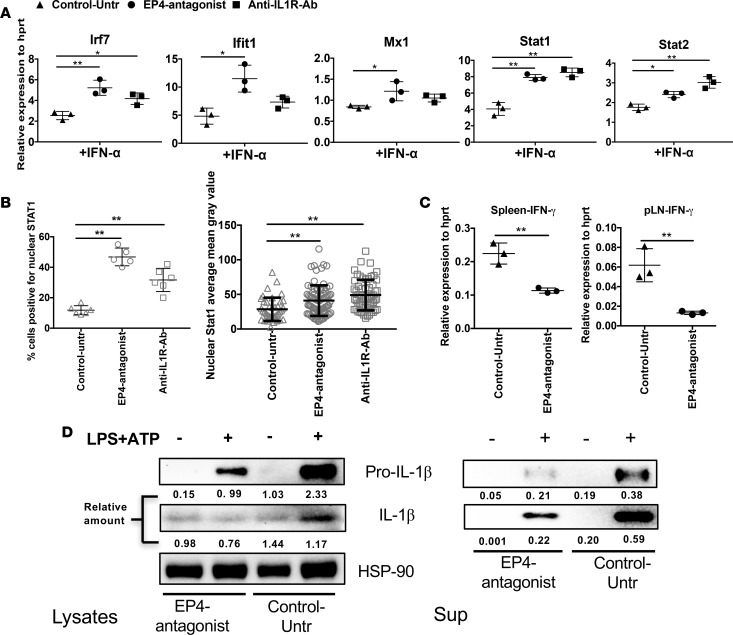
We next sought to elucidate the interplay among IFN-I, IL-1, inflammasome and PGE2 pathways in vivo. Thus, we treated NOD mice in vivo chronically (4–8 weeks) with grapiprant to block PTGER4-mediated inflammatory signals and to determine the effect on IFNAR signaling. We also treated age-matched NOD mice with anti–IL-1R antibody to block IL-1–mediated signaling. Inhibition of PTGER4 or IL-1R starting at 4 weeks significantly improved IFNAR signaling in late prediabetic 8- to 10-week-old NOD mice, as demonstrated by the increased expression of tonic-sensitive Irf7, Ifit1, Mx1, Stat1, and Stat2 in the spleen (Figure 6A). In addition, nuclear STAT1 levels were increased after in vitro stimulation with IFN-α in CD11C+ cells (Figure 6B). Interestingly, compared with blocking IL-1R, PTGER4 antagonist had a greater effect on IFNAR signaling. Next, the effect of PTGER4 antagonist treatment on IL-1 or other inflammatory molecules was tested. As shown in Figure 6C, in vivo PTGER4 blocking significantly inhibited IFN-γ transcript levels in lymph nodes, especially the pancreatic draining lymph node. In enriched CD11c+ DC lysates prepared from in vivo treated mice (with or without PTGER4 inhibitor), PTGER4 inhibition reduced IL-1 protein levels (Figure 6D). Together, these data support the conclusion that PTGER4 signaling contributes to balancing IL-1 and IFN-I signals and blocking PTGER4 corrects some of the innate imbalance we identified in NOD APCs.
Figure 6. In vivo PTGER4 antagonist treatment increases IFNAR signaling but decreases IL-1 and IFN-γ.
(A) Total splenocytes from NOD mice treated with PTGER4 antagonist or anti–IL-1R as indicated were stimulated with IFN-α for 2 hours, and IFN-I–response gene expression was determined by qPCR. (B) IFN-α–stimulated STAT1 nuclear localization. Spleen cells from untreated or treated (PTGER4 antagonist/anti-IL1R-antibody) NOD mice were stimulated with IFN-α for 30 minutes, and nuclear STAT1 localization was determined by confocal microscopy, as in Supplemental Figure 1B. Images (15/slide) were quantified to determine the percentage of CD11c+ cells positive for nuclear STAT1 and the intensity of STAT1 levels within CD11C+ population. (C) IFN-γ transcripts were measured in mixed cells prepared from spleen or pancreatic lymph nodes, 4 weeks after treatment with or without PTGER4 antagonist. (D) Four weeks after treatment with or without PTGER4 antagonist, CD11c+ cells were enriched from the spleen, and IL-1 protein levels were detected by Western blotting. Protein bands were quantified relative to HSP90 (lysate) or total cell number (supernatant). Data are representative of at least 2 independent experiments performed with 3 individual mice per group (A and C) or pooled from 3 mice (B and D), mean ± SD for each of the experiments (A–C). Statistical analysis was performed with 1-way ANOVA with Bonferroni post-test. *P < 0.05, **P < 0.01.
PTGER4 blocking increased the number of healthy islets and decreased autoreactive T cell proliferation in NOD mice.
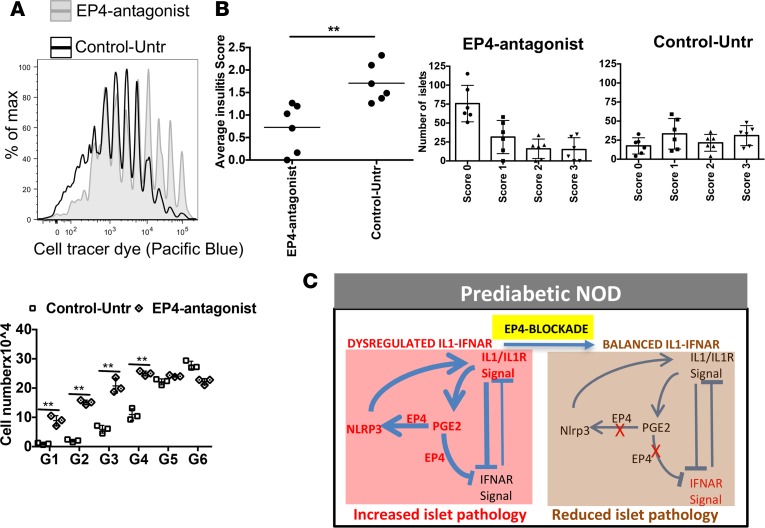
To determine if the PTGER4-mediated shift in innate inflammatory responses altered adaptive autoimmune responses, proliferation of islet-specific T cells in response to endogenous antigen was measured after in vivo blocking of PTGER4. BDC2.5-transgenic CD4 T cells labeled with cell tracer dye were injected into 8-week-old NOD mice treated with grapiprant from week 4–8 or untreated NOD controls. At day 6 after transfer, BDC2.5-transgenic CD4 T cells in the pLNs of grapiprant-treated mice showed significantly less proliferation compared with control mice (Figure 7A). Consistent with these data, PTGER4 inhibition in NOD mice also reduced islet immune infiltrate. Grapiprant treatment substantially reduced autoimmune pathology, as shown by both a statistically significant decrease (P < 0.01) in insulitis score and an increase in the numbers of healthy islets (“0” score) that are free of lymphocytes compared with the untreated mice (Figure 7B). These data suggest that PTGER4 inhibition improves autoimmune pathology by modulating the balance of inflammatory IL-1/IFNAR signaling (Figure 7C) and reducing autoreactive T cell activation and migration.
Figure 7. In vivo EP4 antagonist treatment decreases autoimmune pathology.
(A) NOD mice treated with grapiprant (EP4 antagonist) or untreated (control) for 4 weeks were then injected with cell tracer dye–labeled transgenic BDC2.5 CD4 T cells, and cell proliferation was checked 6 days after transfer. Cell division was compared between EP4 antagonist–treated and untreated control mice by counting cell numbers for each division. (B) Separate group of control or EP4-inhibited mice (did not received BDC2.5 CD4 T cells) were checked for islet lymphocyte infiltration (6 weeks after treatment). Pancreas tissue samples were sectioned for H&E staining, and islet infiltration was ranked as follows: no infiltration, 0; 10%–20%, 1; 20%–50%, 2; and >50%, 3. Data are presented with average score or number of islets with different levels of infiltration with lymphocytes. Data are representative of 2 independent experiments (A and B). Each experiment was performed with 3–4 mice per group. The graph shows mean ± SD for each of the experiments. Statistical analysis was performed with 1-way ANOVA with Bonferroni post-test. (C) Proposed schematic model of altered balance of IL-R/IL-1 and IFNAR signaling in adult prediabetic NOD mice with modulation by PGE2-EP4 and NLRP3, and reestablishment of homeostatic balance after inhibiting EP4 signals. Prediabetic NOD mice showed increased expression of IL1, PGE2, PTGER4, and NLRP3 (red). PGE2-EP4 were key regulators of enhancing NLRP3 and IL1 that was inhibitory to IFNAR signaling (bold lines). After inhibition of EP4, IFNAR signaling increased by reducing IL1.
Discussion
Most studies on the role of IFN-I in the context of autoimmune diabetes have focused on the early events, corresponding to the initial loss of self-tolerance, that occur before the chronic autoimmunity and pathogenic T cell response is established. In this study, we sorted 5 specific subsets of APCs taken from adult prediabetic NOD mice and age-matched diabetes-resistant B6.g7 mice and show downmodulation of IFN-I response. The tonic-sensitive subset of ISGs, which is highly sensitive to canonical IFNAR signaling, drives most of the observed IFN response of downregulation. This alteration of IFNAR signaling in NOD mice affects all 5 APC subsets analyzed, with expression of IFN-related tonic-sensitive genes at about 50% of the level found in controls. Tonic-sensitive gene expression was lower even in NOD Rag–/– mice compared with B6 Rag–/– mice, showing that this innate imbalance is independent of the adaptive autoimmune process and may contribute to the development of pathogenic T cell responses. A blocking IFNAR antibody given after chronic innate immune signals actually accelerates diabetes, which contrasts with prior studies showing that early IFNAR blocking protects from diabetes and suggests a shift in the balance of innate signals.
In addition, at this age, NOD PGE2-driven IL-1–dominant proinflammatory signals increase. PGE2 primarily binds EP2 and PTGER4 on immune cells, and NOD APCs have higher PTGER4 and lower EP2 protein expression compared with B6.g7 mice. Most strikingly, when we block PGE2-PTGER4 signals, multiple cytokines pathways were affected, with decreased IL-1 and IFN-γ and increased IFNAR signaling. Importantly, in vivo inhibition of PGE2-PTGER4 during chronic autoimmunity corrected the impaired IFN-I signals and inhibited pathogenic T cell IFN-γ production and islet infiltration. At this age, blocking IFNAR signaling exacerbates Th1 responses, increasing IFN-γ–secreting T cells, suggesting that IFN-I may limit disease in already established organ-specific autoimmunity. Analysis of human T1D data sets showed parallel innate changes. Together, this suggests that innate changes of T1D may be more similar to those of multiple sclerosis, in which IFN-β is an efficacious treatment (43), and not lupus, in which IFN-I is likely a pathogenic signal even in established disease (44).
IFN-I is critical for the initiation of autoimmunity, as shown by inhibition of diabetes development when NOD mice are treated with anti-IFNAR from 2–4 weeks (5). Early blockade of IFNAR, at a time when the immune cell compartments are developing and chronic stimulation has not yet occurred, increases the number of immature DCs and production of the immunoregulatory cytokines IL-10 and IL-4 (5, 45, 46). IFN-I can regulate T cell function and promotes T cell–mediated IL-10 production (47, 48) but can also limit IL-10 production by activated monocytes under chronic stimulation (49). Thus, the outcomes of IFN-I production or action may change depending on the type of immune cells and its developmental and activation state. Transient increases of IFN-I signatures have been observed early in both humans (at time of autoantibody seroconversion) and mice (at first appearance of autoreactive T cells in the pancreas); these signals can decrease later, as observed in human T1D at the onset of diabetes (7, 8, 38). In this context, our recent study with in vivo CpG-A stimulation in adult prediabetic NOD mice showed that CpG-induced IFN-I responses are muted (lower expression of CD86 and CD80), even as IFN-I levels are increased (6). This likely indicates the appearance of negative regulatory pathways affecting downstream IFN gene expression as a result of chronic IFN-I signaling. Therefore, failure to induce an optimal response to IFN-I could contribute to pathogenesis at this later stage. In fact, once the autoimmune response is established, IFN-I treatment can suppress autoimmune pathology (50, 51). Similarly, we measured more IFN-γ–producing T cells when IFNAR signaling was blocked from 6–8 weeks of age. Our results, together with the previous observations, highlight the importance of IFN-I in reducing autoimmune pathology in the prediabetic NOD mice.
Chronic inflammation is present in NOD mice, can begin as early as 3 weeks of age, and could alter IFN-I responses over time. In other contexts, such as chronic viral infection, the continued presence of elevated IFN-I signal can lead to downmodulation of immune responses (52). The Mathis-Benoist lab recently defined a set of tonic-sensitive ISGs, showing different clusters of ISGs in relation to cell-type specificity, kinetics, and sensitivity to tonic IFN signals (37). IFN-I is produced constitutively at very low levels, even without any external stimulation, and the response to this tonic IFN is critical for normal IFN-I responses (53). In adult prediabetic NOD mice, tonic signal-dependent ISG expression was greatly reduced in all major APC subsets, perhaps as a result of elevated chronic IFN-I levels. When we modeled low-dose chronic IFN treatment in B6.g7 mice by implanting osmotic pumps that provide continuous delivery, a small decrease in IFN response was observed (data not shown). The expression of tonic-sensitive ISGs is impaired less in pDCs compared with other APCs, and pDCs also are not as enriched in proinflammatory pathways as other NOD APCs. This lower inflammatory tone in NOD pDCs could make these cells a good target for antigen-specific tolerance approaches. Although this requires further study, one possibility is that pDCs are more refractory to chronic IFN signals and are one source of the sustained increase in IFN. In contrast, these circuits are downmodulated in other cell types, including APCs that do not produce IFN-I. Because even unfractionated spleen cells displayed impaired IFN responses, this innate alteration may also occur in lymphocytes and may result in downmodulation of regulatory pathways such as IL-10 production.
IFN-I signals can be kept in check by other types of inflammatory signals, so we also focused on pathways that were significantly increased by prediabetic conditions and likely to counterregulate IFN-I, including IL-1 (54). Although IL-1 is associated with T1D pathogenicity (16), blocking IL-1 signal alone does not prevent T1D progression (20). Yet, IL-1 can be an important component of inflammatory immune responses flaring out of control. Recently, investigation on IL-1–IFN-I cross-talk during mycobacterial infection revealed that IL-1 inhibits IFN-I production through eicosanoids (24). IL-1 and IFN-I counterregulate each other, and both signals can be important for immune activation (24, 54–56). Therefore, in some chronic autoimmune environments, less IL-1 but more IFN-I may limit immune pathology. This concept may be relevant to autoimmune diabetes as prediabetic NOD showed significant enrichment with human IL-1–dependent gene signature, and the enrichment was reversed with the blockade of IL-1R. Therefore, our data unveil a complex cytokine interplay in prediabetic NOD mice, in which PGE2 enhances IL-1 production, perhaps via NLRP3 inflammasome activation (12). In contrast, the effect of PGE2 in diabetes-resistant B6.g7 mice is rather inhibitory to NLRP3 inflammasome and IL-1 production. Therefore, PGE2-PTGER4 signaling in NOD mice contributes to increased inflammatory responses in NOD mice.
In vivo blocking of IL-1R by treating NOD mice with anti–IL-1R specific antibody enhances IFNAR signaling. Thus, crosstalk between IL-1 and IFN-I signals occurs in both infectious disease and autoimmune settings but may be regulated differently by PGE2-related signaling. Patients with T1D have increased plasma and urinary PGE2 levels (29, 57), and the PGE2-PTGER4 interaction is directly linked with inflammation (25). Here, we measured increased NOD PGE2 levels and found enhanced levels of PTGER4 expression in NOD APCs, together with other inflammatory molecules, including IL-1, suggesting the establishment of PGE2 and IL-1 as dominant innate signals in prediabetic NOD mice. PGE2-PTGER4 can have divergent effects on cellular response to cytokine production due to the activation of cAMP (58). cAMP acts on multiple cytokine and chemokine signaling pathways, including activation of IFN-γ and Th1 differentiation (59). In T1D, IFN-γR signaling increases diabetogenic CD8+ T cells that selectively destroy β cells (60). Importantly, PTGER4 inhibition in NOD mice resulted in a reduction of the inflammatory burden mediated by IL-1 and IFN-γ but an improvement in IFN-I response and related regulatory pathways. In conclusion, we have shown that the prediabetic stage is associated with diminished tonic-sensitive IFN-I responses that are counterregulated by increased prostaglandin and IL-1 innate mediators. Although blockade of IL-1 signaling alone could enhance IFN-I response, this did not alter autoimmune pathology. Interestingly, PTGER4 inhibition corrects the innate immune dysregulation and results in inhibition of autoreactive T cell proliferation and preserves β cells in the islets. Therefore, modulation of the PGE2/PTGER4 axis may be useful for correcting an imbalance between IFN-I and IL-1 innate signals and for treatment of T1D.
Methods
Mice.
NOD, B6.H2g7 (B6.g7), and RAG1–/– mice in both B6 and NOD backgrounds were purchased from Jackson Laboratories and bred in the NIDDK animal facility. Animals were housed in specific pathogen–free conditions and handled according to the guidelines provided by the Animal Care and Use Committee of NIDDK at the NIH. The B6.g7 congenic strain is on the B6 background but carries the NOD-derived H2g7 haplotype. Age-matched female 8- to 10-week-old mice, with 3–4 mice/group, were used in all experiments except where indicated.
In vivo treatment.
NOD mice at 4 weeks of age were treated with grapiprant (MedChem Express), a member of the piprant class of compounds that is a highly selective antagonist of PTGER4, orally every day until mice were 8 weeks old. Grapiprant was formulated in PBS + 30% DMSO, and mice were fed 10 mg/kg body weight. Anti–IL-1R specific antibody (clone JAMA-147, BioXcell) was injected at 50 μg/kg body weight i.p. daily for 4 weeks in a separate group of NOD mice. Anti-IFNAR–specific antibody (clone MAR1-5A3, BioXcell) or isotype-matched control monoclonal antibody (GIR-208, BioLegend) was injected twice in a week (200 μg/mouse, i.p.) for 2 weeks (week 6–8) in some groups of mice.
Isolation of APCs from lymphoid organs.
Spleens, pLNs, and skin lymph nodes were harvested separately as described by Dong et al. (34). DCs or other APCs were enriched in the mixed cell population from individual spleens and lymph nodes with Collagenase III digestion (Worthington) as described by Guerrero et al. (61). Cells were then strained using a 60-μm filter to obtain a single-cell suspension. Single-cell suspensions were then stained with appropriate antibodies and identified by flow cytometry for cell sorting or analysis of expression of molecules.
Flow cytometry.
Isolated cells were blocked with anti-CD16/32 (Fcγ R III/II, BioLegend) and stained with appropriate antibodies against surface proteins in PBS with 2% FCS as previously described (61).
In some experiments, intracellular proteins were stained after fixation and permeabilization using fixation and permeabilization buffers from eBioscience. For intracellular IFN-γ staining, cells were incubated with brefeldin A (1×, Biolegend) for the last 4 hours of PMA and ionomycin stimulation. Cells were fixed and permeabilized and stained with appropriate antibodies.
The following antibodies were used: anti-CD3 (145-2C11), anti-CD11b (M1/70), anti-CD11c (N418), anti-CD49b (DX5), anti-Ly6C (HK1.4), anti-Ly6G (1A8), anti-Siglec-H (clone 551), anti-CD8, anti-IFN-γ (XMG1.2), and anti-IFNAR (MAR1-5A3) from BioLegend. Brilliant Violet 510–conjugated streptavidin (BioLegend) and Pacific Orange–conjugated streptavidin (Life Technologies) were used to detect biotinylated antibodies, and Aqua Dead Cell Stain (Life Technologies) was used to gate on viable cells. Fluorescence minus one controls include all fluorophores except the color being analyzed. Samples were collected on a BD LSRII flow cytometer with 3 lasers, and analyzed using FlowJo software 9.8.2 (Treestar). Sorting was performed in NHLBI flow core facility at NIH. Flow cytometry gating for 5 different APC subsets, CD8a+, DCIR2+, IAg7, Ly6C, and pDC, was followed as previously described (34). Briefly, spleen lymphocytes were gated first for live and lineage– populations (negative for CD3, CD19, Ly6G, NKP46). From the lineage– population, pDC was gated by using SiglecH+ (specific for pDC). SiglecH– cells were plotted for CD11b and CD11c to separate CD11bhiCD11clo and CD11ChiCD11blo subsets. From this step onward, cells were further separated by following gating: (a) CD11bhiCD11clo cells were separated by using MHCII and Ly6C for sorting MHCII+ (moDCs) and Ly6c+ (monocytes) subsets; (b) CD11ChiCD11blo cells were gated on MHCIIhi (almost all cells are high) and then separated by using DCIR2 and CD8α for sorting DCIR2+ (cDC2) and CD8α+ (cDC1) subsets.
Cells were sorted on a BD FACSAria II cell sorter and immediately lysed with PicoPure RNA extraction buffer. Cell lysates were kept frozen at –80°C. Total RNA was extracted using the Arcturus PicoPure RNA isolation kit (Thermo Fisher Scientific) per the manufacturer’s protocol.
Low-input RNA sequencing libraries were generated from about 300 pg total RNA using the SMART-seq v4 Ultra Low Input RNA kit (Clontech). All samples were subjected to 14 PCR amplification cycles to minimize PCR biases (62). Amplified cDNA libraries were later fragmented through sonication to obtain 200- to 500-bp fragments. Standard Illumina sequencing libraries were prepared using the Rubicon ThruPLEX DNA-seq kit (Rubicon Genomics) according to manufacturer’s protocol. Sample barcoded libraries were sequenced on Illumina’s NextSeq 500 instrumentation using NextSeq 300 Cycle Kit, High Output, V1 reagents (Illumina), and data analysis workflow (bcl2fastq v2.17.1.14) to obtain 150-bp paired-end reads.
Data analysis.
Paired-end RNA sequencing reads were trimmed to remove Illumina’s adapter sequences and were further processed to remove poor quality reads and/or reads mapping to mouse rRNA and tRNA sequences using ArrayStudio package (http://www.omicsoft.com/array-studio). The following criteria was used to remove poor quality reads. First, reads with base quality score (Sanger quality score) <10 were trimmed. Then, reads were filtered out if either trim length is <25 bp, maximal base quality score is <15, average quality score is <10, or poly AGCT rate is ≥ 80%. The pair was filtered out if either read failed the filtering criteria. The sequencing data have been deposited in the NCBI Gene Expression Omnibus under accession GSE104644.
Sequencing reads were aligned to the mouse reference genome (build38) using OSA version 4 (63). To obtain transcript counts data, RSEM package (64) and NCBI mouse RefSeq gene model (released July 2015) annotations were used. Transcripts with 0 counts in more than two-thirds of the samples were discarded from downstream analysis to reduce noise in the expression data. Filtered counts data were later normalized using quantile normalization and differentially expressed transcripts were identified using Limma Voom (65). Batch effects were removed using ComBat (from SVA R package) after conversion to CPM within the Voom function but before calculation of precision weights. Combat was used with defaults, no covariates, and for each subset separately. DE genes were determined using Limma’s eBayes and topTable functions, utilizing the precision weights calculated from Voom normalization. A P value cutoff of 0.05 was used to classify transcripts as differentially expressed in treatment condition. Upstream regulator and pathway analyses on differentially expressed transcripts were performed using QIAGEN’s IPA (https://www.qiagenbioinformatics.com/).
IPA.
IPA analysis was done with the DE genes and sorted the enriched pathways for all the APCs based on P values and Z score. Fold change values were used for IPA analysis for the identification of direction of most enriched pathways, either activated or inhibited. Z score cut off >1.5 and P >0.05 were set as criteria to define which pathways were linked with the prediabetic conditions of NOD mice. Pathway analysis settings, including 140 molecules per network and 25 networks per analysis, a causal network that is related to IFN and inflammation, a mammal species molecular network with stringent filter selected, and DC-like cell types for tissue type, were selected. Multiple and duplicate transcript isoforms for a single gene were resolved by using the isoform with the maximum fold change. Rag–/– RNAseq data analyzed by IPA also used these setting, but with a fold change cutoff of ±0.5.
Promoter motif enrichment analysis.
Differentially expressed transcripts were screened for enrichment of known promoter motifs using Homer v.4.8. Results were obtained using the “findMotifs.pl” function with default settings to include a P value threshold of 0.1 and promoter regions defined as –300 to +50 bp from the TTS. Homer Motif Database, containing over 330 motifs, was the only collection of motifs used for the known enrichment analysis (66). Results reported are all “known motif enrichment” results. Fold enrichment in the motif figure (Figure 1D) depicts the percentage of promoters of transcripts with lower expression in NOD mice than B6.g7 mice that have at least 1 occurrence of the motif divided by percentage of promoters from the remainder of the genome with at least 1 occurrence of the motif.
Motifs enriched in the promoters were taken from the list of genes with expression that was significantly lower in NOD mice than in B6.g7 mice. Motifs that were presented passed the P value cutoff of 0.05 in at least 4 of the DC subsets.
Gene set enrichment analysis.
Gene set enrichment analysis was performed using the Broad Institute’s GSEA software (version 3.0 Beta 2) in combination with a subset of gene sets from the Immunologic Signatures collection of the Molecular Signatures Database (MSigDB database v5.2). The collection was filtered to exclude gene sets that compare one cell type to another cell type and any T cell gene set. Only gene sets comparing different conditions between the same cell type were used. The tonic-sensitive gene set from Mostafavi et al. (37) was added to the collection for the purposes of comparing the enrichment to other gene sets. The GSEA-preranked procedure was used for the analysis using the “classic” enrichment statistic (67, 68). All other parameters were set to default. RefSeq IDs converted to current official MGI gene names and human ortholog HUGO gene names were obtained from MGI website. Genes with the same HUGO gene name (isoforms of same gene) were collapsed/resolved by using the one with the largest fold change.
Prerank metric.
Results from our RNAseq experiment were ranked so that genes with adjusted P values below 0.1 were on the peripheries of the ranked list, while everything in between was sorted based on adjusted P value. The genes on the peripheries of the ranked list were sorted by fold change, with the largest fold change magnitudes furthest from the center of the rank. Two gene sets were generated from data reported by Cabrera et al. (21). The clinical trial expression profiling data were generated after treating recent-onset T1D patients in vivo with either IL-1R antagonist treatment or placebo. Sera collected from these patients was then used to stimulate responder PBMCs. Expression profiling of the PBMCs was then performed. The significant DE microarray probes from IL-1R antagonist treatments (AIDA clinical trial) were separated into probe sets with either increased or decreased detection, as reported by Cabrera et al. (21). Two gene sets were generated from data reported by Chen et al. (41). Their expression profiling data, similar to that of Cabrere et al., was generated after using sera from recent-onset T1D patients and unrelated healthy controls to stimulate healthy responder PBMCs.
For each probe set from data reported by Cabrera et al. (21) and Chen et al. (41), only probes with unambiguous gene annotations (probes mapping only to 1 gene) were used. After removing duplicate HUGO gene names, 4 gene sets resulted: (a) genes with increased expression in recent-onset T1D compared with unrelated healthy controls, (b) genes with decreased expression in recent-onset T1D compared with unrelated healthy controls, (c) genes with increased expression upon IL-1R antagonist treatment, and (d) genes with decreased expression upon IL-1R antagonist treatment. GSEA was performed with these 4 gene sets on our preranked RNAseq data as described in GSEA methods section.
PGE2 ELISA.
PGE2 quantification was based on the forward sequential competitive binding technique in which PGE2 present in a sample competes with HRP-labeled PGE2 for a limited number of binding sites on a mouse monoclonal antibody as described by the manufacturer (R&D Systems). Serum and culture supernatants were diluted 1:5,000 or 1:5, respectively, and added to the wells. Following washings, substrate solution was added and the intensity of the color was recorded at 450 nm. The color was inversely proportional to the concentration of PGE2 in the sample.
Western blot analysis.
Where indicated, total spleen cells or CD11C+ cells (5 × 106) were stimulated in serum-free media with LPS (1 μg/ml) for 4 hours, followed by 30 minutes of PGE2 (1 μM) and, finally, an 1 hour of ATP (31). Where indicated, spleen cells were stimulated with IFN-α (50 U/ml). Protein samples were fractionated on a NuPAGE 4–12% or 12% Bis-Tris gel (Invitrogen) and blotted onto a nitrocellulose membrane (Invitrogen). A panel of different primary antibodies against PTGER2 (LifeSpan Biosciences Inc.); PTGER4 (Novus Biologicals); and STAT1, STAT2, IL-1β, NLRP3, Caspase-1, Lamin B, and HSP90 from Cell Signaling were used in the experiments with species-matched HRP-labeled secondary antibody. Western blots were developed using an enhanced chemiluminescence system according to the manufacturer’s instructions (Amersham Biosciences). The results were visualized by the ChemiDoc Touch Gel and Western Blot Imaging System (Bio-Rad). Data were subjected to densitometric analysis using Image Lab software included with the ChemiDoc.
Confocal microscopy and quantification of nuclear localization of STAT proteins.
Formaldehyde-fixed (4%) (Electron Microscopy Sciences) splenocytes (with or without IFN-α prestimulation for 30 minutes) were air dried on the superfrost microscopic glass slides and blocked with 4% normal goat serum before over night incubation with primary antibodies: rabbit monoclonal STAT1 (D1K9Y, Cell Signaling), rabbit polyclonal STAT2 (Cell Signaling), and hamster CD11c (Biolegend). Species-matched secondary antibodies (conjugated with flurochrome dye Alexa Fluor 594 or Alexa Fluor 488) were added for 2 hours. The slides were washed, air dried, and mounted directly by VECTASHIELD antifade mounting medium with DAPI (Vector Laboratories). Tissue sections treated without the primary antibodies were used as controls.
Images were taken with a Zeiss confocal microscope (Nikon Instruments), and 10–15 fields per condition were analyzed with ImageJ (NIH). An algorithm was developed and used in ImageJ to quantify nuclear STAT1 intensity in cells that were positive for STAT1 and CD11c. CD11c+ cells were defined as cells with a ring of CD11c signal, and nuclear area was defined by DAPI signal. A total of 100–150 CD11c+ cells was analyzed to quantify the nuclear localization of STAT1.
Quantitative gene expression analysis.
For qPCR, total splenocytes or CD11c+ splenocytes were bead-enriched by negative selection using biotinylated antibodies specific for CD3, CD19, NKp46, and Ly6c, followed by anti-biotin magnetic beads according to the manufacturer’s instructions (Miltenyi Biotec). RNA was then extracted immediately using the Qiagen mini prep (Molecular Research Center). RNA was reverse-transcribed into cDNA (Life Technologies), and the cDNA was used as a template for PCR. A quantitative PCR assay was performed using The QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) for Irf7, Stat1, Stat2, Ifih1, Ifit1, Mx1, Il1b, Ptger4, Ifng, and Nlrp3 genes using TaqMan primers/probes from Life Technologies (Irf7, catalog 4331182, Mm00516793_g1; Stat1, catalog 4331182, Mm01257286_m1; Stat2, catalog 4331182, Mm00490880_m1; Ifih1, catalog 4331182, Mm00459183_m1; Ifit1, catalog 4331182, Mm00515153_m1; Mx1, catalog 4331182, Mm00487796_m1; Il1b, catalog 4331182, Mm00434228_m1; Ptger4, catalog 4331182, Mm00436053_m1; Ifng, catalog 4331182, Mm01168134_m1; Nlrp3, catalog 4331182, Mm00840904_m1). Relative gene expression was normalized to the house keeping gene Hprt.
Statistics.
Homoscedastic 2-tailed t tests were performed for most analyses when comparing two independent groups. Where indicated, 1-way ANOVA with Bonferroni post-test was used for multiple group comparisons. P values of less than 0.05 were considered as significant.
Study approval.
All animal procedures were approved by the NIDDK Animal Care and Use Committee under the animal protocol ASP K024-DEOB-13.
Author contributions
MJR and KVT conceived and designed the experiments, interpreted results, and wrote the paper. JAQ, YL, YZ, CHI, AWLK, and TWT performed experiments. KBR analyzed RNAseq data and produced results, VB processed and normalized RNAseq data, and HYS helped analyzing the data. AMWM interpreted results and edited the manuscript text.
Supplementary Material
Acknowledgments
We would like to acknowledge Alice Franks (Diabetes, Endocrinology, and Obesity Branch, NIDDK) for help with mouse husbandry and the NIDDK/NHLBI flow core and Phil McCoy for flow cytometry support. We thank the NHLBI bioinformatics core facility, including Xujing Wang, for gene array support and RNAseq data analysis. We also thank Mark Morris and Merle Elloso from Janssen Pharmaceutical Companies of Johnson & Johnson for scientific discussion and input. We are grateful to Katrin D. Mayer-Barber editing and critical reading of the manuscript and Giorgio Trinchieri and John J. O’Shea for feedback and scientific discussion. This work was supported by the intramural research programs of the NIDDK/NIH and Janssen Research and Development by a collaborative research agreement (CRADA, DK-12-0237).
Version 1. 02/08/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(3):e97843. https://doi.org/10.1172/jci.insight.97843.
Contributor Information
Kameron B. Rodrigues, Email: kameronr@email.arizona.edu.
Juan A. Quiel, Email: juan.quiel@nih.gov.
Yi Liu, Email: yi.liu2@nih.gov.
Vipul Bhargava, Email: vbharga@ITS.JNJ.com.
Yongge Zhao, Email: Yongge.Zhao@nih.gov.
Han-Yu Shih, Email: han-yu.shih@nih.gov.
References
- 1.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32(4):457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol. 2008;180(3):1929–1937. doi: 10.4049/jimmunol.180.3.1929. [DOI] [PubMed] [Google Scholar]
- 3.Delong T, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351(6274):711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulek AM, et al. Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes. Nat Immunol. 2012;13(3):283–289. doi: 10.1038/ni.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Xu B, Michie SA, Rubins KH, Schreriber RD, McDevitt HO. Interferon-alpha initiates type 1 diabetes in nonobese diabetic mice. Proc Natl Acad Sci USA. 2008;105(34):12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman MJ, et al. Despite increased type 1 IFN, autoimmune nonobese diabetic mice display impaired dendritic cell response to CpG and decreased nuclear localization of IFN-activated STAT1. J Immunol. 2016;196(5):2031–2040. doi: 10.4049/jimmunol.1501239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira RC, et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63(7):2538–2550. doi: 10.2337/db13-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panarina M, Kisand K, Alnek K, Heilman K, Peet A, Uibo R. Interferon and interferon-inducible gene activation in patients with type 1 diabetes. Scand J Immunol. 2014;80(4):283–292. doi: 10.1111/sji.12204. [DOI] [PubMed] [Google Scholar]
- 9.Reder AT, Feng X. How type I interferons work in multiple sclerosis and other diseases: some unexpected mechanisms. J Interferon Cytokine Res. 2014;34(8):589–599. doi: 10.1089/jir.2013.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabrera SM, Henschel AM, Hessner MJ. Innate inflammation in type 1 diabetes. Transl Res. 2016;167(1):214–227. doi: 10.1016/j.trsl.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padgett LE, Broniowska KA, Hansen PA, Corbett JA, Tse HM. The role of reactive oxygen species and proinflammatory cytokines in type 1 diabetes pathogenesis. Ann N Y Acad Sci. 2013;1281:16–35. doi: 10.1111/j.1749-6632.2012.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu C, et al. NLRP3 deficiency protects from type 1 diabetes through the regulation of chemotaxis into the pancreatic islets. Proc Natl Acad Sci USA. 2015;112(36):11318–11323. doi: 10.1073/pnas.1513509112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dogan Y, Akarsu S, Ustundag B, Yilmaz E, Gurgoze MK. Serum IL-1beta, IL-2, and IL-6 in insulin-dependent diabetic children. Mediators Inflamm. 2006;2006(1):59206. doi: 10.1155/MI/2006/59206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain MJ, et al. Elevated serum levels of macrophage-derived cytokines precede and accompany the onset of IDDM. Diabetologia. 1996;39(1):60–69. doi: 10.1007/BF00400414. [DOI] [PubMed] [Google Scholar]
- 15.Cabrera SM, Chen YG, Hagopian WA, Hessner MJ. Blood-based signatures in type 1 diabetes. Diabetologia. 2016;59(3):414–425. doi: 10.1007/s00125-015-3843-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferris ST, et al. The islet-resident macrophage is in an inflammatory state and senses microbial products in blood. J Exp Med. 2017;214(8):2369–2385. doi: 10.1084/jem.20170074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11(8):633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradshaw EM, et al. Monocytes from patients with type 1 diabetes spontaneously secrete proinflammatory cytokines inducing Th17 cells. J Immunol. 2009;183(7):4432–4439. doi: 10.4049/jimmunol.0900576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen CM, et al. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007;356(15):1517–1526. doi: 10.1056/NEJMoa065213. [DOI] [PubMed] [Google Scholar]
- 20.Moran A, et al. Interleukin-1 antagonism in type 1 diabetes of recent onset: two multicentre, randomised, double-blind, placebo-controlled trials. Lancet. 2013;381(9881):1905–1915. doi: 10.1016/S0140-6736(13)60023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabrera SM, et al. Interleukin-1 antagonism moderates the inflammatory state associated with Type 1 diabetes during clinical trials conducted at disease onset. Eur J Immunol. 2016;46(4):1030–1046. doi: 10.1002/eji.201546005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21(7):677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlos D, et al. Mitochondrial DNA activates the NLRP3 inflammasome and predisposes to type 1 diabetes in murine model. Front Immunol. 2017;8:164. doi: 10.3389/fimmu.2017.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer-Barber KD, et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511(7507):99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawahara K, Hohjoh H, Inazumi T, Tsuchiya S, Sugimoto Y. Prostaglandin E2-induced inflammation: Relevance of prostaglandin E receptors. Biochim Biophys Acta. 2015;1851(4):414–421. doi: 10.1016/j.bbalip.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Nataraj C, et al. Receptors for prostaglandin E(2) that regulate cellular immune responses in the mouse. J Clin Invest. 2001;108(8):1229–1235. doi: 10.1172/JCI13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrigan E, et al. Csf2 and Ptgs2 epigenetic dysregulation in diabetes-prone bicongenic B6.NODC11bxC1tb mice. Genet Epigenet. 2015;7:5–17. doi: 10.4137/GEG.S29696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benhamou PY, et al. Essential fatty acid deficiency prevents autoimmune diabetes in nonobese diabetic mice through a positive impact on antigen-presenting cells and Th2 lymphocytes. Pancreas. 1995;11(1):26–37. doi: 10.1097/00006676-199507000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Chen SS, Jenkins AJ, Majewski H. Elevated plasma prostaglandins and acetylated histone in monocytes in type 1 diabetes patients. Diabet Med. 2009;26(2):182–186. doi: 10.1111/j.1464-5491.2008.02658.x. [DOI] [PubMed] [Google Scholar]
- 30.Basu S, Larsson A, Vessby J, Vessby B, Berne C. Type 1 diabetes is associated with increased cyclooxygenase- and cytokine-mediated inflammation. Diabetes Care. 2005;28(6):1371–1375. doi: 10.2337/diacare.28.6.1371. [DOI] [PubMed] [Google Scholar]
- 31.Sokolowska M, et al. Prostaglandin E2 inhibits NLRP3 inflammasome activation through EP4 receptor and intracellular cyclic AMP in human macrophages. J Immunol. 2015;194(11):5472–5487. doi: 10.4049/jimmunol.1401343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang EH, Libby P, Vanhoutte PM, Xu A. Anti-inflammation therapy by activation of prostaglandin EP4 receptor in cardiovascular and other inflammatory diseases. J Cardiovasc Pharmacol. 2012;59(2):116–123. doi: 10.1097/FJC.0b013e3182244a12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. 2011;17(9):2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- 34.Dong MB, Rahman MJ, Tarbell KV. Flow cytometric gating for spleen monocyte and DC subsets: differences in autoimmune NOD mice and with acute inflammation. J Immunol Methods. 2016;432:4–12. doi: 10.1016/j.jim.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilliams M, et al. From skin dendritic cells to a simplified classification of human and mouse dendritic cell subsets. Eur J Immunol. 2010;40(8):2089–2094. doi: 10.1002/eji.201040498. [DOI] [PubMed] [Google Scholar]
- 36.Ivashkiv LB1, Donlin LT2. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mostafavi S, et al. Parsing the interferon transcriptional network and its disease associations. Cell. 2016;164(3):564–578. doi: 10.1016/j.cell.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrero JA, Calderon B, Towfic F, Artyomov MN, Unanue ER. Defining the transcriptional and cellular landscape of type 1 diabetes in the NOD mouse. PLoS ONE. 2013;8(3):e59701. doi: 10.1371/journal.pone.0059701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolic T, de Bruijn MF, Lutz MB, Leenen PJ. Developmental stages of myeloid dendritic cells in mouse bone marrow. Int Immunol. 2003;15(4):515–524. doi: 10.1093/intimm/dxg050. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol. 2007;179(11):7577–7584. doi: 10.4049/jimmunol.179.11.7577. [DOI] [PubMed] [Google Scholar]
- 41.Chen YG, et al. Molecular signatures differentiate immune states in type 1 diabetic families. Diabetes. 2014;63(11):3960–3973. doi: 10.2337/db14-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rausch-Derra L, Huebner M, Wofford J, Rhodes L. A prospective, randomized, masked, placebo-controlled multisite clinical study of grapiprant, an EP4 prostaglandin receptor antagonist (PRA), in dogs with osteoarthritis. J Vet Intern Med. 2016;30(3):756–763. doi: 10.1111/jvim.13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yong VW, Chabot S, Stuve O, Williams G. Interferon beta in the treatment of multiple sclerosis: mechanisms of action. Neurology. 1998;51(3):682–689. doi: 10.1212/WNL.51.3.682. [DOI] [PubMed] [Google Scholar]
- 44.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25(3):383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Diana J, et al. Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat Med. 2013;19(1):65–73. doi: 10.1038/nm.3042. [DOI] [PubMed] [Google Scholar]
- 46.Li Q, McDevitt HO. The role of interferon alpha in initiation of type I diabetes in the NOD mouse. Clin Immunol. 2011;140(1):3–7. doi: 10.1016/j.clim.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L, Yuan S, Cheng G, Guo B. Type I IFN promotes IL-10 production from T cells to suppress Th17 cells and Th17-associated autoimmune inflammation. PLoS One. 2011;6(12):e28432. doi: 10.1371/journal.pone.0028432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maine CJ, Teijaro JR, Marquardt K, Sherman LA. PTPN22 contributes to exhaustion of T lymphocytes during chronic viral infection. Proc Natl Acad Sci USA. 2016;113(46):E7231–E7239. doi: 10.1073/pnas.1603738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng X, Yau D, Holbrook C, Reder AT. Type I interferons inhibit interleukin-10 production in activated human monocytes and stimulate IL-10 in T cells: implications for Th1-mediated diseases. J Interferon Cytokine Res. 2002;22(3):311–319. doi: 10.1089/107999002753675730. [DOI] [PubMed] [Google Scholar]
- 50.Brod SA, Malone M, Darcan S, Papolla M, Nelson L. Ingested interferon alpha suppresses type I diabetes in non-obese diabetic mice. Diabetologia. 1998;41(10):1227–1232. doi: 10.1007/s001250051056. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka-Kataoka M, et al. Oral use of interferon-alpha delays the onset of insulin-dependent diabetes mellitus in nonobese diabetes mice. J Interferon Cytokine Res. 1999;19(8):877–879. doi: 10.1089/107999099313398. [DOI] [PubMed] [Google Scholar]
- 52.Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15(4):231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 53.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36(2):166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer-Barber KD, Yan B. Clash of the cytokine titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell Mol Immunol. 2017;14(1):22–35. doi: 10.1038/cmi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku CL, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204(10):2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Page C, Génin P, Baines MG, Hiscott J. Interferon activation and innate immunity. Rev Immunogenet. 2000;2(3):374–386. [PubMed] [Google Scholar]
- 57.Esmatjes E, Levy I, Gaya J, Rivera F. Renal excretion of prostaglandin E2 and plasma renin activity in type I diabetes mellitus: relationship to normoglycemia achieved with artificial pancreas. Diabetes Care. 1987;10(4):428–431. doi: 10.2337/diacare.10.4.428. [DOI] [PubMed] [Google Scholar]
- 58.Vallerie SN, et al. Myeloid cell prostaglandin E2 receptor EP4 modulates cytokine production but not atherogenesis in a mouse model of type 1 diabetes. PLoS One. 2016;11(6):e0158316. doi: 10.1371/journal.pone.0158316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao C, Hirata T, Soontrapa K, Ma X, Takemori H, Narumiya S. Prostaglandin E2 promotes Th1 differentiation via synergistic amplification of IL-12 signalling by cAMP and PI3-kinase. Nat Commun. 2013;4:1685. doi: 10.1038/ncomms2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yi Z, et al. IFN-γ receptor deficiency prevents diabetes induction by diabetogenic CD4+, but not CD8+, T cells. Eur J Immunol. 2012;42(8):2010–2018. doi: 10.1002/eji.201142374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guerrero AD, Dong MB, Zhao Y, Lau-Kilby A, Tarbell KV. Interleukin-2-mediated inhibition of dendritic cell development correlates with decreased CD135 expression and increased monocyte/macrophage precursors. Immunology. 2014;143(4):640–650. doi: 10.1111/imm.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhargava V, Head SR, Ordoukhanian P, Mercola M, Subramaniam S. Technical variations in low-input RNA-seq methodologies. Sci Rep. 2014;4:3678. doi: 10.1038/srep03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu J, Ge H, Newman M, Liu K. OSA: a fast and accurate alignment tool for RNA-Seq. Bioinformatics. 2012;28(14):1933–1934. doi: 10.1093/bioinformatics/bts294. [DOI] [PubMed] [Google Scholar]
- 64.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 68.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.